Abstract
High-level transcription of eps, an operon encoding biosynthesis of an exopolysaccharide virulence factor of the phytopathogen Ralstonia (Pseudomonas) solanacearum, requires the products of at least seven regulatory genes (phcA, phcB, xpsR, vsrA-vsrD, and vsrB-vsrC), which are organized in three converging signal transduction cascades. Because xpsR and the vsrB-vsrC two-component system are the most downstream cascade components required for activation of eps, we explored how these components control transcription from the eps promoter (Peps). Deletion and PCR mutagenesis identified an upstream region of Peps (nucleotides −82 to −62) that is critical for transcription activation by VsrB-VsrC and XpsR and also is required for negative control of Peps by the putative eps regulator EpsR. Using PCR mutagenesis we generated the vsrC1 allele that encodes a response regulator that constitutively activates Peps in the absence of its cognate sensor, VsrB. However, activation of Peps by vsrC1 still required xpsR. Unexpectedly, the amino acid substitution conferring the constitutive phenotype on VsrC1 is 12 residues from its C terminus, outside the known functional domains of response regulators. Finally, a modified DNase I footprinting method was used to demonstrate specific binding of both VsrC1 and VsrC to the −72 to −62 upstream region of Peps.
Ralstonia (Pseudomonas) solanacearum, which causes a lethal wilting disease of solanaceous and many other types of other plants (15, 16), enters hosts via natural openings or wounds in roots and then proceeds to extensively colonize xylem vessels of the vascular system (37, 44). Although secreted plant cell wall-degrading exoenzymes enhance virulence (possibly by facilitating invasion and vascular colonization [23, 24, 37]), it is exopolysaccharide I (EPS I), a large, nitrogen-rich, acidic exopolysaccharide (34), that is the primary virulence factor of R. solanacearum. EPS I is produced in copious amounts and is required for wilting and killing of hosts (8, 29). EPS I apparently causes wilting by restricting water flow through xylem vessels (6). It also markedly enhances the speed and extent of stem colonization (37).
In R. solanacearum, production of EPS I (as well as some exoenzymes) is stringently controlled by a cascading network of more than 10 regulatory genes (5, 11, 20, 41). Inactivation of any of seven genes in this network causes a >85% reduction in transcription from the eps promoter (Peps), leading to loss of EPS I production and the ability to wilt and kill. However, inactivation of all but two of these genes (vsrB and vsrC) can be suppressed by constitutive expression of xpsR from a vector promoter (20). These and other data showed that VsrB, VsrC, and XpsR are the most downstream components in the eps regulatory cascade and suggested that they may directly affect interaction of RNA polymerase with Peps.
The predicted amino acid sequences of VsrB and VsrC imply that they comprise a two-component system in which VsrB is a sensor kinase and VsrC is its cognate response regulator (19, 20). However, no homologs of the very basic XpsR protein have been found. How these three proteins interact to control Peps is not clear. Nonetheless, analogy to other two-component systems (17) implies that, in response to some unknown signal, VsrB phosphorylates VsrC, thereby stimulating it to turn on transcription, possibly via direct binding to Peps. EpsR, a putative DNA-binding protein, is another potential regulator of Peps, since its overproduction strongly represses EPS I synthesis by R. solanacearum strain K60 (18, 25).
Since there was no clear or direct evidence for physical interactions between Peps and VsrC, XpsR, or EpsR, we first used deletion and PCR mutagenesis to define an upstream region of Peps that is absolutely required for its transcription activation by VsrB-VsrC and XpsR. Then we generated a vsrC allele that activated Peps independently of vsrB and used DNase I footprinting to show that both this constitutively active VsrC protein and wild-type VsrC directly bind to the region of Peps that was identified by mutagenesis to be important in transcription activation. However, since the affinity of VsrC for Peps was weak and since the constitutively active VsrC protein was unresponsive to VsrB and still required XpsR for activation of Peps, we speculate that XpsR may facilitate or stabilize binding of VsrC to Peps.
MATERIALS AND METHODS
Bacteria and plasmids.
Most strains and plasmids constructed and used in this work are described in Table 1. R. solanacearum was grown in BG or BSM medium (36) at 30°C, while Escherichia coli was grown in LB (33) at 37°C. The host strain and vectors used for cloning were E. coli DH5α (14) and pTZ19U/18U (31), respectively. pPF12 (7) and pEPS1 (21) were described previously. R. solanacearum strain AW22 was constructed by transposon mutagenesis with Tn3lacZ (7). Derivatives of AW22 were constructed by natural transformation (20) with genomic DNA from AW-R164, AW-C, and AW-MG2 (19). Concentrations (micrograms per milliliter) of antibiotics used to select and maintain plasmids were kanamycin (Km), 50; spectinomycin (Sp), 50; ampicillin (Amp), 100; and tetracycline (Tc), 20.
TABLE 1.
Important R. solanacearum strains and plasmids used
| Strain/plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strain | ||
| AW | Wild-type pathogen, biovar 1 | 38 |
| AW201 | epsA1::npt1 | 21 |
| AW-C | vsrC::cat | 20 |
| AW-R164 | xpsR::nptI | 20 |
| AW22 | eps::Tn3-lacZ | This study |
| AW22B | AW22 vsrB::Ω | This study |
| AW22BC | AW22 vsrC::cat vsrB::Ω | This study |
| AW22RBC | AW22 vsrC::cat vsrB::Ω xpsR::nptI | This study |
| AW19A | epsB::Tn5lacZ | 4 |
| Plasmid | ||
| pXPS1 | 8-kb PstI fragment of pPF12 in pTZ18U | This study |
| pXPS2 | 900-bp BamHI fragment of pEPS1 in pTZ19U | This study |
| pXPS5 | 320-bp SphI fragment of pXPS2 in pTZ18U | This study |
| pTZSZ12 | 176-bp BamHI fragment of pPSZ12 in pTZ18U | This study |
| pPSZ22 | Nucleotides −44 to +23 of Peps fused to lacZ via BamHI site of pRG970 | This study |
| pPSZ15, pPSZ17, pPSZ20, pPSZ12, pPSZ19, pPSZ21 | Same as pPSZ22, but with longer lengths of Peps fused to lacZ (see Fig. 1) | This study |
| pPSZ15-2, pPSZ15-7 | pPSZ15 with −67 A to G and −72 G to T, respectively | This study |
| pEPSM2, pEPSM7 | pPSZ12 with −67 A to G and −72 G to T, respectively | This study |
| pKVC3 | RsaI-BamHI fragment of vsrC in pTZ19U | This study |
| pRKVC3 | vsrC from pKVC3 in pRK415 | This study |
| pVSRC1 | vsrC with H147R and S209L mutations in pRK415 | This study |
| pVSRC2 | vsrC with S209L mutation in pRK415 | This study |
| pVSRCH, pCB92 | vsrC and vsrC1, respectively, in pTrc-His | This study |
| pEPSR | epsR from AW in BamHI and EcoRI sites of pTZ19U | This study |
| pEPSR-T | StyI fragment of epsR in EcoRV site of pTOK2 | This study |
Construction of Peps reporter plasmids.
Peps fragments with various lengths of upstream sequences were made by PCR using primers EPS1 (5′-CCGGATCCCCAACTGTAAATCGTA-3′), EPS2 (5′-CCGGATCCAAACGAAATATGCATT-3′), EPS3 (5′-CCGGATCCTTTCGGTATTGAAGC-3′), EPS4 (5′-CCGGATCCATGCACAACCGTATC-3′), EPS5 (5′-TCGGATCCATCGCCACCGGTACTG-3′), EPS6 (CCGGATCCAGAACGATCCATGTTTC-3′), EPS7 (5′-CCCCGGGTTGGCGTTCTGCCTAT-3′), EPS9 (5′-GAGGATCCAGTTGCAGAAACGGCCA-3′), M13F (TGTAAAACGACGGCCAGT-3′), M13R (5′-AGCGAATAACAATTTCACACAGGA-3′), and T7 (5′-TAATACGACTCACTATAGGG-3′). PCR mixtures (100 μl) contained 10 mM Tris (pH 8.3), 50 mM KCl, 1 mM of each deoxynucleoside triphosphate (dNTP), 2 mM MgCl2, 0.1 nmol of primers, 1 to 5 ng of template, and 2 U of Taq polymerase (Perkin-Elmer). Each amplification cycle consisted of denaturation at 94°C for 1 min, annealing at 50°C for 1 min, and extension at 72°C for 1 min. After 30 cycles, there was a final extension at 72°C for 10 min. The Peps DNA fragments from nucleotides −538 to +23, −337 to +23, and −243 to +23 were amplified using pXPS1 as template and primer pairs EPS6-EPS4, EPS7-EPS4, and EPS9-EPS4, respectively. The Peps DNA fragments from −143 to +23, −101 to +23, −68 to +23, and −44 to +23 were amplified using primer pairs M13F-EPS4, EPS1-EPS4, EPS2-EPS4, and EPS3-EPS4, respectively, and pXPS5 as template. PCR products were digested with BamHI and/or SmaI and ligated into the promoterless lacZ fusion vector pRG970 (43) digested in the same manner. After transformation into E. coli and plating on LB agar with Sp and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), plasmid DNA was isolated from blue colonies, and the orientation and identity of the insert were confirmed.
PCR mutagenesis of Peps and vsrC.
To mutagenize Peps, sequences between nucleotides −143 and +23 were PCR amplified essentially as above using a pTZSZ12 template. However, the dATP concentration was 0.2 mM instead of 1 mM, and MnCl2 was added to 0.5 mM. BamHI-digested PCR products were ligated with BamHI-digested pRG970 and transformed into E. coli, and ∼10,000 blue-colored transformants arising on LB plates with Sp and X-Gal were individually picked and pooled, and plasmid DNA was isolated from them. Pooled plasmids were electroporated into R. solanacearum strain AW201 (epsA1::nptl). After plating on BG agar with Sp and X-Gal, white or pale-blue transformants (i.e., those with decreased LacZ activity) were observed at 0.3%. Plasmids from these colonies were individually isolated, transformed into E. coli, reisolated, and analyzed with restriction enzymes to confirm Peps inserts. Sequence alterations were determined after recloning inserts into pTZ19U.
To mutagenize vsrC, PCRs were performed under mutagenic conditions as above, using pKVC3 as template and an M13F-T7 primer pair. After digestion with HindIII and EcoRI, the PCR products were ligated with similarly digested pTZ19U and transformed into E. coli. Plasmid DNA was isolated en masse from ∼20,000 transformant colonies that were washed from plates. The vsrC alleles in this plasmid pool were released by digestion with HindIII and EcoRI and ligated with similarly digested pRK415 (26), followed by transformation into E. coli. Plasmid DNA was isolated from ∼20,000 pooled Tc-resistant colonies and electroporated into R. solanacearum strain AW22B (vsrB::Ω eps::lacZ). After selection on BG plates with X-Gal and Tc, transformants showing increased expression of the eps::lacZ reporter (i.e., darker-blue color) were analyzed.
To construct the vsrC2 allele, vsrC sequences between nucleotides 523 and 1,194 (GenBank U18134) were amplified using a pRKVC3 template, a T7 primer, and a primer containing vsrC sequences between nucleotides 1,162 and 1,194 but having a C1185T mutation (S209L) and a same-sense C1171G silent mutation that introduced a BamHI site (5′-TCCATGCGCAAGATGATCTGGATCCGCGTGCGC-3′). The resultant PCR product and a plasmid with sequences 1,145 to 1,290 of vsrC were mixed, and PCR amplification was performed with T7 and M13F primers. The 800-bp vsrC PCR product was digested with HindIII and EcoRI and cloned into pRK415. Plasmids from transformants were isolated and screened for the introduced BamHI site. The vsrC allele from one candidate plasmid (pVSRC2) was sequenced to confirm that it contained the S209L mutation.
Purification of His-tagged VsrC proteins.
Wild-type vsrC was PCR amplified using pKVC3 as template and primers M13F and VSRCN (5′-ACGGATCCACGAGCTCGCTGCG-3′; complementary to sequences encoding the N terminus of VsrC). The product was digested with BamHI and cloned in frame to the hexahistidine-encoding tag of BamHI-digested pTrc-HisA (Invitrogen), yielding plasmid pVSRCH. To generate pCB92, the SacI-KpnI fragment of pVSRCH was replaced with the analogous fragment from pVSRC1.
E. coli JM109 (45) cells containing pVSRCH or pCB92 were shaken at 37°C in LB plus Amp until the optical density at 600 nm (OD600) was 0.6. The 100-ml culture was shifted to 28°C, and isopropyl-β-d-thio-galactoside was added to 0.4 mM. After 18 h, cells were harvested, suspended in 10 ml of buffer B (5 mM imidazole, 500 mM NaCl, 20 mM Tris-HCl [pH 8.0]) and sonicated five times for 10 s. The sonicate was centrifuged at 30,000 × g for 25 min, and the supernatant was passed through a 3-ml column containing 1 ml of Ni-nitriloacetic acid resin (Sigma) equilibrated with buffer B. The column was washed with 10 ml of buffer B followed by 10 ml of buffer B with 50 mM imidazole. VsrCs were eluted with buffer B plus 0.5 M imidazole; sodium dodecyl sulfate-polyacrylamide gel electrophoresis indicated >90% purity.
Construction of an epsR null mutant.
The epsR homolog of R. solanacearum strain AW was PCR amplified from genomic DNA as above except that annealing and extension were at 70°C. Primers (EPSRN, 5′-AAGGATCCAGG CGGCGCAGTG-3′ [nucleotides 355 through 381 with an added BamHI site] and EPSRC, 5′-ATGAATTCAGCCCGGCGTGCACGAGGCG-3′ [nucleotides 1,055 through 1,075 with an added EcoRI site]) were based on the sequence of epsR from R. solanacearum strain K60 (GenBank M61197). The resultant PCR product was digested with BamHI and EcoRI and cloned into pTZ18U. DNA sequencing showed that epsR from strain AW is >97% identical to epsR from strain K60.
To construct the epsR null mutant, an internal StyI fragment lacking sequences encoding the first 12 N-terminal residues and last 52 C-terminal residues of EpsR was made blunt ended with T4 DNA polymerase and then cloned into the SmaI site of suicide vector pTOK2 (27). The resultant plasmid (pEPSR-T) was electroporated into R. solanacearum AW, and strains in which the plasmid integrated into epsR by a single recombination event were selected by Tc resistance. Genomic DNA from two strains was prepared and disruption of the genomic copy of epsR was confirmed by PCR; no PCR products could be obtained using EPSRN and EPSRC primers, whereas a PCR product of the predicted size was obtained with wild-type DNA as template. An EPSRN and M13R vector primer gave a PCR fragment of the size predicted for the truncated epsR gene.
Measurement of PglA.
Assay samples were spotted onto nitrocellulose and after blocking for 2 h at 25°C with 5% skim milk (Difco) in TBS (10 mM Tris HCl [pH 7.4], 140 mM NaCl), the membrane was washed three times with TBST (TBS with 0.2% Tween 20) and submerged for 1 h in 10 ml of TBST containing a 1:1,000 dilution of anti-PglA antiserum (39). After three washes with TBST, one with TBST containing 0.85 M NaCl, and one with TBST, bound PglA antibodies were detected with anti-rabbit immunoglobulin G (IgG) conjugated to alkaline phosphatase (Jackson Immunologicals), 5-bromo-4-chloro-3-indolyl-phosphate, and nitroblue tetrazolium (32).
DNase I footprinting.
Target DNA fragments were prepared by PCR essentially as described above and previously (46) using 0.2 mM dNTPs, pPSZ17 as template, and primers *M13L (5′-[6-FAM]-CACGACGTTGTAAAACGACGGCCAGT-3′; PE Applied Biosystems) and T7. The resultant PCR product (containing sequences between −337 and +23 of Peps and labeled at one 5′ end with the 6-FAM fluorescent tag) was gel purified, and 40 ng (10 nM) was used in 10-μl footprinting-reaction mixtures that contained 10 mM Tris-HCl (pH 7.5), 5 mM KCl, 1 mM EDTA, 8% glycerol, and 0.5 to 4 μm (0.4 to 3.5 μg) of purified His-tagged VsrC proteins. The total protein concentration was maintained at 4.5 mg/ml using bovine serum albumin. After 30 min at 30°C, reaction mixtures were placed at 26°C, and 5 μl of RNase-free DNase I (1.2 × 10−5 U/μl, freshly diluted [Boehringer Mannheim]) were added. After 4 min, 15 μl of 0.5 M EDTA (pH 8.0) was added; reactions were extracted with phenol-chloroform and passed through a CENTRI-SEP column (Princeton Separations). Digestion products were vacuum dried, dissolved in 12 μl of deionized formamide, 0.2 μl of GS-500-ROX size standards (PE Biosystems) was added, and the fragmentation patterns were analyzed with an ABI 310 Genetic Analyzer as described previously (46).
General molecular and genetic techniques.
Methods for plasmid isolation from E. coli or R. solanacearum and subsequent transfer into E. coli using the CaCl2-treated competent cells or into R. solanacearum via electroporation were described earlier (21, 22). Restriction enzymes, DNA ligase, Klenow fragment, and other DNA enzymes were used according to the manufacturer's recommendations. DNA sequences were obtained using an ABI 377 sequencer. Other general molecular genetic techniques used are described elsewhere (1, 28).
RESULTS
Two regions of the eps promoter are involved in its transcriptional activation.
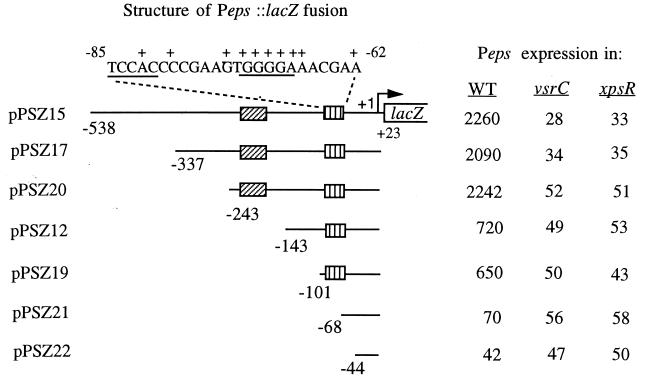
To define the extent of upstream sequences that are required for transcriptional activation of Peps, we constructed a series of reporter plasmids with various lengths of Peps fused to lacZ. Expression of these reporter genes was assayed in an R. solanacearum wild-type strain and in strains lacking key regulators. A reporter plasmid with sequences between −538 and +23 of Peps fused to lacZ(pPSZ15) gave a high level of Peps expression that was strongly dependent on both xpsR and vsrC, since Peps expression was reduced at least 50-fold by inactivation of either regulator (Fig. 1). Deletion of sequences upstream of −337 or −243 did not dramatically alter expression or regulation, indicating that sequences between −243 and +23 are sufficient for wild-type expression from Peps. When sequences between −243 and −143 were deleted (pPSZ12 [Fig. 1]), expression was reduced about threefold, suggesting that a site in the region between nucleotides −243 and −143 stimulates Peps expression. Residual expression from this reporter construct, however, remained strongly dependent on both vsrC and xpsR. Deletion of sequences between −143 and −101 did not significantly reduce transcription below that observed with a reporter having the −143 to +23 region of Peps fused to lacZ. However, when sequences between nucleotides −101 and −68 were deleted, Peps expression was reduced to less than 5% of the levels observed with the −243 to +23 fusion. Expression was only marginally further reduced when Peps sequences down to −44 were deleted (pPSZ22 [Fig. 1]), regardless of the genetic background. These data show that the −101 to −68 region contains sequences that are absolutely required for activation of Peps by these two regulators.
FIG. 1.
Identification of upstream regions involved in transcriptional regulation of Peps by VsrC and XpsR. The eps promoter and various lengths of upstream sequences were fused to lacZ on pRG970, generating plasmids pPSZ12 through pPSZ22. Plasmids were transferred into R. solanacearum wild-type (WT) strain AW, into strain AW-C, a vsrC mutant, and into strain AW-R164, an xpsR mutant. Peps expression (i.e., transcription directed by the Peps fragment) was monitored by measuring LacZ activity (given in Miller units [33]) in cells from cultures grown overnight in BG medium as described previously (4, 20). LacZ activity from cells harboring an empty pRG970 vector was 15. Nucleotide numbering is relative to the transcription start site of eps (21). The hatched boxes indicate the two important regions for activation of eps. The sequence at the top is that of the region identified by PCR mutagenesis as essential for transcription activation by xpsR and vsrC. +, positions of substitution mutations that reduced or eliminated activation of Peps (see Table 2). Underlined sequences, putative palindromic recognition sequences for VsrC. Values are averages from three experiments with <25% variation.
Determination of nucleotides required for activation and regulation of Peps.
To delineate more precisely the sequences that are critical for Peps regulation, a fragment with Peps nucleotides −143 to +23 was subjected to mutagenic PCR, and the products were joined to lacZ on pRG970 to generate transcriptional fusions. The resultant pooled reporter constructs were introduced into wild-type R. solanacearum, and colonies were screened for altered Peps::lacZ expression by using X-Gal. Sixteen mutant plasmids were obtained and characterized further by DNA sequencing and quantitative LacZ assays (Table 2). Peps::lacZ expression from plasmids with single-nucleotide substitutions at Peps nucleotides −74, −71, −70, and −69 was reduced ∼10-fold, almost to the basal levels given by plasmids in which all upstream activation sequences had been deleted (compare to pPSZ22 and pPSZ21 [Fig. 1]). When assayed in xpsR or vsrC mutants of R. solanacearum, expression directed by these mutant promoters was only marginally reduced, indicating that these mutations largely eliminate activation of Peps transcription by VsrC and XpsR. Fusion plasmids with single nucleotide substitutions at Peps nucleotides −82, −79, −72, −68, and −67 were reduced about fivefold. However, when assayed in xpsR or vsrC mutant backgrounds of R. solanacearum, Peps expression from these promoters was reduced an additional three- to fourfold to basal levels, indicating that these mutant promoters are inefficiently activated by VsrC and XpsR. In summary, these results suggest that nucleotides between −82 and −62, and in particular the sequence GTGGGGAA between −74 and −67 (Fig. 1), are important for activation of Peps. It is plausible that this region contains the binding sites for Peps activators, perhaps VsrC and/or XpsR.
TABLE 2.
Expression and regulation of mutant eps promotersa
| Mutation | Peps expression (Miller units) in
|
||
|---|---|---|---|
| WT | vsrC | xpsR | |
| None | 780 | 49 | 53 |
| −82 A to G | 148 | 37 | 48 |
| −79 C to T | 151 | 36 | 42 |
| −74 G to A | 65 | 45 | 52 |
| −72 G to T | 160 | 46 | 37 |
| −71 G to A | 75 | 59 | 65 |
| −70 G to A | 48 | 40 | 37 |
| −69 G to A | 70 | 41 | 39 |
| −68 A to G | 142 | 43 | 58 |
| −67 A to G | 151 | 59 | 55 |
| −62 A to G | 214 | 49 | 51 |
| −38 T to A | 326 | 30 | 32 |
| −32 A to C & −29 T to C | 10,200 | NT | 1,340 |
| −12 T to C | 82 | 31 | 29 |
| −9 A to G | 69 | NT | NT |
| −7 T to C | 45 | 37 | 31 |
Derivatives of Peps::lacZ fusion plasmid pPSZ12 (Fig. 1) containing the indicated mutations were placed in R. solanacearum strains AW (wild-type [WT]), AW-C (vsrC), and AW-R164 (xpsR). Expression (transcription) from Peps was measured by assay of LacZ (given in Miller units [33]) as described in Fig. 1. NT, not tested. Values are averages of three independent determinations with less than 25% variation.
When Peps sequences between nucleotides −538 and −143 were restored in the proper orientation and position to the Peps::lacZ fusion plasmids with substitutions at −67 or −72 and the resultant plasmids placed in wild-type R. solanacearum, Peps expression was largely the same as observed with shorter (−143 to +23) Peps fragments (Table 2 versus Table 3). These results support the conclusion that the primary Peps regulatory sequences required for transcription activation by VsrC and XpsR lie downstream of nucleotide −143, while sequences upstream of −143 can only enhance activation by these regulators.
TABLE 3.
Effect of selected mutations on expression directed by Peps fragments with different upstream lengths
| Plasmid | Peps sequences fused to lacZ | Mutation | Expression from Pepsa |
|---|---|---|---|
| pPSZ15 | −538 to +23 | None | 2,260 |
| pPSZ15-2 | −538 to +23 | −67 A to G | 195 |
| pEPSM2 | −143 to +23 | −67 A to G | 164 |
| pPSZ15-7 | −538 to +23 | −72 G to T | 262 |
| pEPSM7 | −143 to +23 | −72 G to T | 190 |
Three other single-nucleotide substitutions (at nucleotides −12, −9, and −7) also dramatically reduced Peps expression (Table 2). The position and nature of these mutations are consistent with the presumed role of this region as the −10 consensus hexamer of the promoter (21). A single-nucleotide substitution at position −38 reduced Peps expression threefold, while a double mutation changing nucleotides at both −32 and −29 dramatically increased Peps expression (Table 2). These data are consistent with the presumed role of this region as the −35 consensus hexamer that comprises a ς70-type RNA polymerase recognition site (21).
EpsR can inhibit transcription activation of Peps by XpsR and VsrC.
epsR encodes a putative DNA-binding protein that inhibits EPS production by R. solanacearum strain K60, but only when plasmid borne (18, 25, 30). Moreover, the two reported phenotypes of epsR mutants of strain K60 are contradictory (3, 25). Therefore, we needed to explore a possible role for epsR in regulating Peps transcription in our R. solanacearum strain. To do this we transferred plasmid pKL4 containing the K60 epsR gene (25) into our wild-type AW strain harboring a genomic eps::lacZ fusion (AW19A). The addition of pKL4 reduced expression of eps by sevenfold (Table 4). To determine what Peps sequences are required for this effect, pKL4 was transferred into strain AW harboring reporter plasmids with different lengths of Peps sequences fused to lacZ (Fig. 1). Expression from reporters with Peps sequences between −143 and +23 or between −101 and +23 fused to lacZ was specifically reduced by greater than ninefold by the presence of pKL4 (Table 4). However, deletion of Peps sequences between nucleotides −101 and −44 eliminated this effect. When pKL4 was placed in a strain harboring fusion plasmid pEPSM9 or pEPSM3, which cannot be transcriptionally activated due to mutations in the −82 to −62 regulatory region, expression from Peps was not significantly decreased. Thus, epsR affects Peps only if it has been activated, suggesting that epsR interferes with transcriptional activation of Peps mediated by vsrC and xpsR via the −82 to −62 upstream region.
TABLE 4.
Effect of plasmid-borne epsR on expression from wild-type and mutant eps promoters in R. solanacearum
| Straina |
eps expression (Miller units) in presence of:
|
|
|---|---|---|
| pLAFR3 | pKL4 | |
| AW19A (epsB::lacZ) | 1,980 | 253 |
| AW (pPSZ12) [−143 to +23] | 710 | 60 |
| AW (pPSZ19) [−101 to +23] | 568 | 57 |
| AW (pPSZ22) [−44 to +23] | 34 | 31 |
| AW (pEPSM9) [−143 to +23; −70 G to A] | 55 | 44 |
| AW (pEPSM3) [−143 to +23; −74 G to A] | 51 | 37 |
R. solanacearum strains contained either the pLAFR3 vector or pKL4 (pLAFR3 carrying epsR). AW19A is a wild-type strain with a genomic epsB::lacZ reporter. AW strains harbored the indicated Peps::lacZ reporter plasmids (see Fig. 1); numbers in brackets indicate region of Peps fused to lacZ and mutation, if any. eps expression was measured as in Fig. 1.
To further investigate Peps regulation by epsR, we constructed an epsR null mutant of strain AW. Similar to some results with strain K60 (25), inactivation of epsR did not obviously affect EPS production. Moreover, when genomic (eps130::lacZ) or plasmid-borne (pPSZ20 or pSZ21) Peps::lacZ fusions were transferred into the AW strain that lacks EpsR, expression from Peps was the same as in the wild type (data not shown). Thus, epsR affects Peps expression in strain AW only when plasmid-borne, probably due to the 10-fold overproduction of EpsR caused by an elevated copy number (25).
Isolation and characterization of vsrB-independent vsrC alleles.
Previous genetic studies (19, 20) and analogy to other two-component systems (17) suggest that, in response to some signal, VsrB phosphorylates VsrC, converting it into a form that can activate transcription from Peps. However, since XpsR is also required, this assumption is tentative. To explore the dependency of VsrC on the VsrB sensor kinase, as well as XpsR, we set out to isolate vsrC alleles that activate Peps independently of VsrB and/or XpsR. A pool of 20,000 plasmids containing heavily PCR-mutagenized vsrC alleles was transferred into R. solanacearum strain AW22B (eps::lacZ vsrB::Ω), and transformants with elevated eps::lacZ expression were selected. After reisolation and retransformation into AW22B, only one plasmid (pVSRC1) consistently and strongly increased Peps expression in the absence of VsrB. LacZ assays showed that in either AW22B or AW22BC (vsrB vsrC eps::lacZ) double mutants, pVSRC1 caused a greater than ninefold increase in transcription of eps::lacZ (Table 5). In contrast, pRKVC3 with wild-type vsrC only slightly increased eps expression. This suggested that the vsrC1 allele on pVSRC1 harbors a mutation that makes it nearly fully active in the absence of phosphorylation by VsrB. When pVSRC1 was placed in the wild-type reporter strain AW22 or strain AW22C (vsrC eps::lacZ), Peps activation was similar to that observed in strain AW22BC (Table 5), suggesting that the activity of VsrC1 cannot be dramatically increased by VsrB. When pVSRC1 was placed in a strain lacking VsrC, VsrB, and XpsR (AW22RBC), transcription of eps::lacZ showed only a small (less than twofold) increase. Restoring vsrB to this strain (i.e., converting it to a vsrC xpsR mutant) had no effect on its vsrC1-mediated activation of Peps (data not shown). Thus, although VsrC1 protein is very active without VsrB, it still requires xpsR for Peps activation. Not surprisingly, all of our attempts to isolate vsrC alleles that functioned independently of xpsR by screening the pool of mutant vsrC alleles in xpsR mutants were unsuccessful.
TABLE 5.
Activation of eps expression by wild-type and mutant alleles of vsrC in various R. solanacearum regulatory mutants
| Straina |
eps expression (Miller units) in presence of:
|
|||
|---|---|---|---|---|
| No plasmid | pRKVC3 | pVSRC1 | pVSRC2 | |
| AW22B | 3.6 | 5.2 | 32.0 | 28.6 |
| AW22C | 4.1 | 44.0 | 36.1 | 41.4 |
| AW22BC | 3.2 | 4.5 | 29.0 | 24.2 |
| AW22RBC | 3.3 | 3.1 | 5.8 | NT |
| AW22 | 37.2 | 38.3 | 33.0 | NT |
Plasmid pVSRC1 (vsrC1 with H146R and S209L mutations), pVSRC2 (vsrC2 with only an S209L mutation), or pRKVC3 (wild-type vsrC) was placed in R. solanacearum strains harboring a genomic eps::lacZ reporter and various regulatory-gene mutations. Expression of eps was monitored as in Fig. 1. AW22, wild type; AW22B, vsrB mutant; AW22C, vsrC mutant; AW22BC, vsrB vsrC mutant; and AW22RBC, xpsR vsrB vsrC mutant. NT, not tested.
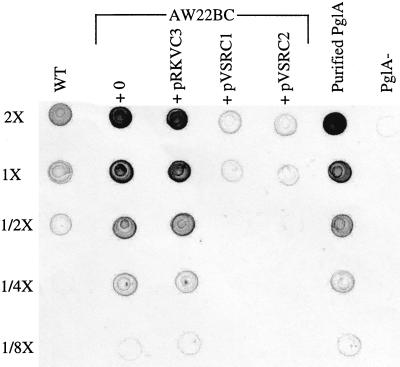
Inactivation of vsrB or vsrC increases PglA production by about sevenfold (19, 20; Fig. 2), indicating that, in addition to positive regulation of eps, the VsrB-VsrC two-component system negatively regulates production of polygalacturonase PglA. Placing the vsrC1 allele in an AW22BC mutant caused its derepressed PglA level to be reduced back to wild-type levels (Fig. 2); in contrast, introduction of wild-type vsrC into the same strain did not affect PglA levels. Thus, vsrC1 exhibits both positive and negative regulation of appropriate targets without the input of VsrB.
FIG. 2.
Regulation of pglA by wild-type vsrC and mutant alleles of vsrC. Concentrated supernatants of 24-h-old BSM cultures of strain AW22 (wild-type background [WT]) or from double mutant strain AW22BC (vsrB vsrC) containing the indicated plasmids were serially diluted twofold in BSM and 5 μl of each dilution (2× through 1/8×) spotted on nitrocellulose. The amount of PglA was assayed using anti-PglA antiserum and alkaline phosphatase-conjugated secondary antibody. pRKVC3 contains wild-type vsrC; pVSRC1 and pVSRC2 contain the vsrB-independent alleles vsrC1 and vsrC2, respectively. PglA−, supernatant from R. solanacearum PG3 (pglA::nptl [39]). Purified PglA (50, 25, 12, 6, and 3 μg, respectively, for each of the 5 dilutions listed at the left) was used for calibration.
DNA sequence analysis of the vsrC1 allele showed two nucleotide substitutions: A996→G and C1185→T, causing substitution of His146 with Arg and of Ser 209 with Leu. To explore the contribution of each amino acid substitution to the vsrC1 phenotype, splice overlap PCR was used to construct a vsrC allele encoding a VsrC with only the S209L substitution. When cloned into pRK415 and assayed in regulatory mutants of R. solanacearum, this allele (vsrC2) had essentially the same effect on expression of eps and pglA as the vsrC1 allele (Table 5 and Fig. 2) indicating that the vsrB-independent phenotype of vsrC1 and vsrC2 is largely the result of the S209L substitution. The position of this altered residue is unexpected and striking, since the analogous region in all other response regulators is outside of the helix-turn-helix DNA-binding domain and other regions implicated in their function (17).
In vitro analysis of VsrC binding to Peps.
Genetic data could not distinguish whether VsrC directly binds to and activates Peps or works via an intermediate, so we investigated the ability of VsrC to bind to Peps in vitro. First we constructed expression plasmids harboring either wild-type vsrC or the constitutively active vsrC1 with their N termini translationally fused in frame to a hexahistidine encoding tag. Both His-tagged alleles were nearly as active as wild-type vsrC; when cloned on pRK415 and placed in an R. solanacearum vsrC mutant, both alleles increased eps::lacZ expression by >10-fold, to ca. 50% of wild-type levels (data not shown). After both His-tagged proteins were purified to >90% homogeneity, up to 20 μg of each was used in gel mobility shift assays with appropriate 32P-labeled Peps fragments. However, no consistent specific mobility shifting was detected under a variety of conditions (data not shown).
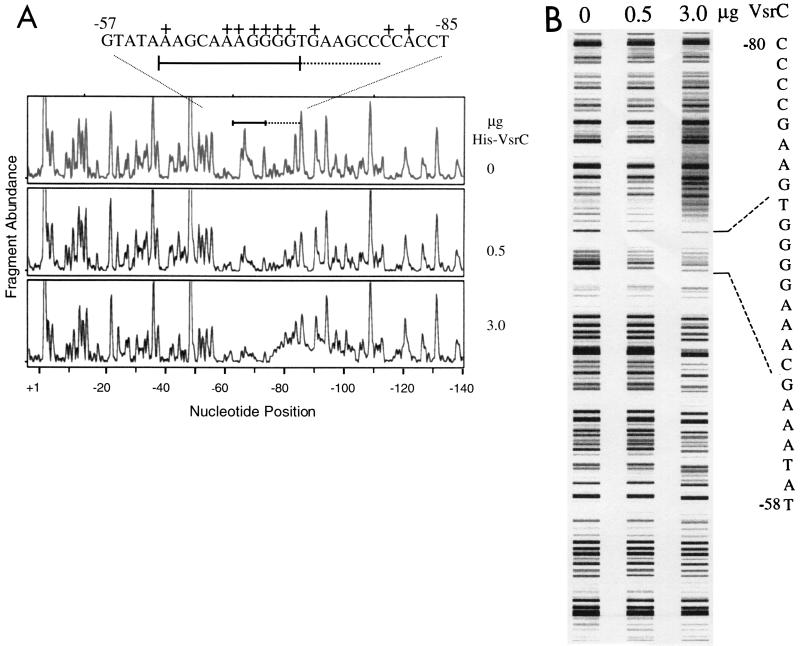
Next we tried a new, rapid footprinting analysis (46) to detect VsrC binding to Peps. A fragment with the −337 to +23 region of Peps that was fluorescently labeled at one 5′-end with 6-FAM was briefly incubated with His-VsrC and then DNase I. Fragmentation patterns were analyzed on an ABI 310 Genetic Analyzer. Run outputs, displayed as electropherograms or “false gel images” (Fig. 3), clearly show an upstream region of Peps where the abundance of certain fragments decreases with increasing amounts of His-VsrC protein in the reaction. This is likely due to a VsrC-specific hindrance of the access of DNase I to this region (i.e., protection). Supporting this, incubation of the Peps fragment with a control protein preparation or incubation of purified His-VsrC with a promoter fragment of a gene not controlled by VsrC did not affect their DNase I fragmentation patterns (data not shown). We also observed that similar to other DNA binding proteins, incubation of the Peps fragment with 3 μg of His-VsrC specifically caused increased DNase I cleavage (i.e., hypersensitivity) adjacent to the protected region (Fig. 3A). Footprinting reactions using the constitutively active His-VsrC1 protein gave essentially the same results (data not shown). From the size of the affected fragments, the positions of nucleotides specifically protected (bound) by VsrC were determined to be between −62 and −72, the same Peps region that harbors many nucleotides important for transcription activation by VsrC and XpsR (Table 2). The hypersensitive region is upstream between nucleotides −76 and −90.
FIG. 3.
DNase I footprinting analysis of VsrC binding to Peps. Reactions were set up, processed, and analyzed using the ABI 310 as described in Materials and Methods and previously (46). (A) Electropherograms from reactions with increasing amounts of His-VsrC. The y-axis gives fluorescence intensity, which is proportional to fragment abundance; the x-axis gives elution position of fragments, which is proportional to their size. Bottom scale gives nucleotide position relative to the eps transcription start site (21) determined using the elution positions of internal size standards. Solid bar, upstream region of Peps that is protected from DNase I digestion by His-VsrC. Dashed line, region made hypersensitive by VsrC. The DNA sequence of the protected region, marked with the same bars, is shown above with nucleotides identified by mutagenesis as critical for transcription activation (see Table 2) marked +. (B) “False gel image” representation of electropherograms in panel A generated by Genotyper 2.5 software, which converts the fluorescence intensity of peaks into proportional gray-scale bands. The sequence of the VsrC-protected region is given on the right.
DISCUSSION
Transcription of eps, an operon encoding biosynthesis of the EPS I virulence factor of R. solanacearum, is controlled by a complex network composed of at least three interacting and environmentally responsive systems (41). We previously found that activation of transcription driven by Peps sequences downstream of nucleotide −143 was completely dependent on xpsR and the vsrB-vsrC two-component system (21). Extending that work here, we showed that sequences downstream of nucleotide −243 are required and sufficient for wild-type Peps expression and regulation. However, when Peps sequences between −243 and −143 were deleted, expression was reduced threefold but remained fully dependent on xpsR and vsrB-vsrC. Thus, the −243 to −143 region enhances the transcription activation that is mediated by these regulators. The global virulence regulator of R. solanacearum, PhcA (5, 41), may play a role in this enhancement, because purified PhcA binds to and protects the −185 to −140 region of Peps from DNase I digestion and because the enhanced activation that requires the −243 to −143 region is absent in phcA mutants (our unpublished data).
Further promoter deletion experiments clearly showed that nucleotides below −101 are absolutely critical for activation of Peps. Subsequent mutagenesis studies (Table 2) revealed that many of the critical nucleotides lie between −82 and −62, in particular within the GTGGGGAA located between −74 and −67. Inactivation of either vsrC or xpsR did not further reduce expression from Peps fragments with mutations in this region, suggesting that VsrC and/or XpsR may directly bind to this site to mediate transcription activation. Using DNase I footprinting we confirmed that VsrC does indeed directly bind to and protect the −74 to −67 region. Recent footprinting experiments (W. Yindeeyoungyeon and M. Schell, unpublished data) have identified another VsrC-protected binding site upstream of a new eps gene. The sequences of these two VsrC-protected sites show extensive similarity and suggest that VsrC may recognize a conserved palindromic consensus sequence found in both regions (TCCNC-N8-GGGGA; Fig. 1). However, in contrast to similar footprinting experiments with another DNA-binding protein (46), a >100-fold excess of wild-type or constitutively active VsrC did not fully protect either site from DNase I, implying that the affinity of VsrC for these sites is relatively low. Perhaps XpsR (or another factor) is required for strong binding. In support of this hypothesis, we found that transcriptional activation by both wild-type and the constitutively active vsrC1 allele still requires xpsR. Although enhancement of DNA binding by an auxiliary protein is not a common property of response regulators (17), another R. solanacearum response regulator, VsrD, also requires an auxiliary protein for its transcriptional regulation (22). Unfortunately, in vitro testing for the effect of XpsR on VsrC binding has not been possible due to the insolubility of our purified XpsR preparation. Alternatively, it is plausible that XpsR may be involved in the phosphorylation status of VsrC; however, the activity of VsrC1 in vivo and in vitro was essentially wild type regardless of the presence of the VsrB sensor kinase. While these data suggest that VsrC1 is fully active in the absence of phosphorylation, other possibilities remain. Site-directed mutagenesis of Asp-58 in VsrC's receiver domain, the presumed site of phosphorylation by VsrB, should better define the role of VsrB and XpsR in phosphorylation and/or activation of VsrC .
The position of the substitution in VsrC that conferred independence from VsrB (residue 209, 12 residues from the C terminus) is interesting and novel, but not surprising considering that vsrC1 was the only bona fide VsrB-independent allele found in a population of >20,000 heavily mutagenized alleles. VsrC, a FixJ-type response regulator (17), is very similar to NarL, the crystal structure of which has been determined (2). Alignment of the C termini of VsrC and NarL (which are 80% similar) suggests that the S209L substitution is located in the middle of helix 10 which follows the helix-turn-helix DNA-binding domain. The region containing helix 10 has not been implicated in transcription activation by response regulators, nor is its amino acid sequence highly conserved. However, circumstantial evidence that the C terminus of a response regulator may interact with RNA polymerase has been reported: deletion of the last two or three residues of BvgA severely inhibited growth of Bordetella pertussis, but this effect was suppressed by mutations affecting the α-subunit of RNA polymerase (42). We have found that vsrC1 also can cause growth inhibition in R. solanacearum. Since genetic and biochemical evidence suggests that interactions between transcriptional regulators and the α-subunit of RNA polymerase are sometimes important in transcriptional activation (35), it is plausible that the C-terminal region of some response regulators (e.g., helix 10) may interact with RNA polymerase to stimulate transcription. Site-directed mutagenesis and chemical cross-linking studies are required to further investigate this possibility.
In contrast, most mutations conferring sensor independence on response regulators are N terminal (e.g., V88L for NarL [10] and some N-terminal deletions [13]). These mutations are thought to mimic changes caused by sensor kinase-mediated phosphorylation; i.e., they cause a conformational change that alleviates occlusion of the C-terminal helix-turn-helix DNA-binding domain (9, 12). Similarly, the substitution at the C terminus of VsrC1 may also alter its conformation in a way that relieves or prevents inhibitory interactions of its helix-turn-helix DNA-binding domain with other parts of the polypeptide that block or reduce its function.
The role of EpsR in Peps regulation and its relationship to xpsR and vsrC remain unclear, because we found that inactivation of epsR did not dramatically affect expression from Peps or EPS I biosynthesis, whereas when present on a multicopy plasmid, epsR did reduce Peps expression by greater than sevenfold. This inhibitory effect occurred only when the VsrC-binding site at Peps was intact. Although indirect evidence for the binding of EpsR to Peps has been reported (3), we were unable to confirm this. Preliminary analyses with lacZ reporters show that elevated levels of EpsR slightly (threefold) reduce expression of xpsR but not of vsrC. Although this implies that environmentally directed overproduction of EpsR could reduce levels of XpsR and hence shut down EPS production, further in vivo and in vitro studies are needed to clarify the physiological role and mechanism of action of EpsR and XpsR with VsrC at the −82 to −62 region of Peps.
ACKNOWLEDGMENTS
This research was supported in part by grant MCB 97-27921 from the National Science Foundation.
The authors thank Tim Hoover and Ellen Neidle for critical reading of the manuscript.
REFERENCES
- 1.Ausubel F, Brent R, Kingston D, Seidman J G, Smith J, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1996. [Google Scholar]
- 2.Baikalov I, Schroder I, Kaczor-Grzeskowiak T, Grzeskowiak K, Gunsalus R P, Dickerson R E. Structure of the Escherichia coli response regulator NarL. Biochemistry. 1996;35:11053–11061. doi: 10.1021/bi960919o. [DOI] [PubMed] [Google Scholar]
- 3.Chapman M R, Kao C C. EpsR modulates production of extracellular polysaccharide in the bacterial wilt pathogen Ralstonia (Pseudomonas) solanacearum. J Bacteriol. 1998;180:27–34. doi: 10.1128/jb.180.1.27-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clough S J, Schell M A, Denny T P. Differential expression of virulence genes and motility in Ralstonia (Pseudomonas) solanacearum during exponential growth. Appl Environ Microbiol. 1997;63:844–850. doi: 10.1128/aem.63.3.844-850.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clough S J, Schell M A, Denny T P. A two-component system in Ralstonia (Pseudomonas) solanacearum modulates production of PhcA-regulated virulence factors in response to 3-hydroxypalmitic acid methyl ester. J Bacteriol. 1997;179:3639–3648. doi: 10.1128/jb.179.11.3639-3648.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denny T P, Carney B F, Schell M A. Inactivation of multiple virulence genes reduces the ability of Pseudomonas solanacearum to cause wilt symptoms. Mol Plant-Microbe Interact. 1990;3:293–300. [Google Scholar]
- 7.Denny T P, Baek S R. Genetic evidence that extracellular polysaccharide is a virulence factor of Pseudomonas solanacearum. Mol Plant-Microbe Interact. 1991;4:198–206. [Google Scholar]
- 8.Denny T P. Involvement of bacterial polysaccharides in plant pathogenesis. Annu Rev Phytopathol. 1995;33:173–197. doi: 10.1146/annurev.py.33.090195.001133. [DOI] [PubMed] [Google Scholar]
- 9.Djordjevic S, Goudreau P N, Xu Q, Stock A M, West A H. Structural basis for methylesterase CheB regulation by a phosphorylation-activated domain. Proc Natl Acad Sci USA. 1998;95:1381–1386. doi: 10.1073/pnas.95.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egan S M, Stewart V. Mutational analysis of nitrate regulatory gene narL in Escherichia coli K-12. J Bacteriol. 1991;173:4424–4432. doi: 10.1128/jb.173.14.4424-4432.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flavier A B, Clough S J, Schell M A, Denny T P. Identification of β-hydroxypalmitic acid methyl ester as a novel autoregulator controlling virulence in Ralstonia solanacearum. Mol Microbiol. 1998;26:251–259. doi: 10.1046/j.1365-2958.1997.5661945.x. [DOI] [PubMed] [Google Scholar]
- 12.Goudreau P N, Stock A M. Signal transduction in bacteria: molecular mechanisms of stimulus-response coupling. Curr Opin Microbiol. 1998;1:160–169. doi: 10.1016/s1369-5274(98)80006-4. [DOI] [PubMed] [Google Scholar]
- 13.Gu B, Lee J H, Hoover T R, Scholl D, Nixon B T. Rhizobium meliloti DCTD, a sigma 54 dependent activator, may be negatively controlled by a subdomain in the C-terminal end of its two-component receiver module. Mol Microbiol. 1994;13:51–66. doi: 10.1111/j.1365-2958.1994.tb00401.x. [DOI] [PubMed] [Google Scholar]
- 14.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 15.Hayward A C. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu Rev Phytopathol. 1991;29:65–87. doi: 10.1146/annurev.py.29.090191.000433. [DOI] [PubMed] [Google Scholar]
- 16.Hayward A C. Hosts of Pseudomonas solanacearum. In: Hayward A C, Hartman G L, editors. Bacterial wilt: the disease and its causative agent Pseudomonas solanacearum. Oxon, United Kingdom: CAB International; 1994. pp. 9–24. [Google Scholar]
- 17.Hoch J A, Silhavy T J. Two-component signal transduction. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- 18.Huang H, Sequeira L. Identification of a locus that regulates multiple functions in Pseudomonas solanacearum. J Bacteriol. 1990;172:4728–4731. doi: 10.1128/jb.172.8.4728-4731.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang J, Denny T P, Schell M A. VsrB, a regulator of virulence genes of Pseudomonas solanacearum, is homologous to sensors of the two-component regulatory family. J Bacteriol. 1993;175:6169–6178. doi: 10.1128/jb.175.19.6169-6178.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang J, Carney B, Denny T P, Weissinger A K, Schell M A. A complex network regulates expression of eps and other virulence genes of Pseudomonas solanacearum. J Bacteriol. 1995;177:1259–1267. doi: 10.1128/jb.177.5.1259-1267.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang J, Schell M A. Characterization of the eps gene cluster of Pseudomonas solanacearum and its transcriptional regulation via a single promoter. Mol Microbiol. 1995;16:977–989. doi: 10.1111/j.1365-2958.1995.tb02323.x. [DOI] [PubMed] [Google Scholar]
- 22.Huang J, Yindeeyoungyeon W, Garg R P, Denny T P, Schell M A. Joint transcriptional control of xpsR, the unusual signal integrator of Ralstonia solanacearum virulence gene regulatory network, by a response regulator and a LysR-type transcriptional activator. J Bacteriol. 1998;180:2736–2743. doi: 10.1128/jb.180.10.2736-2743.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang Q, Allen C. Exo-poly-α-d-galacturonosidase, PehB, is required for wild-type virulence of Ralstonia solanacearum. J Bacteriol. 1997;179:7369–7378. doi: 10.1128/jb.179.23.7369-7378.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang Y, Huang J, Guozhang M, He L-Y, Schell M A. Dramatically reduced virulence of mutants of Pseudomonas solanacearum that are defective in export of extracellular proteins across the outer membrane. Mol Plant-Microbe Interact. 1994;7:370–377. [Google Scholar]
- 25.Kao C C, Gosti F, Huang Y, Sequeira L. Characterization of a negative regulator of exopolysaccharide production by the plant pathogenic bacterium Pseudomonas solanacearum. Mol Plant-Microbe Interact. 1994;7:121–130. doi: 10.1094/mpmi-7-0121. [DOI] [PubMed] [Google Scholar]
- 26.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 27.Kitten T, Willis D K. Suppression of a sensor kinase dependent phenotype in Pseudomonas syringae by ribosomal L35 and L20 proteins. J Bacteriol. 1996;178:1548–1555. doi: 10.1128/jb.178.6.1548-1555.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 29.McGarvey J, Bell C, Denny T P, Schell M A. Analysis of EPS I in culture and in planta using immunological methods: new insights and implications. In: Allen C A, Elphinstone J, Prior P, editors. Bacterial wilt disease: molecular and ecological aspects. Berlin: Springer-Verlag; 1998. pp. 157–163. [Google Scholar]
- 30.McWilliams R, Chapman M, Kowlaczuk M, Hershberger D, Sun J-H, Kao C C. Complementation analyses of Pseudomonas solanacearum extracellular polysaccharide mutants and identification of genes responsive to EpsR. Mol Plant-Microbe Interact. 1995;8:837–844. doi: 10.1094/mpmi-8-0837. [DOI] [PubMed] [Google Scholar]
- 31.Mead D A, Szczesna-Skorupa E, Kemper B. Single-stranded DNA “blue” T7 promoter plasmids: a versatile tandem promoter system for cloning and protein engineering. Protein Eng. 1986;1:67–74. doi: 10.1093/protein/1.1.67. [DOI] [PubMed] [Google Scholar]
- 32.Menon A L, Mortenson L E, Robson R L. Nucleotide sequence and genetic analysis of hydrogenase oxidation (hox) genes in Azotobacter vinelandii. J Bacteriol. 1992;174:4549–4557. doi: 10.1128/jb.174.14.4549-4557.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 34.Orgambide G, Montrozier H, Servin P, Roussel J, Trigalet-Demery D, Trigalet A. High heterogeneity of the exopolysaccharides of Pseudomonas solanacearum strain GMI1000 and the complete structure of the major polysaccharide. J Biol Chem. 1991;266:8312–8321. [PubMed] [Google Scholar]
- 35.Rhodius V A, Busby S J W. Positive activation of gene expression. Curr Opin Microbiol. 1998;1:152–159. doi: 10.1016/s1369-5274(98)80005-2. [DOI] [PubMed] [Google Scholar]
- 36.Roberts D P, Denny T P, Schell M A. Cloning of the egl of Pseudomonas solanacearum and analysis of its role in phytopathogenicity. J Bacteriol. 1988;170:1445–1451. doi: 10.1128/jb.170.4.1445-1451.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saile E, McGarvey J, Schell M A, Denny T P. Role of extracellular polysaccharide and endoglucanase in root invasion and colonization of tomato plants by Ralstonia solanacearum. Phytopathology. 1998;87:1264–1271. doi: 10.1094/PHYTO.1997.87.12.1264. [DOI] [PubMed] [Google Scholar]
- 38.Schell M A. Purification and characterization of an endoglucanase from Pseudomonas solanacearum. Appl Environ Microbiol. 1987;53:2237–2241. doi: 10.1128/aem.53.9.2237-2241.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schell M A, Roberts D P, Denny T P. Analysis of the Pseudomonas solanacearum polygalacturonase encoded by pglA and its involvement in phytopathogenicity. J Bacteriol. 1988;170:4501–4508. doi: 10.1128/jb.170.10.4501-4508.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schell M, Poser E F. Demonstration, characterization, and mutational analysis of NahR protein binding to the nah and sal promoters. J Bacteriol. 1989;171:837–846. doi: 10.1128/jb.171.2.837-846.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schell M A. Regulation of virulence and pathogenicity genes in Ralstonia solanacearum by a complex network. Annu Rev Phytopathol. 2000;38:263–292. doi: 10.1146/annurev.phyto.38.1.263. [DOI] [PubMed] [Google Scholar]
- 42.Stibitz S. Mutations affecting the α-subunit of Bordetella pertussis RNA polymerase suppress growth inhibition conferred by short C-terminal deletions of the response regulator BvgA. J Bacteriol. 1998;180:2484–2492. doi: 10.1128/jb.180.9.2484-2492.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van den Eede G, Deblaere R, Goethals K, Van Montegue M, Holsters M. Broad host range and promoter selection vectors for bacteria that interact with plants. Mol Plant-Microbe Interact. 1992;5:228–234. doi: 10.1094/mpmi-5-228. [DOI] [PubMed] [Google Scholar]
- 44.Vasse J, Pascal F, Trigalet A. Microscopic studies of intercellular infection and protoxylem invasion of tomato roots by Pseudomonas solanacearum. Mol Plant-Microbe Interact. 1995;8:241–251. [Google Scholar]
- 45.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC18. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 46.Yindeeyoungyeon, W., and M. A. Schell. Footprinting with an automated capillary DNA sequencer. Biotechniques, in press. [DOI] [PubMed]