Abstract
Legionella pneumophila, the causative organism of Legionnaires' pneumonia, contains two enzymes with catalatic and peroxidatic activity, KatA and KatB. To address the issue of redundant, overlapping, or discrete in vivo functions of highly homologous catalase-peroxidases, the gene for katA was cloned and its function was studied in L. pneumophila and Escherichia coli and compared with prior studies of katB in this laboratory. katA is induced during exponential growth and is the predominant peroxidase in stationary phase. When katA is inactivated, L. pneumophila is more sensitive to exogenous hydrogen peroxide and less virulent in the THP-1 macrophage cell line, similar to katB. Catalatic-peroxidatic activity with different peroxidatic cosubstrates is comparable for KatA and KatB, but KatA is five times more active towards dianisidine. In contrast with these examples of redundant or overlapping function, stationary-phase survival is decreased by 100- to 10,000-fold when katA is inactivated, while no change from wild type is seen for the katB null. The principal clue for understanding this discrete in vivo function was the demonstration that KatA is periplasmic and KatB is cytosolic. This stationary-phase phenotype suggests that targets sensitive to hydrogen peroxide are present outside the cytosol in stationary phase or that the peroxidatic activity of KatA is critical for stationary-phase redox reactions in the periplasm, perhaps disulfide bond formation. Since starvation-induced stationary phase is a prerequisite to acquisition of virulence by L. pneumophila, further studies on the function and regulation of katA in stationary phase may give insights on the mechanisms of infectivity of this pathogen.
Heme-containing hydroperoxidases were among the first enzymes discovered and have been studied extensively. Three types are found in bacteria: monofunctional catalases, monofunctional peroxidases, and bifunctional catalase-peroxidases. The monofunctional catalases and peroxidases display, respectively, catalatic activity (2H2O2→H2O + O2) and peroxidatic activity (AH2 + H2O2→A + 2H2O), predominantly. Bifunctional catalase-peroxidases display both, but catalatic activity is larger by far, typically a ratio of >100 for catalatic-dianisidine peroxidatic activity, expressed as micromoles of H2O2 reduced per milligram of protein (11, 28, 29, 31).
Studies of the physiological function of monofunctional catalases and bifunctional catalase-peroxidases have focused on the catalatic enzyme activity and an in vivo role in protection from oxidative damage by H2O2 or hydroxyl radical formed from it. Null mutants in aerobic species are often identical to wild type in growth rate but more sensitive to added H2O2 (6, 7, 14, 32, 51, 55). This suggests that these enzymes are important in defense against exogenous H2O2 but that the defense against endogenous H2O2 from aerobic respiration is redundant and that other defenses exist or are induced in the nulls. Sources of exogenous H2O2 include environmental chemical reactions, commensal microorganisms, and the respiratory burst in response to bacterial infection. The peroxidatic activity of bifunctional catalase-peroxidases may be important physiologically (28, 29, 31, 36), but no in vivo peroxidatic cosubstrates have been definitively identified.
Studies of physiological function are more complex when multiple heme hydroperoxidases are present in a single bacterial cell. Escherichia coli K-12 contains a monofunctional catalase, KatE, and a bifunctional catalase-peroxidase, KatG. Differences in regulation are a critical factor in understanding the in vivo roles of these two enzymes. KatG catalase-peroxidase increases during exponential growth in proportion to the rate of endogenous H2O2 production and is induced in response to exogenous H2O2 (21). The principal transcriptional regulator of katG is OxyR (44, 53, 62). KatE monofunctional catalase is important for H2O2 resistance in stationary phase, principally under control of the stress response sigma factor rpoS (25, 35).
Legionella pneumophila, the causative organism of Legionnaires' pneumonia, contains two bifunctional catalase-peroxidases and no monofunctional catalase or peroxidase (6). The fundamental question that we are addressing is whether the two bifunctional enzymes are redundant, overlapping, or independent in their in vivo function. Here we describe cloning and functional studies of the gene encoding KatA. Although KatA function is qualitatively similar to that of L. pneumophila KatB, which we described earlier (6), in macrophage infection, resistance to respiratory and exogenous H2O2, and the growth phase dependence of its expression, the katA gene is required for viability in stationary phase, while inactivation of katB is without effect. We report investigations into this in vivo functional difference and the possible importance of differential subcellular localization in the role of these catalase-peroxidases in stationary-phase phenomena.
MATERIALS AND METHODS
Media and growth conditions.
The cloning strain was E. coli DH5α grown in Luria-Bertani medium (48) and antibiotics (final concentrations: sodium ampicillin, 100, kanamycin sulfate, 50, gentamicin sulfate, 5, and chloramphenicol, 25 μg/ml). For L. pneumophila, AYE broth (30) and CYE plates (16) were generally used with antibiotics as indicated: kanamycin sulfate (25 μg/ml), gentamicin sulfate (5 or 10 μg/ml), and chloramphenicol (5 μg/ml). CAYE broth for stationary-phase survival experiments contained 2 g of yeast extract and 10 g of casamino acids (Difco) per liter and other ingredients identical to those in AYE broth. The parental L. pneumophila strain was wild-type strain JR32. See Table 1 for strains and plasmids used. All culturing was done at 37°C.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| L. pneumophila | ||
| JR32 | Homogenous salt-sensitive isolate of AM511 (Philadelphia-1 Smr r− m+) | H. Shuman (57) |
| PB126 | JR32 katA::Gmr (katA null strain) | This work |
| PB140 | PB126 katB::ΩCm (katA katB double null) | This work |
| E. coli UM383 | HB101 katG17::Tn10(Tet) katE::Tn10(Kan) | P. Loewen |
| Plasmids | ||
| pAM6 | Bluescript KS with a 2.9-kb gentamicin resistance cassette cloned into the HindIII site | A. Marra and H. Shuman |
| pAM6::katA int | 875-bp EcoRI-EcoRI katA internal fragment (codon E67 to codon S358) cloned into the EcoRI site of pAM6 | This work |
| pJBZ281 | lacZ with polylinker upstream for constructing translational fusions, ColE1 ori(Kanr) | M. R. K. Alley (2) |
| pJBZ281::PkatA | 1,761-bp fragment of katA (1,733 nt upstream of ATG start through first nt of codon for Ala-10) cloned using BamHI and PstI into pJBZ281 (katA translational fusion plasmid) | This work |
| pMMB207αB | RSF1010 derivative, IncQ lacIq Cmr Ptac oriT MCS α complementation | L. Wiater and H. Shuman (57) |
| pMMB207αB::katB | katB ORF (288 nt upstream of translational ATG start through 220 nt downstream of TAA stop) cloned using PstI and HindIII into pMMB207αB | 5 |
| pMMB207αB-Km14 | RSF1010 derivative, IncQ lacIq Cmr Ptac oriT MCS α complementations, with kanamycin resistance cassette interrupting mobA at nt 3325 | G. Segal and H. Shuman (45) |
| pMMB207αB-Km14::katA | katA ORF (191 nt upstream of translational ATG start through 267 nt downstream of TAA stop) cloned into BamHI site of pMMB207αB-Km14 in opposite direction from Ptac | This work |
| pNPTS138 | Derivative of pLITMUS 38 cloning vector with nptI, RK2 oriT, and Bacillus subtilus sacB, Kanr | E. Quardokus and Y. Brun (42) |
| pNPTS138::katB::ΩCm | katB null allele (katB::ΩCm) cloned into BamHI site of pNPTS138 | 5 |
MCS, multiple cloning site.
Cloning of L. pneumophila katA.
Southern blots of BamHI-digested genomic DNA showed two bands when hybridized with a fragment of the E. coli katG catalase-peroxidase gene. The entire katA gene was in the 6.7-kb band, and the 5′ end of katB was in the 5.7-kb band (6). A pUC12 library was constructed from the gel-purified 6.7-kb region and patched to a grid. The katA clone was identified by hybridization of blots of BamHI-digested plasmid DNA from progressively smaller pools from the library grid.
PCR methodology.
PCR was used to amplify fragments with restriction sites at their ends suitable for cloning into pJBZ281 to make the katA::lacZ translational fusion vector and into pMMB207αB-Km14 to make the katA complementation vector. The PCR protocol was described previously (6). See Fig. 1 for primer location and sequence.
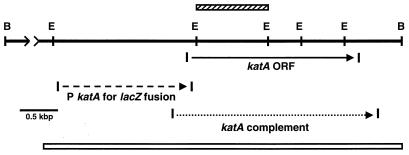
FIG. 1.
Restriction map of the L. pneumophila katA locus. The open horizontal bar at the bottom depicts the region sequenced (5,249 nt). Every nucleotide reported was identified in at least two sequencing runs with different primers. Restriction sites: B, BamHI; E, EcoRI. The distance between BamHI sites is 6.7 kb. The katA ORF and direction of transcription are indicated. The cross-hatched bar above the ORF indicates the internal EcoRI-EcoRI fragment used to make the chromosomal katA disruption. Fragments amplified by PCR are indicated by dashed (lacZ fusion plasmid) and dotted (katA+ complementation plasmid) lines. The 5′ and 3′ PCR primers for the lacZ fusion fragment were TGACAAAGGATCCAGTTTGGCACAAAGCACAACC and GAGTGACTGCAGCAAACAGAGGTATTGTTCGTTTG, respectively. The 5′ and 3′ PCR primers for the complementation plasmid were AAAGGATCAGGTAGGATCCCGTGGCATTAATGAACCGGAGG and AAGCGGTTATCTGGATCCGGTGAACATGAAAGTCGCATGC, respectively. The underlined segments are restriction sites for BamHI, PstI, BamHI, and BamHI, respectively, which replace 6 nt in the L. pneumophila genomic sequence.
Construction of a lacZ fusion vector.
The PCR fragment described in the legend to Fig. 1 (1,733 nucleotides [nt] upstream of the katA ATG to 8 codons downstream of the ATG; 1,861 nt total) was cloned using BamHI and PstI into pJBZ281 (Kmr) to make an in-frame translational fusion. The construct was LacZ+ by X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) in E. coli strain DH5α, indicating that the L. pneumophila katA promoter is recognized in E. coli. After transformation into L. pneumophila strain JR32, Kmr transformants were repeatedly subcultured in the absence of kanamycin. Although the ColE1 origin of pJBZ281 is not well maintained in the absence of drug selection, it was not possible to obtain a chromosomal integrant as was done with a similar pJBZ construct for L. pneumophila katB (6). Therefore, strain JR32 was transformed with pJBZ281::katA, and expression studies were performed with the plasmid fusion with kanamycin in the culture medium.
Construction of a complementing vector.
The PCR fragment described in the legend to Fig. 1 (191 nt upstream of katA ATG to 267 nt downstream of the UAA translational stop; 2,715 nt total) was cloned into the BamHI site of pMMB207αB-Km14 (45). The construct was transformed into L. pneumophila strains by electroporation and maintained by using chloramphenicol in the culture medium.
Construction of null mutants.
A katA null mutant was made by gene disruption using an internal fragment of katA (63). The 919-nt EcoRI-EcoRI fragment from the 5′ end of katA (Fig. 1) was cloned into the EcoRI site of pAM6 Bluescript, with a gentamicin resistance (Gmr) cassette ligated into the HindIII site. L. pneumophila wild-type strain JR32 and katB null strain PB117 (6) were transformed to Gmr by electroporation with this construct. Transformants were subjected to three rounds of overnight culturing in the absence of gentamicin, each followed by plating on CYE with gentamicin (CYE-Gm). Gentamicin-resistant colonies were analyzed by zymogram staining for catalase and peroxidase activities and Southern blotting of BamHI- and PstI-digested genomic DNA probed with the 919-nt katA fragment. These analyses established that the katA and katA katB null mutants arose from the expected gene disruption by single crossover events.
Growth of L. pneumophila in human macrophage-like cell lines.
THP-1 cells were maintained in RPMI 1640 containing 10% fetal calf serum, 2 mM glutamine, and 1 mM sodium pyruvate (46, 52). To promote adherence, 10 pmol of phorbol 12-myristate 13-acetate was added per well (0.5 ml of medium, 3 × 105 cells). After 16 to 20 h, the monolayer was washed twice and then incubated with 0.5 ml of RPMI containing 2 mM glutamine, 1 mM sodium pyruvate, and 20% normal human serum (Gemini Bio-Products). Overnight cultures of L. pneumophila in AYE were diluted in RPMI-normal human serum and added at an initial multiplicity of infection of 0.1. Aliquots were taken from the medium and plated on CYE agar without antibiotics for colony counts. The zero time aliquot was taken 30 to 60 min after infection.
Measurement of resistance to hydrogen peroxide.
Overnight cultures in AYE with appropriate antibiotic (0.1 ml) were spread on CYE plates without antibiotic in 3 ml of 0.8% top agar without nutrients. Whatman 3MM disks (7-mm diameter) received 10 μl of freshly diluted H2O2. Zones of inhibition were measured after 48 h.
Spheroplasting procedure.
Periplasm and cytosol fractions of E. coli were prepared by lysozyme-EDTA treatment (50, 60). Unwashed overnight cultures of E. coli strain UM383 katE katG containing plasmid katA or plasmid katB were treated with 0.1 M Tris-HCl (pH 8.0)–0.25 M sucrose–0.25 mM EDTA–30 μg of lysozyme per ml at room temperature for 30 to 40 min. Spheroplasts were stabilized with 20 mM magnesium chloride, removed by centrifugation, and then lysed by freeze-thawing and sonication.
Enzyme assays.
Supernatants from sonic disruption of cell pellets (49) were quantitatively analyzed for enzyme activities by spectrophotometry. Catalase activity was assayed by monitoring decomposition of 10 mM H2O2 in 50 mM potassium phosphate (pH 7.2) (ɛ240 = 39.4 M−1 cm−1 [1]). Peroxidase activity was assayed in 10 mM potassium phosphate (pH 6.4)–0.17% Triton X-100–1 mM H2O2. Peroxidatic cosubstrates were 0.2 mM NADH or NADPH (ɛ340 = 6.22 × 103 M−1 cm−1), 42 mM pyrogallol (ɛ420 = 2.47 × 103 M−1 cm−1 [36]), or 0.34 mM dianisidine (ɛ460 = 11.3 × 103 M−1 cm−1 [11]). Alkaline phosphatase was assayed by monitoring hydrolysis of 4.8 mM p-nitrophenyl phosphate (ɛ410 = 18.5 × 103 M−1 cm−1) in 100 mM Tris (pH 8.5). Glucose 6-phosphate dehydrogenase was assayed by monitoring oxidation of 3.3 mM glucose 6-phosphate by 0.18 mM NADP+ (ɛ340 = 6.22 × 103 M−1 cm−1) in 50 mM Tris (pH 8.0)–3 mM magnesium chloride. One unit of activity was defined as 1 μmol of substrate (H2O2, peroxidase cosubstrate, p-nitrophenyl phosphate, or glucose 6-phosphate/NADP+) converted per min. β-Galactosidase activity was assayed with o-nitrophenyl-β-d-galactoside as the substrate, and activity was expressed in Miller units (37).
In situ staining of polyacrylamide gels following nondenaturing electrophoresis for catalase and diaminobenzidine peroxidase activity was used for semiquantitative analysis of the separated catalase-peroxidases (12, 23). These methods are based on formation of an achromatic zone by inhibition of H2O2-dependent oxidation of diaminobenzidine by horseradish peroxidase and by formation of a brown zone by H2O2-dependent oxidation of diaminobenzidine, respectively.
Nucleotide sequence accession number.
The GenBank accession number for the katA locus is AF276752. Nucleotide and protein sequences were analyzed using programs of the Genetics Computer Group (University of Wisconsin).
RESULTS
Cloning the gene for KatA catalase-peroxidase.
L. pneumophila katA was cloned by hybridization in a 6.7-kbp genomic BamHI fragment. A total of 5,249 nt were sequenced, including the katA catalase-peroxidase gene (Fig. 1). The katA open reading frame (ORF) encodes 749 amino acids that are 59% identical to the KatB catalase-peroxidase, previously studied by us (6). Nucleotide sequences upstream and downstream of katA showed no homologies to sequences of known function. After we cloned and sequenced katA, Amemura-Maekawa et al. reported the sequence of L. pneumophila katA cloned by PCR amplification from genomic DNA (3). Their sequence of 2,587 nt (GenBank accession number AB017595) corresponds to nt 2,184 to 4,770 in our sequence (AF276752). The sequences are identical except for two differences ≈160 nt before the ATG start codon. Nucleotides G42 and C44 in AB017595 are C2229 and G2231 in AF276752.
Construction of katA null mutants.
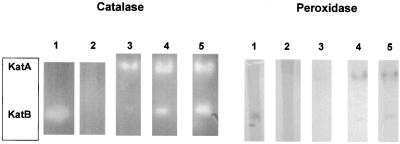
Beginning with wild-type strain JR32 and katB::Cmr null strain pB117, katA and katA katB null strains were made by interruption of katA via a single crossover with an internal fragment of katA (Fig. 1) cloned in a Gmr ColE1 vector. Zymograms for catalatic and diaminobenzidine peroxidase activity demonstrated absence of KatA activity in the katA null and absence of both KatA and KatB activity in the katA katB double null (Fig. 2, lanes 1 and 2). Southern blotting confirmed the expected interruption of wild-type katA (data not shown).
FIG. 2.
Zymogram stains of catalatic and peroxidatic activities. Cell extracts were electrophoresed under nondenaturing conditions and stained for catalatic (left) or diaminobenzidine peroxidatic (right) activity. Lanes 1, strain PB126 (katA null); lanes 2, strain PB140 (katA katB null), 275 μg of cell protein from an overnight culture per lane; lanes 3, 4, and 5, wild-type strain JR32 from, respectively, exponential-phase cultures (optical density at 600 nm, 0.69 and 1.45, respectively) and a 48-h culture (250 μg of cell protein per lane).
Effect of katA null on aerobic growth and H2O2 resistance.
No change in doubling time was observed when either katA or katB alone (6) was inactivated, but the doubling time increased twofold when both genes were inactivated (Table 2). Null mutants of individual catalase-peroxidases were more sensitive to exogenous H2O2 than the wild type, and the katA null was consistently more sensitive than the katB null (Table 2). In nearly all instances, the katA katB double null was not significantly more sensitive to H2O2 than either single null. This suggested that the absence of both catalase-peroxidases elicited a compensatory gene expression not found with either single null which decreased H2O2 sensitivity.
TABLE 2.
Doubling times and H2O2 resistance of mutants
| Strain | Doubling timea (min) | Resistance to indicated concn of H2O2b (zone of clearing, cm)
|
||
|---|---|---|---|---|
| 1 M | 2 M | 3 M | ||
| No plasmid | ||||
| JR32 (wild type) | 118 ± 5 | 3.0 ± 0.07 | 3.4 ± 0.1 | 4.2 ± 0.2 |
| katA null | 128 ± 3 | 3.5 ± 0.2 | 4.0 ± 0.2 | 4.7 ± 0.2 |
| katB null | n.d. | 3.2 | 3.65 ± 0.05 | 4.4 |
| katA katB null | 234 ± 22 | 3.65 ± 0.05 | 3.75 ± 0.05 | 4.3 |
| With pMMB207αB-Km14 | ||||
| katA + katA+ | n.d. | 2.75 ± 0.15 | 3.0 | 3.95 ± 0.15 |
| katA + vector alone | n.d. | 3.4 | 3.75 ± 0.05 | 4.8 ± 0.1 |
In AYE broth. n.d., not determined. Means ± standard deviations are shown.
Disk contained 10 μl of the indicated H2O2 solution. Means ± standard deviations are shown.
Complementation of the katA null with plasmid wild-type katA demonstrated that H2O2 sensitivity in the katA null was due to loss of katA and not to a polar effect on a downstream gene or spontaneous mutation elsewhere in the chromosome (Table 2). With the empty vector, H2O2 sensitivity was the same as in the katA null. With the complementing plasmid, H2O2 sensitivity was less than that of the wild type, presumably due to additional katA from multiple copies of the plasmid. Segal and Shuman demonstrated that inactivation of mobA in pMMB207 (an RSF1010 derivative) rendered the vector suitable for complementation and used pMMB207 mobA::Kmr derivatives to complement icm mutants for defects in infection of human macrophage cell lines and of Acanthamoeba castellanii (45). The pMMB207 mobA+ vector was unsuitable for complementation studies because H2O2 resistance and infectivity were altered by the vector alone without insert (6, 45).
Effect of katA null on growth in human macrophages.
The ability of L. pneumophila to invade, replicate within, and lyse macrophages can be quantified by measuring the increase in titer after addition of L. pneumophila to adherent macrophage-like cell lines, because L. pneumophila is unable to replicate in the tissue culture medium in the absence of macrophages. We previously showed that L. pneumophila was virulent in the absence of KatB and ultimately reached titers comparable to wild-type titers, although the time course of infection was delayed by ∼2 days (6). The katA and katA katB strains constructed in this study were similarly virulent and reproducibly showed a delay of ∼1 day in their infection curves (data not shown). Complementation of the katA null established that this delay phenotype was attributable to loss of katA. With plasmid pMMB207 mobA katA+, the infection curve of the katA null was identical to that of wild-type strain JR32, while the empty vector had no effect on the katA delay phenotype (data not shown). A compensatory change in gene expression in the katA katB double null may account for its 1-day delay phenotype in contrast to the 2-day delay of the katB single null.
Importance of katA in stationary-phase survival.
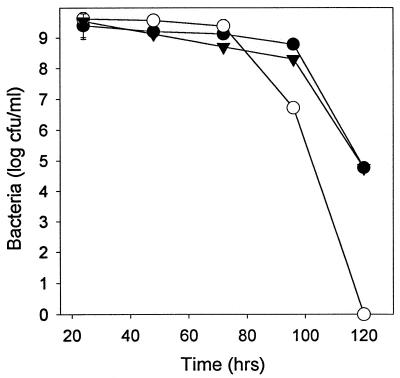
Decreases in stationary-phase plating efficiency of several orders of magnitude have been reported for catalase-peroxidase null mutants of Caulobacter crescentus (51) and Rhodobacter capsulatus (27). A similar deficit in stationary-phase survival was found for the L. pneumophila katA null (Fig. 3). Four days after inoculation, survival was decreased 100-fold. After 5 days no CFU were seen, compared to a titer of >104/ml for the wild type. The survival deficit was completely complemented by the pMMB207 mobA katA+ plasmid, indicating that this phenotype was attributable to katA inactivation and not to polar effects or second-site mutations (Fig. 3). Survival of the katA katB double null in stationary phase was similar to that of the katA null (data not shown), consistent with our earlier observation that inactivation of katB is without effect on this phenotype (6).
FIG. 3.
Stationary-phase survival. Titers were determined on CYE plates at the indicated times following inoculation into CAYE broth of wild-type strain JR32 (●), katA null strain PB126 (○), and katA null strain with complementing vector pMMB207αB-Km14::katA+ (▾). Standard deviations are shown by the error bars or are contained within the symbols.
Regulation of catalase-peroxidase expression.
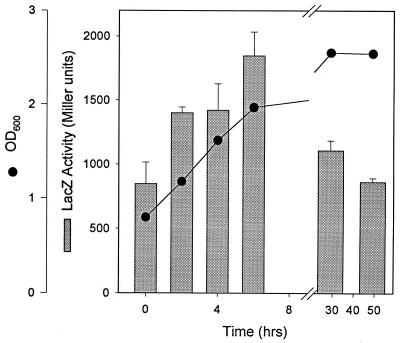
Expression of katA increased during exponential growth and then decreased in stationary phase by ≈50%, as assessed by a plasmid translational lacZ fusion (Fig. 4). This pattern is similar to our previous observations for L. pneumophila katB using a chromosomal lacZ fusion (6). Expression patterns were assessed directly in exponential- and stationary-phase cultures of wild-type L. pneumophila by catalase and diaminobenzidine peroxidase zymograms (Fig. 2, lanes 3 to 5). The zymograms were consistent with the lacZ fusion results in demonstrating an increase in both KatA and KatB during exponential growth. However, the zymograms showed that activity levels of each catalase-peroxidase were higher in stationary than in exponential phase.
FIG. 4.
Expression of katA during growth. Wild-type strain JR32 containing the lacZ translational fusion plasmid pJBZ281::PkatA was cultured in AYE medium, and at the indicated times aliquots were removed for determination of optical density at 600 nm (OD600) (●) and LacZ measurements (bars).
Levels of a gene product may be inaccurately reported by a translational fusion. In the present instance, the discrepancy between fusion (Fig. 4) and zymogram (Fig. 2) data may be due to decreased stability of the chimeric protein and/or decreased export to the periplasm in the stationary phase. (See intracellular location studies below.) We (6) and others (40) observed that peroxidase activity in L. pneumophila stationary-phase cultures is 8 to 10 times higher than during rapid growth. We predicted that KatA was the principal source of peroxidase activity in stationary phase from analysis of katB expression (6). The data in Fig. 2 confirm our prediction. By direct zymogram analysis, KatA peroxidase activity is higher in stationary-phase than in exponential-phase cultures, and KatA, not KatB, is the predominant peroxidase during stationary growth.
To test for induction by exogenous H2O2 in exponential cultures, the katA::lacZ fusion strain in mid-log phase was treated with bolus additions of H2O2 to 15 or 60 μM. LacZ activity increased less than 50%, modest compared to the 20-fold induction seen for C. crescentus katG under comparable conditions (51). No increase in LacZ activity was observed when an L. pneumophila katB::lacZ reporter strain was similarly treated (6).
Substrate specificity of KatA and KatB.
Cloned L. pneumophila katA and katB were expressed in an E. coli strain lacking katE monofunctional catalase and katG catalase-peroxidase. All catalase and peroxidase activity in cell extracts was from the expressed L. pneumophila genes. Activity with four peroxidase cosubstrates was compared to catalatic activity (Table 3). Many bifunctional catalase-peroxidases have NAD(P)H peroxidatic activity, 0.01 to 1% of the catalatic activity, and reduced pyridine nucleotides have been proposed as physiological peroxidatic cosubstrates (28, 31, 36). The peroxidatic activity of L. pneumophila KatA and KatB catalase-peroxidases towards reduced pyridine nucleotides was in this range (0.02% of catalatic activity, Table 3) and was identical for KatA and KatB. KatA and KatB differed in their relative dianisidine peroxidatic activity, with that of KatA being five times higher. This was consistent with our earlier prediction from measurements of activity in L. pneumophila extracts (6). The relative activity of KatA and KatB towards pyrogallol was comparable (Table 3).
TABLE 3.
Enzymatic activitya
| Peroxidase cosubstrates | Relative activity (% of catalase activity)b
|
|
|---|---|---|
| KatA | KatB | |
| H2O2 + dianisidine | 0.28 ± 0.02 | 0.058 ± 0.0004 |
| H2O2 + pyrogallol | 0.29 ± 0.05 | 0.36 ± 0.002 |
| H2O2 + NADH | 0.022 ± 0.001 | 0.022 ± 0.004 |
| H2O2 + NADPH | 0.023 ± 0.005 | 0.019 ± 0.002 |
Enzymatic activities of KatA and KatB were measured in periplasm or spheroplast extracts of E. coli strain UM383 katE katG containing pMMB207αB-Km14 katA or pMMB207αB katB (Table 1). The E. coli strains were grown overnight in LB broth with chloramphenicol.
Activities (in micromoles of H2O2 or peroxidase cosubstrate converted per minute per microliter of extract) are expressed relative to catalase activity, set at 100%. Where appropriate, corrections were made for absorbance changes in extracts of UM383 with the vector pMMB207αB-Km14 alone.
Intracellular localization of KatA and katB in E. coli.
The stationary-phase survival deficit of the L. pneumophila katA null mutant was very similar to what we observed for a null mutant of periplasmic L. pneumophila copper-zinc superoxide dismutase (CuZnSOD [52]). Spheroplasting studies were performed to determine if KatA was also periplasmic. Cytosol and periplasm fractions were cross-contaminated, as judged by their respective markers, glucose 6-phosphate dehydrogenase (Zwf) and alkaline phosphatase (PhoA), when spheroplasting was attempted in L. pneumophila. Therefore, spheroplasting was done with E. coli katE katG strains expressing L. pneumophila katA or katB from a plasmid.
The ratio of enzyme activity in the cytosol to that in the periplasm (Table 4, last column) clearly showed that L. pneumophila KatA was periplasmic, KatB was cytosolic, and the Zwf and PhoA markers localized in their expected subcellular fractions in E. coli. After our spheroplasting studies were completed, Amemura-Maekawa et al. (3) reported that L. pneumophila KatA is periplasmic by expression of a plasmid gene in E. coli. Our results were consistent and additionally demonstrated the cytosolic location of KatB and quantitative data for intracellular locations. The presence of a signal peptide in L. pneumophila katA (3) and independent experiments by ourselves and others (3) establish the periplasmic location of KatA beyond a reasonable doubt.
TABLE 4.
Intracellular locationa
| Strain and enzyme | Activity (U/ml)
|
Activity ratio, C/P | |
|---|---|---|---|
| Cytosol (C) | Periplasm (P) | ||
| katA | |||
| Zwf | 2.28 (±0.03) × 10−4 | 8.1 (±0.1) × 10−5 | 2.8 |
| PhoA | 1.7 × 10−7 | 1.2 (±0.1) × 10−5 | 0.014 |
| KatA | 6.2 (±0.2) × 10−6 | 3.0 (±0.1) × 10−4 | 0.021 |
| katB | |||
| Zwf | 1.1 (±0.2) × 10−3 | 0.35 (±0.13) × 10−3 | 3.1 |
| PhoA | 1.1 (±0.5) × 10−5 | 1.4 (±0.2) × 10−4 | 0.078 |
| KatB | 0.25 (±0.05) | 0.11 (±0.02) | 2.3 |
The values listed are the units of activity per milliliter of original E. coli culture referred to in footnote a of Table 3. KatA activity was assayed by its dianisidine peroxidase activity, and KatB was assayed by its catalase activity. The values for recovery of enzyme activity [(activity in cytosol plus activity in periplasm)/activity in sonic extracts] were 78% for KatA, 34% for glucose 6-phosphate dehydrogenase (Zwf), and 61% for alkaline phosphatase (PhoA) for the katA strain and 83% for KatB, 44% for glucose 6-phosphatase, and 18% for alkaline phosphatase for the katB strain.
DISCUSSION
If a single cell contains multiple enzymes with the same in vitro activity, do the enzymes have redundant, overlapping, or independent functions in bacterial physiology or ecology (4)? We are addressing this question in L. pneumophila, a free-living aquatic bacterium, an intracellular parasite of aquatic amoeba, and a pathogen of pulmonary macrophages (17, 46, 47). L. pneumophila contains two bifunctional catalase-peroxidases, KatA and KatB, ≈60% identical in nucleotide and amino acid sequence (3, 6). Our studies show that, depending on the physiological function, KatA and KatB may be either redundant, overlapping, or independent.
Function of catalase-peroxidases in aerobic growth.
Since L. pneumophila is an obligate aerobe, one might expect that growth would be impaired by eliminating enzymes that defend against reactive oxygen produced in aerobic respiration. However, neither katA nor katB is essential for exponentially growing cultures in complex medium. Null mutants for either have a doubling time identical to that of the wild type. When both catalase-peroxidases are absent, the doubling time increases twofold. These data show that L. pneumophila KatA and KatB are functionally redundant in defense against endogenously produced H2O2. If one catalase-peroxidase is absent, the other—plus any changes in gene expression consequent to the null mutation—compensates to prevent the increased doubling time of the katA katB double null. Since KatA and KatB account for all catalatic and peroxidatic activity in cell extracts, the viability of the katA katB null indicates that neither catalatic nor peroxidatic activity is essential for aerobic survival. Presumably other players in the defense against oxidative stress—superoxide dismutases, alkylhydroperoxide reductase, glutathione, and/or repair mechanisms—can partially compensate for the absence of those activities and sustain aerobic survival, albeit at a reduced growth rate.
When encounters with exogenous H2O2 are mimicked by addition of H2O2 to cultures, katA and katB null mutants are more sensitive than the wild type. In this model of environmental H2O2 exposure, the catalase-peroxidases show overlapping function. Null mutants for either katA or katB are more sensitive to H2O2 than the wild type. The sensitivity of the katA null to H2O2 is greater than that of the katB null, consistent with zymograms showing that KatA is the major catalatic activity in exponential growth and the major peroxidatic activity in stationary phase.
Function of catalase-peroxidases in macrophage infection.
The infectivity of certain bacterial pathogens is attenuated by inactivation of antioxidant enzymes via null mutations or antisense RNA (13, 15, 20, 32, 58, 59). This has been attributed to a compromise of the bacterial defense against reactive oxygen species generated in the respiratory burst following infection. Null mutants in katA or katB catalase-peroxidase (6) or both show a virulence defect akin to some icm and dot mutants of L. pneumophila that display attenuated infectivity, in particular the infection delay seen in the icmF mutant (41, 46, 54). It has been argued that the respiratory burst is not triggered by L. pneumophila because it enters macrophages by the CR1 and CR3 complement receptors (39), whose activation is not ordinarily linked with a burst (61). One explanation of the infection delays in katA and katB nulls is that H2O2 is in fact generated in an L. pneumophila infection despite its mechanism of entry. Alternatively, KatA and KatB may be important for changes in bacterial metabolism related to virulence. Environmental stress—in the form of nutrient deprivation or growth within an amoeba host—enhances the ability of L. pneumophila to infect bone marrow-derived macrophages, peripheral blood monocytes, or cultured macrophage-like cell lines (9, 10, 24). In this study, we observed that katA is a major player in stationary-phase survival, i.e., in surviving the stress of nutrient deprivation. Thus, further studies of the regulation and function of katA may clarify the linkage between environmental stress and L. pneumophila virulence.
Function of catalase-peroxidases in stationary-phase survival.
The most intriguing finding of this study is that inactivation of katA decreases stationary-phase survival by 100- to 10,000-fold, while inactivation of katB is without effect. This clear example of independent physiological roles was investigated by studying the regulation, substrate specificity, and intracellular locations of KatA and KatB. Such a survival deficit is absent from E. coli, in which the maximum decrease in stationary-phase survival in katE, katG, or katE katG nulls is sixfold (34, 38, 51).
Our data indicate that differences in regulation are an unlikely explanation for the different roles of L. pneumophila katA and katB in stationary-phase survival. The katA and katB genes are expressed similarly during growth. Levels of each catalase-peroxidase increase steadily during log phase. Neither is induced by exogenous H2O2 during exponential growth, an adaptive response seen with some other bacterial catalase-peroxidases (22, 51) and catalases (33, 56). In addition, katA was not preferentially induced in stationary phase, even though katA is critical for stationary-phase survival. This contrasts with C. crescentus katG, whose inactivation also decreases stationary-phase survival by >1,000-fold, which is induced 50- to 90-fold in stationary phase (51).
Characterization of the peroxidatic substrate specificity was not revealing about stationary-phase function. The dianisidine peroxidase-catalatic activity of KatA activity is five times greater than that of KatB, but dianisidine is not a physiological substrate. With the potential physiological cosubstrates NADH and NADPH, KatA and KatB catalase-peroxidases have identical ratios of catalatic to peroxidatic activity.
The most compelling clue for understanding the discrete roles of KatA and KatB in stationary-phase survival is their intracellular location, KatA in the periplasm and KatB in the cytosol. Although E. coli KatG is no longer believed to be periplasmic (26), other examples of periplasmic heme hydroperoxidases are known. KatP, a plasmid-encoded periplasmic catalase-peroxidase, is found in the enterohemorrhagic E. coli strain O157:H7 (8), and multiple monofunctional catalases are found in the periplasm of the phytopathogen Pseudomonas syringae (33). Discrete roles for katA and katB in stationary-phase survival define a physiological context for which cytosolic hydroperoxidase activity cannot substitute for periplasmic activity. What periplasm-specific function could be furnished by KatA?
One possibility is decomposition of H2O2 outside the cytosol. Cytosolic catalase activity can defend against external H2O2 because H2O2 readily passes through the inner membrane. If scavenging of H2O2 by KatA is critical in stationary phase, then the outer membrane, periplasm, or outer surface of the inner membrane may contain targets so sensitive to H2O2 that they cannot be adequately protected by cytosolic KatB. In support of this proposal, we found that the mutant lacking periplasmic KatA is in fact more sensitive to external H2O2 than the mutant lacking cytosolic KatB.
Another possibility is that the peroxidatic activity of KatA is critical for maintaining the periplasmic redox potential in stationary phase. The best-characterized periplasmic redox reactions are those in which sulfhydryl groups of periplasmic proteins are oxidized to disulfide bonds via Dsb proteins (43). DsbB, an inner membrane protein, donates electrons to ubiquinone (5), making the electron transport chain the ultimate source of oxidizing power for sustaining periplasmic sulfhydryl oxidation. In stationary phase, electron transport to oxygen is reduced. Therefore, the capacity of electron transport to sustain periplasmic disulfide bond formation will be reduced. Periplasmic KatA may be critical in stationary-phase survival by peroxidatically oxidizing ubiquinol or another periplasmic or inner membrane component using H2O2 as an electron acceptor. This function could maintain periplasmic sulfhydryl oxidation when the oxidizing capacity of the electron transport is diminished.
Periplasmic KatA is one of four antioxidant enzymes known to be essential for stationary-phase survival in gram-negative species. Among the others, L. pneumophila CuZnSOD is also periplasmic (52), but the catalase-peroxidases of C. crescentus (51) and R. capsulatus (18, 19, 27, 29) have not been subjected to localization studies and are not predicted to contain a signal peptide (PSORT Prediction, http://psort.nibb.ac.jp/form.html; Signal P V1.1, http://www.cbs.dtu.dk/services/signalP/). Our studies raise the question of whether a periplasmic location is a sine qua non for the stationary-phase function. This can be studied in L. pneumophila katA by determining if katA without the signal peptide complements the survival deficit of the katA null. The 100- to 10,000-fold deficit in stationary-phase survival is a strong phenotype for identifying suppressors of the katA null. Future studies such as these will give insights on KatA function in stationary phase and on the role of starvation stress in L. pneumophila virulence.
ACKNOWLEDGMENTS
This work was supported by grants MCB-9513076 and MCB-980992 to H.M.S. from the National Science Foundation.
We thank Howard Shuman, Laura Hales, and Gil Segal (Columbia University) for plasmids pMMB207αB-Km14 and pBluescript Gmr (pAM6), Peter Loewen (University of Manitoba) for E. coli strain UM383, and the DNA Sequencing/Synthesis Services Facility at Albert Einstein College of Medicine for DNA sequencing and synthetic oligonucleotides.
REFERENCES
- 1.Aebi H. Catalase in vitro. Methods Enzymol. 1984;108:121–126. doi: 10.1016/s0076-6879(84)05016-3. [DOI] [PubMed] [Google Scholar]
- 2.Alley M R K, Gomes S L, Alexander W, Shapiro L. Genetic analysis of a temporally transcribed chemotaxis gene cluster in Caulobacter crescentus. Genetics. 1991;129:333–342. doi: 10.1093/genetics/129.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amemura-Maekawa J, Mishima-Abe S, Kura F, Takahashi T, Watanabe H. Identification of a novel periplasmic catalase-peroxidase KatA of Legionella pneumophila. FEMS Microbiol Lett. 1999;176:339–344. doi: 10.1111/j.1574-6968.1999.tb13681.x. [DOI] [PubMed] [Google Scholar]
- 4.Aslund F, Beckwith J. The thioredoxin superfamily: redundancy, specificity, and gray-area genomics. J Bacteriol. 1999;181:1375–1379. doi: 10.1128/jb.181.5.1375-1379.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bader M, Muse W, Ballou D P, Gassner C, Bardwell J C. Oxidative protein folding is driven by the electron transport system. Cell. 1999;98:217–227. doi: 10.1016/s0092-8674(00)81016-8. [DOI] [PubMed] [Google Scholar]
- 6.Bandyopadhyay P, Steinman H M. Legionella pneumophila catalase-peroxidases: cloning of the katB gene and studies of KatB function. J Bacteriol. 1998;180:5369–5374. doi: 10.1128/jb.180.20.5369-5374.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bishai W R, Howard N S, Winkelstein J A, Smith H O. Characterization and virulence analysis of catalase mutants of Haemophilus influenzae. Infect Immun. 1994;62:4855–4860. doi: 10.1128/iai.62.11.4855-4860.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunder W, Schmidt H, Karch H. KatP, a novel catalase-peroxidase encoded by the large plasmid of enterohaemorrhagic Escherichia coli O157:H7. Microbiology. 1996;142:3305–3315. doi: 10.1099/13500872-142-11-3305. [DOI] [PubMed] [Google Scholar]
- 9.Byrne B, Swanson M S. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect Immun. 1998;66:3029–3034. doi: 10.1128/iai.66.7.3029-3034.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cirillo J D, Cirillo S L, Yan L, Bermudez L E, Falkow S, Tompkins L S. Intracellular growth in Acanthamoeba castellanii affects monocyte entry mechanisms and enhances virulence of Legionella pneumophila. Infect Immun. 1999;67:4427–4434. doi: 10.1128/iai.67.9.4427-4434.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Claiborne A, Fridovich I. Purification of the o-dianisidine peroxidase from Escherichia coli B: physicochemical characterization and analysis of its dual catalatic and peroxidatic activities. J Biol Chem. 1979;254:4245–4252. [PubMed] [Google Scholar]
- 12.Clare D A, Duong M N, Darr D, Archibald F, Fridovich I. Effects of molecular oxygen on detection of superoxide radical with nitroblue tetrazolium and on activity stains for catalase. Anal Biochem. 1984;140:532–537. doi: 10.1016/0003-2697(84)90204-5. [DOI] [PubMed] [Google Scholar]
- 13.De Groote M A, Ochsner U A, Shiloh M U, Nathan C, McCord J M, Dinauer M C, Libby S J, Vazquez-Torres A, Xu Y, Fang F C. Periplasmic superoxide dismutase protects Salmonella from products of phagocyte NADPH-oxidase and nitric oxide synthase. Proc Natl Acad Sci USA. 1997;94:13997–14001. doi: 10.1073/pnas.94.25.13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeShazer D, Wood G E, Friedman R L. Molecular characterization of catalase from Bordetella pertussis: identification of the katA promoter in an upstream insertion sequence. Mol Microbiol. 1994;14:123–130. doi: 10.1111/j.1365-2958.1994.tb01272.x. [DOI] [PubMed] [Google Scholar]
- 15.Farrant J L, Sansone A, Canvin J R, Pallen M J, Langford P R, Wallis T S, Dougan G, Kroll J S. Bacterial copper- and zinc-cofactored superoxide dismutase contributes to the pathogenesis of systemic salmonellosis. Mol Microbiol. 1997;25:785–796. doi: 10.1046/j.1365-2958.1997.5151877.x. [DOI] [PubMed] [Google Scholar]
- 16.Feeley J C, Gibson R J, Gorman G W, Langford N C, Rasheed J K, Mackel D C, Baine W B. Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. J Clin Microbiol. 1979;10:437–441. doi: 10.1128/jcm.10.4.437-441.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fields B S. The molecular ecology of legionellae. Trends Microbiol. 1996;4:286–290. doi: 10.1016/0966-842x(96)10041-x. [DOI] [PubMed] [Google Scholar]
- 18.Forkl H, Drews G, Tadros M H. Promoter analysis of the catalase-peroxidase gene (cpeA) from Rhodobacter capsulatus. FEMS Microbiol Lett. 1996;137:169–174. doi: 10.1111/j.1574-6968.1996.tb08101.x. [DOI] [PubMed] [Google Scholar]
- 19.Forkl H, Vandekerckhove J, Drews G, Tadros M H. Molecular cloning, sequence analysis and expression of the gene for catalase-peroxidase (cpeA) from the photosynthetic bacterium Rhodobacter capsulatus B10. Eur J Biochem. 1993;214:251–258. doi: 10.1111/j.1432-1033.1993.tb17918.x. [DOI] [PubMed] [Google Scholar]
- 20.Franzon V L, Arondel J, Sansonetti P J. Contribution of superoxide dismutase and catalase activities to Shigella flexneri pathogenesis. Infect Immun. 1990;58:529–535. doi: 10.1128/iai.58.2.529-535.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez-Flecha B, Demple B. Homeostatic regulation of intracellular hydrogen peroxide concentration in aerobically growing Escherichia coli. J Bacteriol. 1997;179:382–388. doi: 10.1128/jb.179.2.382-388.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez-Flecha B, Demple B. Metabolic sources of hydrogen peroxide in aerobically growing Escherichia coli. J Biol Chem. 1995;270:13681–13687. doi: 10.1074/jbc.270.23.13681. [DOI] [PubMed] [Google Scholar]
- 23.Gregory E M, Fridovich I. Visualization of catalase on acrylamide gels. Anal Biochem. 1974;58:57–62. doi: 10.1016/0003-2697(74)90440-0. [DOI] [PubMed] [Google Scholar]
- 24.Hammer B K, Swanson M S. Co-ordination of legionella pneumophila virulence with entry into stationary phase by ppGpp. Mol Microbiol. 1999;33:721–731. doi: 10.1046/j.1365-2958.1999.01519.x. [DOI] [PubMed] [Google Scholar]
- 25.Hengge-Aronis R. Regulation of gene expression during entry into stationary phase. In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1497–1512. [Google Scholar]
- 26.Hillar A, Van Caeseele L, Loewen P C. Intracellular location of catalase-peroxidase hydroperoxidase I of Escherichia coli. FEMS Microbiol Lett. 1999;170:307–312. doi: 10.1111/j.1574-6968.1999.tb13388.x. [DOI] [PubMed] [Google Scholar]
- 27.Hochman A, Figueredo A, Wall J D. Physiological functions of hydroperoxidases in Rhodobacter capsulatus. J Bacteriol. 1992;174:3386–3391. doi: 10.1128/jb.174.10.3386-3391.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hochman A, Goldberg I. Purification and characterization of a catalase-peroxidase and a typical catalase from the bacterium Klebsiella pneumoniae. Biochim Biophys Acta. 1991;1077:299–307. doi: 10.1016/0167-4838(91)90544-a. [DOI] [PubMed] [Google Scholar]
- 29.Hochman A, Shemesh A. Purification and characterization of a catalase-peroxidase from the photosynthetic bacterium Rhodopseudomonas capsulata. J Biol Chem. 1987;262:6871–6876. [PubMed] [Google Scholar]
- 30.Horwitz M A, Silverstein S C. Intracellular multiplication of Legionnaires' disease bacteria (Legionella pneumophila) in human monocytes is reversibly inhibited by erythromycin and rifampin. J Clin Investig. 1983;71:15–26. doi: 10.1172/JCI110744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnsson K, Froland W A, Schultz P G. Overexpression, purification, and characterization of the catalase-peroxidase KatG from Mycobacterium tuberculosis. J Biol Chem. 1997;272:2834–2840. doi: 10.1074/jbc.272.5.2834. [DOI] [PubMed] [Google Scholar]
- 32.Khelef N, DeShazer D, Friedman R L, Guiso N. In vivo and in vitro analysis of Bordetella pertussis catalase and Fe-superoxide dismutase mutants. FEMS Microbiol Lett. 1996;142:231–235. doi: 10.1111/j.1574-6968.1996.tb08435.x. [DOI] [PubMed] [Google Scholar]
- 33.Klotz M G, Hutcheson S W. Multiple periplasmic catalases in phytopathogenic strains of Pseudomonas syringae. Appl Environ Microbiol. 1992;58:2468–2473. doi: 10.1128/aem.58.8.2468-2473.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loewen P C. Isolation of catalase-deficient Escherichia coli mutants and genetic mapping of katE, a locus that affects catalase activity. J Bacteriol. 1984;157:622–626. doi: 10.1128/jb.157.2.622-626.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loewen P C, Hu B, Strutinsky J, Sparling R. Regulation in the rpoS regulon of Escherichia coli. Can J Microbiol. 1998;44:707–717. doi: 10.1139/cjm-44-8-707. [DOI] [PubMed] [Google Scholar]
- 36.Marcinkeviciene J A, Magliozzo R S, Blanchard J S. Purification and characterization of the Mycobacterium smegmatis catalase-peroxidase involved in isoniazid activation. J Biol Chem. 1995;270:22290–22295. doi: 10.1074/jbc.270.38.22290. [DOI] [PubMed] [Google Scholar]
- 37.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. [Google Scholar]
- 38.Mulvey M R, Switala J, Borys A, Loewen P C. Regulation of transcription of katE and katF in Escherichia coli. J Bacteriol. 1990;172:6713–6720. doi: 10.1128/jb.172.12.6713-6720.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Payne N R, Horwitz M A. Phagocytosis of Legionella pneumophila is mediated by human monocyte complement receptors. J Exp Med. 1987;166:1377–1389. doi: 10.1084/jem.166.5.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pine L, Hoffman P S, Malcolm G B, Benson R F, Franzus M J. Role of keto acids and reduced-oxygen-scavenging enzymes in the growth of Legionella species. J Clin Microbiol. 1986;23:33–42. doi: 10.1128/jcm.23.1.33-42.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Purcell M, Shuman H A. The Legionella pneumophila icmGCDJBF genes are required for killing of human macrophages. Infect Immun. 1998;66:2245–2255. doi: 10.1128/iai.66.5.2245-2255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reisenauer A, Mohr C D, Shapiro L. Regulation of a heat shock sigma32 homolog in Caulobacter crescentus. J Bacteriol. 1996;178:1919–1927. doi: 10.1128/jb.178.7.1919-1927.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rietsch A, Beckwith J. The genetics of disulfide bond metabolism. Annu Rev Genet. 1998;32:163–184. doi: 10.1146/annurev.genet.32.1.163. [DOI] [PubMed] [Google Scholar]
- 44.Rosner J L, Storz G. Regulation of bacterial responses to oxidative stress. Curr Top Cell Regul. 1997;35:163–177. doi: 10.1016/s0070-2137(97)80007-6. [DOI] [PubMed] [Google Scholar]
- 45.Segal G, Shuman H A. Intracellular multiplication and human macrophage killing by Legionella pneumophila are inhibited by conjugal components of IncQ plasmid RSF1010. Mol Microbiol. 1998;30:197–208. doi: 10.1046/j.1365-2958.1998.01054.x. [DOI] [PubMed] [Google Scholar]
- 46.Segal G, Shuman H A. Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect Immun. 1999;67:2117–2124. doi: 10.1128/iai.67.5.2117-2124.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shuman H A, Purcell M, Segal G, Hales L, Wiater L A. Intracellular multiplication of Legionella pneumophila: human pathogen or accidental tourist? Curr Top Microbiol Immunol. 1998;225:99–112. doi: 10.1007/978-3-642-80451-9_6. [DOI] [PubMed] [Google Scholar]
- 48.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 49.Steinman H M. Bacteriocuprein superoxide dismutases in pseudomonads. J Bacteriol. 1985;162:1255–1260. doi: 10.1128/jb.162.3.1255-1260.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steinman H M, Ely B. Copper-zinc superoxide dismutase of Caulobacter crescentus: cloning, sequencing, and mapping of the gene and periplasmic location of the enzyme. J Bacteriol. 1990;172:2901–2910. doi: 10.1128/jb.172.6.2901-2910.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steinman H M, Fareed F, Weinstein L. Catalase-peroxidase of Caulobacter crescentus: function and role in stationary-phase survival. J Bacteriol. 1997;179:6831–6836. doi: 10.1128/jb.179.21.6831-6836.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.St. John G, Steinman H M. Periplasmic copper-zinc superoxide dismutase of Legionella pneumophila: a role in stationary phase survival. J Bacteriol. 1996;178:1578–1584. doi: 10.1128/jb.178.6.1578-1584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Storz G, Altuvia S. The OxyR regulon. Methods Enzymol. 1994;234:217–223. doi: 10.1016/0076-6879(94)34088-9. [DOI] [PubMed] [Google Scholar]
- 54.Swanson M S, Isberg R R. Identification of Legionella pneumophila mutants that have aberrant intracellular fates. Infect Immun. 1996;64:2585–2594. doi: 10.1128/iai.64.7.2585-2594.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tichy M, Vermaas W. In vivo role of catalase-peroxidase in Synechocystis sp. strain PCC6803. J Bacteriol. 1999;181:1875–1882. doi: 10.1128/jb.181.6.1875-1882.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Visick K L, Ruby E G. The periplasmic, group III catalase of Vibrio fischeri is required for normal symbiotic competence and is induced both by oxidative stress and by approach to stationary phase. J Bacteriol. 1998;180:2087–2092. doi: 10.1128/jb.180.8.2087-2092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wiater L A, Sadosky A B, Shuman H A. Mutagenesis of Legionella pneumophila using Tn903dIIlacZ: identification of a growth-phase-regulated pigmentation gene. Mol Microbiol. 1994;11:641–653. doi: 10.1111/j.1365-2958.1994.tb00343.x. [DOI] [PubMed] [Google Scholar]
- 58.Wilks K E, Dunn K L, Farrant J L, Reddin K M, Gorringe A R, Langford P R, Kroll J S. Periplasmic superoxide dismutase in meningococcal pathogenicity. Infect Immun. 1998;66:213–217. doi: 10.1128/iai.66.1.213-217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilson T, de Lisle G W, Marcinkeviciene J A, Blanchard J S, Collins D M. Antisense RNA to ahpC, an oxidative stress defense gene involved in isoniazid resistance, indicates that AhpC of Mycobacterium bovis has virulence properties. Microbiology. 1998;144:2687–2695. doi: 10.1099/00221287-144-10-2687. [DOI] [PubMed] [Google Scholar]
- 60.Witholt B, Boekhout M, Brock M, Kingma J, Heerikhuizen H V, Leij L D. An efficient and reproducible procedure for the formation of spheroplasts from variously grown Escherichia coli. Anal Biochem. 1976;74:160–170. doi: 10.1016/0003-2697(76)90320-1. [DOI] [PubMed] [Google Scholar]
- 61.Wright S D, Silverstein S C. Receptors for C3b and C3bi promote phagocytosis but not the release of toxic oxygen from human phagocytes. J Exp Med. 1983;158:2016–2023. doi: 10.1084/jem.158.6.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zheng M, Aslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]
- 63.Zuniga M, Champomier-Verges M, Zagorec M, Perez-Martinez G. Structural and functional analysis of the gene cluster encoding the enzymes of the arginine deiminase pathway of Lactobacillus sake. J Bacteriol. 1998;180:4154–4159. doi: 10.1128/jb.180.16.4154-4159.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]