Abstract
Despite advances in treatment and diagnosis, the prognosis of patients with esophageal squamous cell carcinoma (ESCC) remains poor. MicroRNAs (miRNAs/miRs) are associated with prognosis in esophageal cancer, indicating that they may help guide treatment decisions. The aim of the present study was to explore exosomal miR-185 as a candidate prognostic biomarker and therapeutic target in ESCC, to investigate its biological function and clinical significance, and to ascertain the applicability of circulating exosomal miR-185 for the development of targeted drugs for ESCC treatment. A GeneChip miRNA array was used to compare exosomal miRNA expression in ESCC cell lines under hypoxia with those under normoxia. Exosomal miR-185 expression was then confirmed by reverse transcription-quantitative PCR. Patient background and prognosis were compared between high and low miR-185 expression groups. Functional analyses were performed to evaluate the antitumor effects of miR-185 in ESCC cells. Global Gene Set Enrichment Analysis of The Cancer Genome Atlas data was also performed, and differentially expressed exosomal miRNAs under hypoxia were identified compared to those under normoxia. Hypoxia markedly decreased the expression of exosomal miR-185 in KYSE-960 and T.Tn cell culture media. Overexpression of miR-185 suppressed the migration, invasion and colony-forming abilities of ESCC lines, and also suppressed cell cycle progression and promoted apoptosis after cisplatin treatment. Notably, high miR-185 expression was associated with signaling pathways related to cell death, DNA damage and p53. Furthermore, circulating exosomal miR-185 levels were associated with cN and cStage, and could predict progression-free survival and disease-specific survival of patients with ESCC after initial treatment. In conclusion, miR-185 holds potential as a prognostic biomarker and therapeutic target in ESCC.
Keywords: ESCC, metastasis, cell cycle progression, microRNA-185, prognostic biomarker
Introduction
Esophageal cancer is the eighth most common malignant neoplasm and the sixth leading cause of cancer-related deaths worldwide (1). Despite advances in the treatment of esophageal cancer over recent decades, patient prognosis has shown little improvement (2). Even with the introduction of combination treatment regimens and early diagnostic technology, the survival rate remains unsatisfactory, with a 5-year overall survival rate of 17–20% in Asia (3). Lymph node metastasis is one of the most important prognostic factors of esophageal cancer and generally indicates a poor outcome (4). Therefore, the identification of new prognostic biomarkers and the development of effective treatment methods are imperative for improving the clinical outcomes of patients with esophageal squamous cell carcinoma (ESCC).
Exosomes are a class of extracellular vesicles; that is, lipid bilayer-enclosed carriers of proteins, nucleic acids, lipids and metabolites, which are secreted by cells into the extracellular environment (5). As such, exosomes can transfer bioactive molecules from donor to recipient cells and influence their biological activities (6). Tumor-derived exosomes are released into peripheral blood and their quantity is a prognostic marker of ESCC that alters cellular gene expression and contributes to tumor progression (7). Furthermore, tumor-derived exosomes have been shown to alter cellular gene expression and contribute to tumor progression in ESCC (8). Therefore, a comprehensive understanding of the mechanisms through which circulating exosomal microRNAs (miRNAs/miRs) influence cancer progression is warranted.
Emerging evidence has suggested that hypoxia contributes to ESCC resistance against first-line chemotherapy, such as cisplatin and 5-fluorouracil (9). Our previous study reported that hypoxia-inducible factor (HIF)-1α serves an essential role in hypoxia-induced ESCC progression by maintaining crucial mechanisms, such as epithelial-mesenchymal transition, proliferation, migration/invasion, apoptosis, cell cycle progression and chemoresistance, influencing patient prognosis (10). However, the mechanisms through which hypoxia influences miRNAs in ESCC remain poorly understood.
miRNAs are small noncoding RNAs that regulate gene expression at the post-transcriptional level (11). A growing body of evidence has demonstrated that miRNAs are implicated in the initiation and progression of ESCC by regulating the expression of oncogenes and tumor suppressors (12,13). The prognostic applications of miRNAs in ESCC have recently attracted interest. miR-185 has been identified as a tumor-suppressive miRNA in multiple types of cancer, including hepatocellular carcinoma (14), osteosarcoma (15) and prostate cancer (16), in addition to being associated with the prognosis of colon (17) and gastric cancer (18). However, there is limited literature on the role of exosomal miR-185 in ESCC. The biological roles of miR-185 in cell viability, Ki-67 staining, cell migration, invasion, xenograft model, locoregional staging and molecular mechanisms of ESCC, and the survival of patients with ESCC have been reported (19–21). The present study observed alterations in exosomal miR-185 expression under hypoxic conditions, as well as its association with lymph node metastasis before chemotherapy and the sensitivity of ESCC cells to chemotherapy. Moreover, this study investigated the association of exosomal miR-185 levels with clinicopathological factors and prognosis in ESCC, and explored the mechanism underlying the role of exosomal miR-185 via in vitro experiments using ESCC cell lines and bioinformatics analyses. The findings highlight the importance of exosomal miR-185 as a prognostic biomarker and therapeutic target in ESCC.
Materials and methods
Clinical samples
Plasma samples were collected from 89 patients diagnosed with ESCC at Chiba University Hospital (Chiba, Japan; affiliated with Chiba University Graduate School of Medicine) between May 2011 and April 2017. Patients aged 20–85 years (median, 67 years) with histologically diagnosed ESCC were included, whereas patients with other types of cancer were excluded; however, no other exclusion criteria, such as history of other medical diseases, were applied. All patients were staged according to the Japanese Classification of Esophageal Cancer 11th edition (22). Written informed consent was obtained from all participants. The present study was approved by the Ethics Committee of the Chiba University Graduate School of Medicine (approval no. 1264; Chiba, Japan). Blood examination and sampling were performed before treatment. After receiving patient blood samples, they were centrifuged to obtain plasma at 4°C and 1,690 × g for 10 min, and were stored at −80°C.
Cell lines and cell culture
The KYSE-960 and KYSE-410 human ESCC cell lines were purchased from the Japanese Collection of Research Bioresources Cell Bank, and the T.Tn, TE1, TE6, TE11 and TE14 ESCC cell lines were provided by the Cell Resource Center at Tohoku University (Sendai, Japan). The SCCVII mouse squamous cell carcinoma cell line was kindly provided by Professor Yuta Shibamoto (Department of Quantum Radiology, Nagoya City University, Nagoya, Japan). Immortalized esophageal keratinocyte cells (R2C3), which were established at Chiba University School of Medicine, were used as a control cell line (23). The cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS; cat. no. 10270-106; Gibco; Thermo Fisher Scientific, Inc.) and 100 U/ml penicillin, and were maintained at 37°C in a humidified atmosphere containing 5% CO2. For exosome isolation under normoxic conditions, cells were incubated in DMEM replenished with 10% exosome-free FBS and 1% penicillin/streptomycin at 37°C in 21% O2 and 5% CO2 for 48 h. To simulate physical hypoxia, cells were incubated in 1% O2 and 5% CO2 in a multi-gas incubator for 48 h (cat. no. BL-43MD; TOSC Japan Ltd.).
Exosome isolation from the cell culture medium
A total of 5×106 cells were incubated in medium with 10% exosome-free FBS for 48 h at 37°C and total (10 ml) cell culture medium was harvested. Exosomes were isolated using the total exosome isolation kit (from cell culture media) (cat. no. 4478359; Invitrogen; Thermo Fisher Scientific, Inc.), according to manufacturer's protocol. The same method was performed that was described in our previous study (7).
Profiling of exosomal miRNAs extracted from normoxia/hypoxia culture medium
Microarray analysis was performed on total RNA extracted from exosomes isolated by ultracentrifugation. Prior to exosome isolation, KYSE-960 and T.Tn cells were cultured to 80% confluence, and were cultured in normoxia or hypoxia, as aforementioned. Exosomes were isolated from the cell culture media by ultracentrifugation at 10,000 × g for 90 min at 4°C (Optima TLX Ultracentrifuge; Beckman Coulter, Inc.), as previously reported (8). Exosome extraction was confirmed by transmission electron microscopy (TEM) and nanoparticle tracking analysis (Fig. S1). Nanoparticle tracking analysis was performed using NanoSight NS300 and NTA2.3 software (Fujifilm), according to the manufacturer's protocol, to define the particle size distribution and exosome concentration. Total Exosome RNA & Protein Isolation Kit (cat. no. 4478545; Thermo Fisher Scientific, Inc.) was used to extract total RNA from exosomes. Exosomal RNA from normoxic and hypoxic KYSE-960 and T.Tn cell culture media were analyzed using Affymetrix GeneChip miRNA 4.0 (cat. no. 902412; Affymetrix; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Arrays were incubated in the GeneChip™ Hybridization Oven 645 (Thermo Fisher Scientific, Inc.) at 48°C for 18 h (agitation at 60 rpm) and scanned using the GeneChip Scanner 3000 7G (Thermo Fisher Scientific, Inc.) according to the accompanying manual [GeneChip Command Console (AGCC) 4.0 User Manual]. miRNA expression was calculated using the detection above background algorithm and was normalized by robust multichip analysis. Chip data analysis was performed using R software (4.2.0) (https://www.r-project.org).
Exosome isolation from plasma
Each plasma sample (1–1.5 ml) was centrifuged at 2,000 × g for 20 min at room temperature to remove cells and debris. Exosomes were then isolated using the total exosome isolation kit (from plasma) (cat. no. 4484450; Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The same method was performed that was described in our previous study (7).
TEM
TEM observation was performed using a carbon-coated copper grid (Excel support film; 200 mesh; cat. no. RL26A; Nisshin Em Co., Ltd.) and the negative staining method. Sample preparation was performed according to the method described in a previous report (24). The samples were then subjected to TEM observation (H-7650; Hitachi High-Technologies Corporation) at an acceleration voltage of 80.0 kV.
miRNA and mRNA isolation and detection via reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from the exosomes using the Total Exosome RNA and Protein Isolation Kit (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Total cellular RNA was extracted using the Mini Kit (Qiagen GmbH), according to the manufacturer's protocol. Total RNA was then reverse-transcribed to cDNA using a High-Capacity RNA-to-DNA™ Kit (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. qPCR analysis was performed using TaqMan MicroRNA Assays (Invitrogen; Thermo Fisher Scientific, Inc.) or the SsoFast™ EvaGreen Supermix (Bio-Rad Laboratories, Inc.). The TaqMan primer for hsa-miR-185 (assay ID 002271) was used to detect miR-185 expression; miR-16 (assay ID 000391), which was previously used as a control for cell-free miRNA analysis in our laboratory (25), was used as an internal control for detection of exosomal miR-185 from plasma and cancer cell culture medium, whereas U6 small nuclear RNA (assay ID 001973) was used as an internal control for detection of miR-185 expression in cancer cell lines (all from Applied Biosystems; Thermo Fisher Scientific, Inc.). BCL-2 expression was normalized to β-actin. The thermocycling conditions were as follows: For miRNA detection, samples were incubated at 95°C for 10 min, followed by 40 cycles at 95°C for 15 sec and 60°C for 60 sec; for mRNA detection, samples were incubated for 30 sec at 95°C, followed by 40 cycles at 95°C for 5 sec and 60°C for 10 sec. Relative expression was calculated using the 2−ΔΔCq method (26); BCL-2 and miR-185 specific primers are detailed in Table I.
Table I.
Primer sequences used for reverse transcription- quantitative PCR.
| Name | Sequence, 5′-3′ |
|---|---|
| BCL-2 | F: GGATCCAGGATAACGGAGGC |
| BCL-2 | R: GGCAGGCATGTTGACTTCAC |
| β-actin | F: CATGTACGTTGCTATCCAGGC |
| β-actin | R: CTCCTTAATGTCACGCACGAT |
| hsa-mir-185 stem-loop | AGGGGGCGAGGGAUUGGAGA |
| GAAAGGCAGUUCCUGAUGGUC | |
| CCCUCCCCAGGGGCUGGCUUU | |
| CCUCUGGUCCUUCCCUCCCA | |
| hsa-mir-16-1 stem-loop | GUCAGCAGUGCCUUAGCAGCA |
| CGUAAAUAUUGGCGUUAAGAU | |
| UCUAAAAUUAUCUCCAGUAUU | |
| AACUGUGCUGCUGAAGUAAGG | |
| UUGAC | |
| U6 snRNA stem-loop | GTGCTCGCTTCGGCAGCACATA |
| TACTAAAATTGGAACGATACAG | |
| AGAAGATTAGCATGGCCCCTGC | |
| GCAAGGATGACACGCAAATTC | |
| GTGAAGCGTTCCATATTTT |
F, forward; R, reverse; miR, microRNA; snRNA, small nuclear RNA.
miRNA transfection
KYSE-960 and T.Tn cells were seeded in 6-well plates (2.5×105 cells/well) and, after 24 h, the cells were transfected with miR-185 mimic or negative control (25 pmol/well) using Lipofectamine™ RNAiMAX Transfection Reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The miR-185 mimic and negative control were purchased from Thermo Fisher Scientific, Inc. (mirVana miRNA mimic and negative control; assay ID MC12486). The miR-185 mimic and negative control were transfected into cells for 48 h at 37°C. After 48 h of transfection, the cells were harvested for the subsequent experiments.
Migration, invasion, cell proliferation and colony formation assays
Migration and invasion were detected using Transwell assays. In a 24-well plate, 5×104 transfected KYSE-960 and T.Tn cells/well were seeded in the upper chamber (8-µm pore; cat. no. 354480; Corning BioCoat Matrigel Invasion Chamber; Corning, Inc.) in FBS-free medium. A medium containing 10% FBS was added to the lower chamber. After incubation for 48 h at 37°C, non-invading cells were removed from the upper chamber with a cotton swab, whereas cells on the lower surface were fixed with 99% methanol for 5 sec at room temperature and stained using Diff-Quick Staining (Sysmex Corporation) at room temperature overnight. Images of three random fields from triplicate wells were recorded. Migration assays were performed in the same manner, except that the chambers had no Matrigel coating (8-µm pore; cat. no. 662638; Greiner Bio-One International GmbH), and the incubation time was 24 h at 37°C.
To assess cell proliferation, a total of 5×103 KYSE-960 and T.Tn transfected cells/well were seeded in a 96-well plate. Cell proliferation was assessed using a Cell Counting Kit-8 (CCK-8) assay (Dojindo Laboratories, Inc.). The reagent was added into each well and incubated for a further 2 h at 37°C every 24 h and the absorbance at 450 nm was measured using a microplate reader (Bio-Rad Laboratories, Inc.). The data were statistically analyzed on day 4 using Student's t-test.
For the colony formation assay, 800 cells/well of KYSE-960 and T.Tn cells were plated in a 6-well plate and were maintained in complete culture medium. After 2 weeks, the colonies were stained with a Diff-Quick Stain (Sysmex Corporation) for 15 sec at room temperature. Images of the visible colonies were captured and the number of colonies consisting of >50 cells was counted.
Apoptosis and cell cycle analyses
The IC50 values of cisplatin in KYSE-960 and T.Tn cells were determined using CCK-8 assay. KYSE-960 and T.Tn cells were seeded in 96-well plates (5×103 cells/well) and were incubated at 37°C in a humidified 5% CO2 atmosphere for 24 h. Subsequently, the medium was replaced with fresh medium with or without various concentrations (0–100 µM) of cisplatin (cat. no. P4394; MilliporeSigma). A total of 48 h after cisplatin administration, cell viability was measured using the CCK-8 assay, as aforementioned, and IC50 was calculated. Subsequently, KYSE-960 and T.Tn cell lines were treated with the IC50 of cisplatin for 48 h at 37°C. The cells were then harvested and washed with PBS, resuspended in 100 µl Annexin V Binding Solution, and incubated with 5 µl Annexin V FITC and 5 µl PI solution (Annexin V-FITC Apoptosis Detection Kit; Nacalai Tesque, Inc.) at room temperature for 15 min. Finally, 400 µl Annexin V Binding Solution was added prior to analysis (BD FACSCanto™ II Flow Cytometer; BD Biosciences). The results were analyzed using FlowJo software (FlowJo, 10.8.1; FlowJo, LLC)
For cell cycle analysis, cells were harvested after 48 h of transfection, washed with PBS, harvested and fixed with pre-cooled 70% ethanol at 4°C overnight (18–24 h). After washing, the cells were incubated with 100 µg/ml RNase A (Invitrogen; Thermo Fisher Scientific, Inc.) and 0.1% Triton-100 at 37°C for 5 min at 37°C, and were then stained with 50 µg/ml PI at room temperature for 30 min in the dark. The DNA content was measured on a BD FACSCanto II Flow Cytometer using the MOdfitLT 5.0 software program.
Gene Set Enrichment Analysis (GSEA)
Enrichment analysis of The Cancer Genome Atlas Esophageal Carcinoma Collection (TCGA-ESCA) data (https://gdac.broadinstitute.org/runs/stddata__2016_01_28/data/ESCA/20160128/) was performed using GSEA v4.0.1 (https://www.gsea-msigdb.org/gsea/index.jsp). RNA-seq and miRNA-seq data from a total of 195 patients with esophageal cancer were used to evaluate miRNA expression. Patients were divided into high and low miR-185-5p expression groups based on median miR-185-5p expression.
Statistical analysis
Statistical analysis was performed using SPSS 21 (IBM Corp.) and GraphPad Prism 7.04 (Dotmatics). Data are presented as the mean ± standard deviation. An unpaired Student's t-test and one-way analysis of variance with Tukey's post hoc test were employed for quantitative variables, and χ2 test or Fisher's exact test were employed for qualitative variables to compare the characteristics of each group. The association between exosomal miR-185 expression in cell lines and in cancer cell culture media was determined using Pearson correlation analysis. Progression-free survival (PFS) and disease specific survival (DSS) curves were plotted using the Kaplan-Meier method and results were compared using the log-rank test. PFS was defined as the time-frame between the start of treatment and the date of the first progression, and was used as an indicator of treatment efficacy. DSS was defined as the time-frame between the start of treatment and the date of death due to ESCC, and was used as a prognostic indicator. Each experiment was repeated three times, except for miR microarray analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
miRNA array expression analysis of KYSE-960 and T.Tn cell culture media
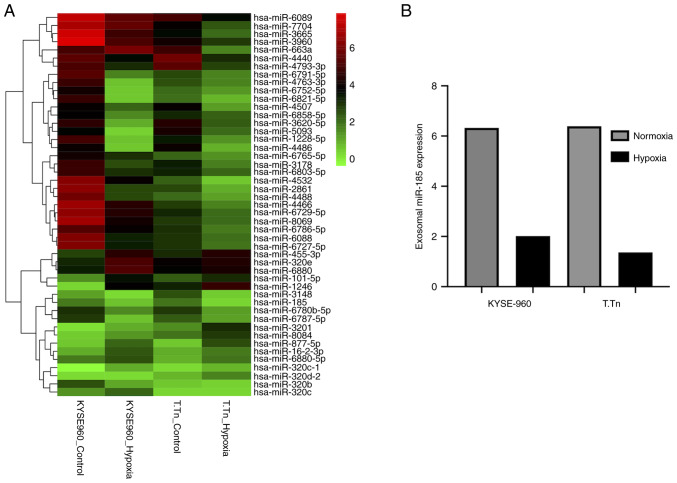
miRNA array analysis was performed to screen differentially expressed exosomal miRNAs in KYSE-960 and T.Tn cells culture medium under normoxic and hypoxic conditions. The expression of 33 exosomal miRNAs exhibited 2-fold downregulation, whereas 12 miRNAs exhibited 2-fold upregulation in normoxic versus hypoxic KYSE-960 cells. In the T.Tn cell line, 33 exosomal miRNAs exhibited 2-fold downregulation, and nine exhibited 2-fold upregulation. Cluster analysis of the intersecting miRNAs is shown in Fig. 1A. The expression levels of exosomal miR-185 were decreased in both KYSE-960 and T.Tn cell culture media under hypoxic conditions (fold change relative to that in KYSE-960 cells: 0.32, fold change relative to that in T.Tn cells: 0.22; Fig. 1B). Different cells respond differently to external stimuli, which may be why inconsistent fold changes occur in the expression levels of miRNAs in KYES-960 and T.Tn cell lines.
Figure 1.
(A) Hierarchical clustering of the differentially expressed miRNAs in ESCC cell culture media. Hypoxia affected miRNA expression in KYSE-960 and T.Tn cell culture media. In the KYSE-960 cell line, 33 miRNAs were downregulated and 12 were upregulated. In the T.Tn cell line, 33 miRNAs were downregulated and nine were upregulated. The color intensity represents the magnitude of expression difference; red indicates miRNAs that were 2-fold upregulated and green indicates miRNAs that were 2-fold downregulated in normoxic cells compared with hypoxic cells. The expression values are log-transformed. (B) Exosomal miR-185 expression, as calculated by detection above background algorithm and normalized by robust multichip analysis, was downregulated in both KYSE-960 and T.Tn cell line culture media (KYSE-960 fold-change: 0.32; T.Tn fold-change: 0.22). miRNA/miR, microRNA.
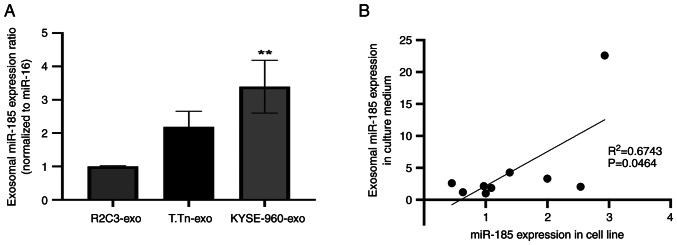
Analysis of miR-185-5p expression in ESCC cells and exosomes
miR-185-5p expression in T.Tn cells was similar to that detected in normal esophageal keratinocytes (R2C3), whereas the expression levels of miR-185-5p were slightly higher in KYSE-960 cells than those in normal esophageal keratinocytes (R2C3), but not statistically significant (Fig. S2). miR-185-5p expression in exosomes derived from T.Tn cell culture medium was two times higher than that in exosomes from normal esophageal keratinocytes, but the difference was not statistically significant. Furthermore, miR-185-5p expression in exosomes derived from KYSE-960 culture medium was significantly higher than that in exosomes from normal esophageal keratinocytes (Fig. 2A). Correlation analysis of miR-185 expression in R2C3, T.Tn, TE1, TE6, TE11, TE14, KYSE-960, KYSE-410 and SCCVII cells, and its expression in exosomes was conducted using data obtained from RT-qPCR (R2=0.6743, P=0.0464; Fig. 2B). A significant positive correlation between miR-185 expression in ESCC cell lines and miR-185 expression in culture medium-derived exosomes was observed.
Figure 2.
(A) R2C3, T.Tn, and KYSE-960 cells were cultured in exosome-free media, and miR-185 expression was measured by RT-qPCR. miR-16 was used as an internal control (**P<0.01 vs. RC23). (B) Correlation analysis of miR-185 expression in exosomes from cancer cell media and in cell lines, quantified by RT-qPCR (R2=0.6743; P<0.05). miR, microRNA; RT-qPCR, reverse transcription-quantitative PCR.
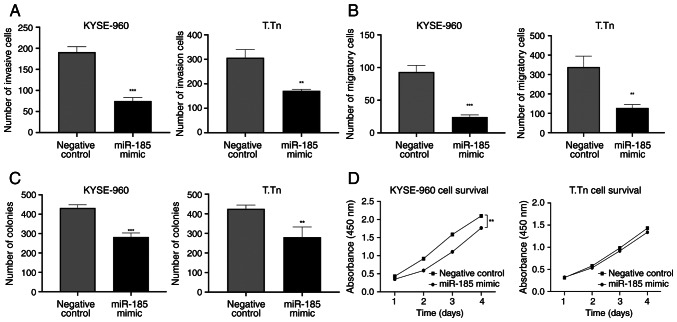
miR-185 overexpression suppresses the invasion, migration and colony formation of ESCC cells in vitro
The statistically significant overexpression of miR-185 in KYSE-960 and T.Tn cells was confirmed by RT-qPCR following miR-185 mimic transfection (>1,000-fold increase in KYSE-960 and T.Tn cell lines, compared with in cells transfected with the negative control mimic; Fig. S3). The invasion and migration of miR-185-overexpressing cells was significantly decreased compared with those in the negative control cells [invasion: KYSE-960, P=0.0001; T.Tn, P=0.0022 (Figs. 3A and S3); migration: KYSE-960, P=0.0003; T.Tn, P=0.0034 (Figs. 3B and S3)]. Cells transfected with the miR-185 mimic exhibited a lower survival rate than those in the negative control group, as determined by the colony formation assay [KYSE-960, P=0.0005; T.Tn, P=0.01 (Figs. 3C and S4)]. The results of the CCK-8 assay indicated that transfection with the miR-185 mimic significantly weakened the proliferative capacity of KYSE-960 cells compared with in the control group (P=0.002); however, the miR-185 mimic was not able to significantly inhibit the proliferative capacity of T.Tn cells (P=0.6) (Fig. 3D).
Figure 3.
Transwell assays were used to detect the (A) invasion and (B) migration of KYSE-960 and T.Tn cells post-transfection. (C) Colony formation assays were performed to assess the colony-forming ability of KYSE-960 and T.Tn cells. (D) Cell Counting Kit-8 assays were used to detect the proliferation of KYSE-960 and T.Tn cells post-transfection. **P<0.01, ***P<0.001 vs. negative control. miR, microRNA.
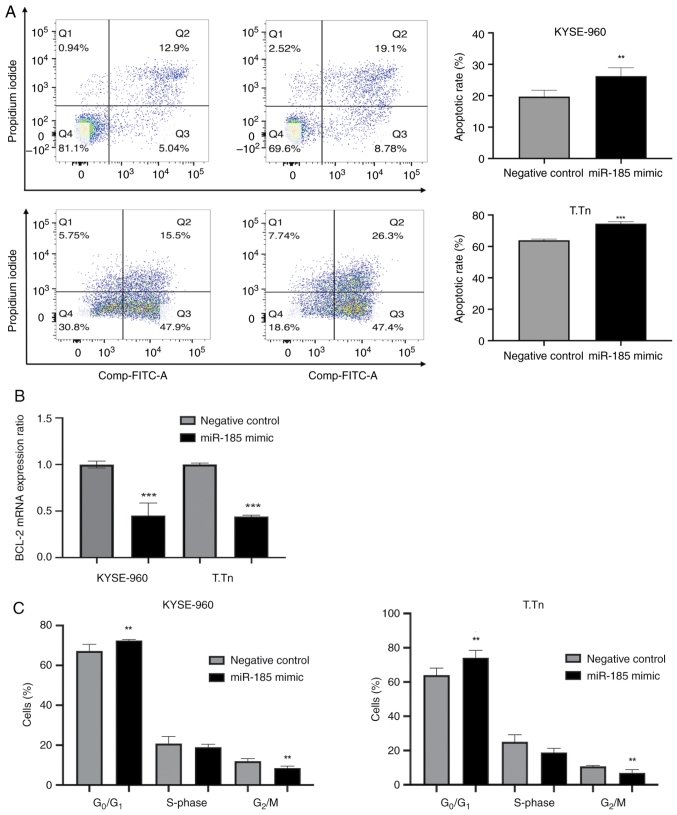
miR-185 regulates apoptosis after cisplatin treatment and cell cycle progression in vitro
The apoptotic rate of miR-185-transfected KYSE-960 and T.Tn cells after cisplatin exposure for 48 h was significantly higher than that of the negative control cells (KYSE-960, P=0.027; T.Tn, P=0.0001; Fig. 4A). In addition, the mRNA expression levels of BCL-2 were reduced in both the KYSE-960 and T.Tn miR-185 mimic groups (Fig. 4B). Cell cycle analyses showed that the percentages of ESCC cells in the G0/G1 phase after miR-185 transfection were significantly higher compared with those in the negative control groups, whereas the percentages of ESCC cells in the G2/M phase were significantly lower (KYSE-960 G0/G1, P<0.05 and G2/M, P<0.02; T.Tn G0/G1, P<0.04 and G2/M, P<0.03; Figs. 4C and S5). Notably, KYSE-960 and T.Tn cells in Fig. 4B and C were not treated with cisplatin.
Figure 4.
(A) Cell apoptosis was evaluated following Annexin V-FITC and propidium iodide staining by flow cytometry. (B) Reverse transcription-quantitative PCR was used to the detect the mRNA expression levels of BCL-2 in KYSE-960 and T.Tn cell lines. β-actin was used as the housekeeping gene. (C) Percentage of cells in each stage of the cell cycle. **P<0.01, ***P<0.001 vs. negative control. miR, microRNA.
miR-185-5p-associated signaling
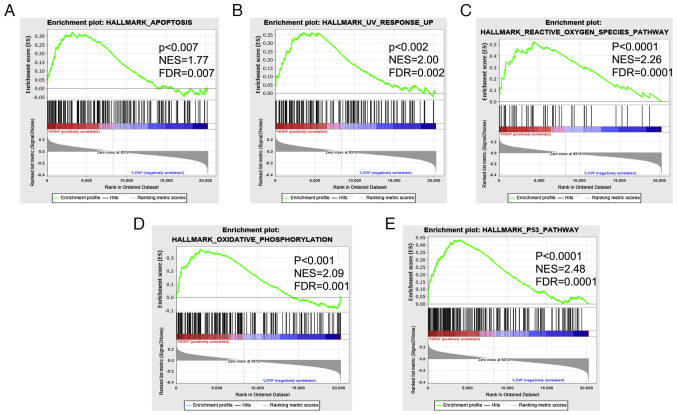
Enrichment analysis of TCGA data was carried out using GSEA, and the results revealed that the high miR-185 expression group was enriched in signaling pathways, such as cell death (apoptosis) and DNA damage (UV response up, reactive oxygen species, oxidative phosphorylation) and p53 signaling (Fig. 5A-E).
Figure 5.
Gene Set Enrichment Analysis was used to analyze the signaling pathways enriched in patients with esophageal cancer and high miR-185-5p expression: (A) Apoptosis, (B) UV response, (C) reactive oxygen species pathway, (D) oxidative phosphorylation and (E) p53 pathway. NES indicates the extent of enrichment per gene set; FDR indicates the significance of enrichment. Gene sets for enrichment analysis were determined based on adjusted P<0.05 and |NES|>1. NES, normalized enrichment score; FDR, false discovery rate.
Characterization of exosomes from patient samples
The present study isolated exosomes from the plasma samples of 89 patients with ESCC. To confirm the successful isolation of exosomes, TEM was employed to characterize their shape in the supernatant (Fig. S6). Notably, plasma exosomes exhibited an elliptical shape.
Relationship between circulating miR-185 levels and clinicopathological characteristics of patients with ESCC
The clinicopathological characteristics of patients are summarized in Table II. The total number of patients was 89, including nine endoscopically treated cases, 55 preoperatively untreated surgical cases and 25 cases treated via neoadjuvant chemotherapy, followed by surgery. Pretreatment staging was performed by whole body analysis, including PET/CT. Patients were divided into high (n=28) and low (n=61) circulating miR-185 groups. A cutoff value of 4.3 was set for the miR-185/miR-16 ratio based on the mean value of miR-185 in patients with esophageal cancer. In patients with high circulating exosomal miR-185, the frequency of lymph node metastasis at preoperative diagnosis was significantly lower (P=0.0045), and cStage was significantly lower in the high circulating exosomal miR-185 group (P=0.0001).
Table II.
Demographics and clinicopathological characteristics of patients with esophageal squamous cell carcinoma.
| Variable | High exosomal miR-185-5p group (n=28) | Low exosomal miR-185-5p group (n=61) | P-value |
|---|---|---|---|
| Mean age ± SD, years | 66.6±8.03 | 66.8±8.32 | 0.923a |
| Sex, n | |||
| Male/Female | 24/4 | 51/10 | >0.999b |
| Histological grade, n | |||
| G1/G2/G3/X | 4/15/7/2 | 11/29/13/8 | 0.853b |
| Location of the tumor | |||
| Ce/Ut/Mt/Lt/Ae | 0/2/15/10/1 | 0/8/27/23/3 | 0.836b |
| cT category, n | |||
| cT1a + 1b/2/3/4a + 4b | 16/7/5/0 | 25/15/21/0 | 0.235c |
| cN category, n | |||
| cN0/1/2/3/4 | 20/2/3/0/3 | 28/11/18/4/0 | 0.0045b |
| cM category, n | |||
| cM0/1 | 28/0 | 61/0 | >0.999b |
| cStage, n | |||
| cStage 0/1/2/3/4a/4b | 7/6/9/3/3/0 | 6/11/19/25/0/0 | 0.0001b |
| Treatment, n | 0.0597b | ||
| Endoscopic | 6 | 3 | |
| Esophagectomy first | 16 | 39 | |
| NAC + esophagectomy | 6 | 19 | |
| pT category, n | |||
| pT0/1a + 1b/2/3/4a | 3/19/2/4/0 | 4/31/10/15/1 | 0.411b |
| pN category, n | |||
| pN0/1/2/3/4/X | 16/3/4/2/0/3 | 31/7/12/8/1/2 | 0.702b |
| pStage, n | |||
| pStage 0/1/2/3/4a/X | 2/11/5/5/2/3 | 1/18/14/15/9/4 | 0.527b |
Student's t-test;
Fisher's exact test;
χ2 test. Ce, cervical esophagus; Ut, upper thoracic esophagus; Mt, middle thoracic esophagus; Lt, lower thoracic esophagus; Ae, abdominal esophagus; NAC, neoadjuvant chemotherapy.
Although the sample size was small, the preoperative patient group with high circulating exosomal miR-185 levels tended to exhibit a slightly higher response rate to chemotherapy than patients with low circulating exosomal miR-185; however, this was not statistically significant (P=0.081; Table III).
Table III.
Pathological response of patients with esophageal squamous cell carcinoma treated with neoadjuvant chemotherapy.
| Variable | High exosomal miR-185-5p group (n=6) | Low exosomal miR-185-5p group (n=19) | P-valuea |
|---|---|---|---|
| Pathological response | |||
| Grade 1a + 1b/2/3 | 3/1/2 | 16/3/0 | 0.081 |
Fisher's exact test.
PFS and DSS of patients with ESCC after initial treatment
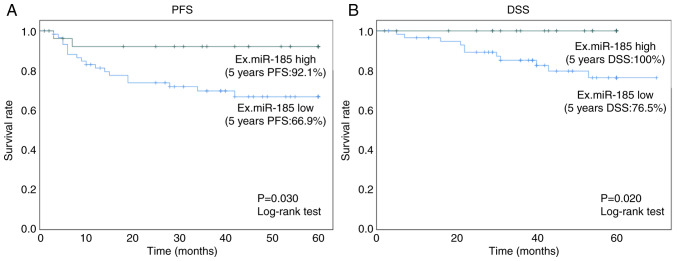
The 5-year PFS rate in the high exosomal miR-185 group was 92.1% (95% CI: 82.0–103.0%), which was significantly higher (P=0.030, log-rank test) than that in the low exosomal miR-185 group (66.9%; 95% CI: 54.0–80.0%) (Fig. 6A). The 5-year DSS rate in the high exosomal miR-185 group was 100%, which was significantly higher (P=0.020, log-rank test) than that in the low exosomal miR-185 group (76.5%; 95% CI: 64.0–89.0%; Fig. 6B).
Figure 6.
Kaplan-Meier survival curves showing the relationship between high or low expression levels of plasma Ex.miR-185-5p with (A) PFS and (B) DSS of patients with esophageal squamous cell carcinoma. DSS, disease-specific survival; Ex., exosomal; miR, microRNA; PFS, progression-free survival.
Discussion
ESCC is one of the most lethal types of cancer and is a public health issue of great concern worldwide (27). Despite advances in its diagnosis and treatment, patient survival rates remain unsatisfactory (28). Therefore, the identification of novel biomarkers and therapeutic targets in ESCC is urgently required.
Exosomal miRNAs can be easily isolated from peripheral blood, which makes them candidate noninvasive biomarkers (29). Their potential to serve as biomarkers in patients with ESCC has been previously reported, along with a possible mechanism of exosomal trafficking (25). Exosomes influence gene expression, and thus, various biological processes (30). However, the influence of exosomal miR-185 levels in plasma on cancer treatment outcomes remains to be fully elucidated. It has been reported that plasma miR-185 levels are decreased in patients with ESCC and that tumor metastasis is suppressed by targeting RAGE (21); the tumor suppressive role of miR-185 in this previous report is consistent with the results of present study. The impact of different storage conditions and treatments on the stability and abundance of individual and total miRNAs in human plasma and plasma exosomes has been investigated; it has been highlighted that exosomal miRNAs have the potential to serve as biomarkers based on their increased stability under various conditions compared with plasma miRNAs (31). Thus, there is a possibility that exosomal miRNAs could serve as novel therapeutic targets for the development of effective methods for the treatment of ESCC. However, to the best of our knowledge, the present study is the first to determine the different profiles of circulating exosomal miR-185 levels in patients with ESCC. The results revealed that exosomal miR-185 was associated with lymph node metastasis and may act as a predictor of the prognosis of patients with ESCC. The data from the current study suggested that exosomal miR-185 may be important for esophageal cancer initiation and progression, and that it could hold promise as a novel suppressor of metastasis in esophageal cancer.
Hypoxia enhances the degree of glycolysis, angiogenesis and other survival responses in tumors, as well as their invasion and metastasis, by activating relevant gene expression through HIFs (32). The expression levels of HIF-1α are elevated in cancer cell lines under hypoxia; notably, HIF-1α drives oncogenic expression in ESCC and is associated with a poor prognosis (10). In the present study, the expression of 33 exosomal miRNAs was downregulated in KYSE-960 and T.Tn cell culture media under hypoxia, whereas certain exosomal miRNAs were upregulated. Notably, exosomal miR-185 expression was decreased under hypoxia in both cell line culture media.
miRNA dysregulation serves a critical role in the initiation and progression of multiple types of human cancer (33,34), by either promoting (35) or suppressing tumorigenesis (36). The characterization of miRNAs involved in ESCC progression and their targets may contribute to the identification of new prognostic markers and therapeutic targets (37). Circulating miRNAs have recently emerged as potential biomarkers for various types of cancer (38). In the present study, exosomal miR-185 was significantly associated with cN and cStage, with high exosomal miR-185 levels in plasma being associated with a good prognosis in patients with ESCC. Upregulation of miR-185 may have a suppressive role in tumor malignancy, paving the way for the development of effective treatment methods in ESCC.
miRNAs are associated with lymph node metastasis in esophageal cancer (22,33,39), which may provide a novel insight into the design of better therapeutic strategies. Furthermore, albeit based on a limited number of cases, the present findings indicated that circulating miR-185 might not be able to influence chemotherapy sensitivity, but that it could predict lymph node metastasis prior to chemotherapy. To provide new insights for designing better therapeutic strategies to treat esophageal cancer in patients with lymph node metastasis and to predict prognosis more accurately, a further study in a larger cohort and a detailed mechanistic investigation are warranted.
Overcoming cisplatin resistance is a major aim in cancer therapy. One of the most widely accepted approaches is through combination with other agents that enhance cisplatin toxicity (40). In the present study, overexpression of miR-185 regulated cancer cell cycle progression and induced apoptosis following cisplatin treatment. The apoptotic rate in a human gastric cancer cell line has been reported to be ~10% without any other intervention except for miR-185 transfection (41). In another study, the apoptotic rate in a breast cancer cell line was reported to be ~20% without any other intervention except for miR-185 transfection (42). In the present study, the negative control group comprised ESCC cells transfected with the control mimic and treated with cisplatin for 48 h, and the target group comprised ESCC cells treated with cisplatin and transfected with the miR-185 mimic. Therefore, considering the previously reported numbers, and the fact that all cells that were treated with cisplatin, it is reasonable that the apoptotic rate detected in the present study was >10%. Tumor cells may utilize several molecular mechanisms to suppress apoptosis and acquire resistance to cytotoxic agents, such as via upregulation of the antiapoptotic protein BCL-2 (43). In the present study, the mRNA expression levels of BCL-2 were suppressed in miR-185-overexpressing cells.
The present study also observed a significant enrichment for hallmark gene sets related to cell death (apoptosis), DNA damage (UV response up, reactive oxygen species and oxidative phosphorylation), and p53 in patients with high versus low miR-185 expression. Such dysregulation is expected to serve a significant role in esophageal cancer cell transformation. These data suggested that miR-185 alters cancer-associated pathway activity.
In the present study, miR-16 was used as an internal control for the clinical evaluation of cell-free miR-185 and U6 was used as an internal reference for evaluation of cellular miR-185; however, a limitation is that these internal controls are not standardized and can only be quantified in relative amounts, which may not accurately reflect the amplification of the primary target and ultimately lead to different Cq value results. Development of accurate internal controls or quantification of absolute value are thus required for further confirmation in future studies. In addition, larger patient cohorts are required for future confirmation of the use of exosomal miR-185 in the prediction of prognosis.
In summary, the data from the current study suggested that miR-185 may be important in ESCC progression and holds promise as a novel suppressor of metastasis in ESCC. Although there are many factors, such as target genes, that can be used as a guide or reference when considering therapeutic efficacy, circulating exosomal miR-185 may act as a potential prognostic biomarker and could have potential as a novel therapeutic target in ESCC.
Supplementary Material
Acknowledgements
The authors are grateful to Ms. Keiko Iida (Department of Frontier Surgery, Chiba University Graduate School of Medicine) for technical advice and assistance.
Glossary
Abbreviations
- ESCC
esophageal squamous cell carcinoma
- RT-qPCR
reverse transcription-quantitative PCR
- HIF
hypoxia-inducible factor
- GSEA
Gene Set Enrichment Analysis
- TCGA
The Cancer Genome Atlas
Funding Statement
This study was supported by JSPS KAKENHI (grant nos. 15K19872, 19K23880 and 21K16414).
Availability of data and materials
The data generated in the present study may be requested from the corresponding author. Microarray analysis data generated in the present study may be found in the Gene Expression Omnibus database under accession number GSE263921 or at the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE263921.
Authors' contributions
YM and HMa conceived the theme, AM and YM designed the study. HW, YM, KKand MK performed the comprehensive analysis of microRNA. AM and HW developed the methodology for in vitro analyses. TS, KM, SI, HMo, TM and YN performed clinical data collection and validation. AM, KM, TT, RO and JH analyzed and interpreted clinical and in vitro data. TS and KM confirm the authenticity of all the raw data. AM and YM wrote the draft of the manuscript under the supervision of MK and HMa. All authors were involved in the planning, data interpretation and core revision of the paper, and read and approved the final version of the manuscript.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of the Graduate School of Medicine, Chiba University (approval no. 1264). Written informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Author's information
Yasunori Matsumoto ORCI: https://orcid.org/0000-0002-6239-6691
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Jin W, Luo W, Fang W, Wang Y, Wang L, Shen Q, Liu W, Zhang H. miR-145 expression level in tissue predicts prognosis of patients with esophageal squamous cell carcinoma. Pathol Res Pract. 2019;215:152401. doi: 10.1016/j.prp.2019.03.029. [DOI] [PubMed] [Google Scholar]
- 3.Chen JG, Chen HZ, Zhu J, Yang YL, Zhang YH, Huang PX, Chen YS, Zhu CY, Yang LP, Shen K, et al. Cancer survival in patients from a hospital-based cancer registry, China. J Cancer. 2018;9:851–860. doi: 10.7150/jca.23039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rice TW, Ishwaran H, Hofstetter WL, Schipper PH, Kesler KA, Law S, Lerut EM, Denlinger CE, Salo JA, Scott WJ, et al. Esophageal Cancer: Associations With (pN+) Lymph Node Metastases. Ann Surg. 2017;265:122–129. doi: 10.1097/SLA.0000000000001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skotland T, Sagini K, Sandvig K, Llorente A. An emerging focus on lipids in extracellular vesicles. Adv Drug Deliv Rev. 2020;159:308–321. doi: 10.1016/j.addr.2020.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Yang H, Fu H, Wang B, Zhang X, Mao J, Li X, Wang M, Sun Z, Qian H, Xu W. Exosomal miR-423-5p targets SUFU to promote cancer growth and metastasis and serves as a novel marker for gastric cancer. Mol Carcinog. 2018;57:1223–1236. doi: 10.1002/mc.22838. [DOI] [PubMed] [Google Scholar]
- 7.Matsumoto Y, Kano M, Akutsu Y, Hanari N, Hoshino I, Murakami K, Usui A, Suito H, Takahashi M, Otsuka R, et al. Quantification of plasma exosome is a potential prognostic marker for esophageal squamous cell carcinoma. Oncol Rep. 2016;36:2535–2543. doi: 10.3892/or.2016.5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsumoto Y, Kano M, Murakami K, Toyozumi T, Suito H, Takahashi M, Sekino N, Shiraishi T, Kamata T, Ryuzaki T, et al. Tumor-derived exosomes influence the cell cycle and cell migration of human esophageal cancer cell lines. Cancer Sci. 2020;111:4348–4358. doi: 10.1111/cas.14660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang P, Zhou J, Liang Z, Yang Y, Luan S, Xiao X, Li X, Zhang H, Shang Q, Zeng X, Yuan Y. Immunotherapy resistance in esophageal cancer: Possible mechanisms and clinical implications. Front Immunol. 2022;13:975986. doi: 10.3389/fimmu.2022.975986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang K, Toyozumi T, Murakami K, Sakata H, Kano M, Endo S, Matsumoto Y, Suito H, Takahashi M, Sekino N, et al. HIF-1α stimulates the progression of oesophageal squamous cell carcinoma by activating the Wnt/β-catenin signalling pathway. Br J Cancer. 2022;127:474–487. doi: 10.1038/s41416-022-01825-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 12.Hemmatzadeh M, Mohammadi H, Karimi M, Musavishenas MH, Baradaran B. Differential role of microRNAs in the pathogenesis and treatment of esophageal cancer. Biomed Pharmacother. 2016;82:509–519. doi: 10.1016/j.biopha.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 13.Harada K, Baba Y, Ishimoto T, Shigaki H, Kosumi K, Yoshida N, Watanabe M, Baba H. The role of microRNA in esophageal squamous cell carcinoma. J Gastroenterol. 2016;51:520–530. doi: 10.1007/s00535-016-1161-9. [DOI] [PubMed] [Google Scholar]
- 14.Niu Y, Tang G. miR-185-5p targets ROCK2 and inhibits cell migration and invasion of hepatocellular carcinoma. Oncol Lett. 2019;17:5087–5093. doi: 10.3892/ol.2019.10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu C, Cai L, Li H. miR-185 regulates the growth of osteosarcoma cells via targeting hexokinase 2. Mol Med Rep. 2019;20:2774–2782. doi: 10.3892/mmr.2019.10534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ostadrahimi S, Abedi Valugerdi M, Hassan M, Haddad G, Fayaz S, Parvizhamidi M, Mahdian R, Fard Esfahani P. miR-1266-5p and miR-185-5p promote cell apoptosis in human prostate cancer cell lines. Asian Pac J Cancer Prev. 2018;19:2305–2311. doi: 10.22034/APJCP.2018.19.8.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang W, Sun Z, Su L, Wang F, Jiang Y, Yu D, Zhang F, Sun Z, Liang W. miRNA-185 serves as a prognostic factor and suppresses migration and invasion through Wnt1 in colon cancer. Eur J Pharmacol. 2018;825:75–84. doi: 10.1016/j.ejphar.2018.02.019. [DOI] [PubMed] [Google Scholar]
- 18.Tan Z, Jiang H, Wu Y, Xie L, Dai W, Tang H, Tang S. miR-185 is an independent prognosis factor and suppresses tumor metastasis in gastric cancer. Mol Cell Biochem. 2014;386:223–231. doi: 10.1007/s11010-013-1860-y. [DOI] [PubMed] [Google Scholar]
- 19.Li BX, Yu Q, Shi ZL, Li P, Fu S. Circulating microRNAs in esophageal squamous cell carcinoma: Association with locoregional staging and survival. Int J Clin Exp Med. 2015;8:7241–7250. [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao ZT, Zhou W, Liu LY, Lan T, Zhan QM, Song YM. Molecular mechanism and effect of microRNA185 on proliferation, migration and invasion of esophageal squamous cell carcinoma. Zhonghua Yi Xue Za Zhi. 2013;93:1426–1431. (In Chinese) [PubMed] [Google Scholar]
- 21.Jing R, Chen W, Wang H, Ju S, Cong H, Sun B, Jin Q, Chu S, Xu L, Cui M. Plasma miR-185 is decreased in patients with esophageal squamous cell carcinoma and might suppress tumor migration and invasion by targeting RAGE. Am J Physiol Gastrointest Liver Physiol. 2015;309:G719–G729. doi: 10.1152/ajpgi.00078.2015. [DOI] [PubMed] [Google Scholar]
- 22.Japan Esophageal Society. Japanese classification of esophageal cancer, 11th edition: Part I. Esophagus. 2017;14:1–36. doi: 10.1007/s10388-016-0551-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sashiyama H, Shino Y, Kawamata Y, Tomita Y, Ogawa N, Shimada H, Kobayashi S, Asano T, Ochiai T, Shirasawa H. Immortalization of human esophageal keratinocytes by E6 and E7 of human papillomavirus type 16. Int J Oncol. 2001;19:97–103. doi: 10.3892/ijo.19.1.97. [DOI] [PubMed] [Google Scholar]
- 24.Jung MK, Mun JY. Sample preparation and imaging of exosomes by transmission electron microscopy. J Vis Exp. 2018;131:56482. doi: 10.3791/56482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeshita N, Hoshino I, Mori M, Akutsu Y, Hanari N, Yoneyama Y, Ikeda N, Isozaki Y, Maruyama T, Akanuma N, et al. Serum microRNA expression profile: miR-1246 as a novel diagnostic and prognostic biomarker for oesophageal squamous cell carcinoma. Br J Cancer. 2013;108:644–652. doi: 10.1038/bjc.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Talukdar FR, di Pietro M, Secrier M, Moehler M, Goepfert K, Lima SSC, Pinto LFR, Hendricks D, Parker MI, Herceg Z. Molecular landscape of esophageal cancer: Implications for early detection and personalized therapy. Ann N Y Acad Sci. 2018;1434:342–359. doi: 10.1111/nyas.13876. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y. Epidemiology of esophageal cancer. World J Gastroenterol. 2013;19:5598–5606. doi: 10.3748/wjg.v19.i34.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manier S, Liu CJ, Avet-Loiseau H, Park J, Shi J, Campigotto F, Salem KZ, Huynh D, Glavey SV, Rivotto B, et al. Prognostic role of circulating exosomal miRNAs in multiple myeloma. Blood. 2017;129:2429–2436. doi: 10.1182/blood-2016-09-742296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 31.Ge Q, Zhou Y, Lu J, Bai Y, Xie X, Lu Z. miRNA in plasma exosome is stable under different storage conditions. Molecules. 2014;19:1568–1575. doi: 10.3390/molecules19021568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu X, Kang Y. Hypoxia and hypoxia-inducible factors: Master regulators of metastasis. Clin Cancer Res. 2010;16:5928–5935. doi: 10.1158/1078-0432.CCR-10-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valeri N, Braconi C, Gasparini P, Murgia C, Lampis A, Paulus-Hock V, Hart JR, Ueno L, Grivennikov SI, Lovat F, et al. MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell. 2014;25:469–483. doi: 10.1016/j.ccr.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR, Yu Y, Chow A, O'Connor ST, Chin AR, et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25:501–515. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, Ngankeu A, Taccioli C, Pichiorri F, Alder H, Secchiero P, et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16:498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Zhang Y, Yang P, Sun T, Li D, Xu X, Rui Y, Li C, Chong M, Ibrahim T, Mercatali L, et al. miR-126 and miR-126* repress recruitment of mesenchymal stem cells and inflammatory monocytes to inhibit breast cancer metastasis. Nat Cell Biol. 2013;15:284–294. doi: 10.1038/ncb2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mao Y, Li L, Liu J, Wang L, Zhou Y. MiR-495 inhibits esophageal squamous cell carcinoma progression by targeting Akt1. Oncotarget. 2016;7:51223–51236. doi: 10.18632/oncotarget.9981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao Y, Song Y, Yao L, Song G, Teng C. Circulating microRNAs: Promising biomarkers involved in several cancers and other diseases. DNA Cell Biol. 2017;36:77–94. doi: 10.1089/dna.2016.3426. [DOI] [PubMed] [Google Scholar]
- 39.Iorio MV, Croce CM. MicroRNAs in cancer: Small molecules with a huge impact. J Clin Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiang Y, Ma N, Wang D, Zhang Y, Zhou J, Wu G, Zhao R, Huang H, Wang X, Qiao Y, et al. MiR-152 and miR-185 co-contribute to ovarian cancer cells cisplatin sensitivity by targeting DNMT1 directly: A novel epigenetic therapy independent of decitabine. Oncogene. 2014;33:378–386. doi: 10.1038/onc.2012.575. [DOI] [PubMed] [Google Scholar]
- 41.Fan L, Tan B, Li Y, Zhao Q, Yuan H, Liu Y, Wang D, Zhang Z. Upregulation of miR-185 promotes apoptosis of the human gastric cancer cell line MGC803. Mol Med Rep. 2018;17:3115–3122. doi: 10.3892/mmr.2017.8206. [DOI] [PubMed] [Google Scholar]
- 42.Değerli E, Torun V, Cansaran-Duman D. miR-185-5p response to usnic acid suppresses proliferation and regulating apoptosis in breast cancer cell by targeting Bcl2. Biol Res. 2020;53:19. doi: 10.1186/s40659-020-00285-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hassan M, Watari H, AbuAlmaaty A, Ohba Y, Sakuragi N. Apoptosis and molecular targeting therapy in cancer. Biomed Res Int. 2014;2014:150845. doi: 10.1155/2014/150845. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in the present study may be requested from the corresponding author. Microarray analysis data generated in the present study may be found in the Gene Expression Omnibus database under accession number GSE263921 or at the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE263921.