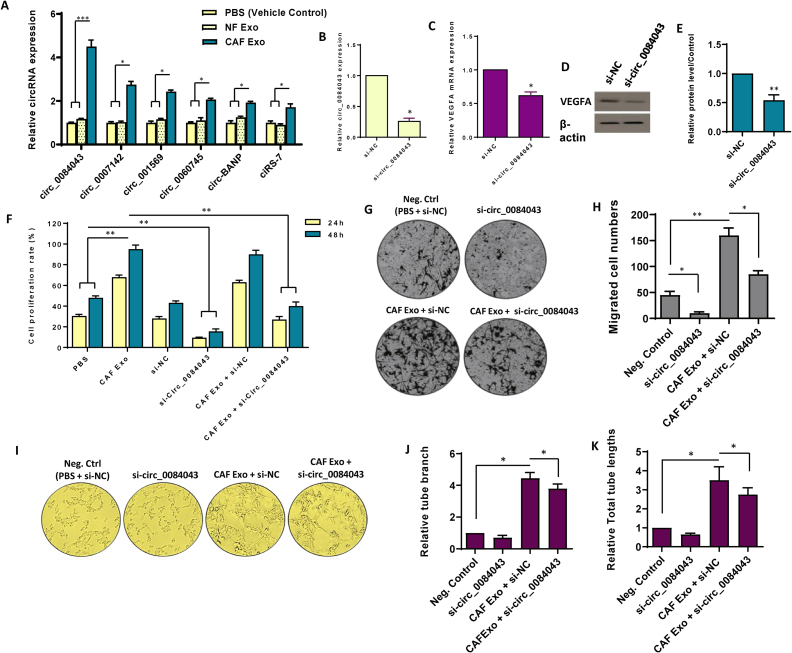
Fig. 4.
Circ_0084043 orchestrates pro-angiogenic effects in endothelial cells induced by exosomes originating from colorectal cancer-associated fibroblasts. A. The differential expression of six candidate circRNAs was assessed in endothelial cells following treatment with 100 μg/mL exosomes derived from cancer-associated fibroblasts (CAF), exosomes derived from normal fibroblasts (NF), and PBS as a vehicle control. Notably, circ_0084043 exhibited the most substantial differences in expression levels among the tested conditions. B. The relative expression of circ_0084043 was examined after treating the cells with siRNA targeting circ_0084043 (si-circ_0084043) for 24 h. C-E. Following the inhibition of circ_0084043 using siRNA, both mRNA (C) and protein (D) levels of VEGFA were evaluated, indicating that circ_0084043 induced VEGFA expression in endothelial cells. The original bots of Fig. 4D are provided in Supplementary Fig. S8. For quantitative analysis, the density of bands was assessed using ImageJ software and represented as relative intensity (E). F. The relationship between the expression levels of CAF-derived exosomal circ_0084043 and cell proliferation in endothelial cells was observed over 24 and 48 h. Upon the inhibition of circ_0084043, cell proliferation decreased. G, H. Microscopic images, along with quantitative analysis, revealed a heightened rate of cell migration in endothelial cells treated with 100 μg/mL CAF-derived exosomes. However, upon inhibition of circ_0084043, cell migration decreased. I. The tube formation assay demonstrated that treating endothelial cells with CAF-derived exosomes (100 μg/mL) increased the formation of tubes, the number of branches, and the overall length. Conversely, inhibiting circ_0084043 significantly decreased the angiogenesis rate in endothelial cells. J, K. Quantitative assessment of the tube length and branch points calculated in three randomly selected fields. The columns represent the means of three independent experiments, with the bars indicating the standard deviation (SD). * p-value <0.05, ** p-value <0.01, *** p-value <0.001.