Abstract
NorM of Vibrio parahaemolyticus apparently is a new type of multidrug efflux protein, with no significant sequence similarity to any known transport proteins. Based on the following experimental results, we conclude that NorM is an Na+-driven Na+/drug antiporter. (i) Energy-dependent ethidium efflux from cells possessing NorM was observed in the presence of Na+ but not of K+. (ii) An artificially imposed, inwardly directed Na+ gradient elicited ethidium efflux from cells. (iii) The addition of ethidium to cells loaded with Na+ elicited Na+ efflux. Thus, NorM is an Na+/drug antiporting multidrug efflux pump, the first to be found in the biological world. Judging from the similarity of the NorM sequence to those of putative proteins in sequence databases, it seems that Na+/drug antiporters are present not only in V. parahaemolyticus but also in a wide range of other organisms.
Drug resistance, especially multidrug resistance, is presently a serious problem in hospitals. Drug efflux from cells is one of the major mechanisms of drug resistance in both prokaryotes and eukaryotes (11, 12, 15, 18, 26). Many drug efflux systems are known to exist in the biological world, and these transporters can be divided into four families: the major facilitator (MF) family, the small multidrug resistance (SMR) family, the resistance nodulation cell division (RND) family, and the ATP binding cassette family (4, 6, 17). Membrane transporters of the MF family possess 12 to 14 transmembrane domains. Transporters of the SMR family are rather small and usually possess four transmembrane domains. Transporters of the RND family require multiple components to function effectively. An electrochemical potential of H+ across cell membranes seems to be the driving force for drug efflux by members of the MF, SMR, and RND families of transporters (13, 18, 28, 29). ATP is utilized as the energy donor in members of the ATP binding cassette family of multidrug efflux pumps (3, 26).
The electrochemical potential of H+ across cell membranes is established mainly by the respiratory chain in aerobic or facultative anaerobic bacteria. The electrochemical potential of H+ across the membrane is converted to that of Na+ by Na+/H+ antiporters (25, 27). Both of the electrochemical potentials of H+ and Na+ across cell membranes can be utilized to drive solute uptake in bacterial cells. Solutes are taken up into cells by an H+/substrate symport mechanism or an Na+/substrate symport mechanism (19). An electrochemical potential of H+ is also utilized to drive extrusion of substrate from cells. Most multidrug efflux pumps in bacteria are driven by H+, which is a mechanism for H+/drug antiport (18). However, no Na+-driven extrusion system for drugs, i.e., no Na+/drug antiporter, has been reported for bacterial cell membranes. Although an Na+/Ca2+ exchanger (16) and an Na+/urea antiporter (9) have been reported for animal cells, no Na+/drug antiporter has been reported for animal cells.
Vibrio parahaemolyticus, a slightly halophilic marine bacterium, is one of the major causes of food poisoning in Japan and many other countries (14). This microorganism requires Na+ for its growth (2). Energy metabolism and energy coupling in membranes of this microorganism are unique (20). Cells of V. parahaemolyticus possess a primary respiratory Na+ pump (24) and Na+-coupled membrane processes, such as an Na+/solute symporter (21, 22, 24) and an Na+-driven flagellar motor (1). We thought that Na+/drug antiporters might exist in this marine organism.
If an Na+/drug antiporter were to exist, it would be anticipated that (i) Na+ would stimulate drug efflux from cells, (ii) an artificially imposed Na+ gradient would elicit drug flux, and (iii) an artificially imposed drug gradient would elicit Na+ flux. Previously, we cloned and sequenced a gene encoding the multidrug efflux protein, NorM, from the chromosome of V. parahaemolyticus (15). The norM gene was expressed in an Escherichia coli mutant, KAM3, which lacks the major drug efflux pump AcrAB, and NorM was characterized (15). Based on its structural features, it was apparent that NorM is a unique transporter, as it shows no sequence similarity to any other known transporters, although it possesses 12 hydrophobic domains, a characteristic of the MF family (15). In this study we investigated whether the NorM multidrug efflux pump of V. parahaemolyticus is an Na+/drug antiporter.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli KAM3 (15) and HIT-1 (8) and V. parahaemolyticus AQ3334 (24) were used in this study. Plasmid pMVP36 is a derivative of pBR322 and carries the norM gene, encoding the NorM multidrug efflux protein, derived from V. parahaemolyticus AQ3334 (15).
Stimulation of energy-dependent ethidium efflux from cells by Na+.
E. coli KAM3 and KAM3/pMVP36 cells were grown separately in L medium (10) supplemented with 40 mM potassium lactate to the late, exponential phase of growth under aerobic conditions at 37°C. The cells were then harvested, washed with a minimal medium (23) (Na+ salts were replaced with K+ salts), and suspended in the same medium. Cells were incubated in the minimal medium supplemented with 25 μM ethidium bromide and 40 μM CCCP (carbonylcyanide-m-chlorophenylhydrazone) at 37°C for 60 min to load ethidium bromide into the cells. Cells were washed three times with a solution containing 100 mM morpholinepropanesulfonic acid (MOPS)–tetramethylammonium hydroxide (pH 7.0), 2 mM MgSO4, and 25 μM ethidium bromide; resuspended in the same buffer (approximately 0.25 mg of protein/ml); and subjected to fluorescence measurement at an excitation wavelength of 500 nm and an emission wavelength of 580 nm. After preincubation at 37°C for 5 min, 10 mM lactate-tetramethylammonium hydroxide (pH 7.0) was added to initiate respiration. Then NaCl, LiCl, or KCl (final concentration, 10 mM) was added to the assay mixture.
Ethidium efflux from cells induced by inwardly directed artificial Na+ gradient.
E. coli KAM3 and KAM3/pMVP36 cells were grown as described above. Cells were harvested at the late-exponential phase of growth and washed twice with 100 mM potassium phosphate buffer (pH 7.0). To load cells with ethidium, the cells were incubated in 100 mM potassium phosphate (pH 7.0) supplemented with 20 μM ethidium bromide, 5 mM 2,4-dinitrophenol, and 5 mM KCN for 60 min at 37°C. The cells were then collected by centrifugation, washed twice with 100 mM potassium phosphate (pH 7.0) supplemented with 20 μM ethidium bromide and 2 mM KCN, and suspended in the same medium (approximately 50 mg of protein/ml). These cells were then subjected to fluorescence measurements. Ethidium efflux from cells was initiated by 100-fold dilution of the cell suspension into 100 mM sodium phosphate (pH 7.0) at 25°C.
Efflux of Na+ induced by addition of ethidium to cell suspension.
E. coli HIT-1/pMVP36 cells were grown in a minimal medium (23) (Na+ salts were replaced with K+ salts) supplemented with 40 mM glycerol at 37°C under aerobic conditions. Cells were harvested at the late-exponential phase of growth, washed twice with a solution containing 100 mM MOPS–tetramethylammonium hydroxide (pH 7.0) and 2 mM MgSO4, and resuspended in the same buffer. A portion (0.5 ml) of this suspension was diluted with 2.5 ml of the same buffer (approximately 25 mg of protein/ml). NaCl was added to yield a final concentration of 100 μM. Cells were incubated at 25°C in a plastic vessel with rapid stirring, and water-saturated N2 gas was introduced continuously to maintain anaerobic conditions. An Na+-electrode (Radiometer, Copenhagen, Denmark) and a reference electrode were put into the vessel. Calibration was carried out by the addition of known amounts of NaCl. An anaerobic solution of serine was added (final concentration, 100 μM) to induce Na+ uptake into cells. Thereafter an anaerobic solution of ethidium bromide was added to the assay mixture (final concentration, 200 μM). Changes in the Na+ concentration of the assay medium were monitored with an Na+ electrode.
Efflux of Na+ induced by addition of ethidium to cell suspension of V. parahaemolyticus.
V. parahaemolyticus AQ3334 cells were grown in a minimal medium (20) supplemented with 40 mM potassium lactate and 1% polypeptone under aerobic conditions at 37°C. Cells were harvested at the late-exponential phase of growth, washed twice with a buffer solution containing 200 mM MOPS–tetramethylammonium hydroxide (pH 7.5) and 5 mM MgSO4, and resuspended in the same buffer (approximately 15 mg of protein/ml). NaCl was added to yield a final concentration of 100 μM. Uptake and efflux of Na+ was monitored with an Na+ electrode as described above.
RESULTS
Effect of Na+ on drug efflux via NorM system.
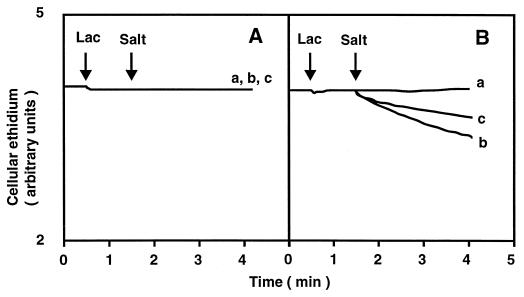
Cells of E. coli KAM3 or KAM3/pMVP36 (pMVP36 carries the norM gene from V. parahaemolyticus) were first loaded with ethidium, a fluorescent substrate for the NorM system (15), under deenergized conditions. Thereafter, an energy donor lactate, a respiratory substrate, was added to initiate drug efflux. In addition, either NaCl, LiCl, or KCl was added to test its effect on drug efflux. Addition of lactate to energy-starved cells of E. coli KAM3 did not cause significant ethidium efflux, and further addition of either KCl, NaCl, or LiCl had no effect (Fig. 1A). Similarly, addition of lactate to energy-starved cells of E. coli KAM3/pMVP36 caused no significant ethidium efflux. However, further addition of NaCl (10 mM) resulted in considerable efflux of ethidium from cells (Fig. 1B, curve b). Addition of LiCl (10 mM) elicited some ethidium efflux (Fig. 1B, curve c). Addition of KCl (10 mM) (Fig. 1B, curve a) caused no ethidium efflux. Thus, ethidium efflux via the NorM system was stimulated by Na+ or Li+.
FIG. 1.
Effect of monovalent cations on energy-dependent ethidium efflux via NorM. Energy-starved and ethidium-loaded cells of E. coli KAM3 and KAM3/pMVP36 were incubated in a buffer containing 100 mM MOPS–tetramethylammonium hydroxide (pH 7.0), 2 mM MgSO4, and 25 μM ethidium bromide. After incubation for 5 min at 37°C, 10 mM lactate–tetramethylammonium hydroxide (pH 7.0) was added to the assay mixture to initiate respiration (first arrow with Lac). At the time point indicated by the second arrow (with Salt), either KCl (curve a), NaCl (curve b), or LiCl (curve c) was added to the assay mixture (final concentration, 10 mM). (A) E. coli KAM3 cells; (B) E. coli KAM3/pMVP36 cells.
Ethidium efflux elicited by artificially imposed Na+ gradient.
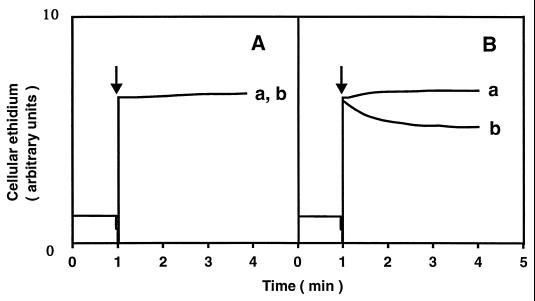
Ethidium was preloaded into energy-starved cells, and no energy source was added in the experiment. No significant ethidium efflux from cells was observed with E. coli KAM3 when an inwardly directed Na+ gradient was imposed (Fig. 2A). However, we observed significant ethidium efflux from KAM3/pMVP36 cells when an inwardly directed Na+ gradient was imposed (Fig. 2B). Imposition of an increasing Na+ gradient increased the initial velocity of ethidium efflux (data not shown).
FIG. 2.
Ethidium efflux from cells elicited by an inwardly directed artificial Na+ gradient. Energy-starved and ethidium-loaded cells of E. coli KAM3 and KAM3/pMVP36 were suspended (approximately 50 mg of protein/ml) in 100 mM potassium phosphate buffer (pH 7.0) containing 2 mM KCN and 20 μM ethidium bromide. An inwardly directed chemical gradient of Na+ was imposed by a 100-fold dilution of the cell suspension into the assay medium indicated below at the time point indicated by the arrow. The assay medium contained either 100 mM potassium phosphate (pH 7.0), 2 mM KCN, and 20 μM ethidium bromide (curve a) or 100 mM sodium phosphate (pH 7.0), 2 mM KCN, and 20 μM ethidium bromide (curve b). The assay was performed at 25°C. (A) E. coli KAM3 cells; (B) E. coli KAM3/pMVP36 cells.
Na+ efflux caused by drug influx.
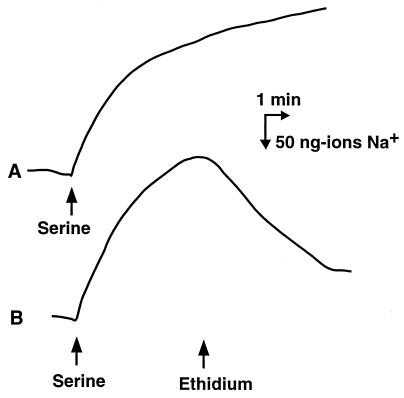
We first prepared Na+-loaded cells. We demonstrated previously that imposition of a chemical gradient of substrate, which is taken up with Na+ via a symport mechanism, elicited Na+ uptake and resulted in accumulation of Na+ in cells (7). In the current experiment, we utilized serine as the substrate and E. coli HIT-1/pMVP36 cells as the test cells. Wild-type E. coli possesses an Na+/serine symporter (7). E. coli HIT-1 cells lack one of the major Na+/H+ antiporters (8). Therefore, the activity of Na+ efflux or Na+ leakage through the antiporter in this strain is weak. The strain is thus suitable for measurement of Na+ flux via another pathway(s) (8). Addition of serine to a cell suspension of HIT-1/pMVP36 under anaerobic conditions induced uptake of Na+ (Fig. 3). When Na+ uptake (upward deflection) reached a plateau level, ethidium was added to the same cell suspension. As anticipated, we observed Na+ efflux caused by the addition of ethidium (Fig. 3). Although HIT-1 control cells showed Na+ uptake elicited by serine, we observed no significant Na+ efflux when ethidium was added (data not shown). Furthermore, we have tested many times whether Na+ efflux takes place with cells possessing NorA or TetA, H+/drug antiporters, or other H+-coupled transporters and we have never observed Na+ efflux (data not shown).
FIG. 3.
Na+ efflux from cells elicited by an inwardly directed ethidium gradient. Energy-starved cells of E. coli HIT-1/pMVP36 were incubated in a buffer consisting of 100 mM MOPS–tetramethylammonium hydroxide (pH 7.0), 2 mM MgSO4, and 100 μM NaCl. Flux of Na+ into and out of cells was measured with an Na+ electrode at 25°C. The first arrow indicates when an anaerobic solution of serine (final concentration, 100 μM) was added to the cell suspension under anaerobic conditions to elicit Na+ uptake into cells. The second arrow indicates when an anaerobic solution of ethidium bromide (final concentration, 200 μM) was added to the assay mixture. Upward deflection represents the uptake of Na+, and downward deflection represents the efflux of Na+.
Na+ efflux elicited by ethidium in V. parahaemolyticus.
We added serine to a cell suspension of V. parahaemolyticus under anaerobic conditions, and Na+ uptake was elicited (Fig. 4A and B). The addition of ethidium to the same cell suspension caused efflux of Na+ (Fig. 4B). This provides solid evidence for the presence of an Na+/ethidium antiporter in V. parahaemolyticus cells.
FIG. 4.
Na+ efflux from V. parahaemolyticus elicited by ethidium influx. Energy-starved cells of V. parahaemolyticus were incubated in a buffer containing 200 mM MOPS–tetramethylammonium hydroxide (pH 7.0), 5 mM MgSO4, and 100 μM NaCl. Flux of Na+ into and out of cells was measured with an Na+ electrode at 25°C. The first arrow indicates the time point when an anaerobic solution of serine (final concentration, 100 μM) was added to the cell suspension under anaerobic conditions to elicit Na+ uptake into cells. At the point indicated by the second arrow, ethidium bromide (final concentration, 200 μM) was added to the assay mixture. Upward deflection represents the uptake of Na+, and downward deflection represents the efflux of Na+.
DISCUSSION
Our experimental results presented here support the idea that NorM mediates Na+/drug antiport. The result that most strongly supports the idea of Na+/drug antiport by NorM is that influx of ethidium, a substrate for NorM, elicited efflux of Na+. So far we have investigated many Na+-coupled transporters and H+-coupled transporters. We have never observed Na+ flux elicited secondarily by H+-coupled transporters. On the other hand, we have observed a slight H+ flux elicited secondarily by Na+-coupled transporters (data not shown). This is likely due to the difference in sensitivity between the H+ electrode and Na+ electrode. The former is much more sensitive than the latter. We tested whether H+ flux takes place with NorM. We have never observed H+ flux due to the NorM system (data not shown). On the other hand, we have detected H+ flux due to NorA and TetA (data not shown). Thus, it is clear that NorM is not an H+/drug antiporter.
NorM of V. parahaemolyticus is believed to be a membrane protein with 456 amino acid residues (15). Very recently we cloned and sequenced, from chromosomal DNA of V. parahaemolyticus, another gene which seems to encode a multidrug efflux pump (unpublished data). This system showed a similar property, requiring Na+ for its activity. Therefore, it is likely that this new system is also an Na+/drug antiporter. We found no sequence similarity between this system and NorM. NorM shares no sequence similarity with any known multidrug efflux protein found in a sequence database (SwissProt) (15). It has been proposed that NorM be classified into a new family of transporters (5). However, NorM shares a high degree of sequence similarity with uncharacterized putative integral membrane proteins deduced from genomic DNA sequences in a wide range of archaeabacteria, eubacteria, and eukaryotes (sequences found in the SwissProt, GenBank, EMBL, and DDBJ databases). One example is YdhE of E. coli (15). Our preliminary result supports the idea that YdhE is also an Na+/drug antiporter (unpublished data). Taken together, these data suggest that Na+/drug antiporters are present not only in a marine bacterium but also in a wide range of other organisms.
ACKNOWLEDGMENTS
We thank Manuel F. Varela of Eastern New Mexico University for critically reading the manuscript.
This study was supported in part by a grant from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Atsumi T, McCarter L, Imae Y. Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces. Nature. 1992;355:182–184. doi: 10.1038/355182a0. [DOI] [PubMed] [Google Scholar]
- 2.Baumann P, Schubert R H W. Family II. Vibrionaceae veron 1965, 5245AL. In: Williams S T, Sharpe M E, Holt J G, editors. Bergey's manual of systematic bacteriology. Vol. 1. Baltimore, Md: Williams & Wilkins; 1984. pp. 516–550. [Google Scholar]
- 3.Bolhuis H, van Veen H W, Molenaar D, Poolman B, Driessen A J, Konings W N. Multidrug resistance in Lactococcus lactis: evidence for ATP-dependent drug extrusion from the inner leaflet of the cytoplasmic membrane. EMBO J. 1996;15:4239–4245. [PMC free article] [PubMed] [Google Scholar]
- 4.Bolhuis H, van Veen H W, Poolman B, Driessen A J, Konings W N. Mechanisms of multidrug transporters. FEMS Microbiol Rev. 1997;21:55–84. doi: 10.1111/j.1574-6976.1997.tb00345.x. [DOI] [PubMed] [Google Scholar]
- 5.Brown M H, Paulsen I T, Skurray R A. The multidrug efflux protein NorM is a prototype of a new family of transporters. Mol Microbiol. 1999;31:394–395. doi: 10.1046/j.1365-2958.1999.01162.x. [DOI] [PubMed] [Google Scholar]
- 6.Gottesman M M, Pastan I. Biochemistry of multidrug resistance mediated by the multidrug transporter. Annu Rev Biochem. 1993;62:385–427. doi: 10.1146/annurev.bi.62.070193.002125. [DOI] [PubMed] [Google Scholar]
- 7.Hama H, Shimamoto T, Tsuda M, Tsuchiya T. Properties of a Na+-coupled serine-threonine transport system in Escherichia coli. Biochim Biophys Acta. 1987;905:231–239. doi: 10.1016/0005-2736(87)90451-2. [DOI] [PubMed] [Google Scholar]
- 8.Ishikawa T, Hama H, Tsuda M, Tsuchiya T. Isolation and properties of a mutant of Escherichia coli possessing defective Na+/H+ antiporter. J Biol Chem. 1987;262:7443–7446. [PubMed] [Google Scholar]
- 9.Kato A, Sands J M. Evidence for sodium-dependent active urea secretion in the deepest subsegment of the rat inner medullary collecting duct. J Clin Investig. 1998;101:423–428. doi: 10.1172/JCI1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lennox E S. Transduction of linked genetic characters of host by bacteriophage P1. Virology. 1955;1:190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- 11.Levy S B. Active efflux mechanisms for antimicrobial resistance. Antimicrob Agents Chemother. 1992;36:695–703. doi: 10.1128/aac.36.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis K. Multidrug resistance pumps in bacteria: variations on a theme. Trends Biochem Sci. 1994;19:119–123. doi: 10.1016/0968-0004(94)90204-6. [DOI] [PubMed] [Google Scholar]
- 13.Mine T, Morita Y, Kataoka A, Mizushima T, Tsuchiya T. Evidence for chloramphenicol/H+ antiport in Cmr (MdfA) system of Escherichia coli and properties of the antiporter. J Biochem (Tokyo) 1998;124:187–193. doi: 10.1093/oxfordjournals.jbchem.a022078. [DOI] [PubMed] [Google Scholar]
- 14.Miwatani T, Takeda Y. Food poisoning due to Vibrio parahaemolyticus in Japan. In: Miwatani T, Takeda Y, editors. Vibrio parahaemolyticus, a causative bacterium of food poisoning. Tokyo, Japan: Saikon Publishing Co.; 1976. pp. 22–25. [Google Scholar]
- 15.Morita Y, Kodama K, Shiota S, Mine T, Kataoka A, Mizushima T, Tsuchiya T. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob Agents Chemother. 1998;42:1778–1782. doi: 10.1128/aac.42.7.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicoll D A, Longoni S, Philipson K D. Molecular cloning and functional expression of the cardiac sarcolemmal Na+-Ca2+ exchanger. Science. 1990;250:562–565. doi: 10.1126/science.1700476. [DOI] [PubMed] [Google Scholar]
- 17.Nikaido H. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science. 1994;264:382–388. doi: 10.1126/science.8153625. [DOI] [PubMed] [Google Scholar]
- 18.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poolman B, Konings W N. Secondary solute transport in bacteria. Biochim Biophys Acta. 1993;1183:5–39. doi: 10.1016/0005-2728(93)90003-x. [DOI] [PubMed] [Google Scholar]
- 20.Sakai Y, Tamao Y, Shimamoto T, Hama H, Tsuda M, Tsuchiya T. Cloning and expression of the 5′-nucleotidase gene of Vibrio parahaemolyticus in Escherichia coli and overproduction of the enzyme. J Biochem (Tokyo) 1989;105:841–846. doi: 10.1093/oxfordjournals.jbchem.a122755. [DOI] [PubMed] [Google Scholar]
- 21.Sarker R I, Ogawa W, Tsuda M, Tanaka S, Tsuchiya T. Characterization of a glucose transport system in Vibrio parahaemolyticus. J Bacteriol. 1994;176:7378–7382. doi: 10.1128/jb.176.23.7378-7382.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarker R I, Ogawa W, Tsuda M, Tanaka S, Tsuchiya T. Properties of a Na+/galactose (glucose) symport system in Vibrio parahaemolyticus. Biochim Biophys Acta. 1996;1279:149–156. doi: 10.1016/0005-2736(95)00252-9. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka S, Lerner S A, Lin E C. Replacement of a phosphoenolpyruvate-dependent phosphotransferase by a nicotinamide adenine dinucleotide-linked dehydrogenase for the utilization of mannitol. J Bacteriol. 1967;93:642–648. doi: 10.1128/jb.93.2.642-648.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsuchiya T, Shinoda S. Respiration-driven Na+ pump and Na+ circulation in Vibrio parahaemolyticus. J Bacteriol. 1985;162:794–798. doi: 10.1128/jb.162.2.794-798.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsuchiya T, Takeda K. Extrusion of sodium ions energized by respiration and glycolysis in Escherichia coli. J Biochem (Tokyo) 1979;86:225–230. [PubMed] [Google Scholar]
- 26.van Veen H W, Konings W N. Drug efflux proteins in multidrug resistant bacteria. Biol Chem. 1997;378:769–777. [PubMed] [Google Scholar]
- 27.West I C, Mitchell P. Proton/sodium ion antiport in Escherichia coli. Biochem J. 1974;144:87–90. doi: 10.1042/bj1440087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yerushalmi H, Lebendiker M, Schuldiner S. EmrE, an Escherichia coli 12-kDa multidrug transporter, exchanges toxic cations and H+ and is soluble in organic solvents. J Biol Chem. 1995;270:6856–6863. doi: 10.1074/jbc.270.12.6856. [DOI] [PubMed] [Google Scholar]
- 29.Zgurskaya H I, Nikaido H. Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc Natl Acad Sci USA. 1999;96:7190–7195. doi: 10.1073/pnas.96.13.7190. [DOI] [PMC free article] [PubMed] [Google Scholar]