Abstract
Formaldehyde is toxic for all organisms from bacteria to humans due to its reactivity with biological macromolecules. Organisms that grow aerobically on single-carbon compounds such as methanol and methane face a special challenge in this regard because formaldehyde is a central metabolic intermediate during methylotrophic growth. In the α-proteobacterium Methylobacterium extorquens AM1, we found a previously unknown enzyme that efficiently catalyzes the removal of formaldehyde: it catalyzes the condensation of formaldehyde and tetrahydromethanopterin to methylene tetrahydromethanopterin, a reaction which also proceeds spontaneously, but at a lower rate than that of the enzyme-catalyzed reaction. Formaldehyde-activating enzyme (Fae) was purified from M. extorquens AM1 and found to be one of the major proteins in the cytoplasm. The encoding gene is located within a cluster of genes for enzymes involved in the further oxidation of methylene tetrahydromethanopterin to CO2. Mutants of M. extorquens AM1 defective in Fae were able to grow on succinate but not on methanol and were much more sensitive toward methanol and formaldehyde. Uncharacterized orthologs to this enzyme are predicted to be encoded by uncharacterized genes from archaea, indicating that this type of enzyme occurs outside the methylotrophic bacteria.
Formaldehyde is a highly reactive chemical that exerts a toxic effect on organisms through its nonspecific reactivity with proteins and nucleic acids (9, 11). All organisms produce low concentrations of formaldehyde as a result of demethylation reactions (5, 13). Methylotrophic bacteria represent an extreme case of formaldehyde handling, as aerobic growth on single-carbon (C1) substrates such as methanol or methane involves formaldehyde as a central metabolic intermediate (22). A typical methylotrophic bacterium with a doubling time of 2 h generates and consumes formaldehyde at a specific rate of 0.5 mmol min−1 g−1 (dry weight), corresponding to 0.1 mmol per min and per ml of cytoplasmic volume (1). Thus, if the consumption of formaldehyde were inhibited, the cytoplasmic formaldehyde concentration would increase to about 100 mM within less than 1 min. Therefore, the metabolism of aerobic methylotrophic bacteria must be constructed and regulated such that the rate of formaldehyde utilization does not become limiting; a limitation would rapidly become detrimental.
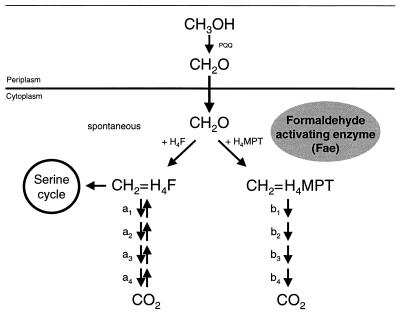
The catabolism of C1 units is best studied in Methylobacterium extorquens AM1. The α-proteobacterium M. extorquens AM1 is a facultative methylotroph that can grow aerobically on methanol and methylamine but also on some multicarbon substrates such as succinate. A scheme of its metabolism during growth on methanol is shown in Fig. 1. Methanol is oxidized to formaldehyde in the periplasm in a reaction catalyzed by a pyrroloquinoline quinone-dependent methanol dehydrogenase. The formaldehyde then crosses the cytoplasmic membrane, and in the cytoplasm it then reacts with the C1 carrier molecules tetrahydrofolate (H4F) and tetrahydromethanopterin (H4MPT) (7). H4MPT is an H4F analogue in methanogenic and sulfate-reducing archaea and was also recently found in most methylotrophic bacteria including methanotrophic bacteria (34). Based on the specificity and activity of the respective C1 unit-converting enzymes, we have proposed that, in M. extorquens AM1, the H4MPT-dependent pathway is the primary formaldehyde oxidation pathway and the H4F-dependent pathway is mainly involved in formaldehyde assimilation (33) which further proceeds by the serine cycle (22) (Fig. 1). A central reaction in the metabolism of methanol in M. extorquens AM1 is the condensation of formaldehyde with H4MPT and H4F, resulting in the formation of the N5,N10-methylene derivatives:
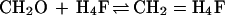 |
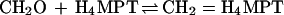 |
These reactions are known to occur spontaneously (8, 16, 20, 25). However, the question arises whether the spontaneous rates are sufficiently high to accommodate the rate of formaldehyde production, as has been discussed before with respect to methylene H4F formation by Attwood and Quayle (1). Studies of nonmethylotrophic organisms suggested that the condensation of formaldehyde and H4F is nonenzymatic (15–17). We have now reinvestigated this problem with a methylotrophic bacterium, which, as pointed out above, must maintain a high rate of formaldehyde consumption inside the cell. We did not find a formaldehyde-H4F condensing activity. However, we discovered and purified an enzyme that catalyzes the condensation of formaldehyde and H4MPT to methylene H4MPT and have shown that it is required for growth of M. extorquens AM1 on methanol and involved in formaldehyde detoxification.
FIG. 1.
C1 metabolism of M. extorquens AM1. Formaldehyde is produced in the periplasm of the cell from methanol by a pyrroloquinoline quinone-dependent methanol dehydrogenase and crosses the membrane. Cytoplasmic formaldehyde reacts with either H4F or H4MPT. The condensation of formaldehyde and H4MPT is catalyzed by Fae (this study). The C1 unit is utilized for further oxidation to CO2 or incorporated via the serine cycle. a1, NADP+-dependent methylene H4F dehydrogenase MtdA (33); a2, methenyl H4F cyclohydrolase FchA (27); a3, formyl H4F synthase; a4, formate dehydrogenase; b1, NAD(P)+-methylene H4MPT dehydrogenases MtdA and MtdB (12, 33); b2, methenyl H4MPT cyclohydrolase Mch (27); b3, formyl methanofuran:H4MPT formyltransferase; b4, formyl methanofuran dehydrogenase.
MATERIALS AND METHODS
Coenzymes.
H4MPT was purified from Methanobacterium thermoautotrophicum Marburg (DSM 2133) (3). H4F was purchased from Sigma. Anoxic stock solutions of H4MPT and H4F were prepared in 50 mM Tricine-KOH (pH 7.0).
Growth of organisms.
M. extorquens AM1 was grown on methanol (100 mM) or succinate (20 mM) at 30°C in minimal medium as described previously (10). The cultures were harvested in the late exponential phase at a cell concentration of 3 g of wet mass/liter. Cells were pelleted by centrifugation at 5,000 × g and stored at −20°C. M. thermoautotrophicum Marburg was grown at 65°C on H2-CO2 (80/20) in a minimal medium (30). Methanosarcina barkeri Fusaro (DSM 804) was grown at 37°C on methanol (18). Cells of the methanogenic archaea were harvested anaerobically in the late exponential phase.
Preparation of cell extracts.
Cells were disrupted in a French press, and cell extracts were prepared as described before (33). Protein concentration was determined by the Bradford assay (2) by using the Bio-Rad reagent with bovine serum albumin as the standard.
Determination of the activity of Fae.
Formaldehyde-activating enzyme (Fae) activity was routinely assayed at 30°C and pH 8.0. The standard assay mixture contained 50 mM Tricine-KOH, 30 mM MgCl2, 50 μM H4MPT (isolated from M. thermoautotrophicum) or 50 μM H4F, enzyme, and 1.6 mM formaldehyde. The reaction was started with formaldehyde. The formation of methylene H4MPT was monitored by measuring the increase in absorbance at 250 nm using a Δɛ250 of 8.5 mM−1 cm−1, and the formation of methylene H4F was monitored by measuring the increase in absorbance at 295 nm using a Δɛ295 of 3.0 mM−1 cm−1. A unit of activity was defined as the formation of 1 μmol of methylene H4MPT from H4MPT and formaldehyde per min minus the spontaneous reaction rate without enzyme added.
Activity of purified Fae was also measured in a coupled assay at 340 nm together with purified methylene H4MPT dehydrogenase (MtdA) (33), which catalyzes the dehydrogenation of methylene H4MPT. Reaction rates of Fae in the coupled assay were calculated indirectly through differences in MtdA activity.
Purification of Fae from M. extorquens AM1.
Cell extracts of M. extorquens AM1 were ultracentrifuged (150,000 × g for 1 h), and the soluble fraction was loaded onto DEAE-Sephacel (26/10; Sigma) columns equilibrated with 50 mM MOPS (morpholine propanesulfonic acid)-KOH, pH 7.0 (buffer A). Protein was eluted with NaCl in buffer A–80 ml of 0 M NaCl–120 ml of 0.1 M NaCl–200 ml of 0.1 to 0.4 M NaCl–100 ml of 0.5 M NaCl. Fae was eluted with about 0.3 M NaCl and applied to a Q-Sepharose (High Performance 16/10; Amersham Pharmacia Biotech) column equilibrated with buffer A. Protein was eluted with a linearly increasing gradient of 0 to 0.3 M NaCl within 450 ml. Fae eluted at 0.13 M NaCl and was subjected to chromatography on hydroxylapatite (16/10; Bio-Rad) equilibrated with 10 mM potassium phosphate, pH 7.0. Protein was eluted with a step gradient of 50, 100, 150, 175, and 200 mM potassium phosphate (25 ml each step). Fae was eluted at 175 mM potassium phosphate. The purification was performed under anoxic conditions. Although the enzyme from M. extorquens AM1 was found to be oxygen tolerant, the anoxic preparation turned out to be more suitable and to yield purer enzyme preparations.
Construction and analysis of a Fae mutant strain.
A 2.0-kb chromosomal region of M. extorquens AM1 containing fae (accession no. AF032114) (7) was amplified using the primers CM-faef, 5′-GTCCCAAATCGATGACGAAG-3′, and CM-faer, 5′-GGTTCACGCGATGTCTCAC-3′. The resulting PCR product was cloned into the pCR2.1 TOPO TA cloning vector (Invitrogen) to yield pCM112. The 2.1-kb BamHI-SphI region of pCM112 was subcloned into pUC19 (36), generating pCM113. The kanamycin resistance gene from pUC4K (32) was then inserted between the two HincII sites found in fae, in the same direction as fae is transcribed, as the direction is known, to create pCM114. The 3.1-kb BamHI-SphI fragment of pCM114 containing the fae::kan allele was then excised, blunted with T4 DNA polymerase, and inserted into the SmaI site of the suicide vector pAYC61 (6) to yield pCM115. Conjugation into M. extorquens AM1 and selection of exconjugates were performed as described previously (6). PCR analysis and antibiotic resistance phenotype confirmed that strain CM115.1 contains a single, interrupted copy of fae generated by allelic exchange. Growth of CM115.1 (fae::kan) on various media was compared to that of a kanamycin-resistant wild-type strain.
A plasmid containing fae under the expression of its own putative promoter, pCM139, was used to complement the fae::kan mutant strain. pCM139 was constructed as follows. The 0.8-kb region containing fae and its putative promoter region was amplified by PCR using the primers CM-Pfaef, 5′-GGATCCTGAGCCTTGGTCCAG-3′, and CM-4010r, 5′-TGACTGCCTCCGATCTAAG-3′. The resulting PCR product was cloned into pCR2.1 TOPO TA (Invitrogen) to generate pCM138. This region was then excised with BamHI and SphI and inserted into a broad-host-range cloning vector recently developed for M. extorquens AM1, pCM62 (C. J. Marx and M. E. Lidstrom, unpublished data), to create pCM139.
RESULTS
Fae activity in cell extracts.
Cell extracts of M. extorquens AM1 grown in the presence of methanol were found to accelerate the reaction of formaldehyde and H4MPT to methylene H4MPT. The activity catalyzing the acceleration of this reaction is designated Fae. A unit of activity was defined as the formation of 1 μmol of methylene H4MPT from H4MPT and formaldehyde per min minus the spontaneous rate without enzyme added. Under standard assay conditions at pH 8.0, the Fae activity of cell extracts from cells grown in the presence of methanol was found to be 1.4 U/mg. Cell extracts of cells grown on succinate exhibited an activity of only 0.3 U/mg, showing that the activity is induced upon growth on methanol.
No acceleration of the spontaneous reaction of formaldehyde and H4F was observed at various pH values and buffers in cell extracts of M. extorquens AM1. Also, no formaldehyde-H4F condensing activity could be observed with purified Fae.
Localization and purification of Fae.
The Fae activity was recovered in the soluble fraction of the cell extract; membrane fractions did not show activity. After three chromatographic steps, preparations contained only one polypeptide, with an apparent molecular mass of 18 kDa, as revealed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Fig. 2) and a specific activity of about 20 U/mg. Purification was about 14-fold with a yield of about 23% (Table 1), indicating that the protein is present in relatively large amounts in the cell, 2% or more.
FIG. 2.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of fractions of Fae upon purification from M. extorquens AM1. Lane M, molecular mass standards (Amersham-Pharmacia Biotech); lane 1, 20 μg of cell extract protein; lane 2, 8 μg of protein after DEAE-Sephacel; lane 3, 6 μg of protein after Q-Sepharose; lane 4, 4 μg of protein after hydroxyapatite.
TABLE 1.
Purification of Fae from M. extorquens AM1 grown on methanola
| Purification step | Protein (mg) | Activity (U) | Sp act (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Soluble fraction | 255 | 357 | 1.4 | 100 | 1 |
| DEAE-Sephacel | 62 | 214 | 3.9 | 60 | 2.8 |
| Q-Sepharose | 11.3 | 122 | 10.8 | 34 | 7.7 |
| Hydroxyapatite | 4.1 | 83 | 20.2 | 23 | 14.4 |
Enzyme activity was determined as described in the text at 30°C.
The activity of purified Fae could also be measured in a coupled assay together with purified methylene H4MPT dehydrogenase (MtdA), which catalyzes the dehydrogenation of methylene H4MPT (33). Reaction rates of Fae were calculated indirectly through differences in MtdA activity and corresponded very well with the activity of Fae determined directly at 250 nm. This experiment confirms that the reaction products formed by spontaneous reaction of formaldehyde and H4MPT as well as the reaction catalyzed by Fae yield the same product, namely, N5,N10-methylene H4MPT. Furthermore, the experiment supports the assumption that in vivo the formation of methylene H4MPT catalyzed by Fae is followed by its dehydrogenation catalyzed by methylene H4MPT dehydrogenase (MtdA or MtdB), which shows high activities of several units per mg in cell extracts (Fig. 1) (12, 33).
Molecular and catalytic properties of Fae.
The apparent molecular mass of Fae was determined by gel filtration on Superdex 200 to be about 60 kDa. Since the subunit molecular mass of Fae was found to be 18 kDa, this finding suggests that Fae has a homotrimeric structure. The UV-visible spectrum of the enzyme was that of a protein lacking a chromophoric prosthetic group.
The spontaneous reaction of formaldehyde with H4MPT and the enzyme-catalyzed reaction exhibited different pH optima (Fig. 3). Whereas the spontaneous reaction proceeded faster in slightly acidic conditions in potassium phosphate buffer, the enzymatic reactions had a more alkaline pH optimum. When a 50 mM Tricine-KOH buffer system was used at pH 8.0, the presence of 30 mM MgCl2 or MgSO4 stimulated the activity about eightfold (Ka = 4 mM). Higher MgCl2 or MgSO4 concentrations did not result in higher reaction rates.
FIG. 3.
pH optimum of the spontaneous reaction of formaldehyde with H4MPT and of the reaction catalyzed by Fae. The assays contained 120 mM potassium phosphate, 40 μM H4MPT, 1.6 mM CH2O, and 5 μg of purified Fae when indicated. Assays were performed at the pH value indicated at 30°C. ●, activity in the absence of enzyme (spontaneous rate); ▴, activity in the presence of enzyme; ▾, activity in the presence of enzyme corrected for the spontaneous rate.
Fae activity depending on the formaldehyde concentration followed Michaelis-Menten kinetics and showed a high affinity of Fae for formaldehyde. Reciprocal plots indicated an apparent Km for formaldehyde of 0.2 mM (Fig. 4A). For the determination of the Km value of H4MPT in the assay, only up to 60 μM H4MPT could be used because of technical reasons. Reciprocal plots indicated an apparent Km value of about 1 mM H4MPT (Fig. 4B). The relatively high apparent Km for H4MPT may be due to the fact that H4MPT from Methanobacterium thermoautotrophicum rather than dephospho-H4MPT from M. extorquens AM1 (33) was used in the assays.
FIG. 4.
Kinetics of Fae with respect to different formaldehyde and H4MPT concentrations. The assay mixtures contained 50 mM Tricine-KOH (pH 8.0), 30 mM MgCl2, and 1.5 μg of purified Fae; the H4MPT concentration was 50 μM (A) or as indicated (B), and the CH2O concentration was as indicated (A) or 1.6 mM (B).
Identification of the gene encoding Fae and similarities to putative proteins from methanogenic archaea.
The N-terminal 40 amino acids of Fae from M. extorquens AM1 were determined by Edman degradation: AKITKVQVGEALVGDGNEVAHIDLIIGPXGSPAETAFXNG. The sequence and predicted molecular mass of 18 kDa were homologous with the gene product previously designated orf18 (7), which is now named fae.
The gene encoding Fae is located within a cluster of genes for proteins with sequence similarities to proteins from methanogenic and sulfate-reducing archaea (7). This cluster contains both the genes for the H4MPT-methanofuran-dependent enzymes involved in the further oxidation of methylene H4MPT to CO2 (Fig. 1) and open reading frames of unknown function (7, 27). Fae exhibits amino acid sequence identities of 50, 49, and 25%, respectively, to the N-terminal portion of putative proteins of unknown function deduced from the genome sequences of Archaeoglobus fulgidus (Orf1305), M. thermoautotrophicum ΔH (Orf1474), and Methanococcus jannaschii (Orf1447) (4, 21, 31). We could not, however, detect a formaldehyde-H4MPT condensing activity in cell extracts of M. thermoautotrophicum Marburg and M. barkeri Fusaro.
Construction of Fae-minus mutants and phenotypical analysis.
In order to assess the physiological importance of Fae activity, a mutant strain with an interrupted fae::kan allele was generated by allelic exchange. The fae::kan mutant strain CM115.1 lacked detectable Fae activity and was unable to grow on methanol but exhibited wild-type growth on succinate (data not shown). Furthermore, growth of the fae::kan mutant strain on succinate-containing solid media was inhibited by the addition of methanol or formaldehyde at MICs of 50 and 200 μM, respectively. These results suggest that the Fae-H4MPT-dependent pathway is the primary formaldehyde detoxification system during growth on both one-carbon and multicarbon compounds. Introduction of pCM139, a plasmid bearing only fae with its putative promoter region, restored wild-type growth to CM115.1, thus confirming that the phenotype is not due to a polar effect on downstream genes.
DISCUSSION
We have shown that M. extorquens AM1 contains a protein, Fae, which catalyzes the condensation of formaldehyde and H4MPT to methylene H4MPT (Fig. 1). From the rates shown in Fig. 3, it can be calculated that the spontaneous rate of formaldehyde condensation with H4MPT at physiological pH is not sufficient to accommodate the formaldehyde production rate but that the consumption rate with Fae would be sufficient. Accordingly, a fae knockout strain was found to be unable to grow on methanol. In addition, the mutant was shown to be formaldehyde sensitive, suggesting an involvement in formaldehyde detoxification of M. extorquens AM1. In this case, it is likely that Fae acts together with the other H4MPT-dependent enzymes to detoxify formaldehyde. The fae gene is indeed clustered with a group of other archaeon-like genes that encode the remainder of the H4MPT-dependent formaldehyde oxidation pathway. It has been previously suggested that this pathway was acquired by horizontal transfer from an ancestral archaeon (7, 34). Our results suggest that Fae may also have been acquired in this way, possibly in the same transfer event.
We did not find a formaldehyde-H4F condensing activity in cell extracts of M. extorquens AM1, nor did purified Fae show this activity. The condensation of formaldehyde with H4F occurs also spontaneously in vitro. The rates are comparable to the rates of condensation of formaldehyde and H4MPT and might be sufficient for methylene H4F formation, which is required for assimilation by the serine cycle and most likely purine and formylmethionine tRNA biosynthesis. The formation of methylene H4F might simply be regulated by the availability of free H4F and formaldehyde. Our hypothesis that the main flux of formaldehyde proceeds via the H4MPT- and not the H4F-dependent pathway is in agreement with an enzyme which accelerates the condensation of formaldehyde with H4MPT.
The only orthologs of Fae are found in methanogenic and sulfate-reducing archaea (Fig. 5). UP to now, the function of these orthologs has been unknown. Their occurrence raises the question as to why methanogens would contain a formaldehyde-H4MPT condensing activity. Formaldehyde is not thought to be a central metabolite of methanogenesis, although formaldehyde can be used as a methanogenic substrate in cell extracts and cell suspensions of methanogenic archaea, albeit only very slowly (14, 24, 26). We could not detect a formaldehyde-H4MPT condensing activity in cell extracts of the methanogenic archaea tested. It is possible, however, that the Fae ortholog in these organisms either is inactive or has activity with a substrate other than H4MPT, formaldehyde, or both. Interestingly, the Fae orthologs in the methanogenic and sulfate-reducing archaea are fused to a second, C-terminal domain that shows identity to 3-hexulose-6-phosphate synthase (HPS, encoded by rmpA) (Fig. 5B) from Methylomonas aminofaciens 77a (35) (Fig. 5B). HPS is the initial formaldehyde-utilizing enzyme found in methylotrophic bacteria that use the ribulose monophosphate pathway for formaldehyde assimilation (35) (Fig. 5B). HPS catalyzes the condensation of formaldehyde with d-ribulose-5-phosphate to generate d-arabino-3-hexulose-6-phosphate, which in ribulose monophosphate pathway methylotrophs is then isomerized to fructose-6-phosphate by 6-phospho-4-hexuloisomerase (PHI, encoded by rmpB) (23, 29). Curiously, HPS and PHI orthologs can be found in a number of genomes of nonmethylotrophic bacteria and nonmethanogenic archaea (Fig. 5B) (19, 28, 37). The HPS and PHI orthologs were recently cloned and purified from Bacillus subtilis, where they apparently function as a formaldehyde detoxification system (37). In the hyperthermophilic archaea Pyrococcus horikoshii and Pyrococcus abyssi, the HPS and PHI orthologs are fused into a single polypeptide (19). The in vivo function of these fusion proteins of fae and rmpA as well as rmpA and rmpB remains to be demonstrated in archaea and may provide new insights into formaldehyde conversion in archaea. The widespread occurrence of orthologs to this group of genes encoding formaldehyde utilization enzymes suggests that formaldehyde may play an unknown but important cellular role in a broad group of prokaryotes.
FIG. 5.
(A) Alignment of the amino acid sequence of Fae from M. extorquens AM1 and putative proteins from the complete genomes of the sulfate-reducing archaeon A. fulgidus (AF1305) (21) and the methanogenic archaeon M. thermoautotrophicum ΔH (Mth1474) (31). (B) Sequence analysis of orthologs of Fae from M. extorquens AM1. Fae from M. extorquens AM1 shows sequence identity to the N-terminal domain of putative proteins from A. fulgidus, M. thermoautotrophicum ΔH, and M. jannaschii (4, 21, 31). The C-terminal domains of these archaeal proteins themselves show sequence identity to 3-hexulose-6-phosphate synthase (HPS; RmpA) from M. aminofaciens (35). A homologue of RmpA is linked to the 6-phospho-4-hexuloisomerase (PHI; RmpB) homologue from M. aminofaciens (29) in Pyrococcus species (19).
ACKNOWLEDGMENTS
This work was supported by the Max Planck Society and by a grant from the NIH (GM36296) to M.E.L.
REFERENCES
- 1.Attwood M M, Quayle J R. Formaldehyde as a central intermediary metabolite of methylotrophic metabolism. In: Crawford R L, Hanson R S, editors. Microbial growth on C1 compounds. Washington, D.C.: American Society for Microbiology; 1984. pp. 315–323. [Google Scholar]
- 2.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 3.Breitung J, Börner G, Scholz S, Linder D, Stetter K O, Thauer R K. Salt dependence, kinetic properties and catalytic mechanism of N-formylmethanofuran:tetrahydromethanopterin formyltransferase from the extreme thermophile Methanopyrus kandleri. Eur J Biochem. 1992;210:971–981. doi: 10.1111/j.1432-1033.1992.tb17502.x. [DOI] [PubMed] [Google Scholar]
- 4.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J-F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrik J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Roberts K M, Hurst M A, Kaine B P, Borodovsky M, Klenk H-P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 5.Case G L, Benevenga N J. Significance of formate as an intermediate in the oxidation of the methionine, S-methyl-L-cysteine and sarcosine methyl carbons to CO2 in the rat. J Nutr. 1977;107:1665–1676. doi: 10.1093/jn/107.9.1665. [DOI] [PubMed] [Google Scholar]
- 6.Chistoserdov A Y, Chistoserdova L V, McIntire W S, Lidstrom M E. Genetic organization of the mau gene cluster in Methylobacterium extorquens AM1: complete nucleotide sequence and generation and characteristics of mau mutants. J Bacteriol. 1994;176:4052–4065. doi: 10.1128/jb.176.13.4052-4065.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chistoserdova L, Vorholt J A, Thauer R K, Lidstrom M E. C1 transfer enzymes and coenzymes linking methylotrophic bacteria and methanogenic archaea. Science. 1998;281:99–102. doi: 10.1126/science.281.5373.99. [DOI] [PubMed] [Google Scholar]
- 8.Escalante-Semerena J C, Rinehart K L, Jr, Wolfe R S. Tetrahydromethanopterin, a carbon carrier in methanogenesis. J Biol Chem. 1984;259:9447–9455. [PubMed] [Google Scholar]
- 9.Feldman M Y. Reactions of nucleic acids and nucleoproteins with formaldehyde. Progr Nucleic Acid Res Mol Biol. 1973;13:1–49. doi: 10.1016/s0079-6603(08)60099-9. [DOI] [PubMed] [Google Scholar]
- 10.Fulton G L, Nunn D N, Lidstrom M E. Molecular cloning of a malyl coenzyme A lyase gene from Pseudomonas sp. strain AM1, a facultative methylotroph. J Bacteriol. 1984;160:718–723. doi: 10.1128/jb.160.2.718-723.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grafstrom R C, Fornace A J, Jr, Autrup H, Lechner J F, Harris C C. Formaldehyde damage to DNA and inhibition of DNA repair in human bronchial cells. Science. 1983;220:216–218. doi: 10.1126/science.6828890. [DOI] [PubMed] [Google Scholar]
- 12.Hagemeier C H, Chistoserdova L, Lidstrom M E, Thauer R K, Vorholt J A. Characterization of a second methylene tetrahydromethanopterin dehydrogenase from Methylobacterium extorquens AM1. Eur J Biochem. 2000;267:3762–3769. doi: 10.1046/j.1432-1327.2000.01413.x. [DOI] [PubMed] [Google Scholar]
- 13.Handler P, Bernheim M L C, Klein J R. The oxidative demethylation of sarcosine to glycine. J Biol Chem. 1941;138:211–218. [Google Scholar]
- 14.Kaesler B, Schönheit P. The role of sodium ions in methanogenesis. Formaldehyde oxidation to CO2 and 2 H2 in methanogenic bacteria is coupled with primary electrogenic Na+ translocation at a stoichiometry of 2-3 Na+/CO2. Eur J Biochem. 1989;184:223–232. doi: 10.1111/j.1432-1033.1989.tb15010.x. [DOI] [PubMed] [Google Scholar]
- 15.Kallen R G, Jencks W P. The dissociation constants of tetrahydrofolic acid. J Biol Chem. 1966;241:5845–5850. [PubMed] [Google Scholar]
- 16.Kallen R G, Jencks W P. The mechanism of the condensation of formaldehyde with tetrahydrofolic acid. J Biol Chem. 1966;241:5851–5863. [PubMed] [Google Scholar]
- 17.Kallen R G, Jencks W P. Equilibria for the reaction of amines with formaldehyde and protons in aqueous solution. J Biol Chem. 1966;241:5864–5878. [PubMed] [Google Scholar]
- 18.Karrasch M, Börner G, Enßle M, Thauer R K. Formylmethanofuran dehydrogenase from methanogenic bacteria, a molybdoenzyme. FEBS Lett. 1989;253:226–230. doi: 10.1016/0014-5793(89)80964-0. [DOI] [PubMed] [Google Scholar]
- 19.Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, Nagai Y, Sakai M, Ogura K, Otsuka R, Nakazawa H, Takamiya M, Ohfuku Y, Funahashi T, Tanaka T, Kudoh Y, Yamazaki J, Kushida N, Oguchi A, Aoki K, Kikuchi H. Complete sequence and gene organization of the genome of a hyperthermophilic archaebacterium, Pyrococcus horikoshii OT3 (supplement) DNA Res. 1998;5:147–155. doi: 10.1093/dnares/5.2.147. [DOI] [PubMed] [Google Scholar]
- 20.Keltjens J T, Caerteling G C, van der Drift C, Vogels G D. Methanopterin and the intermediary steps of methanogenesis. Syst Appl Microbiol. 1986;7:370–375. [Google Scholar]
- 21.Klenk H-P, Clayton R A, Tomb J-F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Peterson S, Reich C I, McNeil L K, Badger J H, Glodek A, Zhou L, Overbeek R, Gocayne J D, Weidman J F, McDonald L, Utterback T, Cotton M D, Spriggs T, Artiach P, Kaine B P, Sykes S M, Sadow P W, D'Andrea K P, Bowman C, Fujii C, Garland S A, Mason T M, Olsen G J, Fraser C M, Smith H O, Woese C R, Venter J C. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 22.Lidstrom M E. The aerobic methylotrophic bacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. New York, N.Y: Springer-Verlag; 1992. pp. 431–445. [Google Scholar]
- 23.Mitsui R, Sakai Y, Yasueda H, Kato N. A novel operon encoding formaldehyde fixation: the ribulose monophosphate pathway in the gram-positive facultative methylotrophic bacterium Mycobacterium gastri MB19. J Bacteriol. 2000;182:944–948. doi: 10.1128/jb.182.4.944-948.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Müller V, Winner C, Gottschalk G. Electron-transport-driven sodium extrusion during methanogenesis from formaldehyde and molecular hydrogen by Methanosarcina barkeri. Eur J Biochem. 1988;178:519–525. doi: 10.1111/j.1432-1033.1988.tb14478.x. [DOI] [PubMed] [Google Scholar]
- 25.Osborn M J, Talbert P T, Huennekens F M. The structure of “active formaldehyde” (N5,N10-methylene tetrahydrofolic acid) J Am Chem Soc. 1960;82:4921–4927. [Google Scholar]
- 26.Poirot C M, Kengen S W M, Valk E, Keltjens J T, van der Drift C, Vogels G D. Formation of methylcoenzyme M from formaldehyde by cell-free-extracts of Methanobacterium thermoautotrophicum. Evidence for the involvement of a corrinoid-containing methyltransferase. FEMS Microbiol Lett. 1987;40:7–13. [Google Scholar]
- 27.Pomper B K, Vorholt J A, Chistoserdova L, Lidstrom M E, Thauer R K. A methenyl tetrahydromethanopterin cyclohydrolase and a methenyl tetrahydrofolate cyclohydrolase in Methylobacterium extorquens AM1. Eur J Biochem. 1999;261:475–480. doi: 10.1046/j.1432-1327.1999.00291.x. [DOI] [PubMed] [Google Scholar]
- 28.Reizer J, Reizer A, Saier M H., Jr Is the ribulose monophosphate pathway widely distributed in bacteria? Microbiology. 1997;143:2519–2520. doi: 10.1099/00221287-143-8-2519. [DOI] [PubMed] [Google Scholar]
- 29.Sakai Y, Mitsui R, Katayama Y, Yanase H, Kato N. Organization of the genes involved in the ribulose monophosphate pathway in an obligate methylotrophic bacterium, Methylomonas aminofaciens 77a. FEMS Microbiol Lett. 1999;176:125–130. doi: 10.1111/j.1574-6968.1999.tb13652.x. [DOI] [PubMed] [Google Scholar]
- 30.Schönheit P, Moll J, Thauer R K. Growth parameters (KS, μmax, YS) of Methanobacterium thermoautotrophicum. Arch Microbiol. 1980;127:59–65. doi: 10.1007/BF00403508. [DOI] [PubMed] [Google Scholar]
- 31.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrokovski S, Church G M, Daniels C J, Mao J-I, Rice P, Nölling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 33.Vorholt J A, Chistoserdova L, Lidstrom M E, Thauer R K. The NADP-dependent methylene tetrahydromethanopterin dehydrogenase in Methylobacterium extorquens AM1. J Bacteriol. 1998;180:5351–5356. doi: 10.1128/jb.180.20.5351-5356.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vorholt J A, Chistoserdova L, Stolyar S M, Thauer R K, Lidstrom M E. Distribution of tetrahydromethanopterin-dependent enzymes in methylotrophic bacteria and phylogeny of methenyl tetrahydromethanopterin cyclohydrolases. J Bacteriol. 1999;181:5750–5757. doi: 10.1128/jb.181.18.5750-5757.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yanase H, Ikeyama K, Mitsui R, Ra S, Kita K, Sakai Y, Kato N. Cloning and sequence analysis of the gene encoding 3-hexulose-6-phosphate synthase from the methylotrophic bacterium, Methylomonas aminofaciens 77a, and its expression in Escherichia coli. FEMS Microbiol Lett. 1996;135:201–205. doi: 10.1111/j.1574-6968.1996.tb07990.x. [DOI] [PubMed] [Google Scholar]
- 36.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 37.Yasueda H, Kawahara Y, Sugimoto S-I. Bacillus subtilis yckG and yckF encode two key enzymes of the ribulose monophosphate pathway used by methylotrophs, and yckH is required for their expression. J Bacteriol. 1999;181:7154–7160. doi: 10.1128/jb.181.23.7154-7160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]