Abstract
Objective:
Systemic lupus erythematosus (SLE) is a highly female-biased systemic autoimmune disease, but the molecular basis for this female bias remains incompletely elucidated. B and T lymphocytes from patients with SLE and female-biased mouse models of SLE exhibit features of epigenetic dysregulation on the X chromosome which may contribute to this strong female bias. We therefore examined the fidelity of dynamic X-chromosome inactivation maintenance (dXCIm) in the pathogenesis of two murine models of spontaneous lupus—NZM2328 and MRL/lpr—with disparate levels of female-bias to determine whether impaired dXCIm contributes to the female bias of disease.
Methods:
CD23+ B cells and CD3+ T cells were purified from age-matched C57BL/6 (B6), MRL/lpr, and NZM2328 male and female mice, activated in vitro, and processed for Xist RNA fluorescence in situ hybridization, H3K27me3 immunofluorescence imaging, qPCR, and RNA sequencing analyses.
Results:
The dynamic relocalization of Xist RNA and the canonical heterochromatin mark, H3K27me3, to the inactive X chromosome was preserved in CD23+ B cells, but impaired in activated CD3+ T cells from the MRL/lpr model (p < 0.01 vs. B6), and even more impaired in the heavily female-biased NZM2328 model (p < 0.001 vs. B6; p < 0.05 vs. MRL/lpr). RNAseq of activated T cells from NZM2328 mice revealed the female-biased upregulation of 32 X-linked genes distributed broadly across the X chromosome, many of which have roles in immune function. Many genes encoding Xist RNA-interacting proteins were also differentially expressed and predominantly downregulated, which may account for the observed mislocalization of Xist RNA to the inactive X chromosome.
Conclusions:
Although evident in T cells from both the MRL/lpr and NZM2328 models of spontaneous SLE, impaired dXCIm is more severe in the heavily female-biased NZM2328 model. The aberrant X-linked gene dosage in female NZM2328 mice may contribute towards the development of female-biased immune responses in SLE-prone hosts. These findings provide important insights into the epigenetic mechanisms contributing to female-biased autoimmunity.
Keywords: X-Chromosome Inactivation, female-biased lupus-like disease, Xist RNA, B cells, T cells, MRL/lpr mice, NZM2328 mice
INTRODUCTION:
Systemic lupus erythematosus (SLE) preferentially afflicts women, with both hormonal and genetic factors contributing to this female bias [1,2]. X-chromosome-linked genetic factors have been heavily implicated in the pathogenesis and female bias of SLE [3], as the X chromosome is enriched for a number of genes with relevant immune functions, including TLR7, CXORF21 (TASL), CD40L, CXCR3, IRAK1, and FOXP3 [4–7]. Furthermore, several SLE susceptibility loci are X-linked [4,5,7], and increased X chromosome copy numbers are associated with SLE disease risk in humans and disease progression in animal models of SLE, presumably through the aberrant dosage of these and other X-linked genes [8–11]. Collectively, these data suggest that X-linked gene dosage is an important contributor to the pathogenesis and female susceptibility of SLE, but the mechanisms underlying the aberrant overexpression of X-linked genes in XX females require further elucidation.
X-Chromosome Inactivation (XCI) is an X-chromosome-specific epigenetic regulatory mechanism that equalizes X-linked gene expression between the sexes by transcriptionally silencing supernumerary X chromosomes. In most XX female somatic cells, transcriptional silencing of the inactive X (Xi) chromosome is maintained through enrichment of Xist RNA, an X-linked long non-coding RNA, and heterochromatic modifications, including H3K27me3, H2AK119-ubiquitin, and DNA methylation, at the Xi [12–15]. Mammalian lymphocytes, however, exhibit “dynamic” XCI maintenance (dXCIm) [16], where naïve lymphocytes lack focal enrichment of Xist RNA and heterochromatic histone marks on the Xi, despite continuous Xist transcription [16–20]. Upon lymphocyte activation, Xist RNA and these heterochromatic marks dynamically relocalize to the Xi, independent of changes in Xist transcription [16–20]. This process of dXCIm is impaired in circulating CD3+ T cells and CD19+ B cells from both pediatric and adult females with SLE, where it is associated with the aberrant expression of X-linked genes [17,20]. Furthermore, B and T lymphocytes from the female-biased NZB/W F1 mouse model of spontaneous lupus-like disease also exhibit impaired dXCIm, specifically in temporal association with the onset of serologic autoimmunity and glomerulonephritis [18]. The impaired relocalization of XIST/Xist RNA to the Xi likely has important functional implications for immune cells, as Xist deletion in murine hematopoietic cells or a human EBV-transformed B cell line (GM12878) results in the overexpression of many X-linked genes, including Tlr7/TLR7, TASL (CXORF21), and Cxcr3/CXCR3 [12,13]. Collectively, these data suggest that impaired dXCIm in lymphocytes is a feature of SLE pathogenesis that may facilitate the expression of proinflammatory X-linked genes from the Xi, thereby contributing to female-biased disease. However, because the NZB/W F1 murine model of spontaneous lupus and human SLE are both heavily female-biased, it is unclear if impaired dXCIm is specific to female-biased models of lupus-like disease, or if it is also observed in models of lupus-like disease that lack a strong female bias. Moreover, both T and B lymphocytes demonstrated impaired dXCIm in these studies, suggesting that disease indiscriminately perturbs dXCIm. Thus, it remains unclear whether impaired dXCIm in a single lymphocyte subset could alter lymphocyte function and contribute to the development of lupus-like disease.
In this study, we sought to determine (1) whether impaired dXCIm is specific to female-biased models of lupus-like disease; and (2) whether simultaneously impaired dXCIm in both T and B cells is a conserved feature across other models of spontaneous lupus-like disease. We examined the epigenetic features of the Xi at the single-cell level in lymphocytes from two murine models of spontaneous lupus-like disease—the NZM2328 model [21], which is heavily female-biased, and the MRL/lpr model, where disease onset and severity are only modestly female-biased [22]. We hypothesized that dXCIm would be impaired in both lymphocyte subsets from the heavily female-biased NZM2328 model of lupus-like disease and intact in lymphocytes from the less female-biased MRL/lpr model, and furthermore that the impairment would occur in temporal association with disease onset.
MATERIALS AND METHODS:
Mouse models
C57BL/6J mice (B6; Strain No. 000664) and MRL/lpr mice (Strain No. 000485) were obtained from the Jackson Laboratory and maintained at the University of Pennsylvania School of Veterinary Medicine. NZM2328 mice were bred and maintained at the University of Southern California. Animals were euthanized using carbon dioxide at three specified age ranges (8–10 weeks, 18–22 weeks, 23–34 weeks) based on the expected timing of disease onset to permit analyses over the course of disease development [21–23]. B6 and MRL/lpr mouse experiments were approved by the University of Pennsylvania Institutional Animal Care and Use Committee (IACUC); approval for NZM2328 animals was obtained through the IACUC at the University of Southern California.
Determination of Disease Activity
Mouse age was used as a proxy for disease activity given the well-established timeframes of disease onset in these spontaneous models [21–23]. To confirm the development of lupus-like disease with mouse age, serum anti-double-stranded DNA antibody concentrations were quantified via enzyme-linked immunosorbent assays performed on serum collected via cardiac puncture at the time of euthanasia, as described previously [18]. Briefly, Immulon 2 high-binding plates (ThermoFisher) were coated with native dsDNA purified from calf thymus (Alpha Diagnostic International) and blocked using bovine serum albumin. Serial dilutions of an anti-dsDNA-antibody-containing stock solution were used to create a standard curve, and sera were sufficiently diluted to ensure that the measured concentrations were within the range of the standard curve. Plates were then treated with HRP-conjugated goat anti-mouse IgG and were subsequently developed using TMB Chromogen Solution and TMB Stop Solution and analyzed at a wavelength of 450 nm.
Isolation and Activation of Splenic B and T Cells
Spleens from B6 and MRL/lpr mice were harvested immediately after euthanasia. Spleens from NZM2328 mice were shipped overnight on ice from the University of Southern California (within 24 hours of euthanasia). CD23+ B cells and CD3+ T cells were isolated from female C57BL/6 (B6), MRL/lpr, and NZM2328 mice over multiple, independent experiments, as described previously [16–18]. Briefly, splenic tissue was mechanically homogenized and subsequently incubated with biotinyated anti-mouse CD23 antibody (BioLegend) and streptavidin microbeads (Miltenyi Biotec). B cells were subsequently purified via MACS® with typically >88% CD19+CD23+ of all live single cells sampled. CD3+ T cells were subsequently purified via negative selection of the resulting CD23- cellular fraction using CD3+ T-cell enrichment columns (R&D Systems; typically >90% CD3+ of all live single cells sampled). Suspensions of purified B and T lymphocytes at time 0 hours (“non-activated”) were allocated for slide preparation for sequential Xist RNA fluorescence in situ hybridization (FISH) and H3K27me3 immunofluorescence (IF), and suspended in TRIzol for qPCR. In vitro activation of lymphocytes was performed for 24 hours (B cells) or for 48 hours (T cells), in the presence of 1 μM CpG (Invivogen) or plate-bound anti-CD3 with soluble anti-CD28 (Bio X Cell), respectively. In vitro-activated cells were then partitioned for sequential Xist RNA FISH and H3K27me3 IF and suspended in TRIzol for qPCR and RNAseq.
Xist RNA Fluorescence in Situ Hybridization (FISH) and Immunofluorescence (IF)
Purified non-activated and in vitro-activated splenic CD23+ B and CD3+ T cells were processed for sequential Xist RNA FISH and H3K27me3 IF as described previously [16,17]. Briefly, Xist RNA FISH was performed using Cy3-labeled oligonucleotide probes for Xist RNA. Nuclei were counterstained using VECTASHIELD® with DAPI. Nuclei were imaged using a Nikon Eclipse Microscope and were assigned one of four Xist RNA localization patterns based on the distribution of Xist RNA transcript locations [17]. The Xist RNA localization score is continuous variable (1–4; 1= Xist RNA is absent or entirely dispersed from the Xi; 4 = Xist RNA is tightly associated with the Xi) calculated as a weighted average of the proportions of four different Xist RNA localization patterns within ≥ 100 nuclei per biological replicate (Supplementary Figure 1). Xist RNA localization patterns for all samples were quantified by one of two reviewers (minimum linearly weighted kappa 0.75). Sequential H3K27me3 IF was performed after Xist RNA FISH using rabbit polyclonal anti-H3K27me3 antibody (Active Motif) and AF488-conjugated goat anti-rabbit IgG. The proportion of nuclei with H3K27me3 foci was determined based on the number of nuclei within the same fields assessed during Xist RNA FISH analysis that exhibited the presence of H3K27me3 IF signal and was quantified across all samples by a single reviewer.
RT-qPCR
RNA was isolated from non-activated and in vitro-activated B and T cells suspended in TRIzol. Briefly, cDNA was synthesized using qScript DNA SuperMix (QuantaBio) and quantified using a NanoDrop™ spectrophotometer. PerfeCTa SYBR Green SuperMix (QuantaBio) was used for qPCR reaction mixtures. Fold-change expression of a gene of interest (2 −ΔΔCT) was determined based on the ΔCT value of that gene in cells (non-activated or 48-hour activated CD3+ T cells, non-activated or 24-hour activated CD23+ B cells) from MRL/lpr or NZM2328 mice relative to the average ΔCT value in matched cell populations from female B6 mice (ΔΔCT), accounting for the expression of the housekeeping gene, Rpl13a, in all animals. The primer pairs (5’−3’) used in this study were: Btk (F: GAGGAGAGGTGAGGAGTCTAGT; R: AGCTCTTCAGTTGGGGAGAAAA); Cd40l (F: ACACGTTGTAAGCGAAGCCAA; R: TCCCGATTAGAGCAGAAGGTG); Cfp (F: TTCACCCAGTATGAGGAGTCC; R: GCTGACCATTGTGGAGACCT); Cxcr3 (F: TACCTTGAGGTTAGTGAACGTCA; R: CGCTCTCGTTTTCCCCATAATC); Il2rg (F: CTCAGGCAACCAACCTCAC; R: GCTGGACAACAAATGTCTGGTAG); Irak1 (F: TCCTCCACCAAGCAGTCAAG; R: AAAACCACCCTCTCCAATCCT); Itm2a (F: TTGCCTCATACTTATGTGCTTCG; R: GCGGAAGGATTTTCGGTTGTTG); Mpp1 (F: TGAGGAAGTTGTTCGGCTCC; R: CTTCTTTGGTGGCCGTGTTG); Msn (F: GGCTTCCCGTGGAGTGAAATC; R: GTCCGGGGCCTTTTTGTCAA); Ogt (F: TTCGGGAATCACCCTACTTCA; R: TACCATCATCCGGGCTCAA); Rps6ka3 (F: TGGCCCTGATGATACTCCAGA; R: GCCGTCAGTCTCTGATGAGG); Tlr7 (F: ATGTGGACACGGAAGAGACAA; R: GGTAAGGGTAAGATTGGTGGTG); Was (F: CCAGCCGTTCAGCAGAACAT; R: GGTTATCCTTCACGAAGCACA); Xist (F: CAGAGTAGCGAGGACTTGAAGAG; R: GCTGGTTCGTCTATCTTGTGGG); Rpl13a (F: AGCCTACCAGAAAGTTTGCTTAC; R: GCTTCTTCTTCCGATAGTGCATC).
RNAseq Sample Preparation and Bioinformatic Analysis
RNA isolated from in vitro-activated CD3+ T cells from 3 B6 female, 5 NZM2328 female, and 3 NZM2328 male mice (18–24 weeks of age) was used for RNAseq. High quality RNA samples (RIN ≥ 8.6) were selected for library construction using the Illumina TruSeq Stranded mRNA Library Preparation kit. Pooled libraries were run on two High Output (75 cycles) flow cells on an Illumina NextSeq 500 sequencer, generating an average read depth of 24 million reads per sample. RNAseq data were analyzed using RStudio (v4.1.1) and Bioconductor 7 [24]. Raw reads were first aligned to the mouse reference genome (EnsDb.Mmusculus.v79) using Kallisto (v0.46.0) and ensembldb (v2.16.4) [25,26]. Transcript counts were generated at the gene level using tximport (v1.20.0) [27]. edgeR (v3.34.1) was used to filter and then normalize the data via the TMM method [28]. Differentially expressed genes were identified using Limma (v3.48.3) [29]. Data have been deposited in the Gene Expression Omnibus (GEO) database (accession number GSE213318). The list of genes encoding Xist RNA interacting proteins was assembled based on previously published work [12,17,30].
Data Visualization
Data were analyzed and visualized using gplots and Prism 9 software (Graphpad). Fluorescence microscopy and image acquisition were performed using Nis-Elements software.
Statistical Analyses
The mean Xist RNA localization scores, percentage of cells with H3K27me3 foci, and log fold-change of gene expression were compared between two groups using the Student’s t-test for normally-distributed data and the Mann-Whitney U tests for non-normally-distributed data. The D’Agostino and Pearson test was used to determine normality for equivocal distributions. Comparisons across three or more groups were performed using a non-equal variance one-way ANOVA with Dunnett’s T3 multiple comparisons testing for normally-distributed data and the Kruskall-Wallis test with Dunn’s multiple comparisons testing for non-normally distributed data. Statistical significance was defined as p < 0.05. For RNAseq analysis, genes exhibiting a log2 fold-change ≥ 0.5 with a Benjamini-Hochberg-adjusted p-value (FDR) < 0.05 were considered differentially expressed. The total number of animals used for each experiment is indicated in the corresponding figure legend. Data are pooled from multiple independent experiments.
RESULTS:
Dynamic Xist RNA Localization is Preserved in Splenic B Cells from Both MRL/lpr and NZM2328 Mice
To determine whether impaired dXCIm in B lymphocytes is a general feature of spontaneous murine lupus independent of a strong female bias, we first characterized the epigenetic features of the Xi in splenic CD23+ follicular B cells from MRL/lpr female mice, and compared them to those from NZM2328 female mice at ages spanning distinct stages of disease (Figure 1A). We observed a positive association between serum anti-dsDNA antibody concentrations and age in both MRL/lpr and NZM2328 mice, in agreement with the expected timing of disease onset (Supplementary Figure 2) [21,31]. Using RNA FISH, we examined the nuclear localization patterns of Xist RNA in naïve, non-activated and in in vitro-activated B cells from female C57BL/6 (B6), MRL/lpr, and NZM2328 mice. Non-activated B cells from age-matched female B6, MRL/lpr, and NZM2328 mice lacked Xist RNA localization at the Xi and exhibited similar Xist RNA localization scores (Figure 1B–C). B cells from B6, MRL/lpr, and NZM2328 mice that were activated in vitro for 24 hours using CpG oligonucleotides exhibited similar levels of dynamic Xist RNA relocalization to the Xi (Figure 1B–C). In contrast to the Xist localization scores from activated B cells from NZB/W F1 mice [18], those from female MRL/lpr and NZM2328 mice did not vary with age/disease status (Supplementary Figure 3). By performing H3K27me3 IF, we found that in vitro-activated B cell nuclei from B6, MRL/lpr, and NZM2328 mice exhibited similar proportions of nuclei with H3K27me3 foci (Figure 1D–E). As with the Xist RNA localization scores, the proportion of H3K27me3 foci in activated B cells was independent of mouse age in all strains (Supplementary Figure 3).
Figure 1. Xist RNA and H3K27me3 localization to the Xi are intact in activated CD23+ B cells from female MRL/lpr and NZM2328 mice.
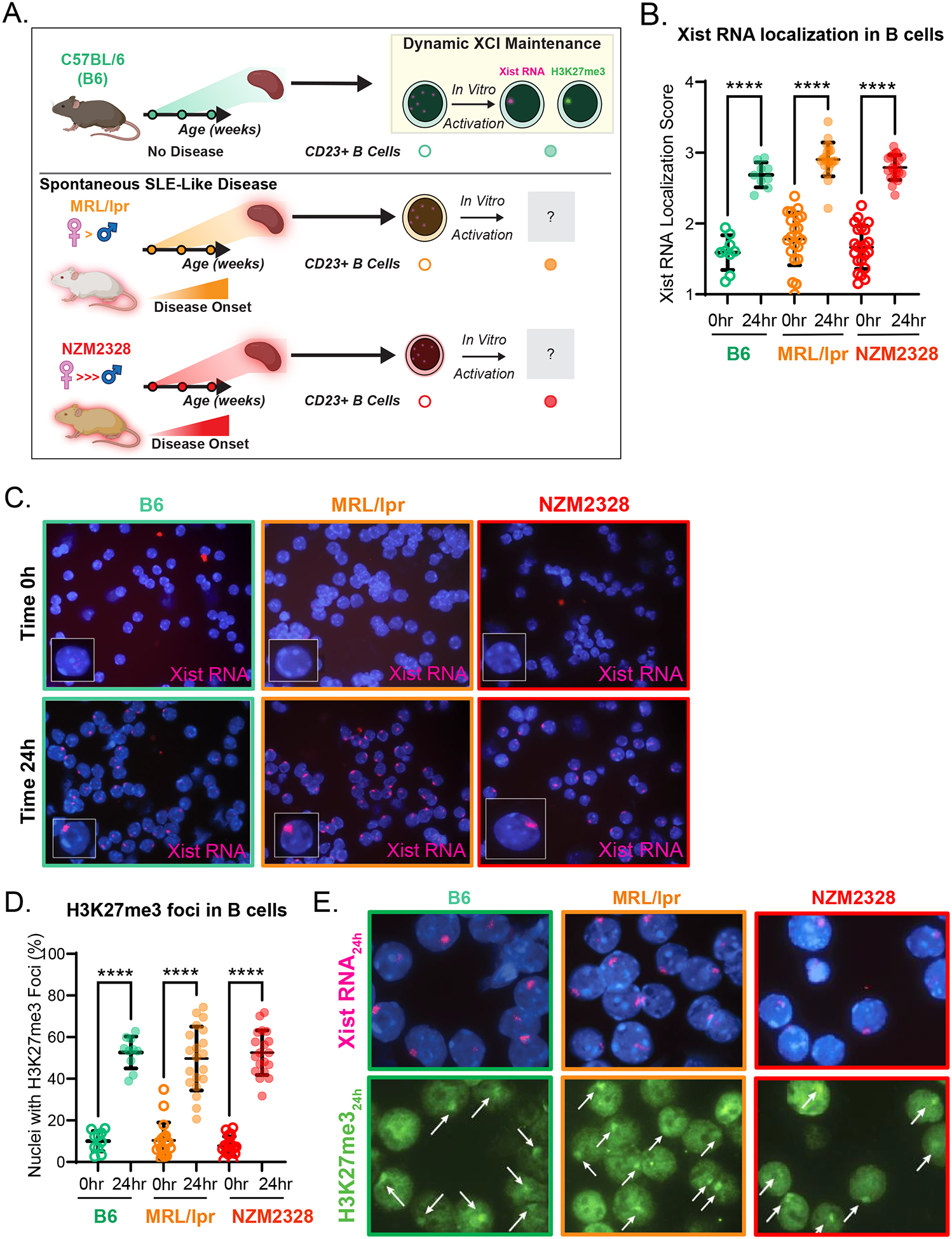
A. Schematic of the experimental approach. B. Mean Xist RNA localization scores ± SD of non-activated (0hr; open circles) and 24-hour-CpG-activated CD23+ splenic B cells (24hr; filled circles) from 8–31 week-old female C57BL/6 (B6) mice (n=9; teal), female MRL/lpr mice (before and during disease; n=21; orange), and female NZM2328 mice (before and during disease; n=21–22; red). C. Representative Xist RNA FISH images from non-activated (Time 0h) and 24-hour-CpG-activated (Time 24h) CD23+ splenic B cells from female B6 (teal border), female MRL/lpr (orange border), and female NZM2328 mice (red border). D. Mean proportion of nuclei ± SD with H3K27me3 foci from non-activated (0hr; open circles) and 24-hour-CpG-activated (24hr; filled circles) CD23+ splenic B cells from 8–31 week-old female B6 mice (n=10; teal), female MRL/lpr mice (n=18–20; orange), and female NZM2328 mice (n=19; red). E. Representative sequential Xist RNA FISH and H3K27me3 IF images for B6 (teal border), MRL/lpr (orange border), and NZM2328 mice (red border). White arrows depict H3K27me3 foci. * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.
XCI Maintenance Remains Intact in Splenic B Cells from MRL/lpr and NZM2328 Mice
Previous work has shown that the localization of Xist RNA in in vitro activated B and T cells from female B6 mice is independent of changes in Xist transcription between the naïve and activated cellular states [16]. Given the robust localization of Xist RNA observed in B cells from female MRL/lpr and NZM2328 mice, we wanted to confirm that this localization was independent of marked increases in Xist transcript levels relative to B6 mice. Using qPCR, we measured the fold-change in Xist expression in female MRL/lpr and NZM2328 mice relative to B6 mice (Figure 2). Xist expression in activated B cells from both MRL/lpr and NZM2328 females was similar to that of B6 females (Figure 2A). The trend towards a modest increase in Xist expression in activated B cells from MRL/lpr, but not NZM2328 mice, suggests that there could be subtle strain-specific differences in Xist expression. To confirm that broader range of Xist expression values in activated B cells from MRL/lpr was not associated with changes in Xi-linked gene expression, we compared the expression of X-linked immunity-related genes broadly distributed across the X chromosome between female (XaXi) and male (XaY) MRL/lpr mice, relative to B6 mice (Figure 2B). This approach provides an indirect approximation of XCI status and Xi-linked gene expression [32,33]. We did not detect any female-biased expression of the X-linked genes, suggesting that X-linked dosage compensation is maintained in MRL/lpr mice, in agreement with the robust localization of Xist RNA to the Xi (Figure 2C). Thus, the overall fidelity of dXCIm is generally preserved in B cells from both models of spontaneous SLE-like disease, regardless of the extent of female-biased disease.
Figure 2. X-linked gene expression is not affected by lupus-like disease in in vitro activated CD23+ B cells from female MRL/lpr mice.
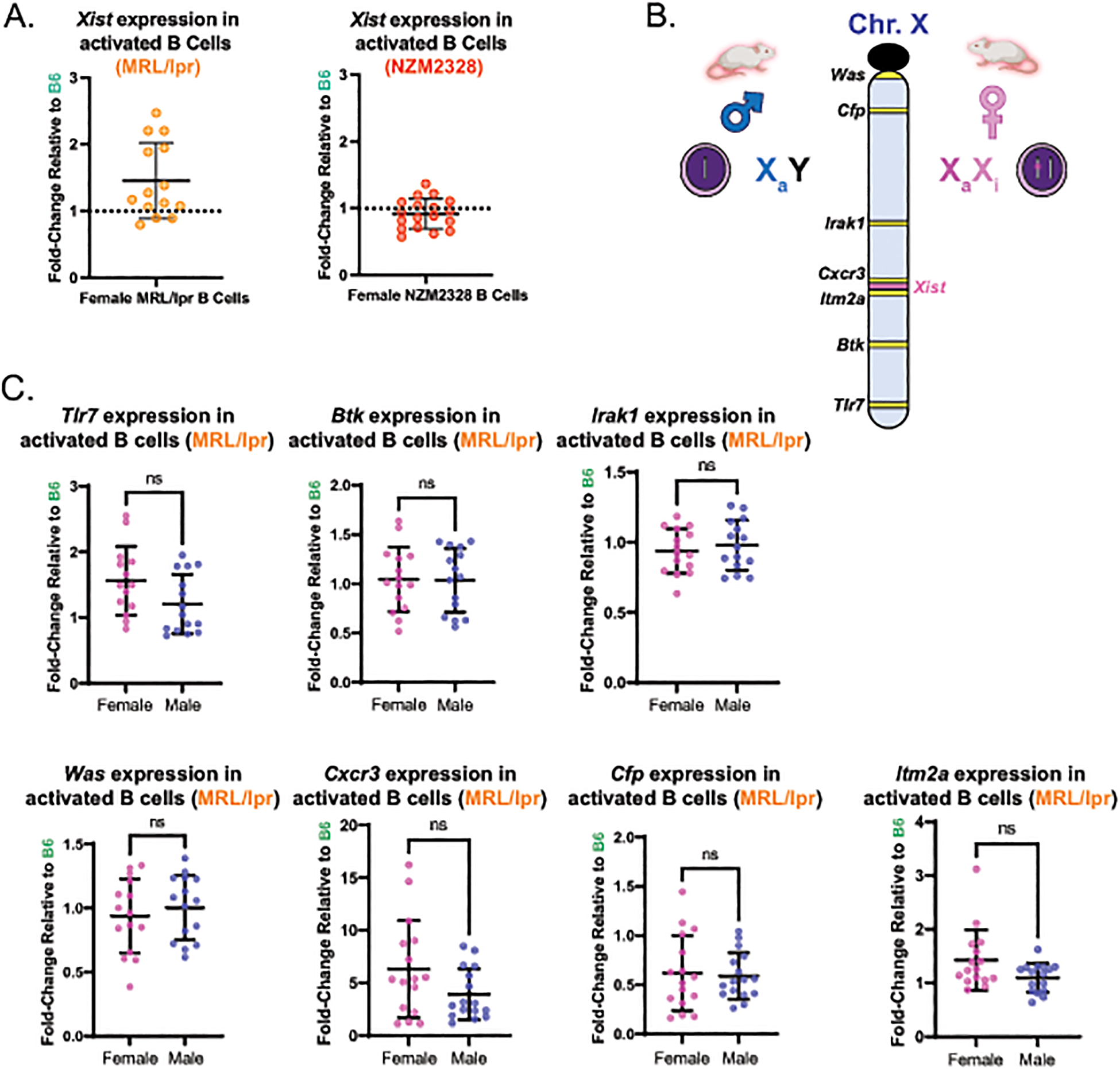
A. Mean fold-change ± SD of Xist expression in CD23+ B cells at 24-hours post-activation with CpG from female MRL/lpr mice (n=14) relative to female B6 mice (n=9) and from female NZM2328 mice (n=19) relative to female B6 (n=10). B. Schematic depicting the approach used to infer gene expression from the Xi. The locations of the X-linked genes used for qPCR analyses in panel C is shown. C. Rpl13a-normalized expression for seven X-linked genes with immune functions and that are distributed broadly over the X chromosome in activated CD23+ B cells from MRL/lpr mice, determined via qPCR. The mean fold-change (± SD) for a given gene was calculated based its expression in 24-hour-CpG-activated CD23+ splenic B cells from 8–31 week-old female MRL/lpr mice (n=14–16) and age-matched male MRL/lpr mice (n=15–16) relative to female B6 mice (n=9–10) using the 2−ΔΔCt approach. ns = not statistically significant.
The Dynamic Localization of Xist RNA to the Xi is Impaired in Activated T Cells from MRL/lpr Mice and is More Dramatically affected in T Cells from NZM2328 Mice, Where It Precedes the Development of Serologic Autoimmunity
Next, we asked whether dXCIm was altered in splenic CD3+ T cells from MRL/lpr and NZM2328 mice. Xist RNA localization scores were similar amongst non-activated CD3+ T cells from B6, MRL/lpr, and NZM2328 female mice (Figure 3A–B). However, the Xist RNA localization scores in in vitro-activated T cells were significantly decreased in MRL/lpr mice relative to B6 mice, and were even more significantly decreased in NZM2328 mice relative to both MRL/lpr and B6 mice (Figure 3A–B). Using sequential RNA FISH/IF, we found that the in vitro-activated T cells from both MRL/lpr and NZM2328 mice also exhibited a lower mean proportion of nuclei containing H3K27me3 foci (Figure 3C–D). As observed in T cells from NZB/W F1 female mice [17], the impairment in Xist RNA localization in activated T cells from MRL/lpr mice was observed in older animals, in temporal association with the development of serologic autoimmunity (Figure 3E). Surprisingly, T cells from NZM2328 mice exhibit impaired Xist RNA localization well before the development of serologic autoimmunity, as females 8–10 weeks of age demonstrated significantly lower Xist RNA localization scores compared to MRL/lpr, B6, and NZB/W F1 mice [21]. Taken together, these data suggest that splenic T cells from NZM2328 female mice possess an intrinsic alteration in the epigenetic modifications at the Xi that precedes the development of lupus-like disease.
Figure 3. Dynamic localization of Xist RNA and H3K27me3 is impaired in in vitro activated CD3+ T cells from both MRL/lpr and NZM2328 female mice.
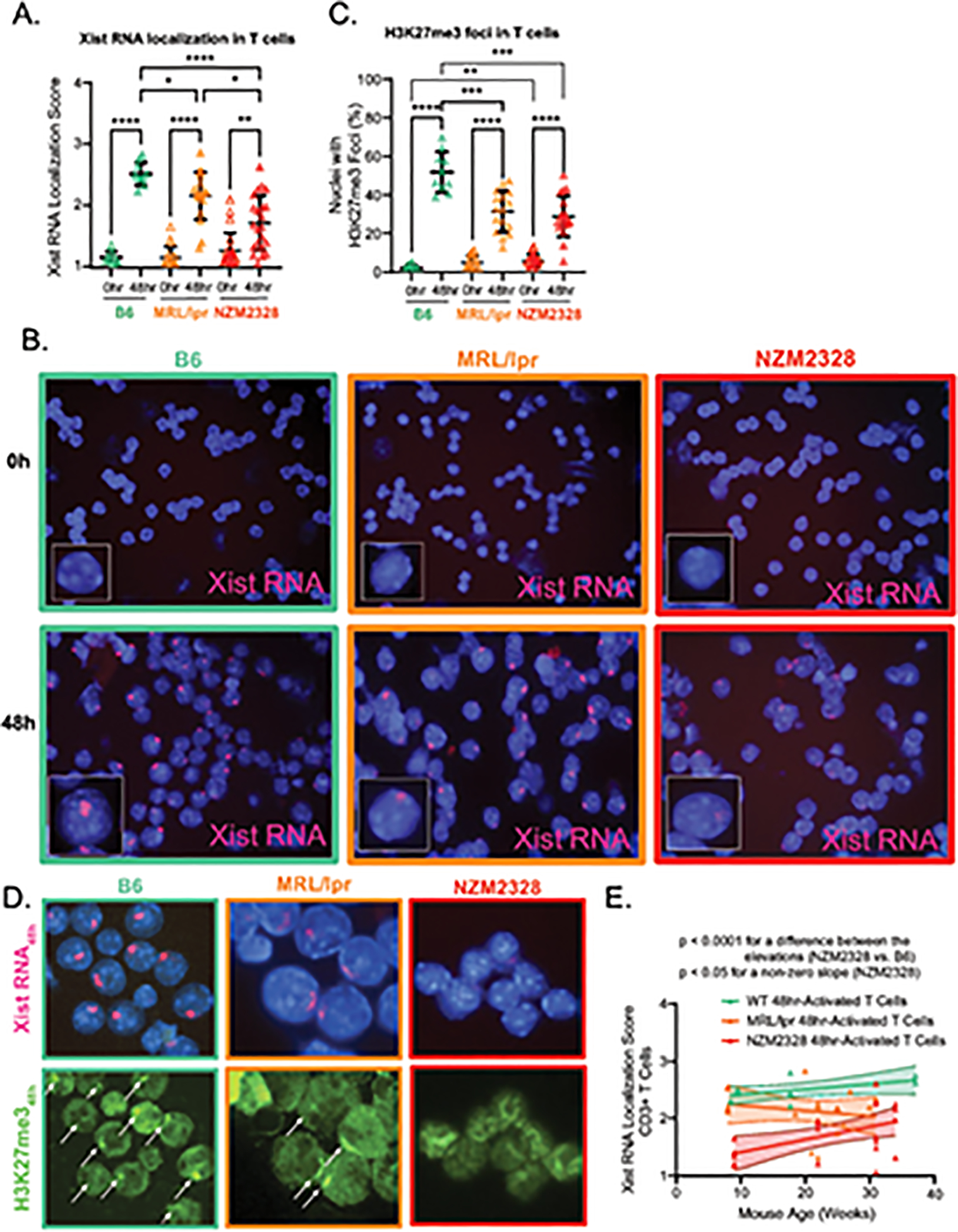
A. Mean Xist RNA localization score ± SD of non-activated (0hr; open triangles) and 48-hour-anti-CD3/CD28-activated CD3+ splenic T cells (48hr; filled triangles) from 8–34 week-old age-matched female B6 (n=10; teal), MRL/lpr (n=18; orange), and NZM2328 mice (n=22; red). B. Representative Xist RNA FISH images from non-activated (Time 0h) and 48-hour-anti-CD3/CD28-activated CD3+ splenic T cells (Time 48h) from female B6 (teal border), MRL/lpr (orange border), and NZM2328 mice (red border). C. Mean proportion of nuclei ± SD with H3K27me3 foci from non-activated (0hr; open triangle) and 48-hour-anti-CD3/CD28-activated CD3+ splenic T cells (48hr; filled triangle) from 8–34 week-old female B6 (n=10; teal), MRL/lpr mice (n=16; orange), and NZM2328 mice (n=19; red). D. Representative sequential Xist RNA FISH and H3K27me3 IF images for B6 (teal border), MRL/lpr (orange border), and NZM2328 (red border) mice. Solid arrows depict H3K27me3 foci. E. Linear regression of mouse age (in weeks) versus Xist RNA localization score for each mouse for stimulated CD3+ T cells from female B6 (teal), MRL/lpr (orange), and NZM2328 mice (red), shown with their 95% confidence intervals. * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.
Activated T Cells from NZM2328 Female Mice Exhibit Aberrant Expression of Immunity Related X-linked Genes
Because in vitro-activated T cells from both MRL/lpr and NZM2328 mice exhibited impaired Xist RNA localization, we hypothesized that X-linked gene expression chromosome-wide may be dysregulated. We first sought to confirm that the impairment in Xist localization was not associated with marked decreases in Xist expression relative to B6 mice. Given the observed relationships between Xist RNA localization and age (Figure 3E), we performed an age-stratified analysis of Xist expression in activated T cells from both MRL/lpr and NZM2328 female mice. In general, we found that Xist expression was either similar to or slightly higher than that observed in B6 females, further supporting the concept of an uncoupling of Xist RNA localization and Xist transcript abundance (Figure 4A). Because Xist RNA localization was moderately impaired specifically within in vitro-activated T cells from older MRL/lpr mice, we compared the expression of several X-linked genes with immune functions relevant to this model and lupus pathogenesis, including Cxcr3 [6,34–36] and Cd40l [37], between activated T cells from older female MRL/lpr and age-matched male MRL/lpr mice (Figure 4B). Notably, we did not identify any female-biased gene expression in these animals, suggesting that the observed magnitude of impairment in Xist RNA localization may not be sufficient to influence the expression of these genes, some of which have exhibited variable escape from XCI in T cells or female-biased expression in T cells from patients with SLE [3,6,37].
Figure 4. Lupus-like disease does not alter X-linked gene expression in in vitro activated CD3+ T cells from female MRL/lpr mice.
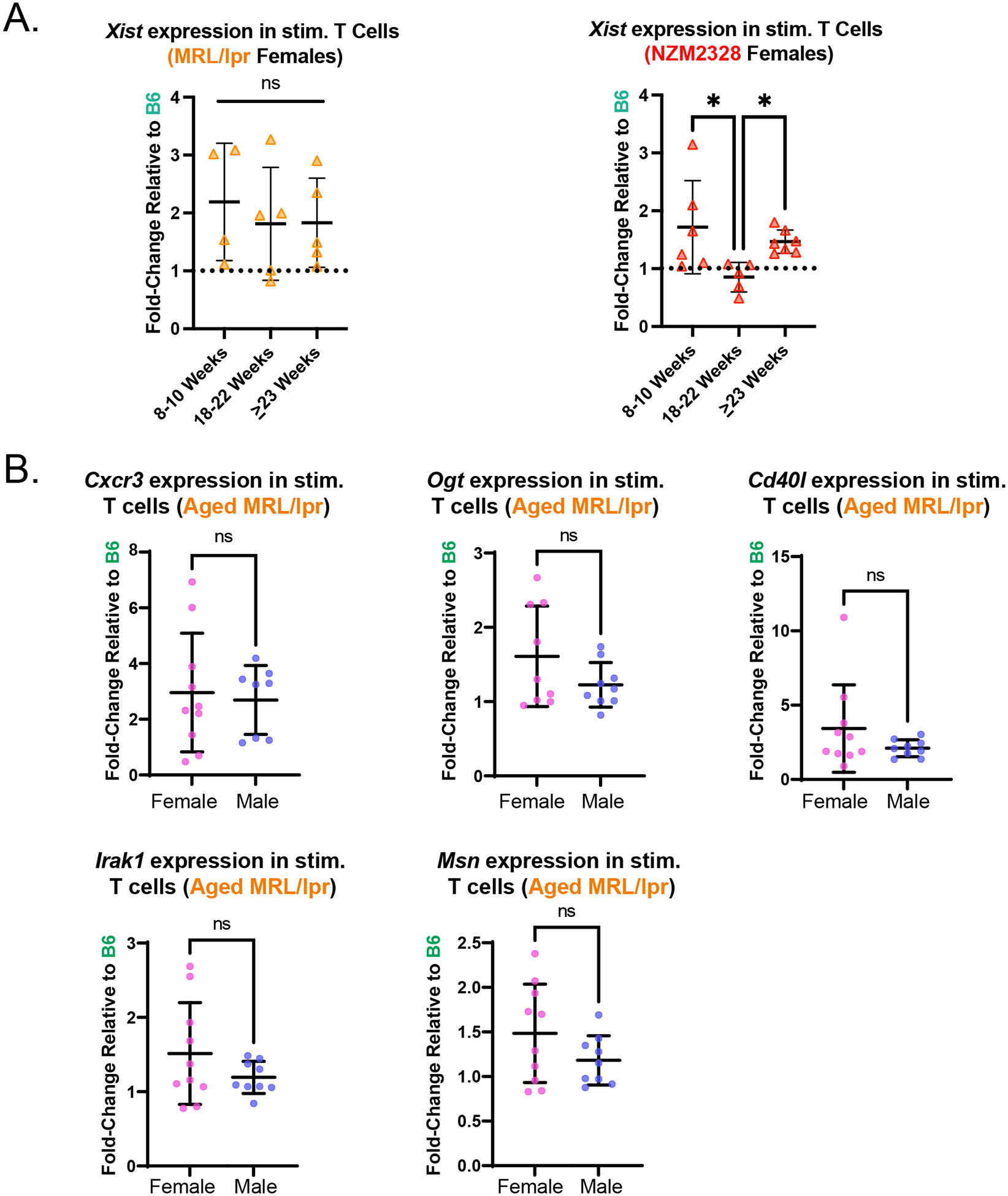
A. Mean fold-change ± SD of Xist expression at 48-hours post-activation with anti-CD3/anti-CD28 relative to 0h (non-activated) for CD3+ T cells from female MRL/lpr (n=14) and NZM2328 mice (n=18), relative to female B6 mice (n=10), stratified by mouse age based on data in Figure 3E. B. Rpl13a-normalized expression for five X-linked genes with relevant immune functions (Cxcr3, Ogt, Cd40l, Irak1, Msn) in 48-hr activated T cells from MRL/lpr mice, determined via qPCR. The mean fold-change (± SD) for a given gene was calculated based on its expression in 48-hour-anti-CD3/CD28-activated CD3+ T cells from 18–31 week-old female MRL/lpr mice (n=9–10) and age-matched male MRL/lpr mice (n=8–9) relative to female B6 mice (n=6). 18–31-week-old mice were specifically selected for this analysis as this age range exhibited impaired Xist RNA localization (Figure 3E). ns = not statistically significant.
Notably, Xist RNA localization was more severely and chronically impaired in activated T cells from the female-biased NZM2328 model. We therefore hypothesized that the magnitude and persistent Xist RNA localization impairment observed in these animals may perturb XCI maintenance and ultimately confer a predisposition towards the development of female-biased disease. To examine X-linked gene expression chromosome-wide, we performed RNAseq on in vitro-activated T cells from NZM2328 female and male mice (aged 18–22 weeks, prior to ages associated with overt disease), and age-matched B6 female mice, and observed differential expression of X-linked genes that clustered according to sex and strain (Figure 5A–B). We identified 32 X-linked genes which were upregulated and 17 X-linked genes which were downregulated in in vitro-activated T cells from NZM2328 females relative to B6 females and NZM2328 males (Figure 5C, gene lists shown in Supplemental Figure 4). Gene Ontology (GO) analysis of the downregulated X-linked genes did not yield any consensus biological processes (data not shown). GO analyses of the differentially upregulated X-linked genes in NZM2328 females revealed that most of these genes were implicated in inflammatory responses (Figure 5D). T cells from NZM2328 mice exhibited female-biased increased expression of a number of X-linked genes with immune functions, including Cxcr3, Foxp3, Tlr8, Il13ra1, as well as the known XCI escape gene Kdm6a (Figure 5C–D, Supplementary Figure 4) [38]. These genes are distributed broadly across the length of the X chromosome, yet the majority are distributed adjacent to the centromere, distal to the Xist locus (Figure 5E). Thus, activated T cells from NZM2328 mice exhibiting a marked reduction in epigenetic modifications at the Xi also demonstrate female-specific aberrant overexpression of a number of X-linked genes with relevant immune functions.
Figure 5. Activated T cells from NZM2328 mice exhibit female-biased expression of X-linked genes with immune functions.
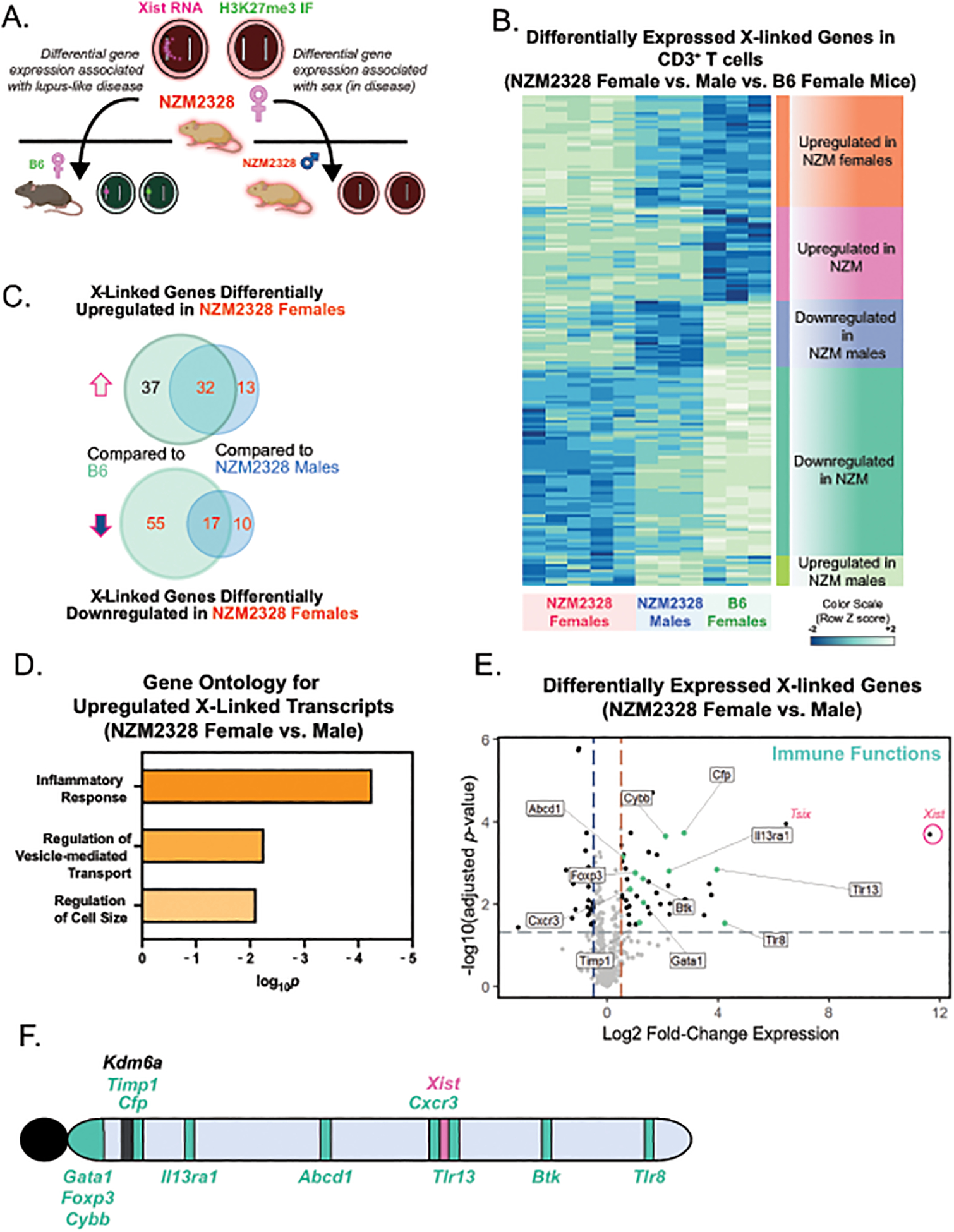
A. Schematic for the experimental approach to determine sex-specific disease expression profiles using in vitro activated T cells from 18–24-week old NZM2328 male (n=3), NZM2328 female (n=5), and B6 female (n=3) mice. B. Heatmap of differentially expressed X-linked genes for in vitro activated T cells from NZM2328 female and male mice and B6 female mice. C. Venn-diagrams showing the number of differentially upregulated and downregulated X-linked genes in in vitro activated T cells from NZM2328 female mice relative to B6 female or NZM2328 male mice. D. Gene Ontology (GO) analyses for Biological Processes of upregulated X-linked genes in in vitro activated T cells comparing NZM2328 female and NZM2328 male mice. E. Volcano plot of differentially expressed X-linked genes in activated T cells from NZM2328 female relative to age-matched NZM2328 male mice. X-linked genes with known immune functions are colored in teal. F. Location of the differentially upregulated immune-related X-linked genes across the X chromosome. The known XCI escape gene, Kdm6a, which is also overexpressed in NZM2328 females, is also shown in black.
Activated Splenic T Cells from NZM2328 Mice Exhibit Aberrant Expression of Genes Encoding Xist RNA-Interacting Proteins
The mislocalization of Xist RNA from the Xi in activated T cells from NZM2328 females may reflect underlying defects in Xist RNA tethering and retention across the Xi. Such functions are mediated via a variety of Xist RNA binding proteins. Work from our lab and others found that deletion or reduced expression of various Xist RNA interacting proteins, including YY1, Ciz1, or hnRNPU, impairs both Xist RNA localization and heterochromatic mark enrichment at the Xi and results in abnormal X-linked gene expression [39–41]. To investigate the possibility that the impaired Xist RNA localization may be due to the aberrant expression of Xist RNA binding proteins, we assembled a list of 354 Xist RNA binding protein (“Xist RNA Interactome”) genes from mouse and human somatic cell types, including fibroblasts, differentiated embryonic stem cells, and a human B cell line [12,30,42,43]. We then compared the expression of these Xist RNA Interactome genes between activated T cells from aged (18–22 weeks) female NZM2328 mice and age-matched female B6 mice. We found that in vitro-activated splenic T cells from NZM2328 females exhibited differential expression of 26 Xist RNA Interactome genes relative to B6 females (Figure 6). Notably, about ~3/4 of these genes were significantly downregulated in T cells from NZM2328 mice, including Trim6, HnrnpA0, and Rad21, all of which have known roles in regulating XCI and Xi-linked gene expression [44–46]. Thus, the aberrant downregulation of Xist RNA Interactome genes may be partially responsible for the observed impairment in dXCIm in T cells from NZM2328 mice.
Figure 6. Activated CD3+ T cells from NZM2328 female mice exhibit aberrant expression of Xist RNA Interactome genes.
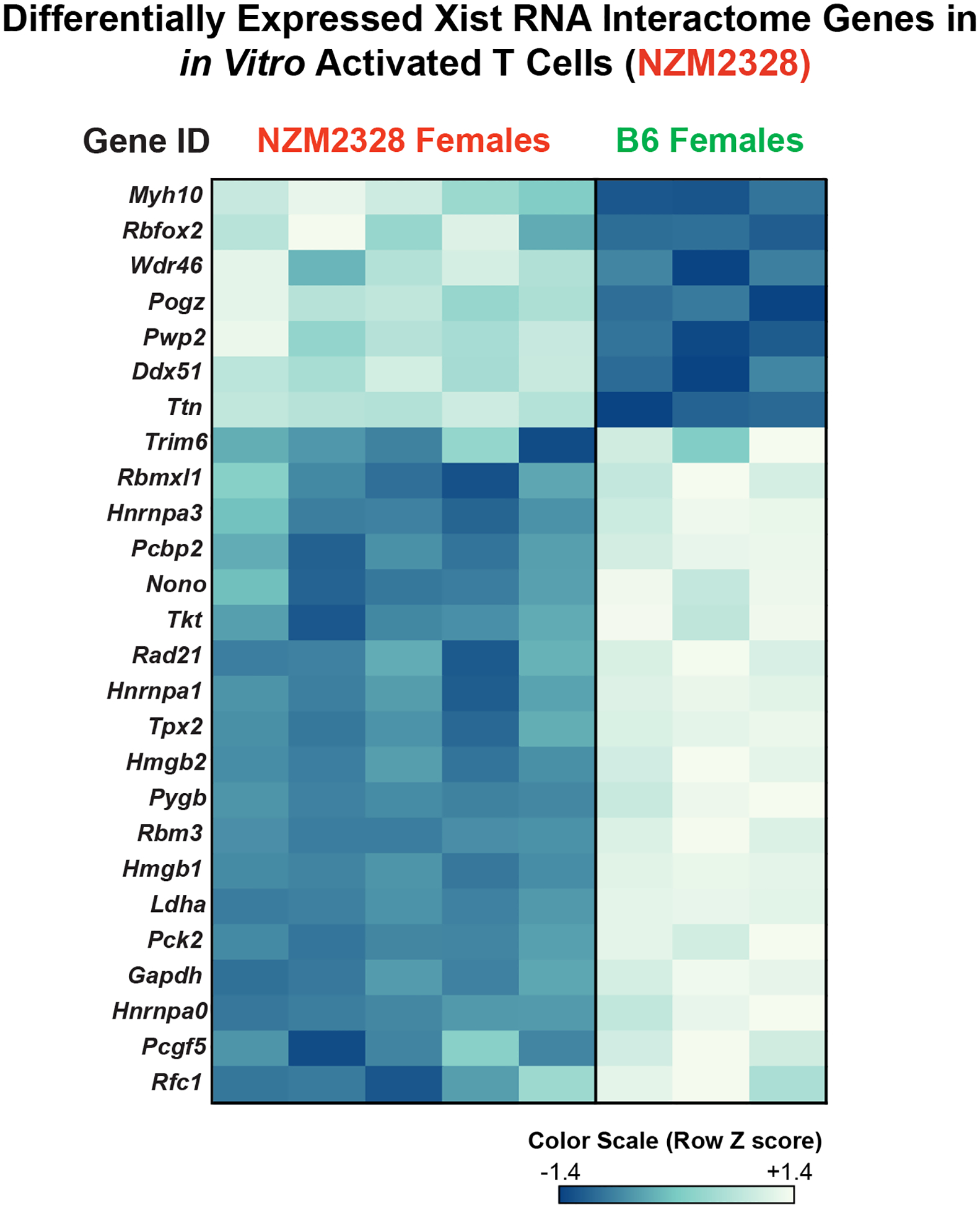
Heatmap of 26 differentially expressed Xist RNA binding protein genes between in vitro activated CD3+ T cells from NZM2328 and age-matched B6 female mice.
DISCUSSION
The molecular mechanisms underlying the female bias of SLE remain incompletely understood. We have previously shown that pediatric and adult patients with SLE exhibit impaired dynamic XCI maintenance in B and T lymphocytes [17,20], but it is not clear from these studies whether impaired dynamic XCI maintenance contributes to the development of female-biased disease, or if it is a consequence of autoimmune disease. By leveraging two murine models of spontaneous lupus-like disease with disparate levels of female bias, we investigated whether impaired dXCIm is a universal secondary feature of lupus-like disease, or whether it is restricted to those models exhibiting female-biased disease onset and severity. We found that splenic CD3+ T cells, but not splenic CD23+ B cells, have significantly reduced enrichment of Xist RNA and H3K27me3 at the Xi in both mouse models of lupus. However, we also found that the reduction in Xist RNA and H3K27me3 enrichment at the Xi was much more striking in activated T cells from the more heavily female-biased NZM2328 model, and furthermore, that this aberrant mislocalization originated before disease onset and persisted into adulthood. Our data suggest that while the impaired localization of Xist RNA and heterochromatic marks in activated T cells is not restricted exclusively to heavily female-biased mouse models of lupus, a sufficient magnitude of impairment may confer a propensity for female-biased disease, as Xist RNA mislocalization was much more severe in the female-biased NZM2328 model of lupus-like disease. Accordingly, we also discovered that in vitro-activated T cells from these mice exhibit female-specific overexpression of X-linked genes with roles in immune function, in conjunction with a predominant downregulation of specific Xist RNA Interactome genes. Collectively, these data suggest that the degree of dXCIm impairment increases with the amount of female-bias in models of lupus-like disease, and furthermore, that impaired dXCIm can occur selectively in specific lymphocyte populations and precede disease onset, thereby potentially conferring a propensity for female-biased disease.
We were surprised to find an impairment in the dynamic localization of Xist RNA in activated T cells from MRL/lpr mice, which is considered to be only modestly female-biased compared to other murine models of SLE-like disease [22]. CD3+ T cells play a critical role in the pathogenesis of the MRL/lpr model, as suggested by (1) their infiltration into target organs including the glomeruli [47]; (2) the delay in disease onset and improved survival upon treatment with anti-CD3 [48] and anti-CD4 monoclonal antibodies [49]; (3) the ability of double-negative (DN) T cells to induce glomerulonephritis in younger MRL/lpr mice [50]; and (4) their expansion within the spleen in temporal association with disease onset [31]. While we observed mislocalization of Xist RNA and heterochromatic marks from the Xi in T cells from MRL/lpr mice, we did not observe female-specific differences in the expression of 5 X-linked genes with immune functions (Figure 4B). It is plausible that the magnitude of impairment in dynamic Xist localization is not sufficient to result in marked changes in gene expression. One possible alternative explanation for this discrepancy is that our indirect approach to query XCI status by comparing female-to-male gene expression for the 5 selected genes was not sufficiently sensitive to detect subtle gene expression changes on the Xi, as the magnitude of Xi-linked expression amongst escape genes is typically lower than that of the active X [32,33,51]. It is also possible that other X-linked genes may be reactivated from the Xi, as we did not perform unbiased RNA sequencing using MRL/lpr samples. Instead, we selected X-linked genes that we considered likely to exhibit female-biased expression, including the chemokine receptor Cxcr3, which exhibits female-biased expression in CD4+ cells from patients with active lupus [6], and has been shown to escape XCI in T cells [34], promote Type 1 immune responses [36], and promote glomerulonephritis in this model [35,52]; and Cd40l, which has also been shown to exhibit female-biased expression in CD4+ T cells from patients with SLE and promotes glomerulonephritis in MRL/lpr mice [37,53,54]. Future studies of single activated CD4+ T cells from MRL/lpr mice investigating the degree of monoallelic and biallelic X-linked gene expression on the chromosome-wide level are necessary to definitively determine which genes may become upregulated from the Xi.
In contrast to the MRL/lpr model, the NZM2328 model exhibits a strong female bias in disease severity and onset. The NZM2328 model is a derivative of the NZB/W F1 model, another female-biased model of spontaneous lupus-like disease [21]. We previously observed impaired dXCIm in CD23+ B cells from female NZB/W F1 mice [18]; thus, we were surprised that dXCIm was largely preserved in CD23+ B cells from female NZM2328 mice. B cells play an important role in lupus pathogenesis in the NZM2328 model, as B cell-deficient NZM2328 mice are protected from glomerular pathology and death [55]. Our findings suggest that the mechanisms regulating dXCIm specifically within CD23+ B cells are not necessary for the development of female-biased lupus-like disease. While we specifically focused on CD23+ B cells in this study, it is possible that other splenic B cell subsets, such as memory B cells, may exhibit impaired dXCIm in NZM2328 mice.
T cells also play an important role in the pathogenesis of spontaneous lupus-like disease in NZM2328 mice, evidenced by the accumulation of activated T cells within diseased glomeruli, the perinephric lymph nodes, and spleens of these animals [23,55,56]. Interestingly, the preferential accumulation of activated T cells in these organs is female-biased and also X-chromosome dependent [11,56]. The mislocalization of epigenetic modifications at the Xi and concomitant female-biased expression of several proinflammatory X-linked genes further support the role of the X chromosome as an important contributor to female-biased disease in the NZM2328 model, and suggest that reductions in Xist RNA and H3K27me3 enrichment at the Xi may permit aberrant gene expression from the Xi in female samples. Notably, we identified a number of X-linked genes with relevant immune functions that were overexpressed in activated T cells from NZM2328 females compared to males. The female-biased expression of Cxcr3 is of particular interest because Cxcr3 can escape XCI in T cells and has a role in promoting both Th1 and Th17 immune responses [34], which are important in the pathogenesis of the NZM2328 model [36,56,57]. Notably, we also observed the female-biased expression of several other X-linked genes with immune functions, including Il13ra1, which is expressed by Th17-polarized cells and can modulate IL17A production [58]. Btk, which has been shown to play an important role in T-cell activation and proliferation and therefore in facilitating autoreactive immune responses [59] also exhibited female-biased expression. Tlr8 expression, which was also female-biased in NZM2328 T cells, has also been detected in murine CD4+ T cells, specifically in a subset of T cells containing regulatory T cells, which is noteworthy given that we also observed female-biased overexpression of Foxp3 [60,61]. Thus, the female-specific increased expression of multiple, broadly distributed X-linked genes with T-cell-related immune functions suggests that aberrant X-linked gene expression in the setting of impaired dXCIm may have functional consequences relevant to female-biased disease development in NZM2328 mice, and may ultimately inform why females rather than males are more likely to develop SLE.
The observed impairment in dXCIm in T cells from female NZM2328 mice likely reflects perturbations within the Xist RNA Interactome. We found 26 autosomal Xist RNA Interactome genes were aberrantly expressed, and often downregulated, in NZM2328 T cells, including several heterogenous nuclear ribonucleoproteins (hnRNPs), including Hnrnpa3, Pcbp2 (Hnrnpe2), Hnrnpa1, and Hnrnpa0 (Figure 6). These nuclear proteins have diverse functions related to RNA processing, including, RNA splicing, RNA trafficking, and the regulation of mRNA translation. Though these Xist binding proteins have not yet been shown to have known roles in specifically tethering Xist RNA to the Xi, they are likely important in preserving dXCIm in female lymphocytes. Depletion of Hnrnpa0 in a human B cell line results in the increased expression of the proinflammatory gene, TLR7, from the Xi, suggesting a regulatory role for XCI maintenance in lymphocytes [12]. It is important to note that the Xist interactome has not been investigated in T cells, nor has it been determined using primary lymphocytes (mouse or human). Thus, it is likely that there are additional, currently unknown, Xist RNA binding proteins specifically in T cells that may be integral for recruiting Xist RNA to the Xi and tethering Xist RNA transcripts across the Xi. The XIST RNA interactome in a human B cell cancer cell line identified 115 XIST RNA binding proteins, only ~70% of which were shared with the XIST RNA Interactome of a human myeloid cell line, suggestive of cell-type-specific XIST RNA Interactome proteins [12]. Thus, future investigations which define the T-cell-specific Xist RNA interactome in primary lymphocytes will likely reveal other important Xist RNA binding proteins whose altered expression may contribute to the development of lupus and lupus-disease, thereby potentially affording new therapeutic targets for this disease.
Finally, it is worth noting that unlike T cells from the female-biased NZB/W F1 model of lupus which develop impaired Xist RNA relocalization with disease onset, T cells from NZM2328 mice exhibit mislocalized Xist RNA well before disease development. Why T cells from NZM2328 females exhibit this defect even early in life is not clear. Of the downregulated Xist interactome genes, only one (Wdr46) was located on a chromosome that harbors one of the four genomic susceptibility loci in the NZM2328 model. Thus, it is possible that these animals possess a currently unidentified, yet heritable, mutation that affects Xist RNA tethering or recruitment to the Xi. In addition, Xist RNA localization at the Xi increased with aging/spontaneous disease development in NZM2328, which did not occur in either NZB/W F1 or MRL/lpr mice. This observation suggests that there may be in vivo selection of T cells with intact XCI maintenance which become the predominant cell population as disease severity increases. Therefore, we propose that impaired dXCIm in T cells is not simply an epiphenomenon of the development of lupus-like disease, and posit that impairments in dXCIm likely contribute to lupus-like disease onset and perhaps disease acceleration. Additional experiments are necessary to establish causality between impaired dXCIm in T cells and the onset/progression of lupus-like disease.
In summary, we found that dXCIm is substantially altered in in vitro-activated T cells from the heavily female-biased NZM2328 model of SLE, in conjunction with the female-biased over-expression of X-linked genes with relevant immune functions. Our data demonstrate that XCI maintenance can be perturbed in specific lymphocyte subsets rather than all lymphocytes, and that the impairments in dXCIm in T cells are more pronounced in the setting of female-biased disease. Collectively, this work suggests that the aberrant X-linked gene dosage arising in the setting of impaired dXCIm in lymphocytes is an important contributor to female-biased autoimmunity, and underscores the need for future studies examining the molecular mechanisms responsible for dXCIm in female-biased autoimmune disease.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Anguera lab for their input and feedback on the data analysis and figures. Figures were created using BioRender.com. This work was supported by grants from the National Institutes of Health (R01-AI134834 to MCA; T32-AR076951-01 to NJ), the Lupus Research Alliance TIL grant (to MCA), the Selena Gomez Fund (to WS), and the Scleroderma Research Foundation (to NJ).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors have no conflicts of interest to disclose.
REFERENCES:
- 1.Tsokos GC. Autoimmunity and organ damage in systemic lupus erythematosus. Nat Immunol [Internet]. (2020);21:605–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramos-Casals M, Brito-Zerón P, Kostov B, Sisó-Almirall A, Bosch X, Buss D, Trilla A, Stone JH, Khamashta MA, Shoenfeld Y. Google-driven search for big data in autoimmune geoepidemiology: Analysis of 394,827 patients with systemic autoimmune diseases. Autoimmun Rev [Internet]. (2015);14:670–9. [DOI] [PubMed] [Google Scholar]
- 3.Jiwrajka N, Anguera MC. The X in seX-biased immunity and autoimmune rheumatic disease. J Exp Med. (2022);219:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Odhams CA, Roberts AL, Vester SK, Duarte CST, Beales CT, Clarke AJ, Lindinger S, Daffern SJ, Zito A, Chen L, Jones LL, Boteva L, Morris DL, Small KS, Fernando MMA, Graham DSC, Vyse TJ. Interferon inducible X-linked gene CXorf21 may contribute to sexual dimorphism in Systemic Lupus Erythematosus. Nat Commun [Internet]. (2019);10:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown GJ, Cañete PF, Wang H, Medhavy A, Bones J, Roco JA, He Y, Qin Y, Cappello J, Ellyard JI, Bassett K, Shen Q, Burgio G, Zhang Y, Turnbull C, Meng X, Wu P, Cho E, Miosge LA, et al. TLR7 gain-of-function genetic variation causes human lupus. Nature. (2022);605:349–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hewagama A, Gorelik G, Patel D, Liyanarachchi P, Joseph McCune W, Somers E, Gonzalez-Rivera T, The Michigan Lupus Cohort, Strickland F, Richardson B. Overexpression of X-Linked genes in T cells from women with lupus. J Autoimmun [Internet]. (2013);41:60–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacob CO, Zhu J, Armstrong DL, Yan M, Han J, Zhou XJ, Thomas JA, Reiff A, Myones BL, Ojwang JO, Kaufman KM, Klein-Gitelman M, McCurdy D, Wagner-Weiner L, Silverman E, Ziegler J, Kelly JA, Merrill JT, Harley JB, et al. Identification of IRAK1 as a risk gene with critical role in the pathogenesis of systemic lupus erythematosus. Proc Natl Acad Sci U S A. (2009);106:6256–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma R, Harris VM, Cavett J, Kurien BT, Liu K, Koelsch KA, Fayaaz A, Chaudhari KS, Radfar L, Lewis D, Stone DU, Kaufman CE, Li S, Segal B, Wallace DJ, Weisman MH, Venuturupalli S, Kelly JA, Pons-Estel B, et al. Rare X Chromosome Abnormalities in Systemic Lupus Erythematosus and Sjögren’s Syndrome. Arthritis Rheumatol. (2017);69:2187–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu K, Kurien BT, Zimmerman SL, Kaufman KM, Taft DH, Kottyan LC, Lazaro S, Weaver CA, Ice JA, Adler AJ, Chodosh J, Radfar L, Rasmussen A, Stone DU, Lewis DM, Li S, Koelsch KA, Igoe A, Talsania M, et al. X Chromosome Dose and Sex Bias in Autoimmune Diseases: Increased Prevalence of 47,XXX in Systemic Lupus Erythematosus and Sjögren’s Syndrome. Arthritis Rheumatol. (2016);68:1290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith-Bouvier DL, Divekar AA, Sasidhar M, Du S, Tiwari-Woodruff SK, King JK, Arnold AP, Singh RR, Voskuhl RR. A role for sex chromosome complement in the female bias in autoimmune disease. J Exp Med. (2008);205:1099–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sasidhar MV, Itoh N, Gold SM, Lawson GW, Voskuhl RR. The XX sex chromosome complement in mice is associated with increased spontaneous lupus compared with XY. Ann Rheum Dis. (2012);71:1418–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu B, Qi Y, Li R, Shi Q, Satpathy AT, Chang HY, Yu B, Qi Y, Li R, Shi Q, Satpathy AT, Chang HY. B cell-specific XIST complex enforces X-inactivation and restrains atypical B cells. Cell [Internet]. (2021);184:1790–1803.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yildirim E, Kirby JE, Brown DE, Mercier FE, Sadreyev RI, Scadden DT, Lee JT. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell [Internet]. (2013);152:727–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Csankovszki G, Nagy A, Jaenisch R. Synergism of Xist RNA, DNA Methylation, and Histone Hypoacetylation in Maintaining X Chromosome Inactivation. J Cell Biol. (2001);153:773–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Payer B, Lee JT. X chromosome dosage compensation: How mammals keep the balance. Annu Rev Genet. (2008);42:733–72. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Syrett CM, Kramer MC, Basu A, Atchison ML, Anguera MC. Unusual maintenance of X chromosome inactivation predisposes female lymphocytes for increased expression from the inactive X. Proc Natl Acad Sci U S A. (2016);113:E2029–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Syrett CM, Paneru B, Sandoval-Heglund D, Wang J, Banerjee S, Sindhava V, Behrens EM, Atchison M, Anguera MC. Altered X-chromosome inactivation in T cells may promote sex-biased autoimmune diseases. JCI Insight. (2019);4:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Syrett CM, Sierra I, Beethem ZT, Dubin AH, Anguera MC. Loss of epigenetic modifications on the inactive X chromosome and sex-biased gene expression profiles in B cells from NZB/W F1 mice with lupus-like disease. J Autoimmun [Internet]. (2020);107:102357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Syrett CM, Sindhava V, Hodawadekar S, Myles A, Liang G, Zhang Y, Nandi S, Cancro M, Atchison M, Anguera MC. Loss of Xist RNA from the inactive X during B cell development is restored in a dynamic YY1-dependent two-step process in activated B cells. PLoS Genet. (2017);13:1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pyfrom S, Paneru B, Knox JJ, Cancro MP, Posso S, Buckner JH, Anguera MC. The dynamic epigenetic regulation of the inactive X chromosome in healthy human B cells is dysregulated in lupus patients. Proc Natl Acad Sci U S A. (2021);118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waters ST, Fu SM, Gaskin F, Deshmukh US, Sung SSJ, Kannapell CC, Tung KSK, McEwen SB, McDuffie M. NZM2328: A new mouse model of systemic lupus erythematosus with unique genetic susceptibility loci. Clin Immunol. (2001);100:372–83. [DOI] [PubMed] [Google Scholar]
- 22.Andrews BS, Eisenberg RA, Theofilopoulos AN, Izui S, Wilson CB, McConahey PJ, Murphy ED, Roths JB, Dixon FJ. Spontaneous Murine Lupus-Like Syndromes - Clinical and Immunopathological Manifestations in Several Strains*. J Exp Med. (1978);148:1198–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agrawal H, Jacob N, Carreras E, Bajana S, Putterman C, Turner S, Neas B, Mathian A, Koss MN, Stohl W, Kovats S, Jacob CO. Deficiency of Type I IFN Receptor in Lupus-Prone New Zealand Mixed 2328 Mice Decreases Dendritic Cell Numbers and Activation and Protects from Disease. J Immunol. (2009);183:6021–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. (2004);5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cunningham F, Allen JE, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, Austine-Orimoloye O, Azov AG, Barnes I, Bennett R, Berry A, Bhai J, Bignell A, Billis K, Boddu S, Brooks L, Charkhchi M, Cummins C, Da Rin Fioretto L, et al. Ensembl 2022. Nucleic Acids Res. (2022);50:D988–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. (2016);34:525–7. [DOI] [PubMed] [Google Scholar]
- 27.Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research. (2015);4:1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. (2009);26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. (2015);43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minajigi A, Froberg JE, Wei C, Sunwoo H, Kesner B, Colognori D, Lessing D, Payer B, Boukhali M, Haas W, Lee JT. A comprehensive Xist interactome reveals cohesion repulsion and an RNA-directed chromosome conformation. Science (80-). (2016);324:89–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J, Karypis G, Hippen KL, Vegoe AL, Ruiz P, Gilkeson GS, Behrens TW. Genomic view of systemic autoimmunity in MRLlpr mice. Genes Immun. (2006);7:156–68. [DOI] [PubMed] [Google Scholar]
- 32.Tukiainen T, Villani AC, Yen A, Rivas MA, Marshall JL, Satija R, Aguirre M, Gauthier L, Fleharty M, Kirby A, Cummings BB, Castel SE, Karczewski KJ, Aguet F, Byrnes A, Gelfand ET, Getz G, Hadley K, Handsaker RE, et al. Landscape of X chromosome inactivation across human tissues. Nature [Internet]. (2017);550:244–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balaton BP, Brown CJ. Escape Artists of the X Chromosome. Trends Genet [Internet]. (2016);32:348–59. [DOI] [PubMed] [Google Scholar]
- 34.Oghumu S, Varikuti S, Stock JC, Volpedo G, Saljoughian N, Terrazas CA, Satoskar AR. Cutting Edge: CXCR3 Escapes X Chromosome Inactivation in T Cells during Infection: Potential Implications for Sex Differences in Immune Responses. J Immunol. (2019);203:789–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takahashi S, Fossati L, Iwamoto M, Merino R, Motta R, Kobayakawa T, Izui S. Imbalance towards Th1 predominance is associated with acceleration of lupus-like autoimmune syndrome in MRL mice. J Clin Invest. (1996);97:1597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Groom JR, Luster AD. CXCR3 in T cell function. Exp Cell Res [Internet]. (2011);317:620–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu Q, Wu A, Tesmer L, Ray D, Yousif N, Richardson B. Demethylation of CD40LG on the Inactive X in T Cells from Women with Lupus. J Immunol. (2007);179:6352–8. [DOI] [PubMed] [Google Scholar]
- 38.Greenfield A, Carrel L, Pennisi D, Philippe C, Quaderi N, Siggers P, Steiner K, Tam PPL, Willard HF, Koopman P. The UTX gene escapes X inactivation in mice and humans. Hum Mol Genet [Internet]. (1998);7:737–42. [DOI] [PubMed] [Google Scholar]
- 39.Hasegawa Y, Brockdorff N, Kawano S, Tsutui K, Tsutui K, Nakagawa S. The matrix protein hnRNP U is required for chromosomal localization of xist RNA. Dev Cell [Internet]. (2010);19:469–76. [DOI] [PubMed] [Google Scholar]
- 40.Jeon Y, Lee JT. YY1 Tethers Xist RNA to the inactive X nucleation center. Cell [Internet]. (2011);146:119–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ridings-Figueroa R, Stewart ER, Nesterova TB, Coker H, Pintacuda G, Godwin J, Wilson R, Haslam A, Lilley F, Ruigrok R, Bageghni SA, Albadrani G, Mansfield W, Roulson JA, Brockdorff N, Ainscough JFX, Coverley D. The nuclear matrix protein CIZ1 facilitates localization of Xist RNA to the inactive X-chromosome territory. Genes Dev. (2017);31:876–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, Sweredoski MJ, Shishkin AA, Su J, Lander ES, Hess S, Plath K, Guttman M. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. (2015);521:232–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chu C, Zhang QC, Da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY. Systematic discovery of Xist RNA binding proteins. Cell. (2015);161:404–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu B, Qi Y, Li R, Shi Q, Satpathy AT, Chang HY. B cell-specific XIST complex enforces X-inactivation and restrains atypical B cells. Cell [Internet]. (2021);184:1790–1803.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kriz AJ, Colognori D, Sunwoo H, Nabet B, Lee JT. Article Balancing cohesin eviction and retention prevents aberrant chromosomal interactions, Polycomb- mediated repression, and X-inactivation Article Balancing cohesin eviction and retention prevents aberrant chromosomal interactions, Polycomb-mediate. Mol Cell [Internet]. (2021);1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luijk R, Wu H, Ward-Caviness CK, Hannon E, Carnero-Montoro E, Min JL, Mandaviya P, Müller-Nurasyid M, Mei H, van der Maarel SM, Beekman M, der Breggen R van, Deelen J, Lakenberg N, Moed M, Suchiman HED, Arindrarto W, van’t Hof P, Jan Bonder M, et al. Autosomal genetic variation is associated with DNA methylation in regions variably escaping X-chromosome inactivation. Nat Commun. (2018);9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo Z, Wang Y, Li R, Huang H, Wang R. Use of laser microdissection in the analysis of renal-infiltrating T cells in murine lupus. Cent Eur J Immunol. (2014);39:285–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henrickson M, Giannini EH, Hirsch R. Reduction of mortality and lymphadenopathy in MRL-lpr/lpr mice treated with nonmitogenic anti-CD3 monoclonal antibody. Arthritis Rheum. (1994);37:587–94. [DOI] [PubMed] [Google Scholar]
- 49.Santoro TJ, Portanova JP, Kotzin BL. The contribution of L3T4+ T cells to lymphoproliferation and autoantibody production in MRL-lpr/lpr mice. J Exp Med. (1988);167:1713–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alexander JJ, Jacob A, Chang A, Quigg RJ, Jarvis JN. Double negative T cells, a potential biomarker for systemic lupus erythematosus. Precis Clin Med. (2020);3:34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cotton AM, Ge B, Light N, Adoue V, Pastinen T, Brown CJ. Analysis of expressed SNPs identifies variable extents of expression from the human inactive X chromosome. Genome Biol. (2013);14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steinmetz OM, Turner J-E, Paust H-J, Lindner M, Peters A, Heiss K, Velden J, Hopfer H, Fehr S, Krieger T, Meyer-Schwesinger C, Meyer TN, Helmchen U, Mittrücker H-W, Stahl RAK, Panzer U. CXCR3 Mediates Renal Th1 and Th17 Immune Response in Murine Lupus Nephritis. J Immunol. (2009);183:4693–704. [DOI] [PubMed] [Google Scholar]
- 53.Russell JQ, Mooney T, Cohen PL, MacPherson B, Noelle RJ, Budd RC. Anti-CD40L Accelerates Renal Disease and Adenopathy in MRL- lpr Mice in Parallel with Decreased Thymocyte Apoptosis. J Immunol. (1998);161:729–39. [PubMed] [Google Scholar]
- 54.Ma J, Xu J, Madaio MP, Peng Q, Zhang J, Grewal IS, Flavell RA, Craft J. Autoimmune lpr/lpr mice deficient in CD40 ligand: spontaneous Ig class switching with dichotomy of autoantibody responses. J Immunol. (1996);157:417–26. [PubMed] [Google Scholar]
- 55.Jacob N, Guo S, Mathian A, Koss MN, Gindea S, Putterman C, Jacob CO, Stohl W. B Cell and BAFF Dependence of IFN-α–Exaggerated Disease in Systemic Lupus Erythematosus-Prone NZM 2328Mice. J Immunol. (2011);186:4984–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bagavant H, Deshmukh US, Wang H, Ly T, Fu SM. Role for Nephritogenic T Cells in Lupus Glomerulonephritis: Progression to Renal Failure Is Accompanied by T Cell Activation and Expansion in Regional Lymph Nodes. J Immunol. (2006);177:8258–65. [DOI] [PubMed] [Google Scholar]
- 57.Jacob N, Yang H, Pricop L, Liu Y, Gao X, Zheng SG, Wang J, Gao H-X, Putterman C, Koss MN, Stohl W, Jacob CO. Accelerated Pathological and Clinical Nephritis in Systemic Lupus Erythematosus-Prone New Zealand Mixed 2328 Mice Doubly Deficient in TNF Receptor 1 and TNF Receptor 2 via a Th17- Associated Pathway. J Immunol [Internet]. (2009);182:2532–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Newcomb DC, Zhou W, Moore ML, Goleniewska K, Hershey GKK, Kolls JK, Peebles RS. A Functional IL-13 Receptor Is Expressed on Polarized Murine CD4 + Th17 Cells and IL-13 Signaling Attenuates Th17 Cytokine Production. J Immunol. (2009);182:5317–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xia S, Liu X, Cao X, Xu S. T-cell expression of Bruton’s tyrosine kinase promotes autoreactive T-cell activation and exacerbates aplastic anemia. Cell Mol Immunol [Internet]. (2020);17:1042–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Caramalho I, Lopes-Carvalho T, Ostler D, Zelenay S, Haury M, Demengeot J. Regulatory T cells selectively express toll-like receptors and are activated by lipopolysaccharide. J Exp Med. (2003);197:403–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tomita T, Kanai T, Fujii T, Okamoto R, Tsuchiya K, Sakamoto N, Akira S, Watanabe M. MyD88-Dependent Pathway in T Cells Directly Modulates the Expansion of Colitogenic CD4+ T cells in Chronic Colitis. J Immunol. (2012);180:5291–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


