Abstract
Protocatechuate (PCA) is the key intermediate metabolite in the lignin degradation pathway of Sphingomonas paucimobilis SYK-6 and is metabolized to pyruvate and oxaloacetate via the PCA 4,5-cleavage pathway. We characterized the 4-carboxy-2-hydroxymuconate-6-semialdehyde (CHMS) dehydrogenase gene (ligC). CHMS is the 4,5-cleavage product of PCA and is converted into 2-pyrone-4,6-dicarboxylate (PDC) by LigC. We found that ligC was located 295 bp downstream of ligB, which encodes the large subunit of the PCA 4,5-dioxygenase. The ligC gene consists of a 945-bp open reading frame encoding a polypeptide with a molecular mass of 34,590 Da. The deduced amino acid sequence of ligC showed 19 to 20% identity with 3-chlorobenzoate cis-dihydrodiol dehydrogenase of Alcaligenes sp. strain BR60 and phthalate cis-dihydrodiol dehydrogenases of Pseudomonas putida NMH102-2 and Burkholderia cepacia DBO1, which are unrelated to group I, II, and III microbial alcohol dehydrogenases (M. F. Reid and C. A. Fewson, Crit. Rev. Microbiol. 20:13–56, 1994). The ligC gene was expressed in Escherichia coli and LigC was purified to near homogeneity. Production of PDC from CHMS catalyzed by LigC was confirmed in the presence of NADP+ by electrospray ionization-mass spectrometry and gas chromatography-mass spectrometry. LigC is a homodimer. The isoelectric point, optimum pH, and optimum temperature were estimated to be 5.3, 8.0, and 25°C, respectively. The Km for NADP+ was estimated to be 24.6 ± 1.5 μM, which was approximately 10 times lower than that for NAD+ (252 ± 3.9 μM). The Kms for CHMS in the presence of NADP+ and NAD+ are 26.0 ± 0.5 and 20.6 ± 1.0 μM, respectively. Disruption of ligC in S. paucimobilis SYK-6 prevented growth with vanillate. Only PCA was accumulated during the incubation of vanillate with the whole cells of the ligC insertion mutant (DLC), indicating a lack of PCA 4,5-dioxygenase activity in DLC. However, the introduction of ligC into DLC restored its ability to grow on vanillate. PDC was suggested to be an inducer for ligAB gene expression.
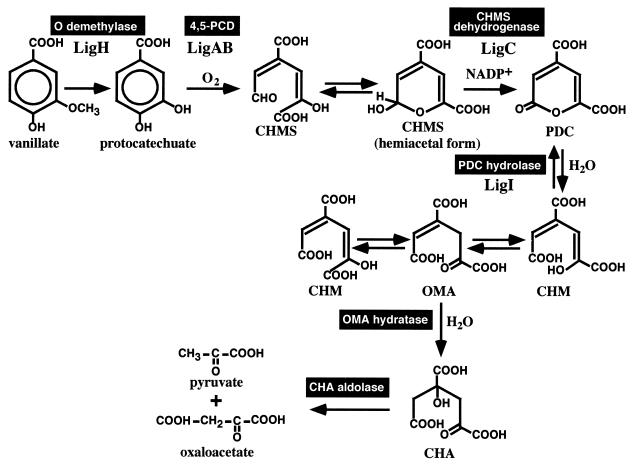
Lignin is a major component of the plant cell wall along with cellulose and hemicellulose, and it is known to be one of the most abundant aromatic compounds in nature. Utilization of this abundant biomass has been expected but has not been established. One of the practical ways to utilize lignin is to degrade it using the enzyme systems of microorganisms to produce commercially valuable compounds. Sphingomonas paucimobilis SYK-6, which has been isolated from pulp effluent, can degrade various dimeric lignin compounds such as β-aryl ether, biphenyl, phenylcoumarane, diarylpropane, and pinoresinol (20). The enzyme genes involved in the degradation of major components of these compounds, β-aryl ether (18, 19, 21, 22) and biphenyl (29, 30), have already been characterized. Vanillate is thought to be a major intermediate metabolite of these dimeric lignin compounds having guaiacyl moieties. As illustrated in Fig. 1, protocatechuate (PCA) is generated following the O demethylation of vanillate (26), and it is metabolized to pyruvate and oxaloacetate via the PCA 4,5-cleavage pathway in S. paucimobilis SYK-6 (20, 23). Three different pathways have been reported for PCA metabolism, the PCA 4,5-cleavage pathway (10, 13–17, 20, 27, 39), the PCA 3,4-cleavage pathway (8), and the PCA 2,3-cleavage pathway (6, 41).
FIG. 1.
Catabolic pathway of vanillate for S. paucimobilis SYK-6 via the PCA 4,5-cleavage pathway. LigA and LigB, small and large subunits of PCA 4,5-dioxygenase (4,5-PCD) (27, 38); LigC, CHMS dehydrogenase (this study); LigI, PDC hydrolase (23); LigH, an essential gene product for vanillate and syringate O demethylations (26).
Numerous genetic and biochemical characterizations of the PCA 3,4-cleavage pathway have been established, while the PCA 4,5- and PCA 2,3-cleavage pathways are poorly understood. Although the PCA 4,5-cleavage pathway was enzymatically characterized by Dagley and coworkers (10, 39) and Maruyama and coworkers (13–17) in the 1980s, no genetic information on the PCA 4,5-cleavage pathway is available except for that included in studies of SYK-6 (23, 27). According to the reports of Kersten et al. (10), PCA is first converted into 4-carboxy-2-hydroxymuconate-6-semialdehyde (CHMS) by the PCA 4,5-dioxygenase in the PCA 4,5-cleavage pathway (Fig. 1). The three-dimensional structure of PCA 4,5-dioxygenase has been elucidated previously (38). The resultant CHMS is in equilibrium between the open form and the cyclic hemiacetal form. The hemiacetal form of CHMS is thought to be oxidized by CHMS dehydrogenase to produce 2-pyrone-4,6-dicarboxylate (PDC). PDC hydrolase transforms PDC to 4-carboxy-2-hydroxymuconate (CHM) or its tautomer, 4-oxalomesaconate (OMA), and the resultant product is converted into 4-carboxy-4-hydroxy-2-oxoadipate (CHA) by a hydratase. Finally, an aldolase cleaves CHA to generate pyruvate and oxaloacetate.
To understand the whole PCA 4,5-cleavage pathway of S. paucimobilis SYK-6, we are planning to characterize all the genes involved in the enzyme reactions and regulation of the PCA 4,5-cleavage pathway. The genes for the PCA 4,5-dioxygenase (ligAB) (27) and PDC hydrolase (ligI) (23) have already been characterized. In this study, we characterize the CHMS dehydrogenase gene, which is located next to ligB and involved in the second step of the PCA 4,5-cleavage pathway. We also suggest that the reaction product of CHMS dehydrogenase, PDC, is an inducer of the ligAB gene expression.
MATERIALS AND METHODS
Strains and plasmids.
The strains and plasmids used in this study are listed in Table 1. S. paucimobilis SYK-6 was grown at 30°C in W minimal salt medium (29) containing 0.2% (wt/vol) vanillate or syringate or in Luria-Bertani (LB) medium (Bacto-Tryptone, 10 g/liter; yeast extract, 5 g/liter; NaCl, 5 g/liter).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| S. paucimobilis | ||
| SYK-6 | Wild type; Nalr Smr | 9 |
| DLC | Mutant derivative of SYK-6; Kmr gene insertion mutant of ligC; Nalr Smr Kmr | This study |
| P. putida PpY1100 | Nalr Smr | 9 |
| E. coli | ||
| JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi Δ(lac-proAB) F′[traD36 proAB+lacIqlacZΔM15] | 42 |
| HB101 | supE44 hsdS20(rB− mB−) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | 3 |
| BL21(DE3) | hsdS gal(λcIts857 ind1 Sam7 nin5 lacUV5-T7 gene 1) | 37 |
| Plasmids | ||
| pUC18 and pUC19 | Cloning vectors; Apr | 42 |
| pBluescript II SK(+) | Cloning vectors; Apr | 36 |
| pET21(+) | Expression vector; Apr T7 promoter | Novagen |
| pKT230 | Broad-host-range vector; Kmr | 2 |
| pVK100 | Broad-host-range cosmid; Kmr Tetr | 11 |
| pBBR122 | Broad-host-range vector; Kmr Cmr | MoBiTec |
| pK19mobsacB | oriT sacB Kmr | 34 |
| pUC4K | Source of Kmr cassette; Apr Kmr | 40 |
| pVA01 | pKT230 with a SYK-6 10.9-kb insert carrying ligABC and ligI | 9 |
| pVAD2 | Deletion plasmid of pVA01; a SalI fragment near the 3′ terminus of the insert was deleted | This study |
| pVAD4 | Deletion plasmid of pVA01; SalI fragments in the middle of the insert were deleted | 23 |
| pHN139F | pUC18 with a 10.5-kb EcoRI fragment of pVA01 carrying ligAB, part of ligC, and ligI | 23 |
| pHN139R | pUC18 carrying the same fragment as pHN139F in the opposite direction | 23 |
| pHNC2 | pET21(+) with a 1.0-kb SmaI-EcoRI fragment carrying part of ligC | This study |
| pSM21 | pET21(+) with a 1.4-kb SmaI-EcoRI fragment carrying ligC | This study |
| pAB16 | pBluescript II SK(+) with a 1.6-kb XbaI-XhoI fragment carrying ligAB | This study |
| pABC25 | pBluescript II SK(+) with a 2.5-kb XbaI-EcoRI fragment carrying ligAB and part of ligC | This study |
| pABC25K | pABC25 with insertion of the Kmr gene of pUC4K replacing a 0.2-kb BsmI fragment | This study |
| pLCD1 | pK19mobsacB with a 3.5-kb XbaI-EcoRI fragment of pABC25K | This study |
| pBBRC1220 | pBBR122 with a 1.4-kb SmaI-HindIII fragment carrying ligC of pSM21 | This study |
| pBBRC1221 | pBBRC1220 with an 0.4-kb PvuII fragment carrying the lac promoter of pUC19 in the SmaI site | This study |
Preparation of substrates.
CHMS was prepared from PCA by using the crude PCA 4,5-dioxygenase (LigAB) prior to the enzyme assay. PCA (1.5 mM) was incubated with 100 μg of crude extract of Escherichia coli JM109 harboring pAB16 carrying ligAB in 20 mM Tris-HCl buffer (pH 8.0) for 1 h at 30°C. The complete conversion of PCA into CHMS was confirmed by electrospray ionization-mass spectrometry (ESI-MS) and gas chromatography-mass spectrometry (GC-MS) with the conditions described below. The resultant CHMS solution was kept at room temperature for 1 h to be equilibrated between the open form and the cyclic hemiacetal forms. Vanillate and other chemicals were purchased from Tokyo Kasei Kogyo Co. (Tokyo, Japan) or Wako Pure Chemical Industries (Osaka, Japan).
DNA manipulations and nucleotide sequencing.
DNA manipulations were carried out essentially as described elsewhere (1, 32). A Kilosequence kit (Takara Shuzo Co., Ltd., Kyoto, Japan) was used to construct a series of deletion derivatives, whose nucleotide sequences were determined by the dideoxy termination method with an ALFexpress DNA sequencer (Pharmacia Biotech, Milwaukee, Wis.). The Sanger reaction (33) was carried out using the Thermosequenase fluorescence-labeled primer cycle sequencing kit with 7-deaza dGTP (Amersham Pharamacia Biotech, Little Chalfont, United Kingdom). Sequence analysis and homology alignment were done using the programs of GeneWorks (IntelliGenetics, Inc., Mountain View, Calif.). The DDBJ databases were employed to search for homologous proteins. Southern hybridization analysis of SYK-6 and its CHMS dehydrogenase gene (ligC) insertion mutants was performed with the DIG system (Boehringer Mannheim Biochemicals, Indianapolis, Ind.).
Enzyme assay.
CHMS dehydrogenase activity was spectrophotometrically determined by measuring the increase in the absorbance at 340 nm derived from the production of NADH (ɛ340 = 6,600 M−1 cm−1; pH 8.0) or NADPH (ɛ340 = 5,070 M−1 cm−1; pH 8.0) from NAD+ or NADP+, respectively, using a DU-7500 spectrophotometer (Beckman, Fullerton, Calif.). Since the reaction product, PDC, has absorbance at 340 nm (ɛ340 = 2,540 M−1 cm−1; pH 8.0) and PDC is produced in an equimolar amount with NADH or NADPH, the increase of absorbance at 340 nm represents the sum of the amount of NADH or NADPH and PDC. We estimated the actual amount of NADH or NADPH by using the sum of molar extinction coefficients for NADH or NADPH and PDC. The 1-ml reaction mixture contained 150 μM CHMS, 200 μM NAD+ or NADP+, and the enzyme in 20 mM Tris-HCl buffer (pH 8.0). The reaction was carried out at 25°C in a cuvette. One unit of the enzyme is defined as the amount that converted 1 μmol of NAD+ or NADP+ to NADH or NADPH, respectively, per min at 25°C. Specific activity was expressed as units per milligram of protein. The optimum pHs were examined in the pH range of 6.0 to 9.0 by using buffers consisting of 20 mM potassium phosphate (pH 6.0 to 8.0) and 20 mM Tris-HCl buffer (pH 7.5 to 9.0). The Km and Vmax values were calculated by the Hanes-Woolf plots obtained from at least three independent experiments.
Enzyme purification.
Enzyme purification was performed according to the method described below using a BioCAD700E apparatus (PerSeptive Biosystems, Framingham, Mass.).
(i) Preparation of cell extract.
The cells were grown in 100 ml of LB medium containing 100 mg of ampicillin per liter at 37°C. The expression of the ligC gene was induced for 3.5 h by adding isopropyl-β-d-thiogalactopyranoside (final concentration, 1 mM) when the optical density (660 nm) of culture reached 0.5. The cells were then pelleted and resuspended in 20 mM Tris-HCl buffer (pH 8.0) (buffer A). Bacterial cells were broken by two passages through a French pressure cell. Crude cell lysate was centrifuged at 15,000 × g for 15 min. Streptomycin (final concentration, 1%) was added to the supernatant that was centrifuged at 15,000 × g for 15 min to remove nucleic acids. The supernatant was recentrifuged at 150,000 × g for 60 min at 4°C and concentrated by ultrafiltration using a Minicon B15 filter (Amicon, Beverly, Mass.) to obtain the crude extract.
(ii) POROS PI anion-exchange chromatography.
The crude extract was applied to a POROS PI (polyethyleneimine) column (7.5 by 100 mm) (PerSeptive Biosystems) previously equilibrated with buffer A. The enzyme was eluted with 88 ml of a linear gradient of 0 to 0.5 M NaCl. The CHMS dehydrogenase was eluted approximately at 0.29 M.
(iii) POROS HQ anion-exchange chromatography.
The fractions containing CHMS dehydrogenase activity eluted from a PI column were pooled, desalted, and concentrated by ultrafiltration using a Minicon B15 filter. The resulting solution was applied to a POROS HQ (quaternized PI) column (4.6 by 100 mm) (PerSeptive Biosystems) previously equilibrated with buffer A. The enzyme was eluted with 33 ml of a linear gradient of 0 to 0.5 M NaCl. The fractions containing CHMS dehydrogenase activity eluting at approximately 0.23 M were pooled.
(iv) POROS PE hydrophobic-interaction chromatography.
The fractions containing CHMS dehydrogenase activity were pooled, desalted, and concentrated as described above. Ammonium sulfate was added to the enzyme solution to a final concentration of 2 M. After the centrifugation at 15,000 × g for 10 min, the supernatant was applied to a POROS PE (phenylether) column (4.6 by 100 mm) (PerSeptive Biosystems) equilibrated with buffer B (buffer A containing 2 M ammonium sulfate). The enzyme was eluted with 25 ml of a linear gradient of 2.0 to 0 M ammonium sulfate. The fractions containing CHMS dehydrogenase activity eluting at approximately 1.62 M were pooled, desalted, and concentrated as described above. Glycerol was added to a final concentration of 10%, and the purified enzyme was stored at −80°C until use.
Analytical methods.
The protein concentration was determined by the method of Bradford (4). The homogeneity of the enzyme preparation was examined by sodium dodecyl sulfate–12% polyacrylamide gel electrophoresis (SDS-PAGE) (12). The molecular mass of the native enzyme was determined by Superdex200 HR10/30 (Pharmacia Biotech) gel filtration column chromatography using a BioCAD700E apparatus. The elution was performed using 50 mM potassium phosphate buffer (pH 7.0) containing 0.15 M NaCl with a flow rate of 0.5 ml/min. The molecular weight was estimated based on the calibration curve of reference proteins.
To determine the N-terminal amino acid sequence, the purified enzyme was subjected to SDS-PAGE (12% polyacrylamide gel) and electroblotted onto a polyvinylidene difluoride membrane (Bio-Rad, Hercules, Calif.). The enzyme band was cut out and analyzed on a PPSQ-21 protein sequencer (Shimadzu, Kyoto, Japan). The isoelectric point of LigC was determined by isoelectric focusing on an Ampholine PAG plate (pH 3.5 to 9.5) (Pharmacia Biotech) using a model Multiphor II electrophoresis system (Pharmacia Biotech).
Identification of the substrate and the reaction products was carried out using a GC-MS (model 5971A; Hewlett-Packard Co., Palo Alto, Calif.) with an Ultra-2 capillary column (50 m by 0.2 mm; Hewlett-Packard Co.) and an ESI-MS (HP1100 series LC-MSD; Hewlett-Packard Co.). For GC-MS analysis, the substrate and the reaction products in the buffer were acidified and extracted with ethylacetate, and then the extract was dried in vacuo and trimethylsilylated (TMS). The analytical conditions for GC-MS were the same as described in the previous study (23). In the analysis by ESI-MS, mass spectra were obtained by negative-mode ESI, with a needle voltage of −3.5 kV and a source temperature of 350°C. The sample was diluted 10-fold with 10 mM Tris-acetate buffer (pH 8.0) and injected into the flow system; the water/methanol ratio was 90:10 (vol/vol), and the flow rate was 0.2 ml/min.
Disruption of the ligC gene.
The 2.5-kb XbaI-EcoRI fragment carrying ligAB and part of ligC was cloned into pBluescript II SK(+) to generate pABC25. The BsmI fragment in the ligC gene of pABC25 was replaced by the 1.2-kb PstI fragment carrying the kanamycin resistance gene from pUC4K. The XbaI-EcoRI fragment of the resultant plasmid pABC25K containing the inactivated ligC gene was cloned into pK19mobsacB (34) to generate pLCD1.
The ligC-disrupted plasmid, pLCD1, was introduced into the SYK-6 cells by electroporation with a Genepulser (Bio-Rad). The colonies resistant to both kanamycin and sucrose were selected as described earlier (23). Southern hybridization analysis of the PstI digests of total DNAs prepared from the candidates for mutants was carried out with the 1.0-kb XhoI-EcoRI and 1.2-kb PstI fragment probes containing ligC and kanamycin resistance genes, respectively.
The metabolites of vanillate by the ligC insertion mutant (DLC) were analyzed. DLC cells grown in 10 ml of LB medium were washed with 10 mM Tris-acetate buffer (pH 8.0). The cells were resuspended in the same buffer and incubated with 12 mM vanillate for 12 h at 30°C. After centrifugation, the supernatant was diluted 20-fold with the buffer and analyzed by ESI-MS as described above.
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper was deposited in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession no. AB035122 and M34835.
RESULTS
Isolation and nucleotide sequence of the CHMS dehydrogenase gene.
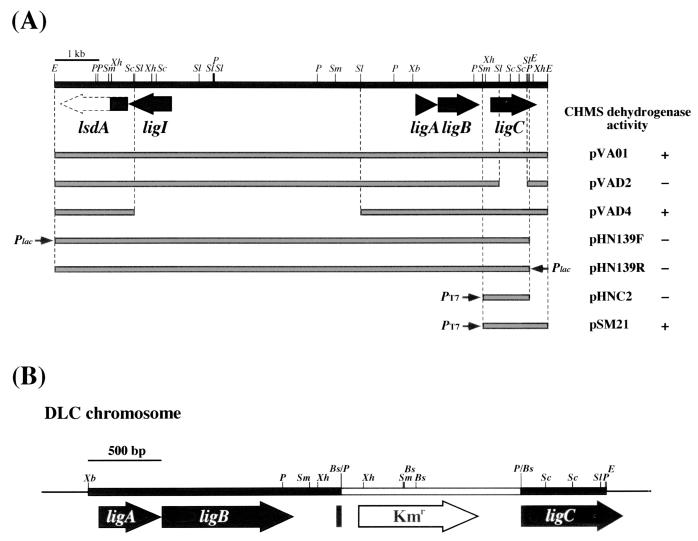
CHMS dehydrogenase activity was detected in the crude extract of Pseudomonas putida PpY1100 harboring pVA01, which contained the PCA 4,5-dioxygenase (ligAB) and PDC hydrolase (ligI) genes. Deletion analysis of pVA01, whose deletions were generated by partial SalI digestion, indicated that the intact CHMS dehydrogenase gene was included in pVAD4 but not in pVAD2 (Fig. 2A). These results suggested that the CHMS dehydrogenase gene was localized downstream of ligB. However, the E. coli transformant harboring pHN139F and pHN139R containing the 10.5-kb EcoRI fragment of pVA01 did not show CHMS dehydrogenase activity (Fig. 2A), and this suggested that the CHMS dehydrogenase gene was truncated at the right end of the 10.5-kb EcoRI fragment. Thus, the 0.4-kb EcoRI fragment located in the right end of pVA01 seemed to contain a part of the CHMS dehydrogenase gene. We determined the nucleotide sequences of the 1.0-kb XhoI fragment and the 0.4-kb EcoRI fragment carrying the partial CHMS dehydrogenase gene. In the 1,608-bp DNA region bounded by the PstI and EcoRI sites, a unique open reading frame (ORF) of 945 bp preceded by a putative ribosome binding site (AGGA) (35) was found and seemed to be the CHMS dehydrogenase gene. This ORF was designated ligC. The 1.4-kb DNA region bounded by SmaI and EcoRI sites carrying the entire ligC gene was cloned in pET21(+) to construct pSM21, and ligC was expressed in E. coli BL21(DE3) under the control of the T7 promoter. Production of the 35-kDa protein was observed by SDS-PAGE (data not shown). The size of this product is close to the molecular mass calculated from the deduced amino acid sequence of ligC (Mr, 34,590). Incubation of CHMS with the crude cell extract containing the ligC gene product (LigC) revealed the decrease in the absorbance at 410 nm derived from CHMS and the increase in the absorbance at 340 nm derived from NADH or NADPH and PDC, which is the reaction product of CHMS (data not shown). These results indicated that LigC actually encodes the CHMS dehydrogenase.
FIG. 2.
Deletion analysis of the CHMS dehydrogenase gene and the insertional inactivation of ligC in S. paucimobilis SYK-6. (A) The CHMS dehydrogenase activities of the cells harboring each subclone are presented. The small arrows indicate the direction of transcription from the lac (Plac) or T7 (PT7) promoters. Large filled arrows indicate the PCA 4,5-cleavage pathway genes, ligI, ligA, ligB, and ligC. A partly filled large arrow represents part of the ORF of the lignostilbene α,β-dioxygenase homolog (lsdA). E. coli JM109, E. coli BL21(DE3), and P. putida PpY1100 were used as host strains for pHN139F and pHN139R; pHNC2 and pSM21; and pVA01, pVAD2, and pVAD4, respectively. E, EcoRI; P, PstI; Sc, SacI; Sl, SalI; Sm, SmaI; Xb, XbaI; Xh, XhoI. (B) Schematic representation of the insertional inactivation of ligC by the kanamycin resistance gene from pUC4K. Bs, BsmI; E, EcoRI; P, PstI; Sc, SacI; Sl, SalI; Sm, SmaI; Xb, XbaI; Xh, XhoI.
A similarity search of the deduced amino acid sequence of ligC revealed 19 to 20% identity with 3-chlorobenzoate cis-4,5-dihydrodiol dehydrogenase (CbaC) of Alcaligenes sp. strain BR60 (25) and phthalate cis-4,5-dihydrodiol dehydrogenases of P. putida NMH102-2 (Pht4) (28) and Burkholderia cepacia DBO1 (OphB) (5). These aromatic dihydrodiol dehydrogenases, whose substrate is a dihydrodiol compound with a carboxyl group, are thought to be a new class of alcohol dehydrogenase, which has little similarity to the group I, II, and III microbial alcohol dehydrogenases (24, 31).
Purification of CHMS dehydrogenase.
In order to characterize the enzyme properties of CHMS dehydrogenase, LigC was purified from the cell extract of E. coli harboring pSM21 by a series of column chromatography procedures with PI, HQ, and PE. LigC was purified approximately 14-fold to near homogeneity with a recovery of 10% (Table 2). N-terminal amino acid sequencing revealed that the first 20 residues completely corresponded to the deduced amino acid sequence of ligC.
TABLE 2.
Purification of CHMS dehydrogenase from E. coli BL21(DE3) harboring pSM21
| Fraction | Total protein (mg) | Total activity (U) | Sp act (U/mg) | Yield (%) | Fold |
|---|---|---|---|---|---|
| Crude extract | 40.7 | 1,040 | 25.6 | 100 | 1 |
| PI | 3.23 | 394 | 122 | 38 | 4.8 |
| HQ | 0.70 | 182 | 260 | 18 | 10 |
| PE | 0.30 | 108 | 360 | 10 | 14 |
Conversion of CHMS into PDC by purified LigC.
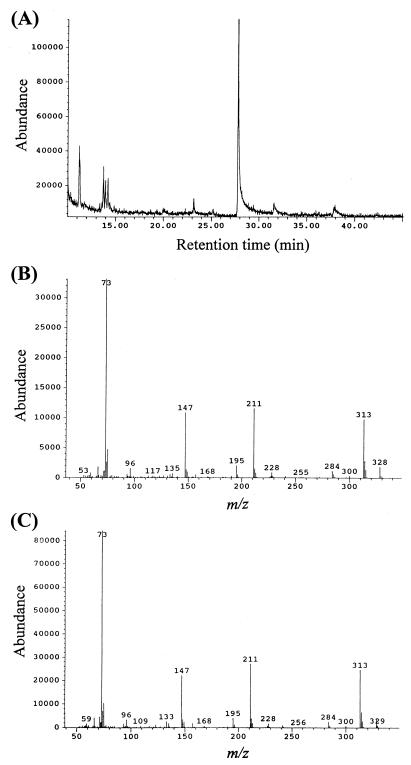
The TMS derivative of CHMS could not be detected by GC-MS, probably due to its instability, so we employed ESI-MS. Negative-ion ESI-MS spectra of CHMS yielded a deprotonated molecular ion at m/z 185.0. When 200 μM CHMS was incubated for 1 h with 0.5 μg of purified LigC and 200 μM NADP+, the ion at m/z 185.0 disappeared completely, and the reaction product ion at m/z 183.0 appeared, which corresponds to the deprotonated molecular ion of PDC. When NADP+ was omitted, the product was not detected. Figure 3 shows the GC and mass spectrum of the TMS derivative of the reaction product of LigC from CHMS. A product peak with a retention time of 27.8 min was observed, and the mass spectrum of the product was identical to that of the authentic PDC. Thus, we concluded that LigC catalyzed the conversion of CHMS into PDC depending on the presence of NADP+.
FIG. 3.
Identification of the reaction product from CHMS catalyzed by LigC. (A) Gas chromatogram of the TMS derivative of the reaction product from CHMS catalyzed by purified LigC. CHMS (100 μM) was incubated with 3 μg of purified LigC for 2 min in the presence of 200 μM NADP+. The reaction product was extracted by ethylacetate and trimethylsilylated (TMS). (B) Mass spectrum of the TMS derivative of the product with a retention time of 27.8 min shown in panel A. (C) Mass spectrum of the authentic TMS-PDC.
Enzyme properties.
Gel filtration column chromatography with a Superdex200 chromatograph demonstrated that the molecular mass of the native LigC was 67 kDa, indicating its dimeric structure. The optimum pH and temperature were estimated to be 8.0 and 25°C, respectively, and the isoelectric point was determined to be 5.3.
Table 3 shows the kinetic constants of LigC. With setting of the initial concentration of CHMS to 150 μM, LigC showed 10-times-higher affinity to NADP+ than to NAD+. Km for NAD+ was estimated to be 252 μM. Vmax for CHMS oxidation with NAD+ showed a value similar to that with NADP+. With adjustment of the initial concentration of a cofactor to 200 μM, Km for CHMS with NAD+ agreed with that with NADP+. However, Vmax for CHMS oxidation with NAD+ did not agree with that with NADP+. The former is two times lower than the latter. These results are probably due to the initial concentration of NAD+ (200 μM) used being lower than the Km value for NAD+ (252 μM).
TABLE 3.
Kinetic constants of the CHMS dehydrogenases
| Strain | Cofactor (μM) | CHMS (μM) | Km (μM)a | Vmax (U/mg) |
|---|---|---|---|---|
| SYK-6 | NAD+ | 150 | 252 ± 3.9 (NAD+) | 449 ± 3.9 |
| NADP+ | 150 | 24.6 ± 1.5 (NADP+) | 363 ± 1.4 | |
| NAD+ (200) | 20.6 ± 1.0 (CHMS) | 175 ± 10 | ||
| NADP+ (200) | 26.0 ± 0.5 (CHMS) | 383 ± 3.8 | ||
| P. ochraceaeb | NAD+ | 158 | 284 (NAD+) | 470 |
| NADP+ | 158 | 17.5 (NADP+) | 470 | |
| NAD+ (110) | 56 (CHMS) | 167 | ||
| NADP+ (110) | 20 (CHMS) | 450 |
The Kms indicated are the values for the compounds shown in parentheses.
The results for the P. ochraceae enzyme have been reported in a previous study (17).
We also examined the influence of sulfhydryl reagents on LigC. Treatments of 5 μg of purified LigC with p-chloromercuribenzoate (10 μM), HgCl2 (10 μM), or 5,5′-dithiobis(2-nitrobenzoate) (100 μM) for 1 h inhibited 78, 83, or 98% of LigC activity, respectively. These results suggested that a cysteine residue might be involved in the enzyme reaction. Neither inhibition nor stimulation of enzyme activity was observed in the presence of 5 mM EDTA.
ligC disruption in S. paucimobilis SYK-6.
In order to clarify the involvement of the ligC gene in the degradation of model lignin compounds, the ligC gene in SYK-6 was disrupted. The insertion mutant of ligC was obtained by the introduction of pLCD1, in which ligC in the 2.5-kb XbaI-EcoRI fragment inserted in pK19mobsacB was inactivated by the insertion of the 1.2-kb kanamycin resistance gene. Southern hybridization analysis of the ligC insertion mutant using the ligC and the kanamycin resistance gene probes revealed that the ligC gene was inactivated by homologous recombination through the double crossover. This mutant strain was designated DLC and used for the following experiments (Fig. 2B). DLC was not able to grow on vanillate, but there was no difference in the growth on syringate between DLC and the wild type, SYK-6.
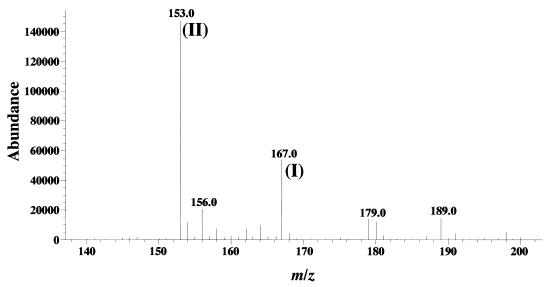
The metabolites of vanillate accumulated during the incubation of the whole cells of DLC and SYK-6 pregrown in LB medium were analyzed by ESI-MS. After 12 h of incubation, only the remaining vanillate and the metabolite, PCA, were detected in the DLC culture, and no CHMS had accumulated (Fig. 4). In the SYK-6 culture, vanillate disappeared thoroughly, and no metabolites were accumulated. When the broad-host-range vector pBBR122 carrying ligC (pBBRC1221) was introduced into the DLC cells, pBBRC1221 restored the ability to grow on vanillate. These results indicated the repression of PCA 4,5-dioxygenase activity in DLC.
FIG. 4.
Identification of the metabolite accumulated in DLC culture from vanillate. Vanillate was incubated with the whole cells of DLC for 12 h. The negative-ion ESI-MS spectrum of a portion of the supernatant of a reaction mixture is shown. The detected ion (II) at m/z 153.0 corresponded to the deprotonated molecular ion of PCA and was a major product from vanillate (ion I). The ion at m/z 185.0 representing CHMS was not detected.
DISCUSSION
The nucleotide sequence of ligC was determined in this study. The deduced amino acid sequence of the ligC gene revealed approximately 20% identity with CbaC of Alcaligenes sp. strain BR60 (25), Pht4 of P. putida NMH102-2 (28), and OphB of B. cepacia DBO1 (5), which are dehydrogenases for carboxylic cis-dihydrodiol compounds involved in the degradation of 3-chlorobenzoate or phthalate. The identities among these enzymes ranged from 45 to 64%, and they do not have any apparent relationship with group I (long-chain zinc-dependent enzyme), group II (short-chain zinc-independent enzyme), or group III (iron-activated enzyme) microbial NAD(P)+-dependent alcohol dehydrogenases (31). Among the enzymes related to CbaC (CbaC family), the amino acid sequence H-(X)11-K-H-V-L-X-E-K-P-X-A was conserved (24). LigC contains the largest part of this conserved sequence, spanning amino acid positions 75 to 96 [H-(X)11-K-H-V-Q-V-E-I-P-L-A]. The CbaC family members also had the consensus sequence G-X-X-G-X-G at their N terminus, which is thought to be the dinucleotide-binding motif for NAD(P)+. However, instead of this motif, LigC had G-X-G-X-X-G. The length of LigC was 315 amino acids, which is much less than those of CbaC (397 amino acids), Pht4 (410 amino acids), and OphB (391 amino acids). Thus, LigC is similar to the CbaC family enzymes, but it does not have an obvious relationship with them.
The ligC gene was located 295 bp downstream of the ligB gene, and the putative ρ-independent terminator was found 24 bp downstream of ligC. This suggested that the transcription of ligC terminates at this terminator. In addition to this sequence, the 21-bp inverted repeat sequence (nucleotide positions 86 to 106 and 213 to 233 in the sequence of AB035122) was found in the intergenic region between ligB and ligC. Further research is needed to address the actual operon structure of ligA, ligB, and ligC.
The ligC gene was expressed in E. coli, and its product was purified to near homogeneity. The reaction product from CHMS catalyzed by purified LigC was determined as PDC by both GC-MS and ESI-MS (Fig. 3). These results are consistent with the reports of Maruyama (13, 14) and Kersten et al. (10). Maruyama et al. initially proposed that the CHMS dehydrogenase would catalyze the conversion of CHMS into CHM, which was not confirmed experimentally (17). Afterwards, Maruyama identified PDC as the reaction product and proposed that this enzyme would be involved in the oxidation of the hemiacetal form to PDC. CHMS seems to be in equilibrium between the open form and the cyclic hemiacetal form (Fig. 1), as suggested by Kersten et al. (10). Such an equilibrium between the open and cyclic hemiacetal forms is also suggested for the naphthalene metabolism by the NAH7 plasmid-encoded enzymes by Eaton and Chapman (7). Based on the chemical structure of PDC as an α-pyrone, it would be produced from the cyclic hemiacetal form of CHMS. Previously, Maruyama et al. (17) determined the CHMS dehydrogenase activity by monitoring the decrease in absorbance at 410 nm, which is the absorption maximum of CHMS (17). In this study, we monitored the generation of NADH and NADPH from NAD+ and NADP+, respectively, during CHMS oxidation to examine the CHMS dehydrogenase activity. The absorbance at 410 nm of CHMS generated from PCA by LigAB gradually decreased; it did not correspond to the amount of CHMS estimated by ESI-MS. When CHMS was prepared from PCA, the amount of CHMS estimated by ESI-MS increased as time went by. However, the absorbance at 410 nm initially increased and then decreased (unpublished results), which indicates that the absorbance at 410 nm represents the amount of the open form of CHMS rather than the total amount of CHMS, and the absorbance at 410 nm is not a good index of the activity of CHMS dehydrogenase, whose substrate was suggested to be a hemiacetal form of CHMS by Kersten et al. (10) and Maruyama (14).
The purified LigC showed features in common with CHMS dehydrogenase of Pseudomonas ochraceae, including the subunit structure, molecular mass, pI, and kinetic parameters (Table 3). Both enzymes showed a much higher affinity to NADP+ than to NAD+, which is a remarkable common kinetic feature. Both were inhibited by the addition of sulfhydryl reagents. This may suggest that the cysteine residue plays a key role in the enzyme reaction. One of the possible roles of cysteine residue is as a ligand to a metal ion. Based on the fact that enzyme activities of both LigC and P. ochraceae CHMS dehydrogenase were not affected by the addition of EDTA, it is clear that the cysteine residue is not involved in the binding of metal ions such as zinc in the group I dehydrogenases (31).
The ligC insertion mutant, DLC, lost the ability to grow on vanillate, indicating that the ligC gene is essential for vanillate catabolism (Fig. 2B). On the other hand, the ligC disruption did not affect the growth on syringate. These results are consistent with those obtained regarding both the ligB (H. Aoshima, E. Masai, S. Nishikawa, Y. Katayama, and M. Fukuda, Abstr. 8th Int. Symp. Microb. Ecol., abstr. 93, 1998) and ligI (23) disruptions. Although PCA 4,5-dioxygenase has the ability to convert 3-O-methylgallate, a metabolic intermediate of syringate, to PDC (10), these results indicated that syringate is mainly metabolized through a pathway in SYK-6 other than the PCA 4,5-cleavage pathway encoded by the ligAB, ligC, and ligI genes.
Interestingly, it was not CHMS but PCA which accumulated from vanillate during the incubation with the ligC insertion mutant DLC (Fig. 4). When pBBRC1221 carrying ligC was introduced into the strain DLC, the resultant transformant recovered the ability to grow on vanillate. We confirmed that the PCA 4,5-dioxygenase activity of the cell extract of E. coli carrying ligAB in response to 100 μM PCA was not inhibited by the equivalent molar amount of CHMS (data not shown). This result suggested that the accumulation of PCA observed in the DLC culture containing vanillate was not due to the product inhibition of PCA 4,5-dioxygenase. Thus, PDC seemed to be an inducing substance for PCA 4,5-dioxygenase encoded by ligAB. In the absence of PDC, PCA 4,5-dioxygenase is not induced in DLC and PCA will be accumulated instead of CHMS. This notion is supported by the results with ligI insertion mutant DLI, which accumulated PDC from vanillate (23). To address the details of regulation of the PCA 4,5-cleavage pathway genes, promoter regions and the regulatory genes governing them should be clarified.
ACKNOWLEDGMENT
This work was supported in part by a Grant-in Aid for Encouragement of Young Scientists (no. 11760057) from the ministry of Education, Science, Sports and Culture, Japan.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1990. [Google Scholar]
- 2.Bagdasarian M, Lurz R, Rückert B, Franklin F C H, Bagdasarian M M, Frey J, Timmis K N. Specific purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010 derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981;16:237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- 3.Bolivar F, Backman K. Plasmids of Escherichia coli as cloning vectors. Methods Enzymol. 1979;68:245–267. doi: 10.1016/0076-6879(79)68018-7. [DOI] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Chang H-K, Zylstra G J. Novel organization of the genes for phthalate degradation from Burkholderia cepacia DBO1. J Bacteriol. 1998;180:6529–6537. doi: 10.1128/jb.180.24.6529-6537.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crawford R L, Bromley J W, Perkins-Olson P E. Catabolism of protocatechuate by Bacillus macerans. Appl Environ Microbiol. 1979;37:614–618. doi: 10.1128/aem.37.3.614-618.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eaton R W, Chapman P J. Bacterial metabolism of naphthalene: construction and use of recombinant bacteria to study ring cleavage of 1,2-dihydroxynaphthalene and subsequent reactions. J Bacteriol. 1992;174:7542–7554. doi: 10.1128/jb.174.23.7542-7554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harwood C S, Parales R E. The β-ketoadipate pathway and the biology of self-identity. Annu Rev Microbiol. 1996;50:553–590. doi: 10.1146/annurev.micro.50.1.553. [DOI] [PubMed] [Google Scholar]
- 9.Katayama Y, Nishikawa S, Nakamura M, Yano K, Yamasaki M, Morohoshi N, Haraguchi T. Cloning and expression of Pseudomonas paucimobilis SYK-6 genes involved in the degradation of vanillate and protocatechuate in P. putida. Mokuzai Gakkaishi. 1987;33:77–79. [Google Scholar]
- 10.Kersten P J, Dagley S, Whittaker J W, Arciero D M, Lipscomb J D. 2-Pyrone-4,6-dicarboxylic acid, a catabolite of gallic acids in Pseudomonas species. J Bacteriol. 1982;152:1154–1162. doi: 10.1128/jb.152.3.1154-1162.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knauf V C, Nester E W. Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid. 1982;8:45–54. doi: 10.1016/0147-619x(82)90040-3. [DOI] [PubMed] [Google Scholar]
- 12.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 13.Maruyama K. Isolation and identification of the reaction product of α-hydroxy-γ-carboxymuconic-ɛ-semialdehyde dehydrogenase. J Biochem. 1979;86:1671–1677. doi: 10.1093/oxfordjournals.jbchem.a132687. [DOI] [PubMed] [Google Scholar]
- 14.Maruyama K. Purification and properties of 2-pyrone-4,6-dicarboxylate hydrolase. J Biochem. 1983;93:557–565. doi: 10.1093/oxfordjournals.jbchem.a134210. [DOI] [PubMed] [Google Scholar]
- 15.Maruyama K. Purification and properties of γ-oxalomesaconate hydratase from Pseudomonas ochraceae grown with phthalate. Biochem Biophys Res Commun. 1985;128:271–277. doi: 10.1016/0006-291x(85)91674-2. [DOI] [PubMed] [Google Scholar]
- 16.Maruyama K. Purification and properties of 4-hydroxy-4-methyl-2-oxoglutarate aldolase from Pseudomonas ochraceae grown on phthalate. J Biochem. 1990;108:327–333. doi: 10.1093/oxfordjournals.jbchem.a123201. [DOI] [PubMed] [Google Scholar]
- 17.Maruyama K, Ariga N, Tsuda M, Deguchi K. Purification and properties of α-hydroxy-γ-carboxymuconic-ɛ-semialdehyde dehydrogenase. J Biochem. 1978;83:1125–1134. doi: 10.1093/oxfordjournals.jbchem.a132002. [DOI] [PubMed] [Google Scholar]
- 18.Masai E, Katayama Y, Kawai S, Nishikawa S, Yamasaki M, Morohoshi N. Cloning and sequencing of the gene for a Pseudomonas paucimobilis enzyme that cleaves β-aryl ether. J Bacteriol. 1991;173:7950–7955. doi: 10.1128/jb.173.24.7950-7955.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masai E, Katayama Y, Kubota S, Kawai S, Yamasaki M, Morohoshi N. A bacterial enzyme degrading the model lignin compound β-etherase is a member of the glutathione-S-transferase superfamily. FEBS Lett. 1993;323:135–140. doi: 10.1016/0014-5793(93)81465-c. [DOI] [PubMed] [Google Scholar]
- 20.Masai E, Katayama Y, Nishikawa S, Fukuda M. Characterization of Sphingomonas paucimobilis SYK-6 genes involved in degradation of lignin-related compounds. J Ind Microbiol Biotechnol. 1999;23:364–373. doi: 10.1038/sj.jim.2900747. [DOI] [PubMed] [Google Scholar]
- 21.Masai E, Katayama Y, Nishikawa S, Yamasaki M, Morohoshi N, Haraguchi T. Detection and localization of a new enzyme catalyzing the β-aryl ether cleavage in the soil bacterium (Pseudomonas paucimobilis SYK-6) FEBS Lett. 1989;249:348–352. doi: 10.1016/0014-5793(89)80656-8. [DOI] [PubMed] [Google Scholar]
- 22.Masai E, Kubota S, Katayama Y, Kawai S, Yamasaki M, Morohoshi N. Characterization of the Cα-dehydrogenase gene involved in the cleavage of β-aryl ether by Pseudomonas paucimobilis SYK-6. Biosci Biotechnol Biochem. 1993;57:1655–1659. doi: 10.1271/bbb.57.1655. [DOI] [PubMed] [Google Scholar]
- 23.Masai E, Shinohara S, Hara H, Nishikawa S, Katayama Y, Fukuda M. Genetic and biochemical characterization of a 2-pyrone-4,6-dicarboxylic acid hydrolase involved in the protocatechuate 4,5-cleavage pathway of Sphingomonas paucimobilis SYK-6. J Bacteriol. 1999;181:55–62. doi: 10.1128/jb.181.1.55-62.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakatsu C H, Providenti M, Wyndham R C. The cis-diol dehydrogenase cbaC gene of Tn5271 is required for growth on 3-chlorobenzoate but not 3,4-dichlorobenzoate. Gene. 1997;196:209–218. doi: 10.1016/s0378-1119(97)00229-1. [DOI] [PubMed] [Google Scholar]
- 25.Nakatsu C H, Wyndham R C. Cloning and expression of the transposable chlorobenzoate 3,4-dioxygenase genes of Alcaligenes sp. strain BR60. Appl Environ Microbiol. 1993;59:3625–3633. doi: 10.1128/aem.59.11.3625-3633.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishikawa S, Sonoki T, Kasahara T, Obi T, Kubota S, Kawai S, Morohoshi N, Katayama Y. Cloning and sequencing of the Sphingomonas (Pseudomonas) paucimobilis gene essential for the O demethylation of vanillate and syringate. Appl Environ Microbiol. 1998;64:836–842. doi: 10.1128/aem.64.3.836-842.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noda Y, Nishikawa S, Shiozuka K, Kadokura H, Nakajima H, Yoda K, Katayama Y, Morohoshi N, Haraguchi T, Yamasaki M. Molecular cloning of the protocatechuate 4,5-dioxygenase gene of Pseudomonas paucimobilis. J Bacteriol. 1990;172:2704–2709. doi: 10.1128/jb.172.5.2704-2709.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nomura Y, Nakagawa M, Ogawa N, Harashima S, Oshima Y. Genes in PHT plasmid encoding the initial degradation pathway of phthalate in Pseudomonas putida. J Ferment Bioeng. 1992;74:333–334. [Google Scholar]
- 29.Peng X, Egashira T, Hanashiro K, Masai E, Nishikawa S, Katayama Y, Kimbara K, Fukuda M. Cloning of a Sphingomonas paucimobilis SYK-6 gene encoding a novel oxygenase that cleaves lignin-related biphenyl and characterization of the enzyme. Appl Environ Microbiol. 1998;64:2520–2527. doi: 10.1128/aem.64.7.2520-2527.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng X, Masai E, Katayama Y, Fukuda M. Characterization of the meta-cleavage compound hydrolase gene involved in degradation of the lignin-related biphenyl structure by Sphingomonas paucimobilis SYK-6. Appl Environ Microbiol. 1999;65:2789–2793. doi: 10.1128/aem.65.6.2789-2793.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reid M F, Fewson C A. Molecular characterization of microbial alcohol dehydrogenases. Crit Rev Microbiol. 1994;20:13–56. doi: 10.3109/10408419409113545. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 35.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementary to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Short J M, Fernandez J M, Sorge J A, Huse W. λZAP: a bacteriophage λ expression vector with in vivo excision properties. Nucleic Acids Res. 1988;16:7583–7600. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 38.Sugimoto K, Senda T, Aoshima H, Masai E, Fukuda M, Mitsui Y. Crystal structure of an aromatic-ring-opening dioxygenase LigAB which is a protocatechuate 4,5-dioxygenase. Struct Fold Des. 1999;15:953–965. doi: 10.1016/s0969-2126(99)80122-1. [DOI] [PubMed] [Google Scholar]
- 39.Tack B F, Chapman P J, Dagley S. Purification and properties of 4-hydroxy-4-methyl-2-oxoglutarate aldolase. J Biol Chem. 1972;247:6444–6449. [PubMed] [Google Scholar]
- 40.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 41.Wolgel S A, Dege J E, Perkins-Olson P E, Juarez-Garcia C H, Crawford R L, Munck E, Lipscomb J D. Purification and characterization of protocatechuate 2,3-dioxygenase from Bacillus macerans: a new extradiol catecholic dioxygenase. J Bacteriol. 1993;175:4414–4426. doi: 10.1128/jb.175.14.4414-4426.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]