Summary
Endolysosomes (EL) are known for their role in regulating both intracellular trafficking and proteostasis. EL facilitate the elimination of damaged membranes, protein aggregates, membranous organelles and play an important role in calcium signaling. The specific role of EL in cardiac atrial fibrillation (AF) is not well understood. We isolated atrial EL organelles from AF goat biopsies and conducted a comprehensive integrated omics analysis to study the EL-specific proteins and pathways. We also performed electron tomography, protein and enzyme assays on these biopsies. Our results revealed the upregulation of the AMPK pathway and the expression of EL-specific proteins that were not found in whole tissue lysates, including GAA, DYNLRB1, CLTB, SIRT3, CCT2, and muscle-specific HSPB2. We also observed structural anomalies, such as autophagic-vacuole formation, irregularly shaped mitochondria, and glycogen deposition. Our results provide molecular information suggesting EL play a role in AF disease process over extended time frames.
Subject areas: Molecular biology, Cell biology, Proteomics, Transcriptomics
Graphical abstract
Highlights
-
•
The EL proteins GAA, Rab7a, CLTB, VPS25, and CCT2 were significantly regulated
-
•
Increased autophagic flux in AF goat model
-
•
Glycogen accumulation and lysosomal-α-glucosidase upregulation
-
•
Increased vesicular trafficking
Molecular biology; Cell biology; Proteomics; Transcriptomics.
Introduction
Atrial fibrillation (AF) is the most common sustained cardiac arrhythmia, accounting for around 14% of all strokes in the UK, and linked to a significantly high risk of developing embolic stroke.1,2 The prevalence of AF in the general population is 2%, although the risk of AF is age-dependent and this figure rises to 3.7–4.2% in ages 60–70 and 10–17% in the over 80s.3 AF is a progressive disease, progressing from paroxysmal to persistent forms, with self-sustaining progression driven by AF-triggered cardiac remodeling4,5 and potentially progression of comorbidities associated with AF.6 Atrial remodeling in AF can be the result of structural7,8 or electrical9 changes, and organelle dysfunction is also observed in AF progression.10
Regional and cell-type specific quantitative proteomics studies have enabled significant progress to be made in the understanding of the proteomic and transcriptomic contribution to AF pathology.11 However, the contribution of organelle remodeling in AF has not been studied extensively, including the potential contribution of changes in acidic organelles12 such as lysosomes and endolysosomes (EL), which play key roles in cellular energy metabolism13 and the trafficking of cellular components.14,15 Lysosomal changes have long been linked to cardiac disease,16 and changes in acidic organelles may be linked to underlying alterations in AF molecular pathways.17 Lysosomes may be linked to AF progression, for example via changes in autophagy.18 Indeed, several studies have recently identified autophagy as a potential mechanism underlying cardiac remodeling in AF progression,18,19 and autophagy has been shown to be increased in AF patients.18 However, little is known of how lysosomal proteins may be altered in AF. Techniques that enable proteomic characterization at the level of individual organelles,20,21 therefore, have the potential to highlight such changes. We have previously developed a modified density gradient method to isolate endolysosomal proteins, which increases the identification of endo-membrane proteins that are trafficked to acidic organelles,21 we applied this endolysosomal isolation method to conduct a proteomic study in a large animal AF model. We combined transcriptomics and proteomics to obtain the mRNA-protein correlation. In addition, we carried out high resolution electron microscopy imaging to confirm structural changes that have previously been reported in goat AF studies22,23 and we specifically look at acidic organelles at the cellular level.
The endolysosomal system consists of a series of membranous vesicles, composed of early endosomes, recycling endosomes, late endosomes and lysosomes. Autophagosomes are responsible for delivering the intracellular contents to lysosomes to complete autophagy. The endocytic pathway consists of acidification of the endosomes, maturation of endosomes to lysosomes accompanied by vesicle trafficking, protein sorting and targeted degradation of mostly sorted cargo. The two opposing sorting systems that are operating in these processes include the endosomal sorting complex required for transport (ESCRT, supports targeted degradation) and the retromer (supports retrograde retrieval of cargo). The EL system is emerging as a central player in a host of neurodegenerative diseases24 and its relevance in other diseases including AF is now being explored.
The present study was conducted using the AF goat as the animal model. The goat model is an ideal substitute for human AF as it has better tolerance than most contemporary animal models for AF, and the goat model is more comparable physiologically to humans compared to other models.25 The goat is a suitable model for conducting long-term cardiac pacing to develop sustained AF,4 and has been successfully utilized for studying electrical, contractile and structural remodeling in sustained AF pathology.26,27,28,29 During prolonged pacing, the AF goat model has been shown to undergo structural remodeling through endomysial fibrosis.28,30 Similar to humans, the AF goat model demonstrates the development of electrical conduction disturbances that give rise to complex activation patterns and endocardial-epicardial dissociation, providing a suitable substrate for the development of atrial arrhythmia. Studies from Wijffels et al.31 and Eijsbouts et al.32 highlight the relevance of the AF goat model’s suitability for antiarrhythmic drug targeted studies.
Our integrated approach combines transcriptomics and proteomics to provide a comprehensive comparative analysis of omics data that includes post-translational data. Dysregulated proteins were identified by performing label-free quantitative mass spectrometric analysis of the AF goat peptides compared to the sham goat models. After identifying dysregulated proteins, molecular pathways were used to understand how these dysregulations potentially affected the lysosomes and acidic organelles. Molecular pathways were analyzed using Cytoscape 3.7.2 with STRING, Kyoto Encyclopedia of Genes and Genomes (KeGG), Gene Ontology (GO), and Reactome pathway annotations to predict the relevant pathways. Furthermore, using GO, an over-representation study was performed. The most significantly regulated proteins identified in the AF goat model were compared against the human proteome, and the highest represented biological processes and cellular components that were altered in the AF goat model were identified based on comparison against human data.
Results
Identifying differential protein expression by quantitative proteomic analysis
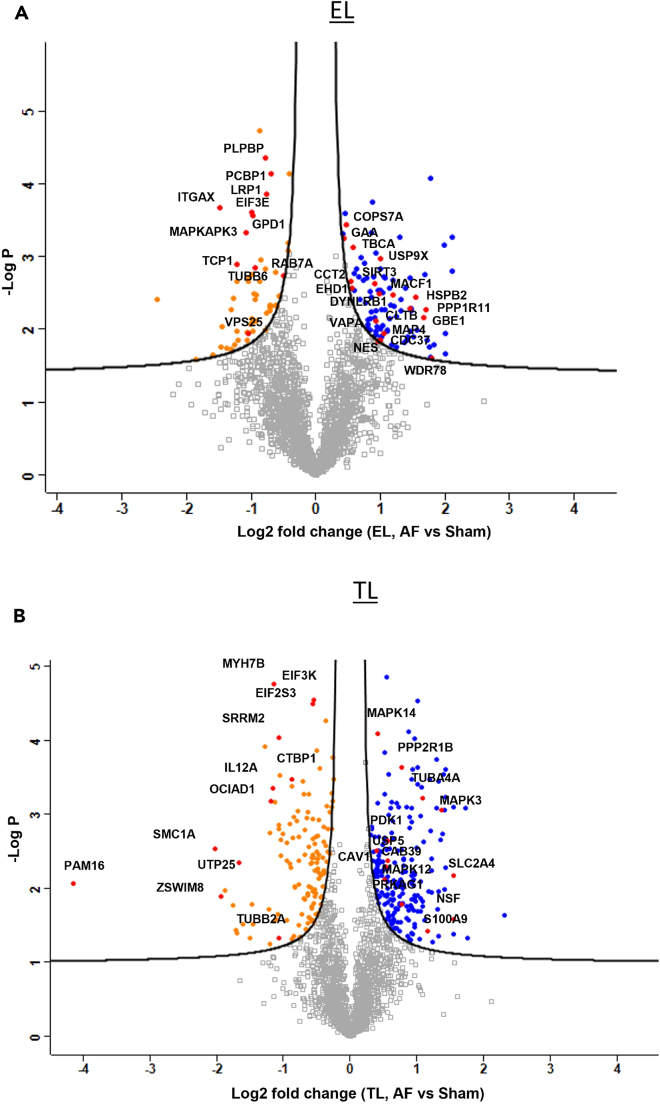
A density gradient approach21 (see STAR methods) was used to isolate fractions corresponding to whole tissue lysate (TL), mitochondria (Mito) and endolysosomal lysate from sham (N = 3) and AF (N = 3) left atrial tissue biopsies from goat hearts. Tissue samples were then prepared as described in STAR methods for liquid chromatography-tandem mass spectrometry (LC-MS). The differential protein-expression levels between AF and sham groups were identified by quantitatively analyzing the mass spectrometric data of TL and EL using the Perseus software platform33 (version 1.6.15.0) (Figure 1A and 1B). The protein intensity values of each biological replicate were separated into protein groups (AF and sham) and imported into Perseus. These data matrixes were filtered by removing proteins with more than two missing values and used for quantitative analysis.
Figure 1.
Volcano plot analysis
(A and B) Volcano plot analysis of Endolysosome fraction (EL) and Tissue Lysate (TL) of the AF vs. sham goat models showing significantly upregulated (Blue) and downregulated (Orange) proteins in the AF (p = 0.05 and FDR = 0.05). Some of the most important proteins discussed are labeled in red. The –log2 transformed p values are plotted against the differentially regulated levels of Proteins in AF and sham.
After the filtration, TL and EL samples each remained with 2104 proteins. Data were log-transformed (log2) and normalized via Z score. Data imputation was not required due to Progenesis reporting signal noise in absence of peptide precursors. Volcano plots were generated for each TL and EL sample by applying a two-way Student’s t test to identify the significant differences in protein regulations between AF and sham conditions (Figures 1A and 1B). The regulation levels were detected using a permutation-based false-discovery rate (FDR) of 5% with 250 randomizations at S0 = 0.1 and 99% confidence level.
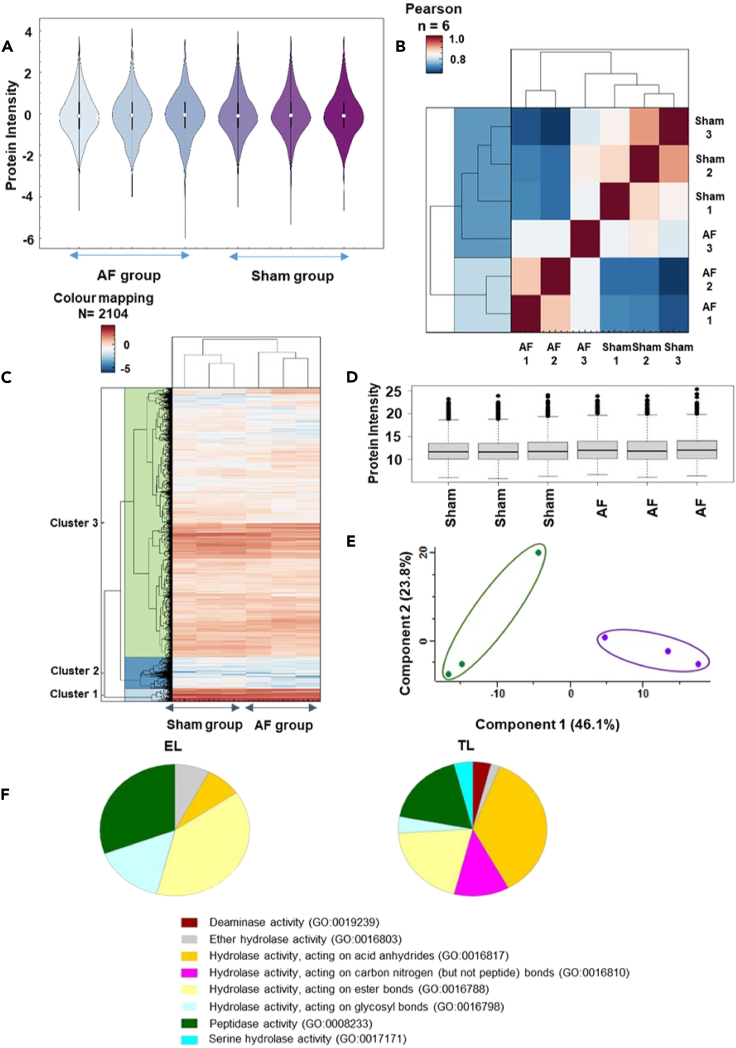
Violin plots (Figure 2A) were created using the kernel density estimation indicating the underlying distribution of the protein intensities between and within sham and AF group biological replicates by samples, color-coded with a gradient of purple, with higher protein intensity values presented as darker than the samples with comparatively lower intensities. Subsequently, Pearson coefficient correlation plots were created to observe the actual correlation between the groups (Figure 2B). Here the correlation ρ represents the interaction between two variables, or the AF and sham biological replicates, on a continuous scale of 1 to −1, where 1, depicts positive correlation, 0 depicts no correlation and −1 depicts negative correlation.34 Heat maps (Figure 2C) were created using Euclidian distance and K-mean clustering of the normalized protein intensities obtained from the quantified protein data matrix. A total of 2104 proteins in EL was observed in the three (EL) protein clusters. The red and blue color codes represent differential protein intensity levels and intensity whisker plots of each biological replicate were created to visualize the protein intensity distribution (Figure 2D). Principal Component Analysis (PCA) (Figure 2E) enabled observation of the vector distribution between and within the sample groups. In the PCA plots (Figure 2E), EL component 1 showed a 46.1% deviation respectively between AF (purple symbols) and sham (green symbols) groups. However, an exceptional segregation of 23.8% was observed in EL first biological replicates of sham groups, demonstrating that the variability between the differential experiment groups is more influential than the variability observed within the biological replicates. The most significantly regulated proteins of the EL fraction (Table 1) were further analyzed in STRING network to identify endolysosomal proteins with their respective structural or functional entities (Figure 3; Table 1).
Figure 2.
Proteomics data quantification
(A) Violin plot shows protein intensity levels in the triplicated AF vs. sham EL samples.
(B) Pearson co-efficient correlation plot explains the direct correlation between the sample protein intensities (1 = highest and 0 = lowest).
(C) Heatmap for the AF and sham EL Samples demonstrating the z scored intensities of the differentially expressed proteins after unsupervised hierarchical clustering.
(D) Whisker plot of the EL AF vs. sham protein intensity distribution shows the median, interquartile and the minimum to maximum outlier distribution.
(E) Principal Component Analysis (PCA) demonstrating the spatial resolution among the averaged vectors of the AF vs. sham goat EL samples, and component one and two variations presents respectively 46.1% and 23.8%.
(F) Gene ontology panther pathway analysis for EL and TL fractions. The molecular function of the endo-lysosome fraction (EL) showed 25% of catalytic activity, compared to the tissue lysate (TL) which showed 26.3% of catalytic activity. The catalytic hydrolase activity was further analyzed, and compared to TL (18%), EL fraction showed higher peptidase activity (30.8%), and hydrolases acting on ester bonds (TL = 20%, EL = 38.5%).
Table 1.
Contains the most significantly regulated endolysosomal (EL) fraction proteins of the AF goat model related to Figure 1A
| Protein ID | Protein name | Gene | Fold change |
|---|---|---|---|
| A0A452F9W2_CAPHI | Prefoldin subunit 4 | PFDN4 | 2.12 |
| A0A452DN27_CAPHI | Single-pass membrane and coiled-coil domain-containing protein 4 | SMCO4 | 2.01 |
| A0A452DY79_CAPHI | Structural maintenance of chromosomes protein | SMC1A | 2.01 |
| A0A452DL17_CAPHI | SAP domain-containing protein | CCAR1 | 1.99 |
| A0A452G2Z7_CAPHI | E3 ubiquitin-protein ligase HECTD3 | HECTD3 | 1.83 |
| A0A452DPX9_CAPHI | WD_REPEATS_REGION domain-containing protein | WDR78 | 1.79 |
| A0A452EJX2_CAPHI | Reticulon | RTN3 | 1.78 |
| A0A452G662_CAPHI | SHSP domain-containing protein | HSPB7 | 1.77 |
| A0A452G8P6_CAPHI | RING finger protein 17 | RNF17 | 1.75 |
| A0A452DM61_CAPHI | Calponin-homology (CH) domain-containing protein | NAV2 | 1.72 |
| A0A452F797_CAPHI | E3 ubiquitin-protein ligase PPP1R11 | PPP1R11 | 1.71 |
| A0A452FPZ5_CAPHI | cAMP-dependent protein kinase inhibitor | PKIG | 1.68 |
| A0A452G133_CAPHI | 1,4-alpha-glucan branching enzyme | GBE1 | 1.67 |
| A0A452F8B4_CAPHI | Cytochrome c oxidase subunit | COX6A2 | 1.56 |
| A0A452G4I2_CAPHI | SHSP domain-containing protein | HSPB2 | 1.55 |
| A0A452F5D8_CAPHI | 2-phospho-D-glycerate hydro-lyase | ENO2 | 1.51 |
| A0A452FLK7_CAPHI | ATP synthase-coupling factor 6, mitochondrial | ATP5PF | 1.47 |
| A0A452DQP7_CAPHI | Actin, alpha cardiac muscle 1 | ACTC1 | 1.47 |
| A0A452EBL3_CAPHI | DNA-directed RNA polymerase III subunit RPC4 | POLR3D | 1.46 |
| A0A452F3N4_CAPHI | Clathrin light chain | CLTB | 1.45 |
| A0A452FRY3_CAPHI | Splicing factor, proline- and glutamine-rich | SFPQ | 1.43 |
| A0A452DRC3_CAPHI | Mitochondrial import inner membrane translocase subunit TIM16 | PAM16 | 1.39 |
| A0A452ERZ9_CAPHI | Complex I-MNLL | NDUFB1 | 1.37 |
| A0A452G711_CAPHI | D-beta-hydroxybutyrate dehydrogenase, mitochondrial | BDH1 | 1.32 |
| A0A452FQ01_CAPHI | Protein cordon-bleu | COBL | 1.25 |
| A0A452E6Y1_CAPHI | Prefoldin subunit 1 | PFDN1 | 1.22 |
| A0A452FJJ3_CAPHI | PRA1 family protein | ARL6IP5 | 1.2 |
| A0A452DY71_CAPHI | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 | MACF1 | 1.19 |
| A0A452FIK7_CAPHI | Lipoma-preferred partner | LPP | 1.18 |
| A0A452F973_CAPHI | Cathelicidin-2 | CATHL2 | 1.16 |
| A0A452EW34_CAPHI | Myosin phosphatase Rho-interacting protein | MPRIP | 1.14 |
| A0A452EQJ4_CAPHI | PDZ domain-containing protein | AHNAK | 1.14 |
| A0A452FP52_CAPHI | Coxsackievirus and adenovirus receptor | CXADR | 1.11 |
| A0A452F158_CAPHI | Protein phosphatase 1 regulatory subunit | PPP1R12C | 1.1 |
| A0A452FDC1_CAPHI | NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial | NDUFS6 | 1.08 |
| A0A452FQ33_CAPHI | Prelamin-A/C | LMNA | 1.07 |
| A0A452G3Z1_CAPHI | Diacylglycerol kinase | DGKD | 1.06 |
| A0A452EIY1_CAPHI | Microtubule-associated protein | MAP4 | 1.05 |
| A0A452FHF8_CAPHI | Prefoldin subunit 6 | PFDN6 | 1.04 |
| A0A452EW73_CAPHI | Autism susceptibility gene 2 protein | AUTS2 | 1.04 |
| A0A452EKC1_CAPHI | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mitochondrial | NDUFB8 | 1.02 |
| A0A452FL98_CAPHI | Ubiquitinyl hydrolase 1 | USP9X | 1 |
| A0A452DMC5_CAPHI | Hsp90 chaperone protein kinase-targeting subunit | CDC37 | 1 |
| A0A452FYK7_CAPHI | BolA-like protein 2 | BOLA2B | 1 |
| A0A452E9A8_CAPHI | TRASH domain-containing protein | RPL24 | 1 |
| A0A452FDZ6_CAPHI | Ubiquitin-fold modifier-conjugating enzyme 1 | UFC1 | 1 |
| A0A452FSE0_CAPHI | Protein-cysteine N-palmitoyltransferase HHAT-like protein | HHATL | 1 |
| A0A452F2G4_CAPHI | Dynein light chain roadblock | DYNLRB1 | 0.98 |
| A0A452G757_CAPHI | IF rod domain-containing protein | NES | 0.96 |
| A0A452G5E3_CAPHI | ATP synthase subunit d, mitochondrial | ATP5PD | 0.96 |
| A0A452EGT0_CAPHI | Complex I-B18 | NDUFB7 | 0.95 |
| A0A452FP74_CAPHI | Gal_mutarotas_2 domain-containing protein | GANAB | 0.94 |
| A0A452ENN3_CAPHI | Leucyl-tRNA synthetase | LARS1 | 0.93 |
| A0A452DKF1_CAPHI | Vesicle-associated membrane protein-associated protein A | VAPA | 0.93 |
| A0A452F4D3_CAPHI | Cystatin domain-containing protein | LOC102186806 | 0.92 |
| A0A452EPW1_CAPHI | NAD-dependent protein deacetylase | SIRT3 | 0.91 |
| A0A452G0B4_CAPHI | Cytochrome b5 heme-binding domain-containing protein | NENF | 0.91 |
| A0A452F760_CAPHI | N-lysine methyltransferase SMYD2 | SMYD2 | 0.9 |
| A0A452DZF9_CAPHI | Matrix-remodeling-associated protein 7 | MXRA7 | 0.87 |
| A0A452FYY0_CAPHI | Short/branched chain specific acyl-CoA dehydrogenase | ACADSB | 0.87 |
| A5JSS3_CAPHI | NADH dehydrogenase (Ubiquinone) 1 alpha subcomplex 4 | NDUFA4 | 0.86 |
| A0A452EKQ0_CAPHI | GRASP55_65 domain-containing protein | GORASP2 | 0.86 |
| A0A452FF94_CAPHI | Glutaredoxin domain-containing protein | GLRX | 0.85 |
| A0A452FEX6_CAPHI | Complex I-B22 | NDUFB9 | 0.85 |
| A0A452FJK2_CAPHI | Fibronectin | FN1 | 0.84 |
| PLMN_CAPHI | Plasminogen (Fragment) | PLG | 0.81 |
| A0A452FQM9_CAPHI | Complex I-B12 | NDUFB3 | 0.81 |
| A0A452G1G3_CAPHI | Helix-destabilizing protein | HNRNPA1 | 0.81 |
| A0A452F1F7_CAPHI | 60S acidic ribosomal protein P2 | RPLP2 | 0.77 |
| A0A452EUX2_CAPHI | Peroxiredoxin-6 | PRDX6 | 0.76 |
| A0A452E9Y7_CAPHI | Myomesin-2 | MYOM2 | 0.74 |
| A0A452GB99_CAPHI | Transgelin | TAGLN2 | 0.71 |
| A0A452F2I2_CAPHI | Epoxide hydrolase | EPHX1 | 0.68 |
| A0A452FAW7_CAPHI | Sodium/potassium-transporting ATPase subunit alpha | ATP4A | 0.67 |
| A0A452ESU7_CAPHI | Heat shock 27 kDa protein | HSPB1 | 0.63 |
| A0A452F8A8_CAPHI | Cerebral dopamine neurotrophic factor | CDNF | 0.6 |
| A0A452EI29_CAPHI | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 6 | NDUFA6 | 0.6 |
| A0A452FI28_CAPHI | Tubulin-specific chaperone A | TBCA | 0.58 |
| A0A452FFI1_CAPHI | EH domain-containing protein 1 | EHD1 | 0.57 |
| A0A452G4D6_CAPHI | CCT-beta | CCT2 | 0.55 |
| A0A452EF68_CAPHI | PCI domain-containing protein | COPS7A | 0.48 |
| A0A452DXZ3_CAPHI | Metaxin-2 | MTX2 | 0.46 |
| A0A452EV92_CAPHI | P-type domain-containing protein | GAA | 0.44 |
| A0A452ETD8_CAPHI | Filamin-B | FLNB | 0.41 |
| A0A452ERC8_CAPHI | Ribonuclease inhibitor | RNH1 | −0.41 |
| A0A452EB34_CAPHI | ADP/ATP translocase 3 | SLC25A6 | −0.43 |
| A0A452FQE3_CAPHI | Malic enzyme | ME1 | −0.43 |
| A0A452DMR1_CAPHI | Ras-related protein Rab-7a | RAB7A | −0.5 |
| A0A452F3X0_CAPHI | Myosin-7B | MYH7B | −0.57 |
| A0A452DSU6_CAPHI | Mediator of ErbB2-driven cell motility 1 | MEMO1 | −0.61 |
| A0A452EI23_CAPHI | Lactamase_B domain-containing protein | ETHE1 | −0.62 |
| A0A452E1G5_CAPHI | Nuclear receptor corepressor 2 | NCOR2 | −0.62 |
| A0A452DVL8_CAPHI | Poly(rC)-binding protein 1 | PCBP1 | −0.7 |
| A0A452G6A1_CAPHI | Dystonin | DST | −0.7 |
| A0A452F3M1_CAPHI | Importin N-terminal domain-containing protein | IPO5 | −0.7 |
| A0A452EH63_CAPHI | Fructose-bisphosphate aldolase | ALDOA | −0.72 |
| A0A452DZ25_CAPHI | NEDD8-activating enzyme E1 regulatory subunit | NAE1 | −0.76 |
| A0A452FYK1_CAPHI | Exosome complex protein LRP1 | LRP1 | −0.76 |
| A0A452FU89_CAPHI | Pyridoxal phosphate homeostasis protein | PLPBP | −0.78 |
| A0A452DU00_CAPHI | PABS domain-containing protein | SMS | −0.79 |
| A0A452E7Y7_CAPHI | PKS_ER domain-containing protein | RTN4IP1 | −0.79 |
| IL6_CAPHI | Interleukin-6 | IL6 | −0.86 |
| A0A452FN18_CAPHI | IF rod domain-containing protein | KRT3 | −0.86 |
| A0A452FAD3_CAPHI | Glyoxylate reductase/hydroxypyruvate reductase | GRHPR | −0.87 |
| A0A452EQN7_CAPHI | CSD_1 domain-containing protein | CARHSP1 | −0.92 |
| A0A452EKB8_CAPHI | Pyridoxal phosphate phosphatase | PDXP | −0.92 |
| A0A452FAZ1_CAPHI | Proteasome subunit beta | PSMB4 | −0.94 |
| A0A452G1A9_CAPHI | Tubulin beta chain | TUBB6 | −0.95 |
| A0A452EXY9_CAPHI | Serine/arginine-rich splicing factor 2 | SRSF2 | −0.95 |
| A0A452FD14_CAPHI | Glycerol-3-phosphate dehydrogenase [NAD(+)] | GPD1 | −0.97 |
| A0A452FCU2_CAPHI | Farnesyl pyrophosphate synthase | FDPS | −0.98 |
| A0A452E698_CAPHI | Chromobox protein homolog 1 | CBX1 | −0.98 |
| A0A452FV62_CAPHI | 4a-hydroxytetrahydrobiopterin dehydratase | PCBD2 | −0.98 |
| A0A452DRD5_CAPHI | Eukaryotic translation initiation factor 3 subunit E | EIF3E | −0.99 |
| A0A452FVJ7_CAPHI | Phospholipase B-like | PLBD1 | −0.99 |
| A0A452FUU5_CAPHI | Perilipin | PLIN3 | −1 |
| A0A452FP83_CAPHI | RRM domain-containing protein | HNRNPC | −1 |
| A0A452ES90_CAPHI | ESCRT-II complex subunit VPS25 | VPS25 | −1.04 |
| A0A452FY19_CAPHI | OCIA domain-containing protein | OCIAD1 | −1.04 |
| A0A452EHJ6_CAPHI | Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial | HADH | −1.05 |
| A0A452EJR1_CAPHI | EGF domain-specific O-linked N-acetylglucosamine transferase | EOGT | −1.06 |
| A0A452DQ55_CAPHI | Protein-synthesizing GTPase | EIF2S3 | −1.07 |
| A0A452EIV8_CAPHI | Non-specific serine/threonine protein kinase | MAPKAPK3 | −1.08 |
| A0A452DKT5_CAPHI | Small nuclear ribonucleoprotein Sm D3 | SNRPD3 | −1.21 |
| A0A452FIX8_CAPHI | Early endosome antigen 1 | EEA1 | −1.21 |
| A0A452EVX4_CAPHI | CCT-alpha | TCP1 | −1.23 |
| A0A452G5M5_CAPHI | Palmdelphin | PALMD | −1.23 |
| A0A452FLJ3_CAPHI | Caveolae-associated protein 1 | CAVIN1 | −1.26 |
| A0A452FNA0_CAPHI | Coronin | CORO1C | −1.28 |
| A0A452G7L4_CAPHI | S-adenosylmethionine synthase | MAT2A | −1.31 |
| A0A452FVB0_CAPHI | Actin-related protein 2/3 complex subunit 3 | ARPC3 | −1.32 |
| A0A452FX49_CAPHI | Hemoglobin subunit beta-C | HBBC | −1.38 |
| Q8WF85_CAPHI | Cytochrome b | cytb | −1.46 |
| A0A452DJR4_CAPHI | SWIM-type domain-containing protein | ZSWIM8 | −1.48 |
| A0A452DS40_CAPHI | VWFA domain-containing protein | ITGAX | −1.48 |
| A0A452F341_CAPHI | Beta-MPP | PMPCB | −1.86 |
| A0A452F1Z3_CAPHI | Argininosuccinate synthase | ASS1 | −2.45 |
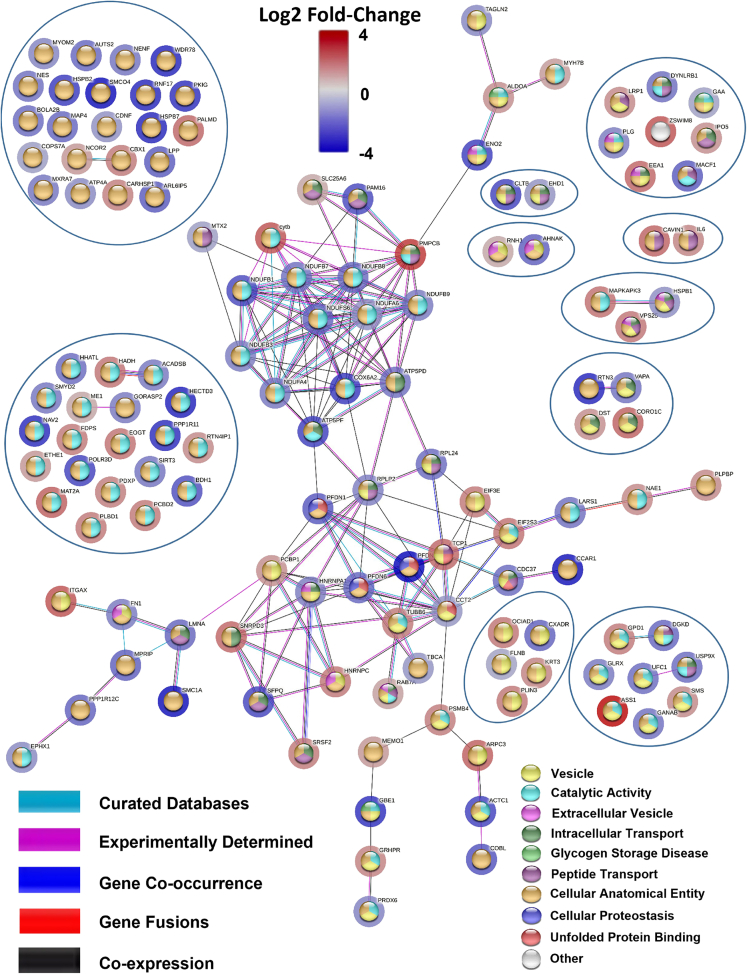
Figure 3.
Using STRING network to study endolysosomal proteins
Edges based on curated databases, experiments, gene co-occurrences, gene fusions, co-expressions and the nodes are colored according to their functional enrichments. The halos around the nodes display the significant log2 fold-change.
Confirmation of selected proteins by western blotting
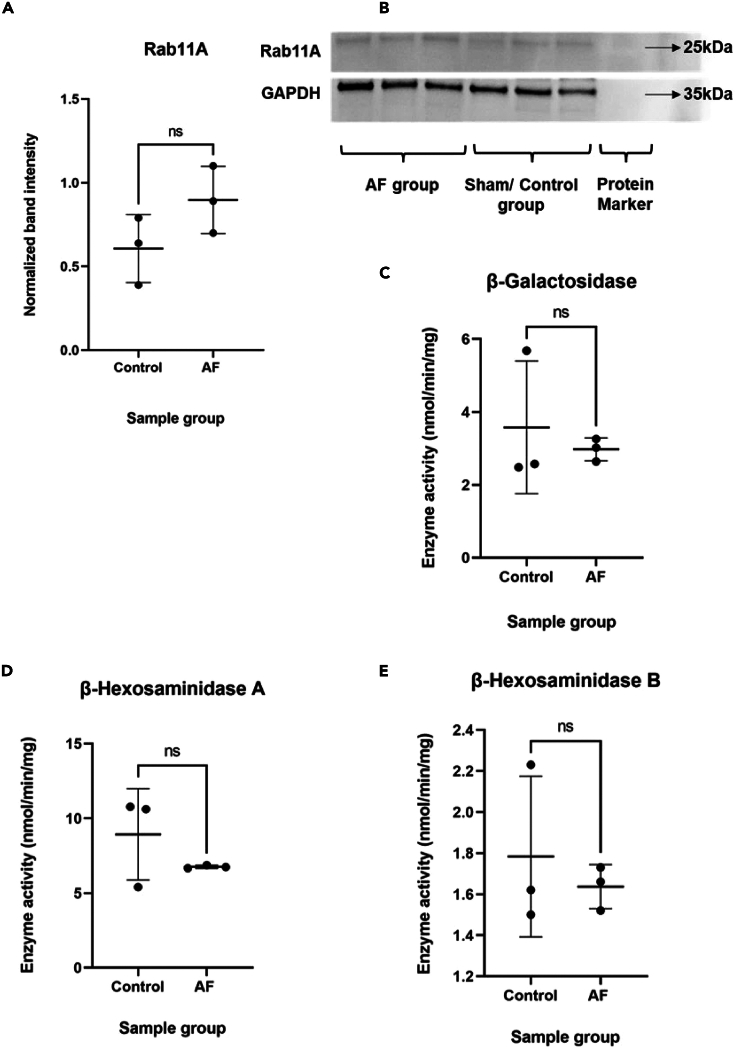
The Ras-related protein Rab-11A (Rab11A) is expressed ubiquitously35,36 and plays important roles in intracellular transport. Rab11A was previously found to be significantly upregulated in the AF goat proteomics model. Western blotting was conducted on the same (N = 3) biological samples retrieved from sham and AF goat models to evaluate this previous finding (Figures 4A and 4B). GAPDH was used as the control protein, and intensities of Rab11A bands were normalized using the GAPDH band intensity. The normalized data are presented as mean ± SD. The normalized band intensity for Rab11A was at 0.61 ± 0.20 for the control group and 0.90 ± 0.20 for the AF group. The Rab11A upregulation in AF was analyzed by conducting a one-way t-test to analyze for upregulation. Although our data did not support a significant upregulation in Rab11A in the AF group, the p-value of 0.07 suggests a trend toward Rab11A upregulation in the AF group when compared to sham controls (N = 3) (Figures 4A and 4B). This difference between our data and data published previously by Lapierre et al.35 is likely the result of the lower power (N = 3) of our data.
Figure 4.
Protein identification using western blotting and lysosomal enzymatic Assays
(A) Western blots were performed on N = 3 biological samples of each sham and AF group. Rab11A (24–25 kDa) and GAPDH control (37 kDa) were detected.
(B) The normalized band intensities of Rab11A data are presented as mean ± SD. The normalized band intensity value for the control group was 0.61 ± 0.20, and for the AF group at 0.90 ± 0.20. After performing a one-way t-test, the normalized protein band intensity showed a trend toward a significant upregulation in the AF group (p = 0.07).
(C) β galactosidase activity is displayed as mean ± SD. The AF group showed an enzymatic activity of 2.64, 3.02 and 3.26 nmol/min/mg, respectively, and the sham group showed 2.48, 2.57 and 5.68 nmol/min/mg. The mean enzymatic activity was 2.57 ± 0.31 for the AF group and 3.57 ± 1.8 for the control group. After performing a Two-way t-test, the β galactosidase activity of the AF group showed no significant regulation (p = 0.60).
(D) β hexosaminidase type-A activity of the AF group (N = 3) showed 6.66, 6.74 and 6.87 nmol/min/mg, respectively, and the sham group showed 5.4, 10.61 and 10.78 nmol/min/mg activity. The normalized mean enzymatic activity was at 8.93 ± 3.1 for the control group and 6.75 ± 0.11 for the AF group, and β hexosaminidase type- A activity of the AF group showed no significant regulation (p = 0.28).
(E) AF group showed (N = 3) 1.52, 1.66 and 1.73 nmol/min/mg of β hexosaminidase type B activity, and the sham group (N = 3) showed 1.5, 1.62 and 2.23 nmol/min/mg of β hexosaminidase type B activity, The mean enzymatic activity was at 1.8 ± 0.39 for the control group and 1.6 ± 0.11 for AF group. No significant regulation was observed in β hexosaminidase type B activity (p = 0.56).
Lysosome hydrolase activity assays
To check for impairment of autophagy, we conducted biochemical assays to investigate whether lysosomal enzyme activity is changed in goat AF. Lysosomal enzyme activity assays were performed on the three most common lysosomal enzymes: β galactosidase, β-hexosaminidase type A, and β hexosaminidase type B (Figures 4C–4E). The enzymatic activities were analyzed using a two-way t-test and no significant difference was detected in any of the 3 lysosomal enzymes studied.
Altered lysosomal enzyme activities are observed in lysosomal storage disorders (LSDs),37 and some patients with LSDs show severe cardiac phenotypes.38 However, we did not observe such significant changes in the AF goat model, indicating little or no impairment in autophagy and this result is reinforced by the western blotting data on LC3I proteins (Figure S2A).
mRNA transcript analysis using next generation sequencing
Distance matrix heatmap
The distances between samples were analyzed using hierarchical clustering to provide an overview of the similarities and dissimilarities between the samples.
Sample distances were displayed using a range of shaded color bar from dark blue to white, 0 being highly similar and displayed as dark blue and 200 being highly dissimilar represented in white color (Figure S3A).
Heatmap
The topmost regulated genes were plotted using hierarchical clustering to determine whether the samples cluster together according to the AF or sham conditions. All the AF and sham goat samples of LA tissue were clustered together, (Figure S3B).
PCA
PCA was performed to analyze the sample distribution and to assess the reliable correlation between the sample triplicates between AF and sham model groups by reduced data dimension for a simpler interpretation. As indicated in Figure S3C, vector deviation of 67.3% was observed at component 1 (PC1) between AF (purple symbols) and sham/control groups (green symbols). An exceptional 11.8% segregation was observed in component 2 (PC2), or the biological replicates within the groups. Since, PC1 value was higher than PC2, it can be concluded that the variance between the two conditions is higher than the variance within a group (Figure S3C).
Whisker plot
The transcript intensity distribution between AF and sham goat model samples were observed using whisker plot (Figure S3D).
The whole tissue lysates (same samples as used in proteomics and Western blotting) were used to isolate mRNA, genomic expression was quantified, and the differential expression plotted as a volcano plot (Figure S4A). These differentially expressed genes were fed into Reactome pathway analysis to study the pathway regulations. We observed a significant upregulation of 235 genes, while 297 genes were significantly down regulated (Table S1.1).
Integrated analysis of transcriptomics and proteomics
The integrated omics analysis highlighted several regulated pathways that were categorized under three confidence levels. These are color coded in gray, black and red which represents non-confident, confident and highly confident pathways, respectively in Figures S4B and S5B. In our analysis and discussion, we do not consider non-confident pathways. Furthermore, the significance of these pathways was determined with a pathway score (PS) to show their enrichment levels in both transcriptomics and proteomics analysis.
Interestingly, our integrated analysis highlighted RHOBTB GTPase, RHOBTB1 and RHOBTB2 GTPase cycles to be significantly enriched in transcriptomics. While the integrated analysis did not highlight a significant enrichment in proteomics. The pathways, mitochondrial ABC transporters, interleukin (IL) 9 signaling, defective POMGNT1 causes MDDGA3, MDDGB3 and MDDGC3, beta-oxidation of pristanoyl-CoA, RUNX3 regulates RUNX1-mediated transcription, RUNX3 Regulates Immune Response and Cell Migration, arachidonate production from DAG, RUNX1 regulates estrogen receptor mediated transcription, abacavir metabolism, coenzyme A biosynthesis, suppression of autophagy, regulation of RUNX1 expression and activity, defective HK1 causes hexokinase deficiency (HK deficiency), activation of BMF and translocation to mitochondria, neurofascin interactions, degradation of GABA, and formation of the active co-factor, UDP-glucuronate are significantly downregulated in proteomics compared to transcriptomics. Some of the downregulated pathways in both the integrated TL proteomics and transcriptomics were, defective PMM2 causes PMM2-CDG (CDG-1a), defective MMAA causes methylmalonic aciduria type cblA, defective MUT causes methyl malonic aciduria mut type, glycogen storage disease types II, IV, XV and 0, defective PGM1 cause of PGM1-CDGII, MET activation of PI3K/AKT signaling, MET activation of PTPNII, inhibition of NO production, and stimulation of the cell death response by PAK-2P34 (labeled in red circles, Figure S4B and Table S2.1).
Like the TL fraction, the integrated analysis of the most significantly up and down regulated EL proteins and genes of transcriptomics analysis (Figure S5B) showed RHOBTB and RHOBTB2 GTPase cycles up regulated. From the integrated analysis, the pathways such as, signaling by PDGFRA transmembrane, juxtamembrane, and kinase domain mutants, signaling by cytosolic FGFR1 fusion mutants, ROBO receptors bind AKAP5, interconversion of 2-oxoglutarate and 2-hydroxyglutarate, IL-6 signaling, defective TPR may confer susceptibility toward thyroid papillary carcinoma (TPC), regulation of Glucokinase by Glucokinase Regulatory Protein, beta-oxidation of pristanoyl-CoA, defective MPI causes MPI-CDG (CDG-1b), suppression of autophagy, MAPK1 (ERK2) activation, phosphor-PLA2 pathway, degradation of GABA, IL-9 signaling, inhibition of NO production, formation of the active cofactor, UDP-glucuronate, and signaling by PDGFRA extracellular domain mutants were downregulated in proteomics compared to transcriptomics. Activation of PPARGC1A (PGC-1alpha) by phosphorylation, defective GFPT1 causes CMSTA1, stimulation of the cell death response by PAK-2P34, MET activation of PI3K/AKT signaling, glycogen storage disorders such as type II, IV, XV, and 0, non-canonical activation of NOTCH3, defective PGM1 cause of PGM1-CDGII, neurofascin interactions, LRR FLII-interacting protein 1 (LRRFIP1) activates type I IFN production, fibronectin matrix formation, mitochondrial ABC transporters, regulation of RUNX1 expression and activity, Tie2 signaling, defective MMAA causes methyl malonic aciduria type cblA, defective MUT causes methyl malonic aciduria mut type and MET activates PTPN11 were the significantly down regulated pathways in, both proteomics and transcriptomics. The regulation of GAP junction activity pathway was downregulated in TL proteomics compared to transcriptomics, (Figure S4B, and Table S2.1).
To identify and understand the overall representation of the pathways related to the EL, acidic organelles, and vesicle trafficking, the enrichment scores from the listed significant pathways of integrated proteomics and transcriptomics analysis were separated (Figure S5A and Table S2.1).
Furthermore, the integrated analysis of the most significantly up or downregulated EL, TL proteins and genes highlighted by the proteomic and transcriptomic analysis showed pathways related to the following as being confidently upregulated (black in color circled; Figures S4B and S5B and Table S2.1); membrane trafficking, vesicle mediated transport, TCA cycle and respiratory pathway, chaperone mediated protein folding, ER to Golgi anterograde transport, ER- Phagosome pathway, lipid metabolism, COPI mediated anterograde transport, while anabolic pathways such as rRNA processing, and cell cycle were among the down regulated pathways with confidence (the most significant list of pathways are summarized in Figures S4B and S5B and the complete list of the pathways are shown in the Table S2.1).
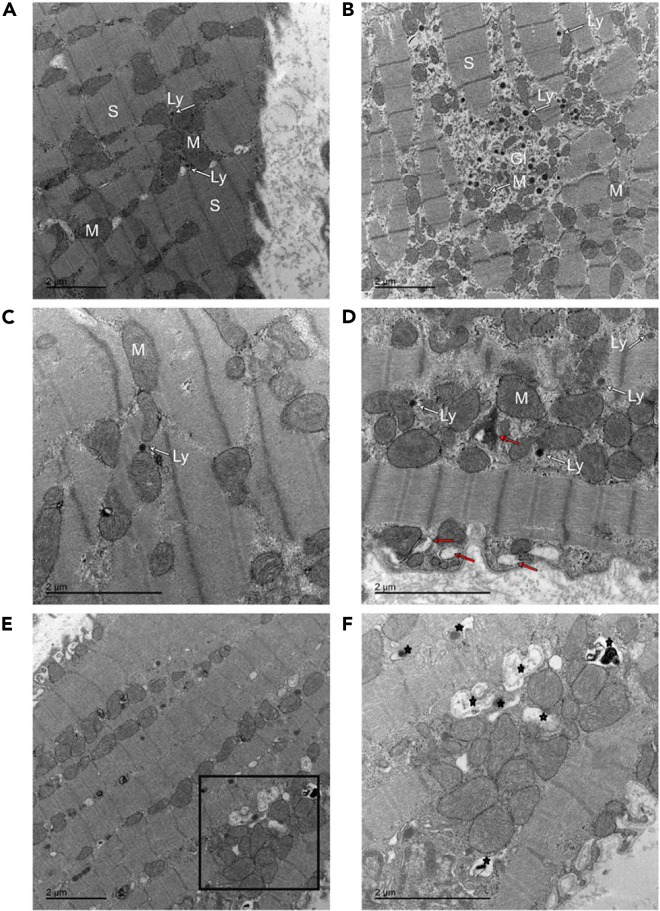
Structural insights using electron microscopy (EM)
We qualitatively analyzed EM images from goat AF and sham control tissue and observed that the sarcomeres were regularly distributed throughout the cytoplasm and there were rows of uniformly sized mitochondria between them in control samples (Figure 5A and 5C panels); similar to observations reported in.39 In AF tissue, we observed increased myolysis (Figure 5B and 5D), with areas depleted in sarcomeres and smaller, irregular mitochondria observed (Figure 5D). EL and autophagic vacuoles and autophagic-lysosomes are more commonly seen in AF tissue (Figure 5E and more zoomed in detail in Figure 5F). Since glycogen is associated with lysosomes or lysosome-like organelles,40 we quantitatively analyzed the amount of glycogen accumulation in our AF and sham tissue samples (Figure S6A). Both manual and automated counting observed increased glycogen levels in AF samples (manual disease mean glycogen count 6354 ± 714.8/nm2 vs. control mean glycogen count 3195 ± 417.9/nm2, p = 0.0023; Automated count disease mean glycogen count 6529 ± 555.0/nm2 vs. control mean glycogen count 4154 ± 415.8/nm2, p = 0.0003. N = 3 for each group. Figure S6B).
Figure 5.
Electron microscopy images of goat left atrial myocardium tissue to identify structural changes
Qualitative electron microscopy (EM) on goat left atrial myocardium tissue samples in sinus rhythm (sham, A and C) and after prolonged sustained atrial fibrillation (B, D, E and F). Panel A shows sarcomeres (S) regularly spaced and surrounded by mitochondria. Glycogen (GL), lysosomes (Ly) and numerous irregular shaped mitochondria observed in atrial fibrillation sample (B). We observe many irregular shaped mitochondria dispersed in myolytic areas, autophagic vacuoles (red arrows) and lysosomes (Ly) in (D). Electron micrographs highlighting increased number of endocytic vesicles and vacuoles including endosomes, autophagosomes, lysosomes in atrial fibrillation samples (E and F), indicated by star symbols). (F) is a higher resolution image of the marked square area in Panel E. Samples for EM were collected from N = 3 for AF and N = 3 for Sinus rhythm control animals.
The overrepresentation of Capra hircus (goat) protein regulation enrichment comparison to total human proteome and to the Cavia porcellus (guinea pig)
The significantly up and down regulated protein/gene names with the respective log2 fold-change values were then uploaded to the Panther–Gene Ontology (GO), and an overrepresentation test was conducted against the reference H. sapiens proteome (Taxonomy ID: 9606) (Figures S1A and S1B). This overrepresentation test was conducted on GO terms, Cellular Component (GOCC) and Biological Process (GOBP). The highest enriched GOCC term included muscle filaments, myofibrils proteasome complex, sarcoplasmic reticulum lumen, endocytic vesicles, and vesicle lumen (Figure S1A). In contrast, the highest enriched GOBP term included the energy metabolism and vesicle-mediated trafficking pathways (Figure S1B).
We performed a Venn analysis to understand the total protein yield of C. hircus TL and EL compared to the C. porcellus protein list that was published previously using this density-gradient based method from Ayagama et al.21 (Figure S1C). These comparisons showed a good overlap between goat and guinea pig data from these separate investigations, with 44.2% shared proteins for TL, and 28.8% shared proteins for EL samples. Due to the higher availability of annotation data for H. sapiens, the UniProt KB identifiers of the most significantly regulated C. hircus proteins identified from the TL volcano plot of the AF compared to the sham samples (Figure 1B) were converted to the H. sapiens identifiers.21
Discussion
Previous studies investigating mechanisms of AF using the goat model have shown structural, electrical, contractile and molecular changes compared with sinus rhythm controls (some examples include Ausma et al.,39 Wijffels et al.4 Allessie et al.,41 van Hunnik et al.,42 and Neuberger et al.43). Proteomics studies have been performed in many cardiac diseases including AF44 and reviewed by45 and have highlighted the need for more multi-omics research to investigate possible implicated molecular pathways in the development of AF. In this study we have focused on changes in EL-related proteins as another factor with a slower time course of development, and their involvement in this disease. Our endolysosomal organelle proteomics approach contributes new insight into regulation and differential functionalities related to these pathways and molecular mechanisms observed in this large animal model of AF. Here, we provide data related to pathway dysregulations in endolysosomal proteins previously not explored. Analysis of such proteins in separated tissue lysate (TL) and endolysosome fractions (EL) increased our ability to uncover EL-specific proteins that were not identified in the TL, such as GAA, CLTB, DYNLRB1, SIRT3, CCT2, and muscle specific HSPB246 (See Table S3.1 for the list of proteins). Combining an integrative multi-omics approach in this study helps us highlight interrelationships of the biomolecules and their functions in this disease and deriving insights into the data we have collected.
Lysosome number and dysfunction has been characterized in several cardiac conditions,47 including congenital atrial septal defects, and AF is a common complication in these patients.48 Further, degenerative changes, including accumulation of lysosomes, have been found to correlate with atrial cellular electrophysiological changes.49 Whilst AF is a common complication in atrial septal defect patients, no published studies have investigated the role of the EL or their corresponding interactive effects on other organelles in AF itself.
The major pathway regulations identified in this study were identified to be influenced from the AMPK signaling pathway (Figures 6 and S7), selective autophagy (aggrephagy and proteasome pathway) (Figures S8 and S9), NADPH oxidase pathway (Figure S10), OXPHOS (Figure S11), gap junction assembly and degradation, ESCRT, protein processing and folding pathway, vesicle-mediated transport, and lysosome vesicle biogenesis (Figures S9–S13). Most of the proteins and interpreted pathways identified in our study are increasingly being recognised in cardiac pathology,17,50 such as gap junctional remodeling,51 mitochondrial-bioenergetics and proteostasis,17 NADPH oxidase,52 and metabolic derangement caused by oxidative stress.53
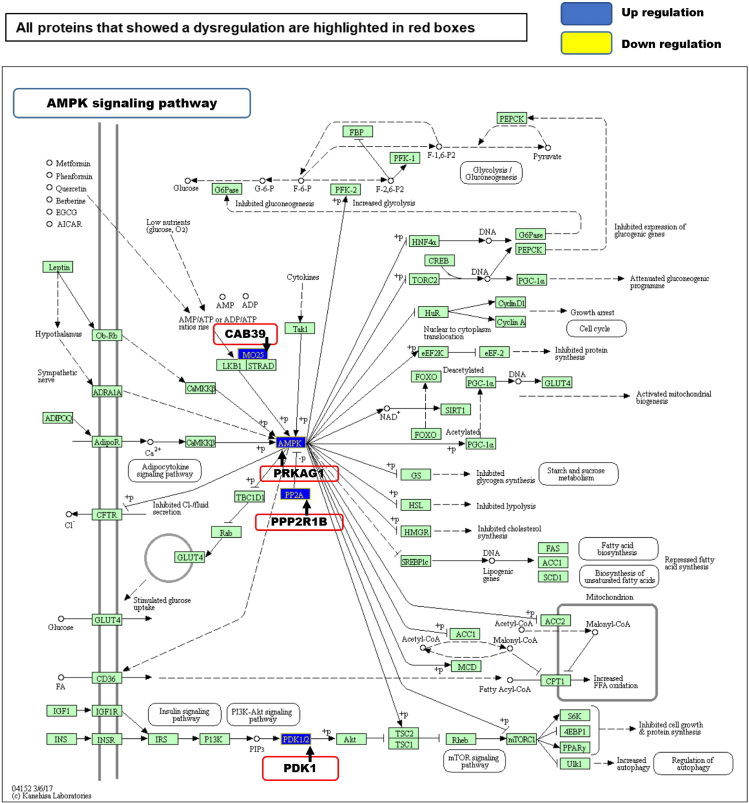
Figure 6.
AMPK signaling pathway depicted using KEGG in tissue lysate fraction
The levels of upregulation of the proteins observed in AMPK pathway are as follows: CAB39/MO25 log2 0.54-fold, PRKAG1/AMPK log2 0.77-fold, PPP2R1B/PP2A log2 0.77-fold and PDK1 log2 0.56-fold. (Arrow = Protein name, protein complex name or protein function. Red bordered boxes = regulated proteins from the query dataset. +P = phosphorylating events. Blue = up regulation (log2 fold-change) and Yellow = down regulation (log2 fold-change)). Pathway analyzed using KEGG database.
LC3s are important biomarkers of autophagy. Comparative analysis of LC3-I and II levels found that in AF tissue, levels of LC3-I were almost completely depleted, while no detection of the lipidated autophagic vacuole (AV)-associated LC3-II was observed. This suggests that high levels of autophagic flux may be occurring in the AF tissue, where the efficient clearance of AVs by lysosomes, outpaces the supply of LC3-I. P70 S6 kinase (p70S6K) is activated in a signaling pathway that includes mTOR and dysregulation of the mTOR pathway is implicated in human disease processes.54 We further tested by Western blotting the levels of p-p70 (p-P70: Total p70) to provide insights into mTOR activity (Figures S2C–S2E). This was measured using a p- P70 (Thr 389): Total p70 ratio, where Thr389 is an mTOR specific epitope. The p-p70 was unchanged, which suggests the heightened autophagy is not mTOR-dependent (Figures S2C and S2D).
The contemporary transcriptomic and proteome of studied samples may vary substantially in terms of overall correlation between transcript and associated protein. In static cell contexts with minimal temporal dynamics, overall correlation can be quite strong,55 but in cells undergoing dynamic response to stimuli or stress, correlation can vary from strongly correlated to almost uncorrelated on a gene by gene basis.56 To integrate across proteomic and transcriptomic results, we adopted the method of Cox and Mann57 to compare changes in terms of pathway up or down regulation relative to a global median across all pathways with identified members. This approach synthesizes pathway changes across all genes/proteins and therefore allowed for more meaningful comparisons of the implication of changes in the transcriptome versus proteome. The approach revealed that, while there was a general downregulation of pathways related to the EL as scored by mRNA levels, this drop was not consistently reflected by changes in the same pathways as scored by proteins. This is partially reflective of the time lag between response at the mRNA level versus response at the protein level, determined by regulation factors, in particular protein translation and turnover rates that vary from protein to protein.
Bioinformatics analysis of our proteomics results suggests that AMPK upregulation is triggered through an ATP depletion path. V-ATPase proton transporters utilize ATP to conduct protons which acidify the lysosomes or EL (Godbey, 2014), which is pivotal for the lysosomal enzymes to be functionally activated. For example, dysfunctional V-ATPases lead to neurodegenerative disorders58 caused by poor substrate digestion in the lysosomes. The increased ATP consumption in tachyarrhythmias,59 such as AF, is expected to create an energy demand in the atria.60 As the cell’s primary energy source, ATP drives active and coupled membrane ion transport61 to maintain cellular ion homeostasis.62
Exploring the functional endolysosomal network proteins in AF
The endolysosomal String network (Figure 3) consists of 134 nodes and 171 edges with an average node degree of 2.55. The functional enrichments in the network were determined using the GO database. 25 proteins from the biological process of intracellular transport and 24 proteins related to peptide transport were identified in the EL network. Six proteins were associated with unfolded protein binding, and 58 proteins were identified for catalytic activity. Furthermore, 48 proteins were components of vesicles, and 11 were components of extracellular vesicles. 118 proteins were part of the anatomical entity of the cell, and 3 proteins were from cellular proteostasis. Notably, 2/8 proteins that regulate the lysosomal storage disorder glycogen storage disease were highlighted in the EL network of AF and 2 more (PYGB and AGL) were significantly upregulated in TL (Figure 3; Table 1).
Endosomal sorting complex required for transport (ESCRT)
Our analysis suggests lower expression levels of proteins involved in the ESCRT pathway in the AF diseased condition (down-regulation of ESCRT-II complex and its cargo). Proteins such as Ras-related protein Rab-7a (RAB7A), and Vacuolar protein-sorting-associated protein 25 (VPS25), were downregulated by −0.50 and −1.04 log2 fold-change respectively (Table 1). RAB7A, a recruiting protein for the tethering molecules,63,64 plays an essential role in the late endosomes to multivesicular bodies (MVB) transition in the ESCRT pathway. Rab-7 is a common modulator/participant in endocytosis and autophagy.65 Moreover, RAB7A participates in the lysosome biogenesis through autophagolysosome formation.66 Vacuolar protein-sorting-associated protein 25 (VPS25) is a leading regulatory component of the ESCRT complex-II that sorts the endosomal cargo proteins to the MVB67 (Figure 3; Table 1).
Lysosomal alpha-glucosidase (GAA) was log2 0.44-fold upregulated in the EL fraction of AF. GAA functions as an enzyme that degrades glycogen in the lysosome68,69,70 (Figure 3; Table 1). Studies have indicated that a substantially high recombinant human GAA level is required to minimize the abnormal glycogen storage in the skeletal and cardiac muscles71,72 (Figure 3; Table 1).
1,4-alpha-glucan-branching enzyme (GBE1) is an enzyme that functions as a vital component in glycogen biosynthesis. GBE1 was upregulated by log2 1.67-fold in AF goat model compared to that of sham model (Figure 3; Table 1). To increase the glycogen molecule solubility, GBE1 generates α-1,6-glucosidic branches from α-1,4-linked glucose chains.73
Lysosome motility and motor-protein based transport
Lysosome motility is mediated by motor proteins74 such as kinesin and dynein and microtubules.75,76 Multiple biological processes, such as degradation of macro molecules,77 cellular homeostasis such as autophagy, signal transduction and metabolic signaling for ATP energy and amino acids, intracellular organelle signaling,78,79,80 require lysosomes to move and position themselves throughout the cytoplasm.81 We identified several motor proteins that are associated with lysosome motility that were upregulated, such as tubulins MACF1 up by log2 1.19-fold, Microtubule-associated protein (MAP4) up by log2 1.05-fold, Tubulin-specific chaperone A (TBCA) up by log2 0.58-fold and we also identified TUBB6 (tubulin beta 6 class V) in EL proteomics fraction and it was found to be downregulated log2 -0.95-fold. Clathrin light chain B (CLTB) was upregulated by log2 1.5-fold in the AF condition. CLTB, along with Vesicle-associated membrane proteins (VAMPs) and adaptor protein complex 1 (AP1) play an essential role in lysosome membrane biogenesis82 (Figure 3; Table 1). Dynein light chain roadblock-type 1 (DYNLRB1) was upregulated by log2 0.98-fold. DYNLRB1 is an essential protein for general dynein-mediated transport and has been shown to be vital for sensory neuron survival.83 Furthermore, DYNLRB1 links dynein with adaptor proteins to regulate dynein and cargo for ideal cellular vesicle transport.84 Simultaneously, Cytoplasmic Dynein 1 acts as a motor protein for the intracellular motility of organelles and retrograde motility of vesicles along the microtubules85 (Figure 3; Table 1). WDR78, a Dynein-f associated motor protein required for the axonemal localization86 was upregulated by log2 1.8-fold. Nestin (NES, upregulated by log2 0.96-fold in AF goat) is an intermediate filament (IFs) involved in vesicle-based communication, vesicle interaction and trafficking.87 Similarly, vesicle-associated membrane protein-associated protein A (VAPA), a protein involved in vesicle trafficking, was upregulated by log2 0.93-fold in the AF goat model88 (Figure 3; Table 1) and is a major ER-lysosome anchor protein.89 Meng Lu et al.90 showed that abolished VAPA-mediated anchoring compromises ER remodeling and significantly increases the speed of lysosome motility. Eps15 homology (EH) domain-containing protein 1 (EHD1) was upregulated by log2 0.6-fold in the EL fraction. EHDs play critical roles in endosome-based membrane protein targeting.91 EHD1 is a retrograde trafficking mediator and a regulator of the cluster of differentiation 44 protein (CD44) that participates in endocytic recycling and lysosomal degradation.92,93 Gudmundsson et al.91 found modulation of EHD expression during myocardial infarction, which suggests that these proteins may play important roles in regulating membrane excitability91 (Figure 3; Table 1).
Intermediate protein networks between the whole cell and endolysosomes - Proteasome and aggresome pathway
The proteasome pathway is part of the selective autophagy process94 and several dysregulated proteins in our EL disease fraction were from this pathway. Protein degradation through ubiquitination is termed as proteasome or aggresome pathway. Aggregated and misfolded proteins are identified by chaperone proteins and tagged by ubiquitination before protein degradation. Proteins related to both aggresome and proteasome formation were found to be up- or downregulated in our AF samples, including; E3 ubiquitin-protein ligase PPP1R11 (PPP1R11) up by log2 1.7, tubulin α chain (TUBA4A) up by log2 1.09, ubiquitin carboxyl-terminal hydrolase 5 (USP5) up by log2 0.39, probable ubiquitin carboxyl-terminal hydrolase FAF-X (USP9X) up by log2 1.00-fold, tubulin β chain (TUBB2A) down by log2 -1.07-fold (Table 1 and Table S1.2). PPP1R11 and USP5 are ubiquitination triggering proteins, and USP9X is both a deubiquitinase, that prevents a protein from the removal of conjugated ubiquitin and a ubiquitin precursor processor95,96(Table 1). The upregulated aggresome formation process that we observed in AF goat model is a well-known factor for the changes in lysosome distribution and its motility.97 Furthermore, Chaperonin containing TPC1 subunit 2 (CCT2) functions independently of ubiquitin and the TRiC complex to facilitate autophagic clearance of solid protein aggregates.98 We detect modestly upregulated levels log2 (0.55) of CCT2 in the EL fraction. In addition to CCT2, we also observe upregulated levels log2 (1.00) of CDC37 and log2 (0.48) COPS7A in the EL fraction. Hsp90-CDC37 complex appear to participate in upstream of autophagy activation for the control of protein quality,99 and COPS7A is a protein from the COP9 signalosome complex (CSN) that mediates de-neddylation to regulate the ubiquitin conjugation pathway and is presumed to participate in structural remodeling that plays a crucial part in the developing stages of AF.44
AMPK upregulation
AMPK pathway is a central regulator of cellular metabolism that is activated mainly by reduced adenosine triphosphate (ATP) levels in the cell (Figures 6 and S7) and cascades a series of downstream chemical reactions in the cell that reprogram metabolism, autophagy,100 cell polarity and growth101 As a result, catabolic pathways are upregulated while inhibiting the anabolic pathways to reduce the cellular ATP consumption level.102
Our Integrated proteomic and transcriptomic data showed a significant upregulation in AMP-activated protein kinase (AMPK) pathway related proteins in AF samples. 5′- AMP-activated protein kinase subunit gamma-1 (PRKAG1/AMPK) log2 0.8--fold, calcium-binding protein 39 (CAB39/MO25) log2 0.54 -fold, Serine/threonine-protein phosphatase 2A regulatory subunit A beta isoform (PPP2R1B/PP2A) log2 0.77-fold and Protein-serine/threonine kinase (PDK1) log2 0.56-fold from AMPK pathway were observed to be upregulated in AF samples (Figures 1B, 6, S7, and Table S1.2). These observed changes suggest a link between AMPK regulatory pathway protein upregulation and AF.19,103,104
The identified AMPK pathway proteins are from AMPK activation through the ATP pathway. CAB39/MO25 is part of the liver kinase B1 (LKB1) and STE-related adaptor protein STRAD protein complex,105 which binds and activates STK11/LKB1. AMPK protein activity is controlled by LKB1, acting as a key upstream regulator for AMPK phosphorylation. PPP2R1B assembles the catalytic subunits of AMPK, and signals from insulin to PKB/AKT1 are transduced by PDK1 through activated phosphorylation. This downstream signaling cascade targets cell survival, glucose and amino acid uptake and glucose storage.106 Therefore, processes such as upregulation of ATP production, activation of glucose intake, inhibition of the cell proliferation and growth, autophagy activation, cytoskeletal remodeling, DNA damage response, and apoptosis102 are regulated by AMPK. The increased expression of the CAB39, AMPK, PP2A and PDK1 proteins provides further support for the upregulation of the AMPK signaling pathway in AF. Of interest is the finding by,107 related to molecular mechanism of electrical modeling, where they showed an increased activity of PP2A (protein phosphatase 2A) results in hypophosphorylation of the calcium channel ICaL (see also Review by.108
Ras-homologous guanosine triphosphatases (Rho GTPases) activation of Nicotinamide adenine dinucleotide phosphate oxidase (NADPH oxidase)
We observed increased levels of proteins from the MAP Kinase signaling network, which represent the RHO-GTPase activation of NADPH Oxidase. These regulated proteins are: Mitogen-activated protein kinase 3 (MAPK3) up by log2 1.37-fold; Mitogen-activated protein kinase 12 (MAPK12) up by log2 0.51-fold; Mitogen-activated protein kinase 14 (MAPK14) up by log2 0.42-fold; and Protein S100-A9 (S100A9) up by log2 1.17-fold (a complete list of the most significantly regulated TL proteins identified in the AF goat model are provided in Table S1.2). MAPK proteins are reported to conduct signaling in mitochondria, Golgi, endoplasmic reticulum (ER) and endosomes.109 Rho family small GTPase proteins activate NADPH Oxidase (NOX), which activates the leading cell stress-response signaling network MAPK (see integrated omics Figures S4B, S5B, S7, and S10). NOX signaling is a cellular stress-responsive mechanism in the cardiovascular system,110,111 causing the production of superoxide, a reactive oxygen species, during cellular stresses initiated by biological, physical or chemical triggers. Cytochrome b-245 heavy chain (CYBB) or NADPH Oxidase 2 is a membrane-bound enzyme that generates superoxide (upregulated by log2 2.06-fold change in our transcriptomics data, a complete list of the most significantly regulated genes identified in the AF goat model mRNA analysis are provided in Table S1.1). The Yoo S et al. 2020 study showed that the oxidative injury by the CYBB/NOX2 caused an electrical remodeling by upregulating the constitutively active acetylcholine dependent potassium current (IKACh) in the canine model.112 Significantly higher NOX-induced superoxide levels have been observed in AF patients, and unlike NOX2, the superoxides produced by dysfunctional NOX had a lesser contribution to electrophysiological remodeling and oxidative injury in the atria of AF patients,52 and these superoxides further worsen the disease condition.113,114,115,116
MAPK3 is a signal transducing protein that regulates transcription, translation,117 and cell cycle-related functions such as the arrangement of the cytoskeleton and cell-cell adhesion during the cell survival state.118,119 Furthermore, MAPK/ERK participate in lysosomal dynamics120 and endosomal recycling.121 MAPK12 is another signal transducing protein that, is triggered by extracellular stress stimuli, such as pro cytokines122 (Figure S10).
MAPK14 is also stimulated by inflammatory triggers123 that conduct the cellular protein turnover for degradation through proteasomes,124 and S100A9 is a calcium- and zinc-binding protein that plays a prominent role in the inflammatory and immune response.125 The observed protein changes in this pathway, indicate increased cellular stress status in our AF goat model (Figure S10) highlighting the relevance of these findings relating to the emerging role of NOX pathways in AF.113
Identifying differential mRNA expression by quantitative transcriptomic analysis
We performed transcriptomics as a confirmatory and supplementary method to our proteomics screen in our goat AF model (Figure S4A). The transcriptome approach has allowed for the detection of unbiased molecular changes in AF.126 There is evidence for inter-related pathways such as oxidative stress, inflammation, thrombogenesis and fibrosis126 and more recently autophagy.127 Our transcriptomic analysis identified upregulation of major ion channel Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (HCN1) by log2 2.80-fold (Table S1.1).128,129 HCN1 gain of function promotes AF. Furthermore, Glycogen debranching enzyme (AGL) was upregulated by log2 2.85-fold change (Table S1.1).
Reactome analysis130 was performed on the differentially expressed genes. The most significantly regulated proteins and genes are presented in a hierarchical visualization of pathways using space filling graphs in Figures S16–S18. KEGG pathways of proteomics as well as transcriptomics and integrated Reactome pathway analysis highlighted AMPK signaling pathways as significantly upregulated (Figure 6, Table S1.1 and a complete list of pathways from the integrated EL, TL proteomics and transcriptomics analysis is in, Table S2.1, and the most significantly highlighted pathways of EL fraction and TL from Reactome analysis are shown in Tables S2.2 and S2.3), and Ribosome biogenesis as significantly down regulated (Figure S14). AMPK (upregulated): These include the following 8 genes SLC2A4, PFKFB2, PRKAA2, STRADB, CAB39L, PRKAG3, PPP2R3A and CCND1. Ribosome biogenesis (downregulated): These include the following 8 genes TRMT112, TBL3, GNL3, GAR1, FBL, NOP56, POP1 and NOB1 (Table S1.1).
Reactome allows us to overlay our quantitative expression data to visualize the extent of change and progression in affected pathways. In the mRNA analysis we found 231 out of 380 identifiers and 885 pathways were identified by at least one identifier (Figure S18). Table S2.4 shows the 25 most relevant pathways sorted according to the p-value.
In summary, the analysis highlights rRNA processing (down regulations), major pathway of rRNA processing in the nucleolus and cytosol (down regulations), unfolded protein response (UPR, down regulations), metabolism of RNA (down regulations), and interferon signaling (up regulations). AMPK, as well as being the master regulator of energy homeostasis, is also a physiological suppressor of UPR.131
There is growing evidence linking AF to metabolic stress and inflammation.132,133 In the cell, ribosomes control translation of proteins and their activity accounts for most of the cells energy consumption.134 Ribosome biogenesis depends on the nutritional and energy status of the cells and is vulnerable to internal and external stress stimuli. Impaired ribosome biogenesis (e.g., in aged tissue) may be protective or a compensatory mechanism.135 AMPK is a central regulator of energy homeostasis136 playing a key role in monitoring cellular energy metabolism and there is growing evidence around the importance of AMPK in the heart.132,137,138 Recently Cao et al.139 showed that γ2-AMPK translocate into the nucleus to suppress pre-rRNA transcription and ribosome biosynthesis during stress, this reduces ER stress and cell death. We identified PRKAG3 (5′-AMP-activated protein kinase subunit gamma-3) among our upregulated genes (upregulated by log2 2.31-fold, Table S1.1). This seems particularly interesting in light of the findings by Cao et al.139 where activation of γ2-AMPK suppresses ribosome biogenesis and protects against myocardial ischemia/reperfusion injury. Our transcriptomics data suggests an adaptation process to the stress conditions created during AF. AMPK signaling is increased, and this consequently would be expected to affect ribosomal RNA transcription, as reflected in the genes that are upregulated (Table S1.1) Su et al.132 highlight a critical role played by AMPK signaling and the resultant alterations in electrophysiological function and structural remodeling in the atria. Additionally, Su et al.132 highlight loss of AMPK affecting the expression of mRNA transcripts encoding gap junction proteins and ion channels in the atrium. Here, our goat model aligns with the notion of an upregulation of AMPK to downregulate energy-consuming processes like ribosome biogenesis.
The datasets obtained, and shared in this study offer a much wider scope for molecular signaling analysis in the goat AF model. Other aspects of AF and aging could be explored in our datasets. As recently was shown in140 that reducing the speed of RNA polymerase II by overexpressing histone components (which counter age-associated changes in nucleosome positioning), extended lifespan in flies and the division potential of human cells. We note in our study POLR2E (major component of Polymerase II complex) downregulation in transcriptomics and histone H3 upregulation was found to be upregulated in proteomics (Tables S1.1 and S1.2). This observation lends weight to our hypothesis that at this six month stage of AF in the goat model, organelle function seems to push in the direction toward compensation mechanisms to favor pro-survival modes.
Glycogen accumulation and lysosomal GAA upregulation
Histological studies in healthy/normal goat by Embi et al. 2014 showed left atrial appendage glycogen levels always exceeded right atrial appendage levels.141 The density and location of glycogen was also distinct and suggested these differences in glycogen were a potential contributory mechanism for the initiation and maintenance of AF, particularly the greater propensity for developing an AF substrate in the left versus the right atrium.141 Studies by Zhang et al. 2015 in pacing-induced AF (dog model), showed AF promoted glycogen deposition.142 In our proteomic data, significantly regulated proteins in the LA of AF goat included GLUT4/SLC2A4, a protein that transports glucose inside the cell (upregulated by log2 1.55) (Tables S1.2 and S2.4). The GLUT4 vesicle translocation to the plasma membrane (Figures S7 and S12) is conducted by the tethering and docking proteins Caveolin (CAV1) and Vesicle-fusing ATPase N-ethylmaleimide-sensitive fusion protein (NSF).143 We observe a significant upregulation of NSF by log2 1.55 (Figure 1B and Table S1.2) and CAV1 by a fold-change of log2 0.57 (Figure S12 and Table S1.2). Moreover, lysosomal-α-glucosidase (GAA), the glycogen degrading enzyme in the lysosomes,68,69 was upregulated by a log2 fold-change of 0.44 in the EL fraction of the AF goat model (Figure 3; Table 1). We chose to quantitatively assess the levels of Glycogenin 1 (GYG1) because in eukaryotes, Glycogenin 1 enzyme initiates glycogen biogenesis by producing an oligosaccharide primer that functions as a substrate for glycogen synthesis in bulk.144 Western blotting was performed on the enzyme GYG1 and a trend towards (p = 0.007) upregulation of GYG1 was observed in the AF samples.
Figures S2F and S2G, provide evidence for increased glycogen synthesis in the AF goat model.141 Here we present specific protein changes that impact on glycogen levels in the cells (Figures S2F and S2G). Reassuringly, the evidence presented from our whole tissue omics analysis (Figures S4B, S5B, and S7), is in keeping with existing structural data (canine142 and goat AF model22) linking glycogen accumulation and fibrosis as factors in the persistent forms of AF.22
Increased autophagic flux in AF goat model
Western blotting was performed on sham and AF groups to identify effects of AMPK upregulation in autophagy (Figures S2A and S2B). For the upregulation of autophagy flux LC3I, and for impaired autophagy flux, LC3II protein markers were blotted. A depletion of LC3I was detected by the absence of LC3I protein in AF compared to the sham goat model, suggesting the upregulation of autophagy flux. Moreover, the LC3II protein was absent in both sham and AF goat models suggesting the absence of impaired autophagic flux instead pointing toward the presence of an overactive flux. This result aligns with a recent study by.18 Network analysis shown in Figure S7, which presents a close network interaction of AMPK with lysosomal and vesicle localized proteins Ras-related GTP-binding protein A (RRAGA), 1,4-alpha-glucan-branching enzyme (GBE1), Glycogen debranching enzyme (AGL), Phosphoglucomutase-1 (PGM1), and Glycogen phosphorylase, brain form (PYGB).
Our integrated omics analysis comparing EL proteomics and transcriptomics highlights important changes occurring in EL proteins involved in this chronic goat AF model. We see a downregulation in suppression of autophagy, MAPK1 activation, inhibition of nitric oxide (NO) production, mitochondrial electron transport chain deregulation, changes in inflammation status (Interleukins), glycogen disease-like pathologies, changes in gap junction activity, stimulation of cell death response by PAK-2p34, upregulation of RhoBTB proteins that are involved in vesicle trafficking processes and retrograde transport from endosomes to the Golgi apparatus (Figure S4B integrated omics). Our observations are relevant and fit with many observations published over the years including the findings that human atrial samples from patients with AF have increased immune cell infiltration compared to those from patients without AF.145 Additionally, NO produced by endothelial NO synthase (eNOS) plays a role in the regulation of cell growth, apoptosis, and tissue perfusion146 and our findings of apoptosis and downregulation of NOS corroborates early findings.147 ER stress and oxidative stress have been highly implicated in many cardiac pathologies148,149 including the pathogenesis of AF.150 Recent studies highlight the importance of PAK2. In Pak2 cardiac deleted mice under stress or overload, there is a defective ER response, cardiac dysfunction, and profound cell death.151
In recent years, a complex relationship between AF, systemic inflammation and oxidative stress has come to light.152 Some evidence suggests that the underlying atrial changes that lead to the development and progression of AF may be inflammatory in nature.153 Histological examination of atrial tissue taken from patients with so-called 'lone' AF refractory to conventional antiarrhythmic treatment showed that two-thirds (66%) had inflammatory changes significant enough to be classified as myocarditis when compared with control samples from patients with Wolff-Parkinson-White syndrome.7 Patients with some systemic autoimmune disorders such as rheumatoid arthritis are at increased risk of developing AF154 and the severity of inflammation as measured by levels of C-reactive protein (CRP) correlates with the incidence of AF in conditions characterized by chronic inflammatory changes.155 It has also been observed that the apogee of the surgical stress response usually occurs at approximately 72 h post-surgery which coincides with the peak post-operative incidence of new-onset AF.156 Moreover, recent work using data from the UK Biobank has a shown strong association between AF and inflammatory indicators in nearly half a million patients.157
Some anti-inflammatory agents have been associated with a reduction in AF burden. One meta-analysis demonstrated a reduction in post-operative AF in patients following cardiac surgery when glucocorticoids were given,158 and in a small trial with patients who had AF catheter ablations, a short course of steroids given after the procedure led to a reduction in early (although not late) recurrence of AF.159 Other agents with anti-inflammatory properties, such as statins and colchicine have also been associated with a protective effect against AF.160,161,162
Our findings lend weight to these conclusions and provide further evidence for the molecular changes in metabolism that may underpin the development of AF. Out of the 2104 proteins, 340 proteins in TL and 148 in EL were significantly changed in AF. We validated Rab11 (Ras-related protein 11) using Western blotting. The LC3I protein was absent in AF, suggesting an increased autophagic flux. The TL fraction, highlighted mitochondrial oxidative-phosphorylation (OXPHOS) and AMPK pathway protein upregulation, indicating a potential increased ATP energy demand in AF. The EL proteins GAA, Rab7a, CLTB, VPS25 and CCT2 were significantly regulated. The upregulation of protein processing suggests increased vesicular trafficking, potentially related to increased metabolic energy demands. Here we use an endolysosomal purification protocol to study atrial specific protein changes in AF and noted changes in EL proteins. We believe this approach allows us to study differences in protein expression more specifically related to EL and globally in health and disease. In order to provide further direct experimental confirmation, our data is expected to provide rationale for future experiments that study links between the EL and AF. These may include viral vector-RNA overexpression in rodent or human induced pluripotent stem cell (iPSC) derived cardiomyocytes targeting the regulated genes and proteins identified in our goat AF omics analysis. As an example, oxidative stress and the AMPK pathway can be induced by hydrogen peroxide and lowering ATP via reduced glucose supplementation to upregulate the AMPK pathway, targeting downstream autophagic pathways and increasing vesicle trafficking of endolysosomal proteins such as GAA, Rab7a, CLTB, VPS25, CCT2 and related proteins. With the availability of AF patient derived cardiomyocytes differentiated from iPSCs,163 electrophysiological and pharmacological approaches might be useful to test any direct or indirect link between AF and EL. Measuring cathepsin activity as a readout of endolysosomal signaling in AF is another experimental option in the future. Other experimental techniques to explore the involvement of lysosomes in AF include live imaging of isolated human atrial cells to study the pH of whole cell or EL in AF cells vs. sinus rhythm controls with ratiometric dyes such as OG488. Endolysosomal related gene knock-in or knock-down (using endolysosomal hits identified in this study) approaches in iPSC cardiac models can also be used to demonstrate whether they can induce functional alterations leading to a cardiac substrate more prone to develop arrhythmias under demanding conditions (e.g., adrenergic stimulation), helping to dissect cellular mechanisms underlying AF.
Conclusion
Studies have shown that prominent differences between paroxysmal AF and sinus rhythm patients relate to changes in expression of proteins involved in metabolic processes.164,165 Ozcan et al. 2015 provided evidence into the role of atrial metabolism for AF substrate evolution, findings that are relevant when considering alternative therapeutic approaches to prevent AF progression.166 Our EL organelle omics approach helps us describe a disease setting where there is increased cellular stress, increased vesicle trafficking, changes in ATP demands, accumulation of glycogen, inflammation, and stimulation of cell death. Our findings uncover new insights linking endolysosomal proteins, ER stress response, RNA biogenesis, and cell apoptosis pathways which may be triggered by failure of protective ER stress response. The current pharmacological therapies for AF are not sufficiently effective to control disease progression and new molecular insights can fuel the development of novel therapeutic strategies. EL in the heart provide a significant contribution to basal calcium transient amplitude and beta-adrenergic responses in both atrial167 and ventricular79 myocytes. Clearly, the multi-functional role of the EL appears to be at play in this disease setting and these results highlight the need for further investigation into the role of endolysosomal pathways in cellular dysfunction and apoptosis in AF. In summary, our endolysosomal proteomics and integrated omics analysis pave the way for future studies focused on identifying suitable EL targets, drug discoveries and biomarker identification. The information provided present a promising option for exploring new pathways for the treatment of AF.
Limitations of the study
The molecular pathways applied on the protein regulations found in this project are interpretations of published literature and databases and provide a fundamental understanding of the disease pathways. Therefore, an establishment of the protein regulation as a complete pathway alteration needs further and extensive investigation. We have used a large animal goat AF model in which chronic AF was maintained by pacing for six months. The data we present is a snapshot of genes and proteins relevant at that point in time of the disease. The multifactorial and heterogeneous nature of AF as a disease25,168,169 pose limitations in the assessment of the occurrence and changes in gene and protein expression especially during the process of disease progression from paroxysmal to chronic AF.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-GYG1 | ThermoFisher Scientific | Cat# PA5-116508; RRID: AB_2901139 |
| Anti- Lamp2 | ThermoFisher Scientific | Cat# PA1-655; RRID: AB_2134625 |
| Anti-COX IV | Abcam | Cat# ab16056; RRID: AB_443304 |
| Anti-Phospholamban | Abcam | Cat# ab85146; RRID: AB_10974942 |
| Polyclonal Goat Anti-Rabbit Immunoglobulins/HRP | Agilent Dako | Cat# P044801-2; RRID: AB_2617138 |
| Biological samples | ||
| Left Atrial tissue from C. hircus Atrial fibrillation models | Maastricht University, Netherlands | |
| Chemicals, peptides, and recombinant proteins | ||
| Lysosome Isolation Buffer | BioVision | K235-50-1 |
| Lysosome Enrichment Buffer | BioVision | K235-50-2 |
| Protease Inhibitor Cocktail | BioVision | K235-50-4 |
| Percoll | Santa Cruz Biotechnology | sc-500790 |
| Sucrose | Fisher Scientific | 15503022 |
| 4-Methylumbelliferyl N-acetyl-b-D-glucosaminide | Merck (Sigma-Aldrich) | CAS 37067-30-4 |
| Na2CO3 | Merck (Sigma-Aldrich) | CAS 497-19-8 |
| Bolt™ 4 to 12%, Bis-Tris, 1.0 mm, Mini Protein Gel, 10-well | ThermoFisher Scientific | NW04120BOX |
| MES SDS Running Buffer | Life Technologies | NP0002 |
| Transfer Buffer | ThermoFisher Scientific | NP0006 |
| Westar Supernova detection substrate | Cyanogen | XLS3,0020 |
| Triton X-100 solution | Merck (Sigma-Aldrich) | CAS 9002-93-1 |
| Sodium Acetate Buffer | Merck (Sigma-Aldrich) | CAS 126-96-5 |
| Tris HCL | Merck (Sigma-Aldrich) | CAS 1185-53-1 |
| Iodoacetamide | Merck (Sigma-Aldrich) | CAS 144-48-9 |
| Dithiothreitol | Merck (Sigma-Aldrich) | CAS 3483-12-3 |
| Urea | Merck (Sigma-Aldrich) | CAS 57-13-6 |
| Deposited data | ||
| Mass spectrometry proteomics data, Proteome Xchange via PRIDE partner repository, dataset identifier PRIDE: PXD041056 |
This paper | http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD041056 |
| mRNA sequencing Transcriptomics data, GEO Accession code: GSE228289 |
This paper | |
| A modified density gradient proteomic-based method to analyze endolysosomal proteins in cardiac tissue | Ayagama et al., 202121 | https://www.cell.com/iscience/fulltext/S2589-0042(21)00917-2 |
| Experimental models: Organisms/strains | ||
| C.hircus | Maastricht University, Netherlands | N/A |
| Software and algorithms | ||
| Progenesis QI software platform (version 4.2) | Waters™ Cooperation | www.nonlinear.com |
| Perseus software platform (version 1.6.15.0) | Tyanova et al., 2016170 | http://coxdocs.org/doku.php?id=perseus:start |
Resource availability
Lead contact
Rebecca-Ann Burton. r.a.b.burton@liverpool.ac.uk.
Materials availability
This study did not generate new unique reagents or codes.
Data and code availability
Mass Spectrometry data has been deposited in PRIDE and genomics data in GEO. See resource availability for accession numbers. Data can be requested by contacting the lead contact. This study did not generate new unique codes.
Experimental model and study participant details
Animals
AF was induced and maintained in farm reared female goats (C. hircus) aged between 24 and 34 months (weight 72 ±8 kg) for 6 months (AF goat model was created as conducted in,42 followed by an open chest sacrifice experiment (N = 4 AF and N = 4 sham controls, N = each animal, n = the number of replicates from each condition). (The goat study was carried out in accordance with the principles of the Basel declaration and regulations of European directive 2010/63/EU, and the local ethical board for animal experimentation of the Maastricht University approved the protocol).
Method details
Tissue homogenization
Frozen left atrial tissue biopsies of AF and sham goat were thoroughly cleaned using phosphate buffered solution (PBS) and weighed. A minimum of 100 mg tissue is weighted in order to perform proteomics. Each atrium biopsy sample was cut using sterile scalpels and gently homogenized using a 7 mL Dounce homogenizer in Lysosome isolation buffer (LIB) [Containing 1:500 protease inhibitor cocktail (PIC) and phosphatase inhibitor (PHI) (Bio vision), (PhosSTOP Roche)]. Preparations were further homogenized in a 1 mL Dounce homogeniser and transferred to chilled 1.5 mL ultracentrifugation tubes (Beckmann coulter). Sample preparations were mixed at a ratio of 1:1.5 Lysosome enrichment buffer [(LEB) (Biovision, containing 1:500 PIC)] to homogenate by inverting tubes, and were stored on ice for 5 min until the centrifugation.21
Tissue lysate (TL)
Samples were centrifuged at 13,000 g × 2 min at 4°C (TLX Beckmann Coulter Ultra Centrifuge) and the supernatant or the TL, was collected.21
Endo-lysosome fraction (EL)
The collected supernatant was retained and repeated for a further centrifugation step at 29,000 g × 30 min at 15°C (500 μL of under-laid 2.5 M sucrose with over-laid 500 μL Percoll). The supernatant above the sucrose and Percoll intermediate was collected for further fractionation. Firstly, ultracentrifuge tubes were underlaid with 2.5 M sucrose and overlaid with a series of Percoll dilutions (1.11 g/mL – 1.04 g/mL in ddH2O). The ultracentrifuge tubes were centrifuged at 67,000 g × 30 min at 4°C. The fraction at 1.04 g/mL was collected and labeled as the endolysosomal fraction (EL).21 N = 3 biological replicates for each AF and sham conditions were used in proteomic analysis.
Liquid chromatography-tandem mass spectrometry analysis
5 μL of 200 mM dithiothreitol (30 min at room temperature) was added to each sample to reduce and proceeded with alkylation using 20 μL of 200 mM iodoacetamide (30 min at room temperature), followed by methanol-chloroform precipitation. The sample proteins were pelleted at this stage and was re-suspended in 6 M urea in 400 mM Tris-HCl, pH 7.8. The 6M urea was then diluted to 1 M with 400 mM Tris-HCl at pH 7.8, and the proteins were digested in trypsin at a ratio of 1:50 (overnight at 37C°). The trypsin digested samples were then processed at the Target Discovery Institute, Oxford.
Samples were then acidified to a final concentration of 1% formic acid, and the samples were desalted on Sola HRP SPE cartridges (Thermo Fisher Scientific) and dried down using a SpeedVaccum centrifuge. Dried down protein Samples were further desalted online (PepMAP C18, 300 μm × 5 mm, 5 μm particle, Thermo Fisher Scientific) for 1 min (flow rate of 20 μL/min and separated on an EASY-Spray column) (PepMAP C18, 75 μm × 500 mm, 2 μm particle, ES803, Thermo Fisher Scientific) over 60 min using a gradient of 2–35% acetonitrile in 5% DMSO/0.1% formic acid at 250 nL/min. Separation and analysis were performed on a Dionex Ultimate 3000 RSLC system coupled to an Orbitrap Fusion Lumos platform (both Thermo Fisher Scientific) using standard parameters (Universal Method).171
MS scans were acquired at a resolution of 120,000 between 400 and 1,500 m/z. An AGC target of 4.0E5 and MS/MS spectra detection was carried out using rapid scan mode in the linear ion trap at a 35% collision energy after collision-induced dissociation fragmentation (CIDF). An AGC target of 4.0E3 for up to 250 ms, employing a maximal duty cycle of 3 s, prioritising the most intense ions and injecting ions for all available parallelisable time. Selected precursor masses were excluded for 30 s.171
Mass spectrometry data were analyzed quantitatively with the Progenesis QI software platform (WatersTM Cooperation, www.nonlinear.com) (version 4.2), and database searches were carried out against the UniProt C. hircus database (UP000291000). Automatic processing was selected. All runs in the experiment were adjusted to the function suitability, and runs were aligned automatically. The peak picking was selected between 10 and 75 min. The group runs option was set to conditions, and relative quantitation using Hi-N was selected. Finally, proteins were grouped.
Quantification and statistical analysis
Quantification and statistical analysis of mass spectrometry data
Quantitative analysis for significant differences of protein regulation between the AF and sham conditions of TL and EL samples and data visualization were performed using the Perseus software platform33 (version 1.6.15.0). Using protein intensity values of biological replicates, the protein groups were created and uploaded as a data matrix in Perseus with the respective protein abundances as main columns. The data matrix was reduced by filtering based on categorical columns to remove proteins where more than two intensity values were absent from six biological replicates, and remaining data with no more than 2 missing values were quantitatively analyzed. A total of 2,104 proteins in TL and EL remained after filtering. Groups of biological replicates for TL and EL fractions were defined in categorical annotation rows. Data were log transformed (log2) and normalised via Z score.
Data imputation was not required due to Progenesis reporting signal noise in absence of peptide precursors (Tables S4 and S5). Therefore, principal component analysis (PCA) was performed on 100% valid values. A volcano plot was generated based on normalised intensities applying two-way Student’s t test to probe for significant difference of protein regulation between AF and sham conditions of each TL and EL samples. A permutation-based false-discovery rate (FDR) was determined with 250 randomizations and S0 = 0.1 (default). The quantified proteins were accounted with 99% confidence level at 5% FDR.
Principal component analysis (PCA)
Principal component analysis is a reduced data dimension-interpretation of the protein groups’ distribution between the sample groups and it is one of the most popular multivariate statistical techniques. A PCA plot can provide a window to a large dataset by identifying the common vectors, therefore summarizing the variation.172 As indicated in Figure 2E the respective component 1 and component 2 vector deviations were observed between AF (purple symbols) and sham (green symbols) EL groups demonstrating that the variability is driven by the differential experiment groups rather than the variability within the sample group.
Violin plot analysis
The violin plots were produced using the InstantClue omics tool.173 The protein intensities used were transformed to log2, normalised by z scoring, and AF vs. sham groups were colour-coded; a gradient of purple for the distribution of protein intensities (Figure 2A). This Violin plot represents a kernel density estimation of the underlying protein intensity distribution (The kernel estimated density distribution is the nonparametric representation of the probability density function of a random variable (www.mathworks.com). The quantified protein intensity matrix used for the violin plot is created in Perseus 1.6.15.0 33.
Heatmap analysis
The heat or clustering maps were produced in InstantClue omics tool using Euclidian distance and K-mean clustering of the normalised protein intensities. The intensity values are obtained from the quantified matrix created in Perseus 1.6.15.0. A total of 148 proteins in EL were categorized into protein clusters according to the protein intensities (EL = 3 clusters). The regulation level variations are displayed using color codes (red and blue), where red represents the up regulation of the protein clusters and blue represents the down regulation of these clusters (Figure 2C).
Statistics for network analysis
The string network edges of EL and TL were created based on the interaction evidence, which are experiments, gene fusions, databases, co-occurrences and co-expressions. The minimum required interaction score was set to default or medium confidence (0.4), and only the query proteins were used and external interactors were excluded.
Sample preparation for transcriptomics
Frozen AF and sham LA goat tissue were collected without the RNase contamination, and samples were thawed using an RNAlater stabilisation solution (AM7020, Invitrogen). Samples were then homogenised using bead disruption, and in a 1 mL of Trizol (15596026, Thermo Fischer), approximately 50–100 mg of tissue were solubilised. The supernatant was collected from lysates after incubating them in RT for 5 min and centrifuging at 12,000 × g for 5 min at 4°C. Furthermore, it was followed with a re-centrifugation at 12,000 g at 4°C for 15 min, after vigorously shaking with 200 μL chloroform. The upper aqueous phase was separated, 1:1 volume ice-cold isopropanol was added, and gently mixed. This procedure was followed by centrifugation at 12,000 g for 30 min at 4°C, and the collected RNA pellet was washed with 75% ethyl alcohol (10048291, Thermo Fisher) and dried.
Qualification and quantification of mRNA
The isolated RNA was screened for contaminants and degradation levels using 1% agarose, and a NanoPhotometer spectrophotometer (IMPLEN, CA, USA) was used to check the purity (RNA Nano 6000 Assay Kit of the Bio-analyser 2100 system (Agilent Technologies, CA, USA) was used to detect the RNA integrity and assess quantitation.
Sequence libraries were prepared using the NEBNext Ultra TM RNA Illumina (NEB, USA) Library Prep Kit. Furthermore, using a PE Cluster Kit cBot-HS all the samples were clustered, and sequenced using an Illumina.
Fastp was used to remove the poly-N, and adapter reads from the raw data and process. The reads Q score over 50% was considered low quality, and Q score at 20 and 30 were identified as clean. Genome web browser National Center for Biotechnology Information/European Molecular Biology Laboratories-European Bioinformatics Institute (NCBI/Ensembl-EBI) was used as the reference genome with HISAT2 program (daehwankimlab.github.io/hisat2/manual/).
Integrated analysis of proteomics and transcriptomics statistics
The proteomic data from Progenesis and the transcript data were analyzed in R v4.1.2. Both proteomic intensities and transcript counts were normalised by Variance Stabilizing Normalisation (VSN).174 Due to the poor annotation status of Capra hircus gene, protein and pathway IDs, the data were 'humanised'; human protein-coding homolog gene IDs were assigned to matchable Goat IDs using the getLDS function in the biomaRt package.175 In total, 27552 IDs across the proteomic and transcriptomic datasets were mapped to 18658 human IDs. Further analysis was performed on the human IDs.
Removal of sample outliers
The outliers of AF and sham samples were identified by conducting principal component analysis on the individual and shared proteomics and transcriptomics data, then the outliers were removed to increase the confidence of the study (Please refer to the PCA plot showing the outliers of combined proteomics and transcriptomics data in the Figure S15).
For each AF vs. Control comparison, t-statistics were calculated for the AF versus control comparison for each gene. For all human Reactome pathways containing genes found in the dataset, we calculated rank-based 2D enrichment MANOVA p-values after57 for all pathways with at least 2 genes found on both sides of each paired comparison, over the three proteomics datasets and the transcriptomics dataset. Missing values for genes that had non-missing value at least one in any proteomics dataset were treated as ranked last; conversely, genes with no non-missing values except in the transcriptomics dataset were treated as missing (and excluded from ranking) in the proteomics datasets. To correct for multiple testing, pathway p-values were converted to q-values using the fdr tool package.176 We set a threshold of 1% FDR for significance.
Protein quantitation
Sample fractions EL or TL were mixed at a ratio of 1:1 with radio-immunoprecipitation (RIPA) buffer (Thermo Scientific). Protein concentrations of EL fractions and TL were determined using the Bicinchoninic acid assay (BCA Protein Assay Kit, Thermo Scientific). Bovine serum albumin was used as a protein standard, and serial dilutions were prepared from the initial stock concentration of 2 mg/mL to prepare a standard curve. To ensure accuracy and reproducibility, protein assays were performed in triplicate. Absorbance values were measured at 562 nm. Protein concentrations were calculated by linear regression analysis.21
SDS/PAGE gel preparation and Western blotting
Sample fractions EL and TL were solubilised, and proteins denatured using SDS/PAGE loading buffer (bio rad) and 2-mercaptoethanol (Sigma-Aldrich). Proteins were separated by gel electrophoresis (NW04120BOX, NuPAGE 4%–12% Bis-Tris protein gels, 20X MES buffer). The gel was transferred to nitrocellulose membrane (NC) (Bio-Rad) for protein transfer (X-cell-II blot module, Thermo Fisher Scientific). NC membrane was incubated in 5% skimmed milk. The primary antibodies anti-GYG1 (1:1000, sc-271109, Santa Cruz), anti-Rab11 (1:1000, ab65200, abcam) were incubated. Goat anti-rabbit antibody (1:2,500, Dako P0448) was used as the secondary antibody to detect the protein markers. The secondary antibodies were detected via chemiluminescence using Westar Supernova (XLS3, 0020, Cyanogen) and the protein bands were visualized in a ChemiDoc XRS + imager (Bio-rad with image Lab software).21 Un-paired t test was performed on the comparative band intensity values obtained from the Western blots to understand the significant changes of the protein levels between AF and sham/control goat groups.
Preparation of atrial tissue extracts for Westernblot analysis
Frozen left atrial regions were weighed and homogenised at a concentration of 20 mg wet weight/mL of RIPA buffer (10 mM Tris-HCl, pH 8, 150 mM NaCl, 0.5% IGEPAL-CA630, 0.5% sodium deoxy-cholate, 0.1% SDS) containing protease inhibitors (5 mM EDTA, 1 mM EGTA, 5 mg/mL leupeptin, 5 mg/mL aprotinin, 2 mg/mL pepstatin, 120 mg/mL Pefabloc, 2 mM 1,10-phenanthroline), using 10 strokes of a Dounce homogenizer connected to an overhead stirrer (Wheaton, NJ) set to speed 4. The resulting homogenate was then centrifuged at 10,000 x g at 4°C for 10 min, and the supernatant was collected. Total protein content of samples was estimated using a bicinchoninic acid protein assay kit (Pierce) and equalised to 1 mg/mL using RIPA buffer. Samples were frozen as 1 mL aliquots at −80°C or boiled for 3 min at 100°C in Laemmli tris-glycine sample buffer (62.5 mM Tris-HCl, pH 6.8, 10% v/v glycerol, 2% w/v SDS, 0.01% w/v bromophenol blue) and subsequently used for Western immunoblot.
SDS-PAGE and Westernblot analysis
14% polyacrylamide tris-glycine gels were used immunoblot detection of LC3-I and II (Cell Signaling Technology, UK), as well as phospho- and total p70S6 kinase (Cell Signaling Technology, UK). Tris-glycine gel solutions were made from a 30% T (total w/v %), 2.6% C (cross-linker w/v %) acrylamide/bisacrylamide stock solution (Sigma, UK) and run on a triple-wide gel system (C.B.S. Scientific, USA). 50 μg of protein from each sample was loaded onto gels and run at 150 V. Proteins were transferred onto Protran reinforced nitrocellulose membrane (0.2 μm pore size, Amersham, UK) at 400 mA for 2 h at 4°C. Uniform transfer of proteins onto nitrocellulose was confirmed by reversible staining with Ponceau S (0.1% w/v, 1% acetic acid, Sigma). Membranes were then blocked for 1 h at room temperature in 5% skimmed milk/TBS-T (Tris-buffered saline solution containing 0.1% Tween 20), then washed (three times for 5 min) in TBS-T, and incubated with appropriate primary antibodies in a 1% BSA/TBS-T solution for 2 h at room temperature or overnight at 4°C. Membranes were washed (three times for 5 min) in TBS-T before horseradish peroxidase-conjugated secondary antibodies (sheep anti-mouse HRP (Li-Cor Biosciences, UK) and donkey anti-rabbit (Li-Cor Biosciences, UK) made in 2.5% milk/TBS-T) were added for 1 h at room temperature. Membranes were washed (three times for 20 min) in TBS-T, and the proteins of interest were visualized using chemiluminescent substrates (Pierce). Fuji Super RX film (FujiFilm, Dusseldorf, Germany) band intensities were quantified using Image Studio Lite software (Li-Cor Bioscience, UK).
Lysosomal hydrolase activity assays
To fluorometrically measure the lysosome enzyme levels, artificial sugar substrates containing the fluorophore 4-methylumbelliferone (4-MU) were used. For measuring β-hexosaminidase activity, 3 mM 4-MU N-acetyl-β-D-glucosaminide (Sigma Aldrich) in 200 mM sodium citrate buffer, pH 4.5 and 0.1% Triton X-100 was used as substrate. For β-galactosidase activity, 1 mM 4-MU β-D-galactopyranoside (Sigma Aldrich) in 200 mM sodium acetate buffer, pH 4.3, 100 mM NaCl, and 0.1% Triton X-100 was used as substrate. The reaction was stopped by adding chilled 0.5 M Na2CO3, and the released fluorescent 4-MU was measured in a Clariostar OPTIMA plate reader (BMG Labtech, Ortenberg, Germany) with an excitation at 360 nm and emission at 460 nm. A standard curve for free 4-MU was used to calculate the enzyme activity. Results were calculated as total Units of enzyme activity (nmol/h) and normalised with respect to protein content.21
Sample preparation for electron microscopy (EM)
Left atrial samples (from N = 3 sham animals and 3 AF animals) were prepared by chemical fixation. Approximately 1 mm3 pieces of tissue from the left atria were rapidly dissected and fixed in Karnovsky fixative containing paraformaldehyde 4%, glutaraldehyde 5%, cacodylate buffer 80 mM, pH 7.4,177 and were embedded in Spurr’s resin.178 Sections were cut to approximately 70–80 nm thickness (Reichart Ultracut) then post-stained with 2% aqueous uranyl acetate and Reynolds lead citrate for contrast. Images were obtained using a transmission electron microscope (Thermo Fisher Tecnai T12 TEM, operated at 120 kV, using a Gatan OneView camera). Extensive quantitative ultra-structure studies have been published on the goat AF model. We performed unbiased qualitative EM analysis to cross check our results with that of the goat AF studies performed by Ausma et al.39
Manual glycogen EM image analysis
Analysis and measurements were made using Gatan DigitalMicrograph and ImageJ software. Raw EM images were analyzed using ImageJ (Version 2.3.0/1.53s). The area of interest was manually selected using the box tool, avoiding cellular organelles such as Golgi, lysosome, mitochondria etc. 5–7 boxes per image were selected in order to cover most of the cytosolic space. Box sizes in pixels were generated by ImageJ. Glycogen quantification was evaluated manually using the multi-point tool, one point represents one glycogen. The overall number of points was recorded as an output by ImageJ. The results are presented as glycogen counts per nmˆ2.
Automated glycogen EM image analysis
We performed independent automated image analysis to cross-check the manual counts performed. This removes user bias. Images were processed to obtain glycogen count estimates from pre-selected regions of interest. The image processing pipeline begins by cropping out each area of interest from the larger image. Noise in the cropped image is then reduced by applying a Gaussian filter to smooth the image and a min filter to accentuate dark areas. Following this, the image is binarized at a color threshold that separates the dark glycogen shapes from the background. Foreground shapes are extracted from the binary image and counted if their area is larger than a minimum size. For larger foreground shapes which contain multiple glycogen, the count is estimated as the maximum number of glycogen deposits which can fit into the area. The parameters for filtering foreground shapes by size and the threshold for binarizing images were selected by visually inspecting results from the image processing pipeline. All image analysis was conducted in Python 3.10.11 with opencv-python 4.7.0 (source code available at https://doi.org/10.5281/ZENODO.7892313).
Additional resources
All the required resources are provided in the STAR methods and supplementary data.
Acknowledgments
R.A.B.B. is funded by a Sir Henry Dale Wellcome Trust and Royal Society Fellowship (109371/Z/15/Z) and R.A.B.B. acknowledges support from The Returning Carers’ Fund (Oxford University, Medical Sciences Division). T.A. acknowledges the Global Challenges Research Fund, University of Oxford. R.A.B.B. is a Senior Research Fellow of at Linacre College. R.A.B.B. acknowledges research funds from the Ellis T Davies Fellowship Endowment, University of Liverpool. S.J.B. is funded by the British Heart Foundation. R.A.C. is funded by the Wellcome Trust and Royal Society. F.M.P. is a Wellcome Trust Investigator in Science and a Royal Society Wolfson merit award holder. D.A.P. was funded by the Mizutani Foundation. A.G. is a Wellcome Trust Senior Investigator and a Principal Investigator of the British Heart Foundation Center of Research Excellence at the University of Oxford. G.M. is a Wellcome Trust Senior Research Fellow. N.H. is a British Heart Foundation Senior Clinical Research Fellow (FS/SCRF/20/32005). We would like to thank the Galione, Parrington, and Tammaro Groups, Department of Pharmacology, University of Oxford for access to scientific equipment. We thank Dr Errin Johnson and Raman Dhaliwal from the Dunn School of Pathology for Electron Microscopy technical support. The goat model work was supported by the Netherlands Heart Foundation (CVON2014-09, RACE V Reappraisal of Atrial Fibrillation: Interaction between hypercoagulability, Electrical remodeling, and Vascular Destabilisation in the Progression of AF, and Grant number 01-002-2022-0118, EmbRACE: Electro-Molecular Basis and the theRapeutic management of Atrial Cardiomyopathy, fibrillation and associated outcomEs), the European Union (CATCH ME: Characterizing Atrial fibrillation by Translating its Causes into Health Modifiers in the Elderly, grant number 633196; MAESTRIA: Machine Learning Artificial Intelligence Early Detection Stroke Atrial Fibrillation, grant number 965286). This research was funded in whole, or in part, by the Wellcome Trust [109371/Z/15/Z]. For the purpose of Open Access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission.
Author contributions
R.A.B.B. conceived and designed the study; S.V. and U.S. performed the goat AF experiments and provided the tissue for this study. T.A., G.B., R.F., and H.K. performed LC-MS/MS; T.A. and B.B. performed western blots; T.A. and D.P. performed enzyme assays; R.A.B.B., T.A., R.A.C., S.J.B., and F.P. contributed intellectually to the project and R.A.B.B., T.A., and S.J.B. drafted the paper. T.A. performed statistical analysis of proteomics and pathways analysis. A.P.C. and T.A. performed statistical analysis, on transcriptomics. P.D.C. performed integrated analysis of proteomics versus transcriptomics. S.J.B. performed the species comparison study. R.A.C., D.A., Q.S., and K.A. analyzed EM data. Q.S., G.R.M., and K.A. performed the manual and automated glycogen counts. T.A. created the figures related to proteomics. T.A. and A.P.C. created figures related to transcriptomics. P.D.C. and T.A. created integrated proteomics and transcriptomics analysis figures. G.R.M. created code for analyzing EM images for glycogen. B.B. performed western blotting studies on LC3 and p-p70. N.H., F.M.P., and A.G. contributed intellectually and provided technical access to equipment for this study. L.H. contributed intellectually relating to metabolism aspects of this study. All authors contributed to writing of the paper.
Declaration of interests
The authors declare no competing interests.
Published: March 28, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109609.
Supplemental information
References
- 1.Samuthpongtorn C., Jereerat T., Suwanwela N.C. Stroke risk factors, subtypes and outcome in elderly Thai patients. BMC Neurol. 2021;21:322. doi: 10.1186/s12883-021-02353-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Workman A.J., Kane K.A., Rankin A.C. Cellular bases for human atrial fibrillation. Heart Rhythm. 2008;5:S1–S6. doi: 10.1016/j.hrthm.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zoni-Berisso M., Lercari F., Carazza T., Domenicucci S. Epidemiology of atrial fibrillation: European perspective. Clin. Epidemiol. 2014;6:213–220. doi: 10.2147/CLEP.S47385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wijffels M.C., Kirchhof C.J., Dorland R., Allessie M.A. Atrial fibrillation begets atrial fibrillation. A study in awake chronically instrumented goats. Circulation. 1995;92:1954–1968. doi: 10.1161/01.cir.92.7.1954. [DOI] [PubMed] [Google Scholar]
- 5.Botto G.L., Luzi M., Sagone A. Atrial fibrillation: the remodelling phenomenon. Eur. Heart J. Suppl. 2003;5:H1–H7. [Google Scholar]
- 6.Heijman J., Linz D., Schotten U. Dynamics of Atrial Fibrillation Mechanisms and Comorbidities. Annu. Rev. Physiol. 2021;83:83–106. doi: 10.1146/annurev-physiol-031720-085307. [DOI] [PubMed] [Google Scholar]
- 7.Frustaci A., Chimenti C., Bellocci F., Morgante E., Russo M.A., Maseri A. Histological substrate of atrial biopsies in patients with lone atrial fibrillation. Circulation. 1997;96:1180–1184. doi: 10.1161/01.cir.96.4.1180. [DOI] [PubMed] [Google Scholar]
- 8.Ausma J., Coumans W.A., Duimel H., Van der Vusse G.J., Allessie M.A., Borgers M. Atrial high energy phosphate content and mitochondrial enzyme activity during chronic atrial fibrillation. Cardiovasc. Res. 2000;47:788–796. doi: 10.1016/s0008-6363(00)00139-5. [DOI] [PubMed] [Google Scholar]
- 9.Lee H.C. Electrical remodeling in human atrial fibrillation. Chin. Med. J. 2013;126:2380–2383. [PubMed] [Google Scholar]
- 10.Pool L., Wijdeveld L.F.J.M., de Groot N.M.S., Brundel B.J.J.M. The Role of Mitochondrial Dysfunction in Atrial Fibrillation: Translation to Druggable Target and Biomarker Discovery. Int. J. Mol. Sci. 2021;22:8463. doi: 10.3390/ijms22168463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doll S., Dreßen M., Geyer P.E., Itzhak D.N., Braun C., Doppler S.A., Meier F., Deutsch M.A., Lahm H., Lange R., et al. Region and cell-type resolved quantitative proteomic map of the human heart. Nat. Commun. 2017;8:1469. doi: 10.1038/s41467-017-01747-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim H., Kang M.G., Park H.W., Park J.R., Hwang J.Y., Kim K. Anderson-Fabry disease presenting with atrial fibrillation as earlier sign in a young patient: A case report. World J. Clin. Cases. 2021;9:4823–4828. doi: 10.12998/wjcc.v9.i18.4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halcrow P.W., Lynch M.L., Geiger J.D., Ohm J.E. Role of endolysosome function in iron metabolism and brain carcinogenesis. Semin. Cancer Biol. 2021;76:74–85. doi: 10.1016/j.semcancer.2021.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bose S.J., Ayagama T., Burton R.A.B. In: Proteolytic Signaling in Health and Disease. Zelanis A., editor. Academic Press; 2022. Chapter 3 - Lysosomal proteases and their role in signaling pathways Proteolytic Signaling in Health and Disease; pp. 41–61. [DOI] [Google Scholar]
- 15.Halcrow P.W., Kumar N., Afghah Z., Fischer J.P., Khan N., Chen X., Meucci O., Geiger J.D. Heterogeneity of ferrous iron-containing endolysosomes and effects of endolysosome iron on endolysosome numbers, sizes, and localization patterns. J. Neurochem. 2022;161:69–83. doi: 10.1111/jnc.15583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wildenthal K., Decker R.S. The role of lysosomes in the heart. Adv. Myocardiol. 1980;2:349–358. [PubMed] [Google Scholar]
- 17.Papathanasiou K.A., Giotaki S.G., Vrachatis D.A., Siasos G., Lambadiari V., Iliodromitis K.E., Kossyvakis C., Kaoukis A., Raisakis K., Deftereos G., et al. Molecular Insights in Atrial Fibrillation Pathogenesis and Therapeutics: A Narrative Review. Diagnostics. 2021;11:1584. doi: 10.3390/diagnostics11091584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan Y., Zhao J., Gong Y., Wang D., Wang X., Yun F., Liu Z., Zhang S., Li W., Zhao X., et al. Autophagy exacerbates electrical remodeling in atrial fibrillation by ubiquitin-dependent degradation of L-type calcium channel. Cell Death Dis. 2018;9:873. doi: 10.1038/s41419-018-0860-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia L., Verdejo H.E., Kuzmicic J., Zalaquett R., Gonzalez S., Lavandero S., Corbalan R. Impaired cardiac autophagy in patients developing postoperative atrial fibrillation. J. Thorac. Cardiovasc. Surg. 2012;143:451–459. doi: 10.1016/j.jtcvs.2011.07.056. [DOI] [PubMed] [Google Scholar]
- 20.Au C.E., Bell A.W., Gilchrist A., Hiding J., Nilsson T., Bergeron J.J. Organellar proteomics to create the cell map. Curr. Opin. Cell Biol. 2007;19:376–385. doi: 10.1016/j.ceb.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Ayagama T., Bose S.J., Capel R.A., Priestman D.A., Berridge G., Fischer R., Galione A., Platt F.M., Kramer H., Burton R.A.B. A modified density gradient proteomic-based method to analyze endolysosomal proteins in cardiac tissue. iScience. 2021;24:102949. doi: 10.1016/j.isci.2021.102949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ausma J., Wijffels M., Thoné F., Wouters L., Allessie M., Borgers M. Structural changes of atrial myocardium due to sustained atrial fibrillation in the goat. Circulation. 1997;96:3157–3163. doi: 10.1161/01.cir.96.9.3157. [DOI] [PubMed] [Google Scholar]
- 23.Ausma J., Litjens N., Lenders M.H., Duimel H., Mast F., Wouters L., Ramaekers F., Allessie M., Borgers M. Time course of atrial fibrillation-induced cellular structural remodeling in atria of the goat. J. Mol. Cell. Cardiol. 2001;33:2083–2094. doi: 10.1006/jmcc.2001.1472. [DOI] [PubMed] [Google Scholar]
- 24.Hu Y.B., Dammer E.B., Ren R.J., Wang G. The endosomal-lysosomal system: from acidification and cargo sorting to neurodegeneration. Transl. Neurodegener. 2015;4:18. doi: 10.1186/s40035-015-0041-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schüttler D., Bapat A., Kääb S., Lee K., Tomsits P., Clauss S., Hucker W.J. Animal Models of Atrial Fibrillation. Circ. Res. 2020;127:91–110. doi: 10.1161/CIRCRESAHA.120.316366. [DOI] [PubMed] [Google Scholar]
- 26.Schotten U., Duytschaever M., Ausma J., Eijsbouts S., Neuberger H.R., Allessie M. Electrical and contractile remodeling during the first days of atrial fibrillation go hand in hand. Circulation. 2003;107:1433–1439. doi: 10.1161/01.cir.0000055314.10801.4f. [DOI] [PubMed] [Google Scholar]
- 27.Verheule S., Tuyls E., van Hunnik A., Kuiper M., Schotten U., Allessie M. Fibrillatory conduction in the atrial free walls of goats in persistent and permanent atrial fibrillation. Circ. Arrhythm. Electrophysiol. 2010;3:590–599. doi: 10.1161/CIRCEP.109.931634. [DOI] [PubMed] [Google Scholar]
- 28.Verheule S., Tuyls E., Gharaviri A., Hulsmans S., van Hunnik A., Kuiper M., Serroyen J., Zeemering S., Kuijpers N.H.L., Schotten U. Loss of continuity in the thin epicardial layer because of endomysial fibrosis increases the complexity of atrial fibrillatory conduction. Circ. Arrhythm. Electrophysiol. 2013;6:202–211. doi: 10.1161/CIRCEP.112.975144. [DOI] [PubMed] [Google Scholar]
- 29.Maesen B., Zeemering S., Afonso C., Eckstein J., Burton R.A.B., van Hunnik A., Stuckey D.J., Tyler D., Maessen J., Grau V., et al. Rearrangement of atrial bundle architecture and consequent changes in anisotropy of conduction constitute the 3-dimensional substrate for atrial fibrillation. Circ. Arrhythm. Electrophysiol. 2013;6:967–975. doi: 10.1161/CIRCEP.113.000050. [DOI] [PubMed] [Google Scholar]
- 30.Eckstein J., Maesen B., Linz D., Zeemering S., van Hunnik A., Verheule S., Allessie M., Schotten U. Time course and mechanisms of endo-epicardial electrical dissociation during atrial fibrillation in the goat. Cardiovasc. Res. 2011;89:816–824. doi: 10.1093/cvr/cvq336. [DOI] [PubMed] [Google Scholar]
- 31.Wijffels M.C., Dorland R., Allessie M.A. Pharmacologic cardioversion of chronic atrial fibrillation in the goat by class IA, IC, and III drugs: a comparison between hydroquinidine, cibenzoline, flecainide, and d-sotalol. J. Cardiovasc. Electrophysiol. 1999;10:178–193. doi: 10.1111/j.1540-8167.1999.tb00660.x. [DOI] [PubMed] [Google Scholar]
- 32.Eijsbouts S., Ausma J., Blaauw Y., Schotten U., Duytschaever M., Allessie M.A. Serial cardioversion by class IC Drugs during 4 months of persistent atrial fibrillation in the goat. J. Cardiovasc. Electrophysiol. 2006;17:648–654. doi: 10.1111/j.1540-8167.2006.00407.x. [DOI] [PubMed] [Google Scholar]
- 33.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 34.Dytham C. 3 Edition. John Wiley & Sons; 2011. Choosing and Using Statistics : A Biologist's Guide. [Google Scholar]
- 35.Lapierre L.A., Dorn M.C., Zimmerman C.F., Navarre J., Burnette J.O., Goldenring J.R. Rab11b resides in a vesicular compartment distinct from Rab11a in parietal cells and other epithelial cells. Exp. Cell Res. 2003;290:322–331. doi: 10.1016/s0014-4827(03)00340-9. [DOI] [PubMed] [Google Scholar]
- 36.Khvotchev M.V., Ren M., Takamori S., Jahn R., Südhof T.C. Divergent functions of neuronal Rab11b in Ca2+-regulated versus constitutive exocytosis. J. Neurosci. 2003;23:10531–10539. doi: 10.1523/JNEUROSCI.23-33-10531.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Platt F.M., d'Azzo A., Davidson B.L., Neufeld E.F., Tifft C.J. Lysosomal storage diseases. Nat. Rev. Dis. Primers. 2018;4:27. doi: 10.1038/s41572-018-0025-4. [DOI] [PubMed] [Google Scholar]
- 38.Nair V., Belanger E.C., Veinot J.P. Lysosomal storage disorders affecting the heart: a review. Cardiovasc. Pathol. 2019;39:12–24. doi: 10.1016/j.carpath.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 39.Ausma J., van der Velden H.M.W., Lenders M.H., van Ankeren E.P., Jongsma H.J., Ramaekers F.C.S., Borgers M., Allessie M.A. Reverse structural and gap-junctional remodeling after prolonged atrial fibrillation in the goat. Circulation. 2003;107:2051–2058. doi: 10.1161/01.CIR.0000062689.04037.3F. [DOI] [PubMed] [Google Scholar]
- 40.Geddes R., Stratton G.C. The influence of lysosomes on glycogen metabolism. Biochem. J. 1977;163:193–200. doi: 10.1042/bj1630193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allessie M., Ausma J., Schotten U. Electrical, contractile and structural remodeling during atrial fibrillation. Cardiovasc. Res. 2002;54:230–246. doi: 10.1016/s0008-6363(02)00258-4. [DOI] [PubMed] [Google Scholar]
- 42.van Hunnik A., Zeemering S., Podziemski P., Simons J., Gatta G., Hannink L., Maesen B., Kuiper M., Verheule S., Schotten U. Stationary Atrial Fibrillation Properties in the Goat Do Not Entail Stable or Recurrent Conduction Patterns. Front. Physiol. 2018;9:947. doi: 10.3389/fphys.2018.00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neuberger H.R., Schotten U., Blaauw Y., Vollmann D., Eijsbouts S., van Hunnik A., Allessie M. Chronic atrial dilation, electrical remodeling, and atrial fibrillation in the goat. J. Am. Coll. Cardiol. 2006;47:644–653. doi: 10.1016/j.jacc.2005.09.041. [DOI] [PubMed] [Google Scholar]
- 44.Liu B., Li X., Zhao C., Wang Y., Lv M., Shi X., Han C., Pandey P., Qian C., Guo C., Zhang Y. Proteomic Analysis of Atrial Appendages Revealed the Pathophysiological Changes of Atrial Fibrillation. Front. Physiol. 2020;11:573433. doi: 10.3389/fphys.2020.573433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Souza A.I., Camm A.J. Proteomics of atrial fibrillation. Circ. Arrhythm. Electrophysiol. 2012;5:1036–1043. doi: 10.1161/CIRCEP.112.973008. [DOI] [PubMed] [Google Scholar]
- 46.Sugiyama Y., Suzuki A., Kishikawa M., Akutsu R., Hirose T., Waye M.M., Tsui S.K., Yoshida S., Ohno S. Muscle develops a specific form of small heat shock protein complex composed of MKBP/HSPB2 and HSPB3 during myogenic differentiation. J. Biol. Chem. 2000;275:1095–1104. doi: 10.1074/jbc.275.2.1095. [DOI] [PubMed] [Google Scholar]
- 47.Chi C., Riching A.S., Song K. Lysosomal Abnormalities in Cardiovascular Disease. Int. J. Mol. Sci. 2020;21:811. doi: 10.3390/ijms21030811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kottmeier C.A., Wheat M.W. Myocardial lysosomes in experimental atrial septal defects. Circ. Res. 1967;21:17–24. doi: 10.1161/01.res.21.1.17. [DOI] [PubMed] [Google Scholar]
- 49.Mary-Rabine L., Albert A., Pham T.D., Hordof A., Fenoglio J.J., Malm J.R., Rosen M.R. The relationship of human atrial cellular electrophysiology to clinical function and ultrastructure. Circ. Res. 1983;52:188–199. doi: 10.1161/01.res.52.2.188. [DOI] [PubMed] [Google Scholar]
- 50.Sygitowicz G., Maciejak-Jastrzębska A., Sitkiewicz D. A Review of the Molecular Mechanisms Underlying Cardiac Fibrosis and Atrial Fibrillation. J. Clin. Med. 2021;10:4430. doi: 10.3390/jcm10194430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van der Velden H.M., Ausma J., Rook M.B., Hellemons A.J., van Veen T.A., Allessie M.A., Jongsma H.J. Gap junctional remodeling in relation to stabilization of atrial fibrillation in the goat. Cardiovasc. Res. 2000;46:476–486. doi: 10.1016/s0008-6363(00)00026-2. [DOI] [PubMed] [Google Scholar]
- 52.Kim Y.M., Guzik T.J., Zhang Y.H., Zhang M.H., Kattach H., Ratnatunga C., Pillai R., Channon K.M., Casadei B. A myocardial Nox2 containing NAD(P)H oxidase contributes to oxidative stress in human atrial fibrillation. Circ. Res. 2005;97:629–636. doi: 10.1161/01.RES.0000183735.09871.61. [DOI] [PubMed] [Google Scholar]
- 53.Manna P., Jain S.K. Obesity, Oxidative Stress, Adipose Tissue Dysfunction, and the Associated Health Risks: Causes and Therapeutic Strategies. Metab. Syndr. Relat. Disord. 2015;13:423–444. doi: 10.1089/met.2015.0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen Z.Y., Liang K., Qiu R.X. Targeted gene delivery in tumor xenografts by the combination of ultrasound-targeted microbubble destruction and polyethylenimine to inhibit survivin gene expression and induce apoptosis. J. Exp. Clin. Cancer Res. 2010;29:152. doi: 10.1186/1756-9966-29-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lundberg E., Fagerberg L., Klevebring D., Matic I., Geiger T., Cox J., Algenäs C., Lundeberg J., Mann M., Uhlen M. Defining the transcriptome and proteome in three functionally different human cell lines. Mol. Syst. Biol. 2010;6:450. doi: 10.1038/msb.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buccitelli C., Selbach M. mRNAs, proteins and the emerging principles of gene expression control. Nat. Rev. Genet. 2020;21:630–644. doi: 10.1038/s41576-020-0258-4. [DOI] [PubMed] [Google Scholar]
- 57.Cox J., Mann M. 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinf. 2012;13:S12. doi: 10.1186/1471-2105-13-S16-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song Q., Meng B., Xu H., Mao Z. The emerging roles of vacuolar-type ATPase-dependent Lysosomal acidification in neurodegenerative diseases. Transl. Neurodegener. 2020;9:17. doi: 10.1186/s40035-020-00196-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nesheiwat Z., Goyal A., Jagtap M. StatPearls Publishing; 2022. Atrial Fibrillation in: StatPearls. [Google Scholar]
- 60.Rennison J.H., Li L., Lin C.R., Lovano B.S., Castel L., Wass S.Y., Cantlay C.C., McHale M., Gillinov A.M., Mehra R., et al. Atrial fibrillation rhythm is associated with marked changes in metabolic and myofibrillar protein expression in left atrial appendage. Pflugers Arch. 2021;473:461–475. doi: 10.1007/s00424-021-02514-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.HASSELBACH W. Atp-driven active transport of calcium in the membranes of the sarcoplasmic reticulum. Proc. R. Soc. Lond. B Biol. Sci. 1964;160:501–504. doi: 10.1098/rspb.1964.0064. [DOI] [PubMed] [Google Scholar]
- 62.Grant A.O. Cardiac ion channels. Circ. Arrhythm. Electrophysiol. 2009;2:185–194. doi: 10.1161/CIRCEP.108.789081. [DOI] [PubMed] [Google Scholar]
- 63.Gutierrez M.G., Munafó D.B., Berón W., Colombo M.I. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J. Cell Sci. 2004;117:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- 64.Vanlandingham P.A., Ceresa B.P. Rab7 regulates late endocytic trafficking downstream of multivesicular body biogenesis and cargo sequestration. J. Biol. Chem. 2009;284:12110–12124. doi: 10.1074/jbc.M809277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hyttinen J.M.T., Niittykoski M., Salminen A., Kaarniranta K. Maturation of autophagosomes and endosomes: a key role for Rab7. Biochim. Biophys. Acta. 2013;1833:503–510. doi: 10.1016/j.bbamcr.2012.11.018. [DOI] [PubMed] [Google Scholar]
- 66.Press B., Feng Y., Hoflack B., Wandinger-Ness A. Mutant Rab7 causes the accumulation of cathepsin D and cation-independent mannose 6-phosphate receptor in an early endocytic compartment. J. Cell Biol. 1998;140:1075–1089. doi: 10.1083/jcb.140.5.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Im Y.J., Wollert T., Boura E., Hurley J.H. Structure and function of the ESCRT-II-III interface in multivesicular body biogenesis. Dev. Cell. 2009;17:234–243. doi: 10.1016/j.devcel.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zirin J., Nieuwenhuis J., Perrimon N. Role of autophagy in glycogen breakdown and its relevance to chloroquine myopathy. PLoS Biol. 2013;11:e1001708. doi: 10.1371/journal.pbio.1001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Adeva-Andany M.M., González-Lucán M., Donapetry-García C., Fernández-Fernández C., Ameneiros-Rodríguez E. Glycogen metabolism in humans. BBA Clin. 2016;5:85–100. doi: 10.1016/j.bbacli.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Roig-Zamboni V., Cobucci-Ponzano B., Iacono R., Ferrara M.C., Germany S., Bourne Y., Parenti G., Moracci M., Sulzenbacher G. Structure of human lysosomal acid α-glucosidase-a guide for the treatment of Pompe disease. Nat. Commun. 2017;8:1111. doi: 10.1038/s41467-017-01263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Van den Hout J.M., Reuser A.J., de Klerk J.B., Arts W.F., Smeitink J.A., Van der Ploeg A.T. Enzyme therapy for pompe disease with recombinant human alpha-glucosidase from rabbit milk. J. Inherit. Metab. Dis. 2001;24:266–274. doi: 10.1023/a:1010383421286. [DOI] [PubMed] [Google Scholar]
- 72.Zhu Y., Li X., Kyazike J., Zhou Q., Thurberg B.L., Raben N., Mattaliano R.J., Cheng S.H. Conjugation of mannose 6-phosphate-containing oligosaccharides to acid alpha-glucosidase improves the clearance of glycogen in pompe mice. J. Biol. Chem. 2004;279:50336–50341. doi: 10.1074/jbc.M409676200. [DOI] [PubMed] [Google Scholar]
- 73.Froese D.S., Michaeli A., McCorvie T.J., Krojer T., Sasi M., Melaev E., Goldblum A., Zatsepin M., Lossos A., Álvarez R., et al. Structural basis of glycogen branching enzyme deficiency and pharmacologic rescue by rational peptide design. Hum. Mol. Genet. 2015;24:5667–5676. doi: 10.1093/hmg/ddv280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bandyopadhyay D., Cyphersmith A., Zapata J.A., Kim Y.J., Payne C.K. Lysosome transport as a function of lysosome diameter. PLoS One. 2014;9:e86847. doi: 10.1371/journal.pone.0086847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cabukusta B., Neefjes J. Mechanisms of lysosomal positioning and movement. Traffic. 2018;19:761–769. doi: 10.1111/tra.12587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Levi V., Serpinskaya A.S., Gratton E., Gelfand V. Organelle transport along microtubules in Xenopus melanophores: evidence for cooperation between multiple motors. Biophys. J. 2006;90:318–327. doi: 10.1529/biophysj.105.067843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.de Duve C. The lysosome turns fifty. Nat. Cell Biol. 2005;7:847–849. doi: 10.1038/ncb0905-847. [DOI] [PubMed] [Google Scholar]
- 78.Aston D., Capel R.A., Ford K.L., Christian H.C., Mirams G.R., Rog-Zielinska E.A., Kohl P., Galione A., Burton R.A.B., Terrar D.A. High resolution structural evidence suggests the Sarcoplasmic Reticulum forms microdomains with Acidic Stores (lysosomes) in the heart. Sci. Rep. 2017;7:40620. doi: 10.1038/srep40620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Capel R.A., Bolton E.L., Lin W.K., Aston D., Wang Y., Liu W., Wang X., Burton R.A.B., Bloor-Young D., Shade K.T., et al. Two-pore Channels (TPC2s) and Nicotinic Acid Adenine Dinucleotide Phosphate (NAADP) at Lysosomal-Sarcoplasmic Reticular Junctions Contribute to Acute and Chronic β-Adrenoceptor Signaling in the Heart. J. Biol. Chem. 2015;290:30087–30098. doi: 10.1074/jbc.M115.684076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wong Y.C., Kim S., Peng W., Krainc D. Regulation and Function of Mitochondria-Lysosome Membrane Contact Sites in Cellular Homeostasis. Trends Cell Biol. 2019;29:500–513. doi: 10.1016/j.tcb.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pu J., Guardia C.M., Keren-Kaplan T., Bonifacino J.S. Mechanisms and functions of lysosome positioning. J. Cell Sci. 2016;129:4329–4339. doi: 10.1242/jcs.196287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saftig P., Klumperman J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat. Rev. Mol. Cell Biol. 2009;10:623–635. doi: 10.1038/nrm2745. [DOI] [PubMed] [Google Scholar]
- 83.Terenzio M., Di Pizio A., Rishal I., Marvaldi L., Di Matteo P., Kawaguchi R., Coppola G., Schiavo G., Fisher E.M.C., Fainzilber M. DYNLRB1 is essential for dynein mediated transport and neuronal survival. Neurobiol. Dis. 2020;140:104816. doi: 10.1016/j.nbd.2020.104816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vaughan K.T., Vallee R.B. Cytoplasmic dynein binds dynactin through a direct interaction between the intermediate chains and p150Glued. J. Cell Biol. 1995;131:1507–1516. doi: 10.1083/jcb.131.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ansar M., Ullah F., Paracha S.A., Adams D.J., Lai A., Pais L., Iwaszkiewicz J., Millan F., Sarwar M.T., Agha Z., et al. Bi-allelic Variants in DYNC1I2 Cause Syndromic Microcephaly with Intellectual Disability, Cerebral Malformations, and Dysmorphic Facial Features. Am. J. Hum. Genet. 2019;104:1073–1087. doi: 10.1016/j.ajhg.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang Y., Chen Y., Zheng J., Wang J., Duan S., Zhang W., Yan X., Zhu X. Vertebrate Dynein-f depends on Wdr78 for axonemal localization and is essential for ciliary beat. J. Mol. Cell Biol. 2019;11:383–394. doi: 10.1093/jmcb/mjy043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lasič E., Trkov Bobnar S., Wilhelmsson U., de Pablo Y., Pekny M., Zorec R., Stenovec M. Nestin affects fusion pore dynamics in mouse astrocytes. Acta Physiol. 2020;228:e13399. doi: 10.1111/apha.13399. [DOI] [PubMed] [Google Scholar]
- 88.Weir M.L., Xie H., Klip A., Trimble W.S. VAP-A binds promiscuously to both v- and tSNAREs. Biochem. Biophys. Res. Commun. 2001;286:616–621. doi: 10.1006/bbrc.2001.5437. [DOI] [PubMed] [Google Scholar]
- 89.Zhao Y.G., Liu N., Miao G., Chen Y., Zhao H., Zhang H. The ER Contact Proteins VAPA/B Interact with Multiple Autophagy Proteins to Modulate Autophagosome Biogenesis. Curr. Biol. 2018;28:1234–1245.e4. doi: 10.1016/j.cub.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 90.Lu M., van Tartwijk F.W., Lin J.Q., Nijenhuis W., Parutto P., Fantham M., Christensen C.N., Avezov E., Holt C.E., Tunnacliffe A., et al. The structure and global distribution of the endoplasmic reticulum network are actively regulated by lysosomes. Sci. Adv. 2020;6:eabc7209. doi: 10.1126/sciadv.abc7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gudmundsson H., Hund T.J., Wright P.J., Kline C.F., Snyder J.S., Qian L., Koval O.M., Cunha S.R., George M., Rainey M.A., et al. EH domain proteins regulate cardiac membrane protein targeting. Circ. Res. 2010;107:84–95. doi: 10.1161/CIRCRESAHA.110.216713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu Y., Song Y., Cao M., Fan W., Cui Y., Cui Y., Zhan Y., Gu R., Tian F., Zhang S., et al. A novel EHD1/CD44/Hippo/SP1 positive feedback loop potentiates stemness and metastasis in lung adenocarcinoma. Clin. Transl. Med. 2022;12:e836. doi: 10.1002/ctm2.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang J., Naslavsky N., Caplan S. EHDs meet the retromer: Complex regulation of retrograde transport. Cell. Logist. 2012;2:161–165. doi: 10.4161/cl.20582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kraft C., Peter M., Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat. Cell Biol. 2010;12:836–841. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 95.Dupont S., Mamidi A., Cordenonsi M., Montagner M., Zacchigna L., Adorno M., Martello G., Stinchfield M.J., Soligo S., Morsut L., et al. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell. 2009;136:123–135. doi: 10.1016/j.cell.2008.10.051. [DOI] [PubMed] [Google Scholar]
- 96.Zhang Y., Duan C., Yang J., Chen S., Liu Q., Zhou L., Huang Z., Xu Y., Xu G. Deubiquitinating enzyme USP9X regulates cellular clock function by modulating the ubiquitination and degradation of a core circadian protein BMAL1. Biochem. J. 2018;475:1507–1522. doi: 10.1042/BCJ20180005. [DOI] [PubMed] [Google Scholar]
- 97.Zaarur N., Meriin A.B., Bejarano E., Xu X., Gabai V.L., Cuervo A.M., Sherman M.Y. Proteasome failure promotes positioning of lysosomes around the aggresome via local block of microtubule-dependent transport. Mol. Cell Biol. 2014;34:1336–1348. doi: 10.1128/MCB.00103-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang Z., Klionsky D.J. CCT2, a newly identified aggrephagy receptor in mammals, specifically mediates the autophagic clearance of solid protein aggregates. Autophagy. 2022;18:1483–1485. doi: 10.1080/15548627.2022.2083305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Calderwood S.K., Murshid A., Prince T. The shock of aging: molecular chaperones and the heat shock response in longevity and aging--a mini-review. Gerontology. 2009;55:550–558. doi: 10.1159/000225957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li Y., Chen Y. AMPK and Autophagy. Adv. Exp. Med. Biol. 2019;1206:85–108. doi: 10.1007/978-981-15-0602-4_4. [DOI] [PubMed] [Google Scholar]
- 101.Shackelford D.B., Shaw R.J. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat. Rev. Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mihaylova M.M., Shaw R.J. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 2011;13:1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen M.C., Chang J.P., Wang Y.H., Liu W.H., Ho W.C., Chang H.W. Autophagy as a mechanism for myolysis of cardiomyocytes in mitral regurgitation. Eur. J. Clin. Invest. 2011;41:299–307. doi: 10.1111/j.1365-2362.2010.02410.x. [DOI] [PubMed] [Google Scholar]
- 104.Czegledi A., Tosaki A., Gyongyosi A., Zilinyi R., Tosaki A., Lekli I. Electrically-Induced Ventricular Fibrillation Alters Cardiovascular Function and Expression of Apoptotic and Autophagic Proteins in Rat Hearts. Int. J. Mol. Sci. 2019;20:1628. doi: 10.3390/ijms20071628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zeqiraj E., Filippi B.M., Deak M., Alessi D.R., van Aalten D.M.F. Structure of the LKB1-STRAD-MO25 complex reveals an allosteric mechanism of kinase activation. Science. 2009;326:1707–1711. doi: 10.1126/science.1178377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pullen N., Dennis P.B., Andjelkovic M., Dufner A., Kozma S.C., Hemmings B.A., Thomas G. Phosphorylation and activation of p70s6k by PDK1. Science. 1998;279:707–710. doi: 10.1126/science.279.5351.707. [DOI] [PubMed] [Google Scholar]
- 107.Christ T., Boknik P., Wöhrl S., Wettwer E., Graf E.M., Bosch R.F., Knaut M., Schmitz W., Ravens U., Dobrev D. L-type Ca2+ current downregulation in chronic human atrial fibrillation is associated with increased activity of protein phosphatases. Circulation. 2004;110:2651–2657. doi: 10.1161/01.CIR.0000145659.80212.6A. [DOI] [PubMed] [Google Scholar]
- 108.Schotten U., Verheule S., Kirchhof P., Goette A. Pathophysiological mechanisms of atrial fibrillation: a translational appraisal. Physiol. Rev. 2011;91:265–325. doi: 10.1152/physrev.00031.2009. [DOI] [PubMed] [Google Scholar]
- 109.Mor A., Philips M.R. Compartmentalized Ras/MAPK signaling. Annu. Rev. Immunol. 2006;24:771–800. doi: 10.1146/annurev.immunol.24.021605.090723. [DOI] [PubMed] [Google Scholar]
- 110.Jiang F., Zhang Y., Dusting G.J. NADPH oxidase-mediated redox signaling: roles in cellular stress response, stress tolerance, and tissue repair. Pharmacol. Rev. 2011;63:218–242. doi: 10.1124/pr.110.002980. [DOI] [PubMed] [Google Scholar]
- 111.Schröder K., Weissmann N., Brandes R.P. Organizers and activators: Cytosolic Nox proteins impacting on vascular function. Free Radic. Biol. Med. 2017;109:22–32. doi: 10.1016/j.freeradbiomed.2017.03.017. [DOI] [PubMed] [Google Scholar]
- 112.Yoo S., Pfenniger A., Hoffman J., Zhang W., Ng J., Burrell A., Johnson D.A., Gussak G., Waugh T., Bull S., et al. Attenuation of Oxidative Injury With Targeted Expression of NADPH Oxidase 2 Short Hairpin RNA Prevents Onset and Maintenance of Electrical Remodeling in the Canine Atrium: A Novel Gene Therapy Approach to Atrial Fibrillation. Circulation. 2020;142:1261–1278. doi: 10.1161/CIRCULATIONAHA.119.044127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Youn J.Y., Zhang J., Zhang Y., Chen H., Liu D., Ping P., Weiss J.N., Cai H. Oxidative stress in atrial fibrillation: an emerging role of NADPH oxidase. J. Mol. Cell. Cardiol. 2013;62:72–79. doi: 10.1016/j.yjmcc.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Antoniades C., Demosthenous M., Reilly S., Margaritis M., Zhang M.H., Antonopoulos A., Marinou K., Nahar K., Jayaram R., Tousoulis D., et al. Myocardial redox state predicts in-hospital clinical outcome after cardiac surgery effects of short-term pre-operative statin treatment. J. Am. Coll. Cardiol. 2012;59:60–70. doi: 10.1016/j.jacc.2011.08.062. [DOI] [PubMed] [Google Scholar]
- 115.Mighiu A.S., Recalde A., Ziberna K., Carnicer R., Tomek J., Bub G., Brewer A.C., Verheule S., Shah A.M., Simon J.N., Casadei B. Inducibility, but not stability, of atrial fibrillation is increased by NOX2 overexpression in mice. Cardiovasc. Res. 2021;117:2354–2364. doi: 10.1093/cvr/cvab019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dudley S.C., Hoch N.E., McCann L.A., Honeycutt C., Diamandopoulos L., Fukai T., Harrison D.G., Dikalov S.I., Langberg J. Atrial fibrillation increases production of superoxide by the left atrium and left atrial appendage: role of the NADPH and xanthine oxidases. Circulation. 2005;112:1266–1273. doi: 10.1161/CIRCULATIONAHA.105.538108. [DOI] [PubMed] [Google Scholar]
- 117.Sha Q.Q., Dai X.X., Dang Y., Tang F., Liu J., Zhang Y.L., Fan H.Y. A MAPK cascade couples maternal mRNA translation and degradation to meiotic cell cycle progression in mouse oocytes. Development. 2017;144:452–463. doi: 10.1242/dev.144410. [DOI] [PubMed] [Google Scholar]
- 118.Pullikuth A.K., Catling A.D. Scaffold mediated regulation of MAPK signaling and cytoskeletal dynamics: a perspective. Cell. Signal. 2007;19:1621–1632. doi: 10.1016/j.cellsig.2007.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shi Y., Xia Y.Y., Wang L., Liu R., Khoo K.S., Feng Z.W. Neural cell adhesion molecule modulates mesenchymal stromal cell migration via activation of MAPK/ERK signaling. Exp. Cell Res. 2012;318:2257–2267. doi: 10.1016/j.yexcr.2012.05.029. [DOI] [PubMed] [Google Scholar]
- 120.Takemasu S., Nigorikawa K., Yamada M., Tsurumi G., Kofuji S., Takasuga S., Hazeki K. Phosphorylation of TMEM55B by Erk/MAPK regulates lysosomal positioning. J. Biochem. 2019;166:175–185. doi: 10.1093/jb/mvz026. [DOI] [PubMed] [Google Scholar]
- 121.He H., Kogut M.H. CpG-ODN-induced nitric oxide production is mediated through clathrin-dependent endocytosis, endosomal maturation, and activation of PKC, MEK1/2 and p38 MAPK, and NF-kappaB pathways in avian macrophage cells (HD11) Cell. Signal. 2003;15:911–917. doi: 10.1016/s0898-6568(03)00100-1. [DOI] [PubMed] [Google Scholar]
- 122.Enslen H., Raingeaud J., Davis R.J. Selective activation of p38 mitogen-activated protein (MAP) kinase isoforms by the MAP kinase kinases MKK3 and MKK6. J. Biol. Chem. 1998;273:1741–1748. doi: 10.1074/jbc.273.3.1741. [DOI] [PubMed] [Google Scholar]
- 123.Xu P., Derynck R. Direct activation of TACE-mediated ectodomain shedding by p38 MAP kinase regulates EGF receptor-dependent cell proliferation. Mol. Cell. 2010;37:551–566. doi: 10.1016/j.molcel.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Qi X., Pohl N.M., Loesch M., Hou S., Li R., Qin J.Z., Cuenda A., Chen G. p38alpha antagonizes p38gamma activity through c-Jun-dependent ubiquitin-proteasome pathways in regulating Ras transformation and stress response. J. Biol. Chem. 2007;282:31398–31408. doi: 10.1074/jbc.M703857200. [DOI] [PubMed] [Google Scholar]
- 125.Simard J.C., Girard D., Tessier P.A. Induction of neutrophil degranulation by S100A9 via a MAPK-dependent mechanism. J. Leukoc. Biol. 2010;87:905–914. doi: 10.1189/jlb.1009676. [DOI] [PubMed] [Google Scholar]
- 126.Steenman M. Insight into atrial fibrillation through analysis of the coding transcriptome in humans. Biophys. Rev. 2020;12:817–826. doi: 10.1007/s12551-020-00735-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Çubukçuoğlu Deniz G., Durdu S., Doğan Y., Erdemli E., Özdağ H., Akar A.R. Molecular Signatures of Human Chronic Atrial Fibrillation in Primary Mitral Regurgitation. Cardiovasc. Ther. 2021;2021:5516185. doi: 10.1155/2021/5516185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rivolta I., Binda A., Masi A., DiFrancesco J.C. Cardiac and neuronal HCN channelopathies. Pflugers Arch. 2020;472:931–951. doi: 10.1007/s00424-020-02384-3. [DOI] [PubMed] [Google Scholar]
- 129.Fenske S., Krause S.C., Hassan S.I.H., Becirovic E., Auer F., Bernard R., Kupatt C., Lange P., Ziegler T., Wotjak C.T., et al. Sick sinus syndrome in HCN1-deficient mice. Circulation. 2013;128:2585–2594. doi: 10.1161/CIRCULATIONAHA.113.003712. [DOI] [PubMed] [Google Scholar]
- 130.Reactome Pathway Database Home - Reactome Pathway Database. 2024. www.reactome.org
- 131.Leclerc G.M., Leclerc G.J., Kuznetsov J.N., DeSalvo J., Barredo J.C. Metformin induces apoptosis through AMPK-dependent inhibition of UPR signaling in ALL lymphoblasts. PLoS One. 2013;8:e74420. doi: 10.1371/journal.pone.0074420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Su K.N., Ma Y., Cacheux M., Ilkan Z., Raad N., Muller G.K., Wu X., Guerrera N., Thorn S.L., Sinusas A.J., et al. Atrial AMP-activated protein kinase is critical for prevention of dysregulation of electrical excitability and atrial fibrillation. JCI Insight. 2022;7:e141213. doi: 10.1172/jci.insight.141213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gutierrez A., Van Wagoner D.R. Oxidant and Inflammatory Mechanisms and Targeted Therapy in Atrial Fibrillation: An Update. J. Cardiovasc. Pharmacol. 2015;66:523–529. doi: 10.1097/FJC.0000000000000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Turi Z., Lacey M., Mistrik M., Moudry P. Impaired ribosome biogenesis: mechanisms and relevance to cancer and aging. Aging (Albany NY) 2019;11:2512–2540. doi: 10.18632/aging.101922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jung M., Jin S.G., Zhang X., Xiong W., Gogoshin G., Rodin A.S., Pfeifer G.P. Longitudinal epigenetic and gene expression profiles analyzed by three-component analysis reveal down-regulation of genes involved in protein translation in human aging. Nucleic Acids Res. 2015;43:e100. doi: 10.1093/nar/gkv473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kim J., Yang G., Kim Y., Kim J., Ha J. AMPK activators: mechanisms of action and physiological activities. Exp. Mol. Med. 2016;48:e224. doi: 10.1038/emm.2016.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Dyck J.R.B., Lopaschuk G.D. AMPK alterations in cardiac physiology and pathology: enemy or ally? J. Physiol. 2006;574:95–112. doi: 10.1113/jphysiol.2006.109389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Arad M., Seidman C.E., Seidman J.G. AMP-activated protein kinase in the heart: role during health and disease. Circ. Res. 2007;100:474–488. doi: 10.1161/01.RES.0000258446.23525.37. [DOI] [PubMed] [Google Scholar]
- 139.Cao Y., Bojjireddy N., Kim M., Li T., Zhai P., Nagarajan N., Sadoshima J., Palmiter R.D., Tian R. Activation of γ2-AMPK Suppresses Ribosome Biogenesis and Protects Against Myocardial Ischemia/Reperfusion Injury. Circ. Res. 2017;121:1182–1191. doi: 10.1161/CIRCRESAHA.117.311159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Debès C., Papadakis A., Grönke S., Karalay Ö., Tain L.S., Mizi A., Nakamura S., Hahn O., Weigelt C., Josipovic N., et al. Ageing-associated changes in transcriptional elongation influence longevity. Nature. 2023;616:814–821. doi: 10.1038/s41586-023-05922-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Embi A.A., Scherlag B.J., Ritchey J.W. Glycogen and the propensity for atrial fibrillation: intrinsic anatomic differences in glycogen in the left and right atria in the goat heart. N. Am. J. Med. Sci. 2014;6:510–515. doi: 10.4103/1947-2714.143282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhang L., Huang B., Scherlag B.J., Ritchey J.W., Embi A.A., Hu J., Hou Y., Po S.S. Structural changes in the progression of atrial fibrillation: potential role of glycogen and fibrosis as perpetuating factors. Int. J. Clin. Exp. Pathol. 2015;8:1712–1718. [PMC free article] [PubMed] [Google Scholar]
- 143.Kanzaki M., Pessin J.E. Insulin signaling: GLUT4 vesicles exit via the exocyst. Curr. Biol. 2003;13:R574–R576. doi: 10.1016/s0960-9822(03)00478-0. [DOI] [PubMed] [Google Scholar]
- 144.Chaikuad A., Froese D.S., Berridge G., von Delft F., Oppermann U., Yue W.W. Conformational plasticity of glycogenin and its maltosaccharide substrate during glycogen biogenesis. Proc. Natl. Acad. Sci. USA. 2011;108:21028–21033. doi: 10.1073/pnas.1113921108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bonilla I.M., Sridhar A., Györke S., Cardounel A.J., Carnes C.A. Nitric oxide synthases and atrial fibrillation. Front. Physiol. 2012;3:105. doi: 10.3389/fphys.2012.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dimmeler S., Haendeler J., Nehls M., Zeiher A.M. Suppression of apoptosis by nitric oxide via inhibition of interleukin-1beta-converting enzyme (ICE)-like and cysteine protease protein (CPP)-32-like proteases. J. Exp. Med. 1997;185:601–607. doi: 10.1084/jem.185.4.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Feng Q., Song W., Lu X., Hamilton J.A., Lei M., Peng T., Yee S.P. Development of heart failure and congenital septal defects in mice lacking endothelial nitric oxide synthase. Circulation. 2002;106:873–879. doi: 10.1161/01.cir.0000024114.82981.ea. [DOI] [PubMed] [Google Scholar]
- 148.Binder P., Nguyen B., Collins L., Zi M., Liu W., Christou F., Luo X., Hille S.S., Frey N., Cartwright E.J., et al. Pak2 Regulation of Nrf2 Serves as a Novel Signaling Nexus Linking ER Stress Response and Oxidative Stress in the Heart. Front. Cardiovasc. Med. 2022;9:851419. doi: 10.3389/fcvm.2022.851419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Dhalla N.S., Temsah R.M., Netticadan T. Role of oxidative stress in cardiovascular diseases. J. Hypertens. 2000;18:655–673. doi: 10.1097/00004872-200018060-00002. [DOI] [PubMed] [Google Scholar]
- 150.Li J., Solus J., Chen Q., Rho Y.H., Milne G., Stein C.M., Darbar D. Role of inflammation and oxidative stress in atrial fibrillation. Heart Rhythm. 2010;7:438–444. doi: 10.1016/j.hrthm.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Binder P., Wang S., Radu M., Zin M., Collins L., Khan S., Li Y., Sekeres K., Humphreys N., Swanton E., et al. Pak2 as a Novel Therapeutic Target for Cardioprotective Endoplasmic Reticulum Stress Response. Circ. Res. 2019;124:696–711. doi: 10.1161/CIRCRESAHA.118.312829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Boos C.J., Lip G.Y.H. Inflammation and atrial fibrillation: cause or effect? Heart. 2008;94:133–134. doi: 10.1136/hrt.2007.119651. [DOI] [PubMed] [Google Scholar]
- 153.Korantzopoulos P., Letsas K.P., Tse G., Fragakis N., Goudis C.A., Liu T. Inflammation and atrial fibrillation: A comprehensive review. J. Arrhythm. 2018;34:394–401. doi: 10.1002/joa3.12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lazzerini P.E., Capecchi P.L., Laghi-Pasini F. Systemic inflammation and arrhythmic risk: lessons from rheumatoid arthritis. Eur. Heart J. 2017;38:1717–1727. doi: 10.1093/eurheartj/ehw208. [DOI] [PubMed] [Google Scholar]
- 155.Baek Y.S., Kim T.H., Uhm J.S., Kim J.Y., Pak H.N., Lee M.H., Joung B. Prevalence and the clinical outcome of atrial fibrillation in patients with Autoimmune Rheumatic Disease. Int. J. Cardiol. 2016;214:4–9. doi: 10.1016/j.ijcard.2016.03.083. [DOI] [PubMed] [Google Scholar]
- 156.Dobrev D., Aguilar M., Heijman J., Guichard J.B., Nattel S. Postoperative atrial fibrillation: mechanisms, manifestations and management. Nat. Rev. Cardiol. 2019;16:417–436. doi: 10.1038/s41569-019-0166-5. [DOI] [PubMed] [Google Scholar]
- 157.Yang X., Zhao S., Wang S., Cao X., Xu Y., Yan M., Pang M., Yi F., Wang H. Systemic inflammation indicators and risk of incident arrhythmias in 478,524 individuals: evidence from the UK Biobank cohort. BMC Med. 2023;21:76. doi: 10.1186/s12916-023-02770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Liu C., Wang J., Yiu D., Liu K. The efficacy of glucocorticoids for the prevention of atrial fibrillation, or length of intensive care unite or hospital stay after cardiac surgery: a meta-analysis. Cardiovasc. Ther. 2014;32:89–96. doi: 10.1111/1755-5922.12062. [DOI] [PubMed] [Google Scholar]
- 159.Kim Y.R., Nam G.B., Han S., Kim S.H., Kim K.H., Lee S., Kim J., Choi K.J., Kim Y.H. Effect of Short-Term Steroid Therapy on Early Recurrence During the Blanking Period After Catheter Ablation of Atrial Fibrillation. Circ. Arrhythm. Electrophysiol. 2015;8:1366–1372. doi: 10.1161/CIRCEP.115.002957. [DOI] [PubMed] [Google Scholar]
- 160.Salih M., Smer A., Charnigo R., Ayan M., Darrat Y.H., Traina M., Morales G.X., DiBiase L., Natale A., Elayi C.S. Colchicine for prevention of post-cardiac procedure atrial fibrillation: Meta-analysis of randomized controlled trials. Int. J. Cardiol. 2017;243:258–262. doi: 10.1016/j.ijcard.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 161.Lennerz C., Barman M., Tantawy M., Sopher M., Whittaker P. Colchicine for primary prevention of atrial fibrillation after open-heart surgery: Systematic review and meta-analysis. Int. J. Cardiol. 2017;249:127–137. doi: 10.1016/j.ijcard.2017.08.039. [DOI] [PubMed] [Google Scholar]
- 162.Liu T., Li L., Korantzopoulos P., Liu E., Li G. Statin use and development of atrial fibrillation: a systematic review and meta-analysis of randomized clinical trials and observational studies. Int. J. Cardiol. 2008;126:160–170. doi: 10.1016/j.ijcard.2007.07.137. [DOI] [PubMed] [Google Scholar]
- 163.Benzoni P., Campostrini G., Landi S., Bertini V., Marchina E., Iascone M., Ahlberg G., Olesen M.S., Crescini E., Mora C., et al. Human iPSC modelling of a familial form of atrial fibrillation reveals a gain of function of If and ICaL in patient-derived cardiomyocytes. Cardiovasc. Res. 2020;116:1147–1160. doi: 10.1093/cvr/cvz217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ghezelbash S., Molina C.E., Dobrev D. Altered atrial metabolism: an underappreciated contributor to the initiation and progression of atrial fibrillation. J. Am. Heart Assoc. 2015;4:e001808. doi: 10.1161/JAHA.115.001808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chiang D.Y., Zhang M., Voigt N., Alsina K.M., Jakob H., Martin J.F., Dobrev D., Wehrens X.H.T., Li N. Identification of microRNA-mRNA dysregulations in paroxysmal atrial fibrillation. Int. J. Cardiol. 2015;184:190–197. doi: 10.1016/j.ijcard.2015.01.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ozcan C., Battaglia E., Young R., Suzuki G. LKB1 knockout mouse develops spontaneous atrial fibrillation and provides mechanistic insights into human disease process. J. Am. Heart Assoc. 2015;4:e001733. doi: 10.1161/JAHA.114.001733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Collins T.P., Bayliss R., Churchill G.C., Galione A., Terrar D.A. NAADP influences excitation-contraction coupling by releasing calcium from lysosomes in atrial myocytes. Cell Calcium. 2011;50:449–458. doi: 10.1016/j.ceca.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 168.Nattel S., Dobrev D. Electrophysiological and molecular mechanisms of paroxysmal atrial fibrillation. Nat. Rev. Cardiol. 2016;13:575–590. doi: 10.1038/nrcardio.2016.118. [DOI] [PubMed] [Google Scholar]
- 169.Xi Y., Cheng J. Dysfunction of the autonomic nervous system in atrial fibrillation. J. Thorac. Dis. 2015;7:193–198. doi: 10.3978/j.issn.2072-1439.2015.01.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 171.Davis S., Charles P.D., He L., Mowlds P., Kessler B.M., Fischer R. Expanding Proteome Coverage with CHarge Ordered Parallel Ion aNalysis (CHOPIN) Combined with Broad Specificity Proteolysis. J. Proteome Res. 2017;16:1288–1299. doi: 10.1021/acs.jproteome.6b00915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yacine A.-S., Dacheng X. Principal Component Analysis of High Frequency Data. Chicago Booth Research Paper. 2017;114:287–303. doi: 10.2139/ssrn.2645886. [DOI] [Google Scholar]
- 173.Nolte H., MacVicar T.D., Tellkamp F., Krüger M. Instant Clue: A Software Suite for Interactive Data Visualization and Analysis. Sci. Rep. 2018;8:12648. doi: 10.1038/s41598-018-31154-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Huber W., von Heydebreck A., Sültmann H., Poustka A., Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18:S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 175.Durinck S., Spellman P.T., Birney E., Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Strimmer K. fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics. 2008;24:1461–1462. doi: 10.1093/bioinformatics/btn209. [DOI] [PubMed] [Google Scholar]
- 177.Karnovsky M.J., Karnovsky M.L. A formaldehyde-glutaraldehyde fixative of high osmolality for use in electron-microscopy. JCB (J. Cell Biol.) 1965;27:137. [Google Scholar]
- 178.Spurr A.R. A low-viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruct. Res. 1969;26:31–43. doi: 10.1016/s0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass Spectrometry data has been deposited in PRIDE and genomics data in GEO. See resource availability for accession numbers. Data can be requested by contacting the lead contact. This study did not generate new unique codes.