Abstract
Background:
Nosocomial pathogens are known to exacerbate morbidity and mortality in contemporary critical healthcare. Hospital fomites, which include inanimate surfaces, have been identified as “breeding grounds” for pathogens that cause nosocomial infections. This systematic review aimed to deliver incisive insights on nosocomial pathogens in intensive care units (ICUs) and the role of fomites as potential reservoirs for their transmission.
Method:
An extensive exploration of electronic databases, including PubMed and Scopus, from 1990 to 2023, was carried out between 25th and 29th May 2023, per standard PRISMA guidelines. Information were extracted from articles that reported on fomites in the ICU. Studies that did not quantitatively report the fomite contamination, and those that exclusively took samples from patients in the ICU were excluded from the analysis.
Results:
About 40% of the total samples collected on fomites from all the studies yielded microbial growth, with species of Staphylococcus being the most predominant. Other prevalent microbes were Acinetobacter baumannii, Escherichia coli, Klebsiella pneumoniae, Candida spp., Enterococcus sp., and Enterobacter sp. The neonatal intensive care unit (NICU) had the highest proportion of contaminated fomites. Among known fomites, the sphygmomanometer exhibited a 100% detection rate of nosocomial pathogens. This included E. aerogenes, Staphylococcus aureus, coagulase-negative Staphylococci (CoNS), E. coli, and K. pneumoniae. Multidrug-resistant (MDR) bacteria, such as methicillin-resistant S. aureus (MRSA), vancomycin-resistant Enterococci (VRE), extended-spectrum beta-lactamase (ESBL)-producing E. coli, and MDR Pseudomonas aeruginosa were commonly isolated on fomites in the ICUs.
Conclusion:
Many fomites that are readily used in patient care in the ICU harbour nosocomial pathogens. The most common fomite appeared to be mobile phones, sphygmomanometers, and stethoscopes, with Staphylococcus being the most common contaminant. Consequently, the need for rigorous disinfection and sterilization protocols on fomites in the ICU cannot be overemphasized. Additionally, heightened awareness on the subject among health professionals is crucial to mitigating the risk and burden of nosocomial infections caused by drug-resistant bacteria.
Keywords: Fomite, nosocomial, pathogen, intensive care unit, Staphylococcus, A. baumannii, critical care, multidrug resistance, hospital, public health
Introduction
Nosocomial infections, also known as healthcare-associated infections (HAIs), are those infections that are acquired during the process of receiving healthcare, and include occupational infections incurred by staff, health professionals, patients, and visitors.1 -3 Pathogens that are responsible for nosocomial infections are termed nosocomial pathogens, and they include a wide range of bacterial, viral, and fungal species. These pathogens and their infections pose significant problems, requiring urgent attention worldwide. 4 Critically ill patients in the intensive care unit (ICU) are often immunocompromised, and they are at a high risk for nosocomial infections than are patients in other areas of the hospital. 5 Nosocomial infections cause significant morbidity and mortality in contemporary critical care medicine, as some nosocomial pathogens are increasingly becoming multidrug-resistant.6,7 Multidrug resistance in nosocomial infections complicates patient management, extends treatment duration, and heightens economic burden with excessive healthcare costs. 8 Nosocomial infections have prevalence rates of 1.6% to 45.8% or higher in less developed countries,9,10 6.5% in the European Union, and 3.2% in the United States, resulting in a cost of more than $4.5 billion for the latter. Since the global burden of HAIs is uncertain due to inadequate surveillance systems, it is probable that the prevalence of nosocomial infections is significantly higher worldwide. 11 The extent to which the hospital environment serves as a reservoir of nosocomial pathogens, however, remains a subject of debate, amidst limited information. 12
Inanimate objects and surfaces can serve as reservoirs of nosocomial pathogens in the ICU. 13 Such objects are commonly referred to as fomites, a term defining objects that when contaminated with infectious agents, can transfer these agents to a new host.14 -16 In an ICU, fomites can include medical equipment, surfaces, and other inanimate objects.17 -19 The role of hospital fomites in the transmission of nosocomial organisms is still a topical issue, but there is no clear consensus on the matter. 20 Several studies have identified fomites in the ICU to harbour nosocomial pathogens and be a contributor to their outbreak in the unit.21 -24 Disturbingly, several nosocomial pathogens are drug-resistant, and in some cases, extensively drug-resistant or multidrug-resistant due to their exposure to numerous antibiotics in the hospital setting.25 -31 Medical equipment, such as ventilators, catheters, faucet aerators, and dialysis machines, as well as other inanimate surfaces, can become contaminated with pathogens and serve as reservoirs for infections.5,24,32,33 Proper cleaning and disinfection of equipment are essential to preventing the spread of the infections. Surfaces such as bed rails, doorknobs, pens, and countertops also harbour pathogens, and must be regularly cleaned and disinfected.34,35 The ability of pathogens to persist on reservoirs is a significant challenge in the prevention and control of nosocomial infections in the ICU. Also, the persistence of bacteria, viruses, and fungi on inanimate surfaces vary accordingly. 36
As it is nearly impossible to eliminate the use of equipment and other fomites in the ICU, compliance with standards and guidelines can help reduce or manage HAIs. 37 With current technological advancements and increased expectations for high-quality healthcare services, it is crucial to analyze the frequency and causes of nosocomial infections, especially, in ICUs. 4 Therefore, it is necessary to identify key inanimate reservoirs in ICUs, the common pathogens they harbour, and how long these pathogens persist on them. This would aid in devising effective infection control programmes in hospitals and help develop a reliable and sustainable plan in controlling infections in critical care units. The lack of precise information on the role of fomites in the spread of nosocomial pathogens makes it difficult to implement control plans, resulting in increased costs for both healthcare systems and patients.4,38,39 This systematic review, therefore, aimed at providing a comprehensive analysis on fomites and their associated pathogens, as well as antibiotic resistance and persistence of these pathogens on fomites within the ICU. Its focus encompasses the neonatal intensive care unit (NICU), pediatric intensive care unit (PICU), surgical intensive care unit (SICU), burns intensive care unit (BICU), and the medical intensive care unit (MICU).
Method
Search strategy
The systematic review was conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guidelines. 40 Between 25th and 29th May 2023, we carried out an extensive exploration of electronic databases, including PubMed and Scopus, spanning 1990 to 2023. In order to ensure that our search was comprehensive, the following search terms were used: ““intensive care unit” and fomite”, “fomite in intensive care unit,” “fomite and pathogen in intensive care unit,” “intensive care unit,” “fomite and infection,” “nosocomial infection,” “intensive care unit,” “inanimate surface,” and “pathogen persistence on fomites in ICU.” Moreover, “fomite” and “intensive care unit” were included in the following search queries: nosocomial infections, persistence, bacteria, fungi, and viruses. Furthermore, the citations of each study identified during the primary search were evaluated for possible relevance, as were similar articles that appeared with the search results on PubMed.
Inclusion and exclusion criteria
Based on the research keywords, we incorporated studies that presented both qualitative and quantitative data on nosocomial pathogens present on fomites, as well as their prevalence on inanimate surfaces in the ICU. The types of studies used included cross-sectional, longitudinal, prospective, and outbreak studies. In the case of outbreak studies, we included those that collected samples from patients and fomites and further reported on the organisms that were recovered from the fomites; in such instances, we only included the fomite part of the outbreak reports. Our selection was limited to articles that were accessible to us, available in full text, and published in the English language. Publications excluded from the review were reports, case-control studies, commentaries, and letters to editors. Published review articles and textbooks were also excluded. Besides, studies that reported on pathogens that were not associated with fomites were excluded. Moreover, studies that did not specify sample size for various fomites and did not report on the number of positive samples were also excluded, as were preprints and studies whose sample sizes were each less than 10.
Study selection
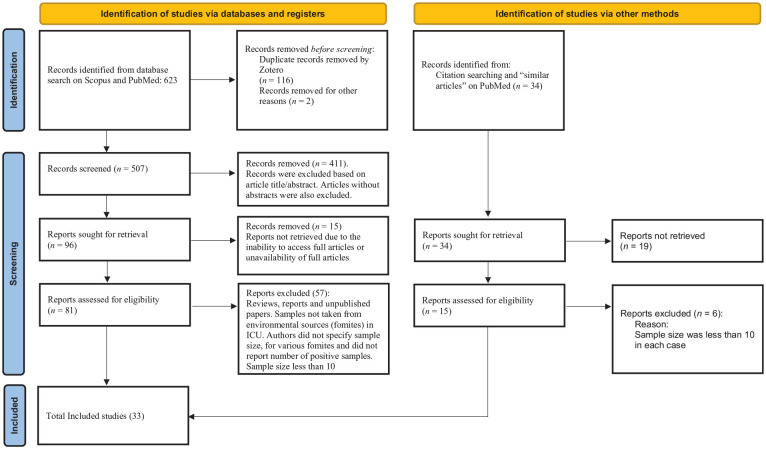
The Scopus and PubMed searches yielded 623 articles, which were screened using the Zotero reference tool (Version 6.0.30, made by Corporate for Digital Scholarship), to identify and download articles that are available for free. After eliminating duplicates using Zotero, the resulting 507 records were examined based on their titles and abstracts in relation to the inclusion criteria. Subsequently, 41 potential articles were each subjected to careful independent evaluations, with only articles published in the English language considered. Finally, 33 articles were included in this study after a systematic evaluation was carried out on the complete texts of the remaining 81 studies to ascertain their eligibility per the criteria specified in the study, as shown in Figure 1.
Figure 1.
A flow diagram of our systematic review process (PRISMA guide).
Quality assessment
This study was established on the basis of previously published research articles with observational evidence. All duplicates were carefully inspected and eliminated in order to retain the quality of the review. The abstracts of searched articles were thoroughly checked and verified before analysis to ensure that quality and relevant information in the literature were included in the review process. The quality of the articles selected was evaluated independently by authors A.-H.O., S.D. and A.O.
Data extraction
Data were extracted by A.-H.O. and E.S.D. from individual studies using a form and database developed for the purposes of this review in the Microsoft Excel 2013 software. The form captured data such as authors’ names, title of the study, year of publication, study setting, keywords, fomites assessed, sampling method, microbial identification method, pathogens assessed, prevalence of nosocomial pathogens, and the duration of pathogens on the fomites. For studies that reported their findings in percentages, the percentages were converted to whole numbers to ensure uniformity.
Results
Overview
In accordance with the inclusion and exclusion criteria and the PRISMA checklist, 40 we selected 33 articles that investigated reservoirs of pathogens in ICUs. The studies were carried out across a diverse range of 17 countries, spanning various regions globally. Three articles were on outbreak or post-outbreak studies41 -43; two targeted human adenoviruses/rotavirus.44,45 Additionally, two other studies46,47 specifically targeted Staphylococcus aureus and SARS-CoV-2, respectively. The computer, computer mouse, the space bar on the computer keyboard, and other parts of the computer were collectively labeled as “Computer and its parts” for the purpose of uniformity in this review. Similarly, the sink, sink outlet, and drain were labeled as “Sink”. The total number of fomites identified in this study was 29, as shown in Table 1. Overall, about 40% of the total samples collected on these fomites yielded microbial growth, and Staphylococcus was the most isolated genus of bacteria. S. aureus was the most predominant species identified. Of the 33 studies, 27 isolated bacteria6,19,23,32,41 -43,46,48 -65, three isolated viruses,44,45,47 and only one study examined fungi. 66 Two studies67,68 isolated both bacteria and fungi on fomites in the ICU. Among the studies that reported on bacteria, fifteen6,19,32,43,46,49,53,56 -58,60,61,63,64 reported on the antibiotic susceptibility profiles of the isolates. None of the studies assessed the longevity or persistence of nosocomial pathogens on a fomite in the ICU. However, the persistence of common nosocomial pathogens on inanimate surfaces has been studied and reviewed in other reports.69,70 Thus, the persistence of the common nosocomial pathogens in our review were inferred from these reports and other similar studies for discussion purposes.
Table 1.
Summary of individual studies methodologies, bacterial identification methods, and quantitative reports.
| Fomites | Site of Study | Organism Isolated | Number of Collected Samples | Number of Positive Samples | Percent Positive (%) | Sampling Method | Organism Identification Method | Article/Source |
|---|---|---|---|---|---|---|---|---|
| Incubators and their Door Locks | ICU | Human adenovirus (HAdV) | 48 | 3 | 6 | A minimum of 50% of fomite surfaces were scraped using swabs. | Genomic quantification and sequencing | Ganime et al. 45 |
| Mattresses and Pillows | ICU | MRSA, VRE, ESBL producers (52% of all isolates were MDR) | 11 | 8 | 73 | Cutting out a segment of material using sterile gloves | PCR, Vitek2 GPS-IX or Vitek 2 AST-N149 cards | Hu et al. 58 |
| Gowns/Coats | ICU | S. aureus, Acinetobacter baumannii, Stenotrophomonas maltophilia, Klebsiella pneumoniae, and Serratia rubidae; 22.2% of the Gram-negative bacteria were MDR. | 31 | 15 | 48 | Use of moistened swab on 4 cm2 areas of cuffs and abdominal regions of gowns. | Automated identification tests (Vitek System) | Pilonetto et al. 64 |
| Yankauer Catheters and Suction Machines | ICU | S. aureus (15% were MRSA), Pseudomonas aeruginosa and Escherichia coli, Candida spp., CoNS, and Enterococcus spp. (10% were VRE) | 20 | 16 | 80% | Tip soaking (Submerging 3 cm catheter tips in 8 mL thioglycollate broth) | Standard microbiological techniques | Brown & Willms et al. 32 |
| Trolleys/ Trays | S. aureus (20.6% were MDR) | 82 | 10 | 12 | Swabbing | PCR and standard microbiological methods | Veloso et al. 46 | |
| ICU | Enterococcus spp., S. aureus, Gram-negative rods, and moulds | 174 | 8 | 5 | A sterile rayon-tipped swab (D2-Tupfer; Heinz Herenz, Hamburg) moistened with sterile saline solution was used to sample the surfaces. | Microbiological testing | Hartmann et al. 67 | |
| Thermometers | ICU | CoNS and MDR S. haemolyticus | 18 | 18 | 100 | Cotton swab moistened with sterile normal saline | Standard microbiological methods and biochemical tests | Sued et al. 19 |
| NICU | MDR Klebsiella spp. | 12 | 1 | 8 | Swabbing | Identification by API 20E identification system, and DNA fingerprinting | Macrae et al. 43 | |
| Parenteral Nutrition (PN) or Expressed Breast Milk | NICU | MDR Klebsiella spp. | 32 | 5 | 16 | Swabbing | Identification by API 20E identification system, and DNA fingerprinting | Macrae et al. 43 |
| Pulse Oximetry | NICU | MDR Klebsiella spp. | 12 | 4 | 33 | Swabbing | Identification by API 20E identification system, and DNA fingerprinting | Macrae et al. 43 |
| Surrounding Air | ICU | Penicillium spp., Aspergillus spp., Curvularia spp., Alternaria spp., Paecelomyces spp., Zygomycetes, Fusarium spp., Cladosporium spp., and sterile mycelium | 40 | 43 | 83 | Sedimentation plate method | Visualization of the macro- and micro-morphology characteristics of the growing colonies | Gonçalves et al. 66 |
| Mobile Phones | ICU | ESBL-producing Enterobacter spp., ESBL-producing Klebsiella pneumoniae, Klebsiella oxytoca, and E. coli; 53.3% of all these were MDR. | 491 | 104 | 21 | A sterile cotton swab moistened with trypticase soy broth was rotated on the entire surface of each phone. | Microbiological procedures | Loyola et al. 60 |
| ICU | Staphylococcus epidermidis, Staphylococcus hominis, Bacillus spp., S. aureus, Staphylococcus warneri, S. haemolyticus, Streptococcus mitis, and Streptococcus oralis | 130 | 93 | 72 | Surface swabbing of each mobile phone’s buttons, using a sterile gel swab that is moistened with saline. | Microbiological methods and MicroScan | Al-beeshi et al. 48 | |
| ICU | SARS-CoV-2 | 51 | 2 | 4 | Nylon FLOQ Swab | RT-PCR and viral culture | Espinoza et al. 47 | |
| ICU | Streptococcus spp., MRSA, CoNS, Enterococcus spp., Non-fermentative Gram-negative bacteria, coliforms, moulds, and yeasts | 200 | 189 | 95 | A sterile swab moistened by saline was rotated on the surfaces. | Biochemical tests | Ulger et al. 68 | |
| ICU | CoNS, S. aureus, Sarcina spp., Bacillus spp., Corynebacterium spp., and Neisseria spp. | 50 | 40 | 80 | A sterile saline-moistened swab (Copan S.p.A, Brescia, Italy) was rotated across both sides of mobile phones’ surfaces. | Standard microbiological methods and biochemical tests | Kotris et al. 59 | |
| ICU | CoNS, MRSA, Micrococci, and ESBL-producing E. coli | 55 | 48 | 87 | Saline-wet-sterile swab sticks were rubbed over the entire surface area of each phone. | Standard microbiological techniques | Anupriya et al. 49 | |
| ICU | CoNS, Bacillus spp., and MRSA, Acinetobacter spp., moulds, Paenibacillus spp., Streptococcus viridians, and Aerococcus spp. | 50 | 50 | 100 | Used “E-Swab COPAN” | Standard microbiological methods and biochemical tests | Galazzi et al. 56 | |
| ICU | CoNS, Streptococcus viridans, S. aureus (1.4% were MRSA), Micrococcus, E. coli, Diphtheroids, Bacillus spp., Pantoea spp., Moraxella osloensis, Pseudomonas stutzeri, Sphingomonas paucimobilis, Acinetobacter lwoffii, and A. baumannii | 213 | 157 | 74 | Sides, backs and screens of mobile phones, and in some cases, phone covers, were swabbed using sterile swabs. | Microbiological methods | Heyba et al. 57 | |
| ICU | P. aeruginosa, Acinetobacter spp., MRSA, VRE, and MDR Enterococcus spp. | 491 | 107 | 22 | A sterile cotton swab moistened with trypticase soy broth was rotated on the entire surface of each phone. | Standard microbiological methods and biochemical tests | Loyola et al. 61 | |
| ICU |
S. epidermidis, S. aureus, E. coli, K. pneumoniae, P. aeruginosa, Acinetobacter spp., Bacillus spp., Proteus, and Streptococcus. None of the isolates were sensitive to sulfamethoxazole-trimethoprim, tetracycline and ampicillin. |
56 | 53 | 95 | Saline-wet-sterile swab sticks (Sterilin, UK) were rubbed over the entire surface area of each phone. | Standard bacteriological procedures | Nwankwo et al. 62 | |
| Faucets and Aerators | ICU | S. paucimobilis, P. aeruginosa, C. meningosepticum, Achromobacter xylosoxidans, Burkholderia cepacia, and S. maltophilia | 162 | 54 | 33 | Sterile cotton swabs were used on the inner surfaces of the faucet aerators. | Standard biochemical methods | Wang et al. 23 |
| ICU | S. aureus (MRSA), Enterobacter, and Enterococcus | 64 | 7 | 11 | Rolling of saline-moistened-sterile rayon-tipped swab (Baxter Healthcare Corporation, Deerfield, Ill), on the entire surface being tested | VITEK system and API 20E, pulsed-field gel electrophoresis | Bures et al. 6 | |
| NICU | P. aeruginosa | 28 | 18 | 64 | Swab and first-flush cold water | qPCR | Bédard et al. 41 | |
| Pens | ICU | CoNS and Micrococcus spp. | 20 | 17 | 85 | Swabbing | Standard microbiological techniques | Wolfe et al. 35 |
| Sphygmomanometers | ICU | E. aerogenes, S. aureus (58.7% were MDR), CoNS (28.3%), E. coli (72.7%), and K. pneumoniae | 18 | 18 | 100 | Sterile cotton-tipped applicator sticks, moistened with sterile normal saline, were employed. | Colony morphology, Gram staining and biochemical tests | Darge et al. 53 |
| ICU | CoNS and MDR S. haemolyticus | 24 | 24 | 100 | Cotton swab moistened with sterile normal saline | Standard microbiological methods and biochemical tests | Sued et al. 19 | |
| Bedside Tables | ICU | S. aureus (58.7% were MDR), CoNS (28.3%), E. coli (72.7%), and C. freundii (20%) | 19 | 19 | 100 | Sterile cotton-tipped applicator sticks, moistened with sterile normal saline, were employed. | Colony morphology, Gram staining, and biochemical tests | Darge et al. 53 |
| ICU | Rotavirus A (RVA) and human adenovirus (HAdV) | 120 | 52 | 43 | A minimum of 50% of fomite surfaces were scraped using swabs | Genomic quantification and sequencing | Ganime et al. 45 | |
| Computer and its Parts | ICU | Rotavirus A (RVA) and human adenovirus (HAdV) | 60 | 22 | 37 | A minimum of 50% of fomite surfaces were scraped using swabs. | Genomic quantification and sequencing | Ganime et al. 45 |
| ICU | MRSA, Enterobacter, and Enterococcus | 80 | 19 | 24 | Rolling of saline-moistened-sterile rayon-tipped swabs (Baxter Healthcare Corporation, Deerfield, Ill), on the entire surface being tested | VITEK system and API 20E, pulsed-field gel electrophoresis | Bures et al. 6 | |
| ICU | Enterococcus spp., S. aureus, Gram-negative rods, and moulds | 444 | 26 | 6 | A sterile rayon-tipped swab (D2-Tupfer; Heinz Herenz, Hamburg) moistened with sterile saline solution was used to sample the surfaces. | Microbiological testing | Hartmann et al. 67 | |
| Hand Sanitizer Dispensers | SICU | CoNS, S. aureus, Micrococcus spp., Bacillus spp., Diphtheroids, Aerobic Actinomycetes, non-lactose-fermenting non-enterics, and lactose-fermenting enterics | 17 | 17 | 100 | Sterile cotton-tipped swabs moistened with sterile saline | Biochemical test and Vitek 2 | Eiref et al. 55 |
| ICU | Human adenoviruses | 14 | 8 | 57 | A minimum of 50% of fomite surfaces were scraped using swabs | Genomic quantification and sequencing | Ganime et al. 44 | |
| Stethoscopes | ICU | CoNS, S. aureus, K. pneumoniae, E. coli, C. freundii, E. aerogenes, and P. vulgaris | 61 | 61 | 100 | Sterile cotton-tipped applicator sticks, moistened with sterile normal saline, were employed. | Colony morphology, Gram staining, and biochemical tests | Darge et al. 53 |
| ICU | Normal flora, A. iwoffi, A. baumannii, MRSA, and Acinetobacter radioresistens | 46 | 15 | 33 | Swabbing of diaphragm and bell of stethoscopes using sterile cotton bud moistened with sterile saline | Standard protocols | Whittington et al. 65 | |
| ICU | CoNS | 18 | 18 | 100 | Cotton swab moistened with sterile normal saline | Standard microbiological methods and biochemical tests | Sued et al. 19 | |
| Identity Badges and Common Access Cards (CACs) | BICU | CoNS, Micrococcus spp., Gram-positive rods, S. aureus, Stomatococcus spp., and Streptococcus viridans | 118 | 89 | 75 | Swabbing | Standard microbiological methods | Caldwell et al. 50 |
| Door Knobs and Handles | ICU | Human Adenovirus | 21 | 8 | 38 | A minimum of 50% of fomite surfaces were scraped using swabs. | Genomic quantification and sequencing | Ganime et al. 44 |
| ICU | S. aureus (20.6% MDR of total S. aureus isolates) | 36 | 6 | 17 | Swabbing | PCR and standard microbiological methods | Veloso et al. 46 | |
| Infusion Pumps | ICU | S. aureus and moulds | 214 | 2 | 1 | A sterile rayon-tipped swab (D2-Tupfer; Heinz Herenz, Hamburg) moistened with sterile saline solution was used to sample the surfaces. | Microbiological testing | Hartmann et al. 67 |
| Ventilators | ICU | Enterococcus spp., S. aureus, Gram-negative rods, and moulds | 222 | 8 | 4 | A sterile rayon-tipped swab (D2-Tupfer; Heinz Herenz, Hamburg) moistened with sterile saline solution was used to sample the surfaces. | Microbiological testing | Hartmann et al. 67 |
| Medical Charts (Records Books) | ICU | CoNS, S. aureus (MRSA), E. faecalis, Streptococcus viridans, A. baumannii, Corynebacterium spp., Bacillus spp., E. coli, S. paucimobilis, MRSA, P. aeruginosa, Pantoea spp., and K. pneumoniae | 422 | 272 | 64 | Swabbing | Microbiological, biochemical laboratory techniques, and automated methods | Chen et al. 51 |
| Patient Files | ICU | P. aeruginosa (32.3% were MDR), MRSA, S epidermidis, K. pneumoniae (14.7% were MDR), A. baumannii (13.7% were MDR), and S. marcesens (0.9% were MDR) | 102 | 87 | 85 | Swabbing | Biochemical tests, API 20E, and API 20 NE | Panhotra et al. 63 |
| Accompanying Arm Chairs | ICU | Rotavirus A (RVA) and human adenovirus (HAdV) | 96 | 49 | 51 | A minimum of 50% of fomite surfaces were scraped using swabs. | Genomic quantification and sequencing | Ganime et al. 45 |
| Cardiac Monitor Keyboards | NICU | Rotavirus A (RVA) and human adenovirus (HAdV) | 36 | 4 | 11 | A minimum of 50% of fomite surfaces were scraped using swabs. | Genomic quantification and sequencing | Ganime et al. 45 |
| Sink, Outlet and Drains | ICU | MDR P. aeruginosa | 76 | 37 | 49 | Swabbing | Biochemical tests and matrix-assisted laser desorption/ ionization time-of-flight (MALDI-TOF) mass spectrometry | De Jonge et al. 54 |
| ICU | S. maltophilia | 12 | 3 | 25 | Swabs and preflush water | Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry | Guyot et al. 42 | |
| NICU | P. aeruginosa | 28 | 25 | 89 | Swabbing | Culture, qPCR | Bédard et al., 41 | |
| ICU | S. aureus (20.6% were MDR) | 36 | 8 | 22 | Swabbing | PCR and standard microbiological methods | Veloso et al. 46 | |
| Tap water | ICU | P. aeruginosa | 233 | 81 | 35 | Collection of first 250 mL of flush of water | API20 NE identification system | Coppry et al 52 |
| NICU | P. aeruginosa | 28 | 14 | 50 | Swab and first-flush cold water in sterile polypropylene bottle | qPCR | Bédard et al., 41 | |
| Companion Chairs | ICU | Human adenoviruses | 19 | 5 | 26 | A minimum of 50% of fomite surfaces were scraped using swabs. | Genomic quantification and sequencing | Ganime et al. 44 |
Abbreviations: MDR, multidrug-resistant; MRSA, methicillin-resistant Staphylococcus aureus; VRE, vancomycin-resistant Enterococcus; ESBL, extended-spectrum beta-lactamase; CoNS, coagulase-negative Staphylococci; ICU, intensive care unit; NICU, neonatal intensive care unit; BICU, burns intensive care unit; SICU, surgical intensive care unit; PCR, polymerase chain reaction; DNA, deoxyribonucleic acid.
Sampled surfaces
The sampling of surfaces designated as fomites or potential fomites varied extensively across the different studies. Furthermore, the scope presented diversity in terms of the inclusion of various types of inanimate surfaces during the sampling process. Among the pool of inanimate surfaces sampled, parenteral nutrition (PN), 71 pulse oximetry, 72 and curtains 73 had relatively lower sample sizes of less than 10, and were, therefore, excluded from further analysis in order to ensure a higher statistical power of this study. 74 Overall, of the 29 fomites, the 3 most sampled in the ICU included mobile phones,47 -49,56,57,59-62,68 sinks,41,42,46,54 and faucets6,23,41 (Table 1).
Laboratory methods
Culture analysis was used in all the studies, except for those of Ganime et al. 44 and Ganime et al. 45 who employed molecular techniques. All the selected studies utilized moisturized swabs for collecting samples on single surfaces, except Gonçalves et al. 66 who used the sedimentation plate method and other means32,52,58,66 (Table 1). With regard to pre-culture media, nine studies used broths such as Letheen broth, 64 thioglycollate broth,6,32 trypticase soy broth51,60,tryptone soya broth, 58 and brain heart infusion broth. De Jonge et al. 54 used an unspecified selective broth in their study. Most of the studies commonly used culture media such as MacConkey agar, mannitol salt agar, blood agar, and chocolate agar, but Sued et al. 19 and Chen et al. 51 did not clearly indicate the media used. Organisms other than bacteria were cultured via different means such as Vero cells, Dulbecco’s modified eagle’s medium, and cysteine-lactose electrolyte-deficient (CLED) plates42,44,45,47 (Table 2). Reported incubation temperatures ranged from 18 °C to 38 °C for 15 hours to 168 hours. However, the majority of the studies had their incubation temperatures ranging between 35 °C and 38 °C. Four studies42,44,45,51 did not report on incubation time and temperature (Table 2).
Table 2.
Details of methodologies and geographical location of the individual studies.
| Articles | Media Used | Broth Used | Incubation Temperature and Duration | Country |
|---|---|---|---|---|
| Al-Beeshi et al. 48 | Sheep blood agar and MacConkey agar | NA | 37 °C for 48 h | Saudi Arabia |
| Anupriya et al. 49 | Nutrient agar, blood agar and MacConkey’s agar | NA | 37 °C for 24 h | India |
| Bédard et al. 41 | Reasoner’s 2A agar | NA | 22 °C for 24-48h | Canada |
| Brown & Willms et al. 32 | Thioglycollate broth, blood agar, chocolate agar, colistin-nalidixic acid blood agar, and MacConkey agar | Thioglycollate broth | 35 °C to 37 °C for 24, 48, and 72 h | United States |
| Bures et al. 6 | Trypticase soy agar, blood agar MacConkey agar, and Columbia colistin-nalidixic acid agar | Thioglycollate broth | 37 °C for 48 h | United States |
| Caldwell et al. 50 | Trypticase soy agar with 5% sheep blood (Beckton, Dickinson, and Company), MacConkey II agar, and fluid thioglycollate medium | NA | 35 to 37 °C for up to 48 h | United States |
| Chen et al. 51 | Sheep blood agar and eosin-methylene blue agar | Trypticase soy broth | Not stated | Taiwan |
| Coppry et al. 52 | Cetrimide agar plates | NA | 37 °C for 24 and 48 h | France |
| Darge et al. 53 | Blood agar, MacConkey agar, and mannitol salt agar | NA | 37 °C for 24 h | Ethiopia |
| De Jonge et al. 54 | MacConkey agar | Selective broth; not specified | 38 °C for 15-18 h | Netherland |
| Espinoza et al. 47 | Vero cells | NA | NA | Brazil |
| Eiref et al. 55 | Trypticase soy agar and blood agar | NA | 35 °C for 24-72 h | United States |
| Galazzi et al. 56 | Brain heart infusion agar plus 5% sheep blood | NA | 35 ± 2 °C for 48 h | Italy |
| Ganime et al. 44 | Dulbecco’s modified eagle’s medium | NA | NA | Brazil |
| Ganime et al. 45 | Dulbecco’s modified eagle’s medium | NA | NA | Brazil |
| Gonçalves et al. 66 | Trypticase soy agar and Chapman agar | NA | 25 °C for 7 d (168 h) | Brazil |
| Guyot et al., 42 | Cysteine-lactoseelectrolyte-deficient (CLED) agar | NA | NA | United Kingdom |
| Hartmann et al. 67 | Blood agar | NA | 36 °C for 48 h | Germany |
| Heyba et al. 57 | Blood agar and chocolate agar | NA | 37 °C for 48 h | Kuwait |
| Hu et al., 58 | Horse blood agar | Tryptone soya broth | 37 °C-37 °C for 18-48h | United Kingdom |
| Kotris et al. 59 | Blood agar | NA | 35 °C ± 2 for 18-24 h | Croatia |
| Loyola et al. 60 | MacConkey agar | 3 mL trypticase soy broth | 35 °C for 18-24 h | Peru |
| Loyola et al. 61 | MacConkey agar, mannitol salt agar, and blood agar | Trypticase soy broth | 18-24 h at 35°C | Peru |
| Macrae et al. 43 | MacConkey agar agar | NA | Not stated | United Kingdom |
| Nwankwo et al. 62 | MacConkey and blood agar | NA | 37 °C for 18-24 h | Nigeria |
| Pilonetto et al. 64 | Letheen broth, MacConkey agar, XLD agar, cetrimide agar, and mannitol salt agar | Letheen Broth | 35 °C for 48 h | Brazil |
| Panhotra et al. 63 | Blood agar and MacConkey agar | NA | 37 °C for 48 h | Saudi Arabia |
| Sued et al. 19 | 5% sheep blood agar and Mueller Hinton agar | NA | 35 °C for 48 h | Brazil |
| Ulger et al. 68 | Blood agar supplemented with 5% defibrinated sheep blood and eosin methylene blue agar | NA | 37 °C for 48 h | Turkey |
| Veloso et al. 46 | Mannitol salt agar | Brain Heart Infusion broth | 35-37 °C for 24 and 48 h | Brazil |
| Wang et al. 23 | Sheep blood agar | NA | 37 °C for 3-5 d | Taiwan |
| Wolfe et al. 35 | 10% sheep blood agar | NA | 35 °C for 24 h and 48 h | United States |
| Whittington et al. 65 | Blood agar and MacConkey agar | NA | 37 °C for 24 h | United Kingdom |
Abbreviation: NA, not available.
The method for bacterial identification varied across the studies. All the articles reported standard microbiological techniques, including colony morphology, Gram stain reaction, microscopic morphology, biochemical reactions, molecular microbial methods, and modern automated identification techniques such as the MALDI-TOF mass spectrometry in the identification of microbes (Table 1). The most common identification methods included biochemical tests and conventional automated identification machines. Metagenomic analysis was also adopted in identifying and further analyzing the processed samples from the fomites.19,23,41 -47,58,61
Frequency of contamination and microbial presence
A significant number of the fomites showed contamination of more than 40% across all studies. However, contamination frequency varied from study to study. Interestingly, certain surfaces displayed a higher percentage of contamination despite being sampled less frequently. For instance, the sphygmomanometer, although subjected to lower sampling frequency, exhibited a 100% detection rate of organisms commonly associated with nosocomial infections, including E. aerogenes, S. aureus, CoNS, E. coli, and K. pneumoniae.19,53 This was also the case for the thermometers19,43, Yankauer catheters and suction machines, 32 and mattresses and pillows 58 which showed a higher percentage of contamination with nosocomial pathogens. Mobile phones emerged as the fomite with the most extensive body of research, garnering significant attention in numerous studies. Notably, the sampling frequency of mobile phones within each study was consistently high and it equally yielded a significant load of nosocomial pathogens, revealing its substantial capacity to harbour nosocomial pathogens47 -49,56,57,59-62,68 (Table 1).
Microorganisms that have the potential of causing nosocomial infections were isolated on various surfaces in the ICU, with the NICU emerging as the predominant unit. Data were presented as a percentage of positive sampling based on the frequency of positive results from the number of surfaces sampled in Table 1. The examined literature demonstrated a high prevalence of nosocomial pathogens, particularly for CoNS, S. aureus, and MRSA, in the ICUs (Tables 1 and 3). Though CoNS are considered normal flora in healthy individuals, S. epidermidis and S. haemolyticus (the most common species in CoNS) are common causes of infections associated with invasive procedures, indwelling devices or implanted foreign bodies, and among the immunocompromised. Infections from these pathogens include bacteraemia, urethritis, and endocarditis, among others. 75
Table 3.
Studies that reported the total number of isolates and the number of various bacteria isolated.
| Studies that reported the most prevalent isolates | TOTAL NUMBER OF ISOLATES | Staphylococcus spp. (CoNS & MRSA) | Bacillus spp. | A. Baumannii | E. Coli | P. aeruginosa | Klebsiella spp. | Streptococcus spp. | Others (Proteus mirabilis, S. paucimobili, Candida, Yeast, Achromobacter, C. meningosepticum,Serratia Rubedia, C. freundii, other Enterobacter |
|---|---|---|---|---|---|---|---|---|---|
| Al-Beeshi et al. 48 | 159 | 104 | 9 | NA | NA | NA | NA | 6 | 40 |
| Brown & Willms et al. 32 | 25 | 9 | NA | NA | NA | NA | NA | NA | 9 |
| Bures et al. 6 | 33 | 16 | NA | NA | NA | NA | NA | NA | 8 |
| Chen et al. 51 | 409 | 245 | 17 | 14 | 23 | 6 | 9 | 20 | 52 |
| Darge et al. 53 | 171 | 93 | NA | NA | NA | NA | 10 | NA | 13 |
| Loyola et al. 60 | 105 | NA | NA | NA | 105 | NA | 23 | NA | 48 |
| Nwankwo et al. 62 | 97 | 38 | 18 | 3 | 8 | 11 | 4 | 8 | 7 |
| Panhotra et al. 63 | 87 | 24 | NA | 14 | NA | 33 | 15 | NA | 1 |
| Pilonetto et al. 64 | 18 | 11 | NA | 2 | NA | NA | 2 | NA | 3 |
| Wang et al. 23 | 66 | NA | NA | NA | NA | 14 | NA | 47 | |
| Ulger et al. 68 | 307 | 231 | NA | NA | NA | NA | NA | 12 | 64 |
| Wolfe et al. 35 | 20 | 17 | NA | NA | NA | NA | NA | NA | NA |
Abbreviations: NA, not available.
All the ESKAPE pathogens, including Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp., were encountered on fomites such as patient files, medical charts (records books), stethoscopes, sphygmomanometers, bedside tables, and many more fomites (Table 1). These ESKAPE pathogens are a group of bacteria that have the ability to “escape” the effects of commonly used antibiotics, posing a significant challenge to healthcare systems worldwide. Other organisms commonly associated with nosocomial infections detected in various ICUs included E. coli, Candida sp., Enterococcus sp., S. haemolyticus, and Pantoea spp. (Tables 1 and 3).
Nosocomial pathogens and their prevalence on the fomites
Although nosocomial pathogens were identified across all studies, a notable presence of specific pathogens was consistently associated with certain fomites, suggesting a potential role of these fomites as reservoirs within the ICU. Notably, S. aureus was found on the majority of the fomites examined, emphasizing its widespread distribution. Additionally, Klebsiella spp. and P. aeruginosa were particularly prominent in sink and taps samples. Alongside Staphylococcus sp., P. aeruginosa, and K. pneumoniae were frequently associated with mobile phones, reinforcing their significance as potential carriers of these organisms. Furthermore, P. aeruginosa exhibited a common association with sink outlets, faucets, and aerators. Nosocomial pathogens were widely isolated from Yankauer catheters and suction machines in one study. A number of viral and fungal isolates were reported in some studies, but in relatively lower amounts compared with bacteria. Mobile phones, 47 bedside tables, 45 bed rails, 45 companion chairs, 44 and incubators and its door locks 45 harboured viruses and in relatively fewer amounts. Twelve studies6,23,32,35,48,51,53,60,62 -64,68 further provided the total number of bacteria isolates, making it possible to determine the most prevalent contaminants of fomites in the ICU. Staphylococcus sp., comprising mainly CoNS, S. aureus, and MRSA were predominant, and were detected in all the 12 studies, except two23,60 (Table 3). Further, P. aeruginosa and Klebsiella sp. were randomly isolated across all studies in moderate amounts.
In the studies that reported on antimicrobial susceptibility, mattress and pillow appeared to harbour multiple drug-resistant bacteria such as MRSA, VRE, and ESBL producers. 58 Similarly, bedside table, which is often proximal to patients, appeared to harbour numerous drug-resistant bacteria, showing 100% contamination in the study of Darge et al. 53 . Mobile phones also randomly harboured ESBL-producing E. coli, ESBL-producing Enterobacter sp, and ESBL-producing K. pneumoniae. 60 The sink, outlet, and drain were contaminated with MDR P. aeruginosa. 53
Discussion
In recent years, a plethora of evidence has emerged regarding the colonization of nosocomial pathogens on inanimate surfaces within hospital settings.76,77 The literature further provides compelling evidence that microorganisms present in the healthcare environment are a source of nosocomial infections. This corresponds to the fact that patients in ICUs are vulnerable to fomite-associated nosocomial infections and, thus, necessitates the need to frequently evaluate fomites in critical care units.78,79 A comprehensive review that quantifies the prevalence of these pathogens within the ICU is yet to be conducted. Consequently, we undertook this systematic review to address that significant gap in literature. In this present study, we generally observed mobile phones, aerators and faucets, the stethoscope, and the sphygmomanometer to be the most potentially contaminated fomites in the ICU. The prevalence of the different nosocomial pathogens in the overall samples varied greatly, but S. aureus led the charts. The microbiological methods employed by all the studies for sampling and microbial identification in the ICU are capable of effectively recovering microbes from fomites. It is worth mentioning that the utilization of modern techniques, such as MALDI-TOF, would have been more efficient in recovering and identifying isolates at the species level in studies that relied only on biochemical tests. 80
We observed that some studies focused on specific organisms and utilized techniques tailored to isolate only those targeted organisms. The fact that 40% of the total samples collected from fomites yielded high positive cultures in this regard suggests that the occurrence of positive cultures on fomites in the ICU would surpass 40% if all nosocomial pathogens were targeted for recovery in all the studies. The generally high prevalence of nosocomial pathogens in ICUs reported here is consistent with other findings.81,82 The observed variation of nosocomial pathogens on the fomites is also comparable with those in the reports of Abubakar et al. 81 and Bhatta et al. 83 which noted variable pathogens in ICU and other hospital settings, respectively. Such variations could be attributed to the fact that these pathogens persist under different conditions such as temperature, humidity, and the characteristics of the fomite they contaminate. Some microbes, such as Acinetobacter sp., are capable of surviving on both dry and wet surfaces for a long period of time (several weeks) in a wide range of temperatures and pH. A study by Kramer et al. 36 and others 84 reported that Gram-positive bacteria, such as Enterococcus spp. (including VRE strains), S. aureus (including MRSA strains), and S. pyogenes survive for months on dry surfaces. The authors further found that Gram-negative species, such as Acinetobacter sp., E. coli, Klebsiella sp., P. aeruginosa, Serratia marcescens, and Shigella sp., can thrive on inanimate surfaces over months. In another study that aimed at determining the longevity of pathogens on objects made of cotton, wool, silk, and cotton-polyester, S. aureus, E. coli, P. aeruginosa, and A. baumannii persisted for weeks. 85
In this present study, bacteria dominated on the studied fomites; only a few fomites harboured viral and fungal organisms. Viral infections have been associated with many infectious outbreaks, 86 but often receive less attention and are somewhat overlooked compared to bacteria, despite their significant impact. 87 Less frequent groups of organisms like Candida, although rare, have a high mortality rate among immunocompromised patients, 88 and most drugs for their treatment have significant side effects. 89
In 2019, the World Health Organization recognized 6 pathogens as significant in nosocomial infections: P. aeruginosa, A. baumannii, E. coli, S. pneumoniae, K. pneumoniae, and S. aureus.48,90,91 At least, one or more of these nosocomial pathogens have been isolated from at least one fomite, although there seems to be no record regarding their isolation from either of companion chairs, accompanying armchairs, incubators and its door locks, and cardiac monitor keyboards. The alarming distribution of these nosocomial pathogens across the fomites in this study is consistent with the findings of Muhammad et al. 82
Out of the 29 fomites identified in this study, S. aureus, the most commonly isolated organism, was present on 19. Furthermore, studies that reported the number of isolates show that Staphylococcus sp., comprising mainly CoNS and MRSA, are the most predominant across all reported studies, except in the case of Loyola et al. 60 and Wang et al. 23 This observation is similar to those of a number of studies4,81,90,91 focusing on ambulances and other parts of hospital settings. Some fomites were constantly 100% contaminated across all studies, as observed in regard to the sphygmomanometer and stethoscope. These are instruments commonly used in measuring blood pressure and listening to internal sounds of patients’ bodies in hospital settings, and as a result, are highly exposed to multiple contacts between clinicians and patients.92,93 Sphygmomanometers recorded 100% contamination in two studies and the isolates were MDR bacteria associated with nosocomial infections, especially among immunocompromised patients.43,94 The sphygmomanometer has several parts, but a notable part capable of harbouring organisms is the cuff, whose physical features make it might be highly conducive to harbouring microbes. The cuff is usually in direct contact with patients and often rubs around their upper arm. 95 The high contamination rate in this present study corroborates the findings of Zargaran et al. 96 who reported a 85% contamination rate of sphygmomanometer cuffs in clinical settings. We also observed that the stethoscope harboured Staphylococcal species, such as S. aureus (MRSA) and CoNS, in all the studies alongside other nosocomial pathogens such as E. coli and A. baumannii. Some fomites, such as taps, persistently harboured P. aeruginosa, which is reported to be effective in biofilm formation, an attribute that enhances their longevity in water and moist surfaces (including the surface of soaps and in liquid soap).13,97,98
ESKAPE pathogens, which are known for their ability to “escape” the effects of commonly used antibiotics were commonly distributed on many fomites. A considerable number of drug-resistant bacteria were reported on several fomites. Mattresses and pillows, which are in direct contact with hospitalized patients, tend to be contaminated with pathogens such as MRSA, VRE, and ESBL producers. All these pathogens have been previously reported in outbreaks in ICUs.99 -101 As a result, mattresses and pillows may be involved in the cross-transmission of pathogens among critically ill patients.102,103 Furthermore, mobile phones appeared to be a potential reservoir of MDR nosocomial pathogens, such as ESBL-producing Enterobacter sp. and ESBL-producing Klebsiella sp. This report aligns with the predictions and findings of Tekerekoğlu et al. 104 and Olsen et al., 105 but contradicts the findings of Muhammad et al 82 who reported no MDR pathogens on mobile phones in hospitals. The absence of MDR pathogens in Muhammad et al.'s 82 report could be attributed to their strict reporting on only the healthcare workers’ mobile phones. The detection and reports of MDR pathogens associated with nosocomial infections on these commonly used inanimate surfaces in the proximity of immunocompromised patients need prompt attention.
One goal of this study was to report fomite contamination based on the specific ICU types, but this information was available for only seven fomites. Among these fomites, the NICU recorded both the highest prevalence of nosocomial pathogens and the largest number of fomites, encompassing six distinct inanimate surfaces: thermometers, PN, pulse oximetry, faucets and aerators, cardiac monitor keyboard, sink, and tap water. The high prevalence of nosocomial pathogens and the abundance of fomites in the NICU pose significant risks to neonates, especially considering their vulnerable and still-developing immune systems. These factors can potentially compromise the health and well-being of these fragile infants. Clostridium difficile is also a common nosocomial pathogen reportedly associated with numerous hospital-acquired infections. It was anticipated to be reported due to its recognized longevity and perseverance, with the ability to survive for extended periods on inanimate surfaces. 106 However, none of the studies reported here identified C. difficile contamination on any of the fomites in the ICU.
Given the challenges associated with isolating or recovering certain clinically relevant organisms, such as viruses and C. difficile, metagenomic analysis emerges as a valuable tool for investigating and characterizing microbial communities present on inanimate surfaces in the ICU. In this present study, the articles that implemented metagenomics in identifying bacteria observed a remarkable organism recovery from the fomites involved which may have escaped traditional microbiological methods.19,41,43 -45 Specifically, recovering viruses for microbiological study presents a significant challenge; however, metagenomic analysis yielded positive results in recovering SARS-CoV-2 and human adenoviruses on fomites.44,45,47
The use of this approach further provided insights into the diversity, abundance, and potential pathogenicity of bacteria found on these fomites and helped in tracing a potential source of bacterial outbreak by analyzing the genomics of the samples on potential fomites and the clinical isolate.42,43,46 Likewise, through genome analysis, Wang et al. 23 found C. meningosepticum from faucet cultures to be similar to the C. meningosepticum isolates recovered from four different patients located in different units. Similarly, Hu et al. 58 who performed metagenomics and subsequent phylogenetic analysis, revealed that, closely related microbiomes contaminate similar categories of fomites. Tracking the presence of antibiotic resistance genes in bacteria is necessary to deduce appropriate measures to avoid their spread via horizontal or vertical gene transfer107 -112 or other factors of interest; ESBL-producing bacteria isolated from mobile phones of healthcare workers were found to harbour bla genes which have long been linked to antibiotic resistance. 61
The identification of these nosocomial pathogens in the ICUs is a pressing issue that demands swift action. Interventions such as the use of copper-silver alloy coats (which possess antibacterial activity) on commonly touched surfaces, such as tap handles and door handles, could be employed.113 -116 It is also important to develop suitable disinfection methods that will help to reduce or eliminate nosocomial agents in ICUs. Several research studies have reported the use of disinfectants, UV irradiation, and phages to curb infections as a result of contaminated fomites.117 -120 Furthermore, the efficacy of disinfection of fomites in the ICU depends on several factors, including concentration of disinfectants, fomite pathogenic load, the frequency of disinfection, and the type of pathogens present on the fomites.121 -123 Hence, efficient methods for disinfection and elimination of nosocomial pathogens in ICU fomites are needed, especially those that can remediate against a broad range of resistant nosocomial pathogens.
This systematic review had some limitations, including the difference in study periods across all the articles analyzed. Some studies were conducted during or after an outbreak, and some only targeted organisms that matched the interests of the invstigators, and these may have introduced unintended bias in the results. Moreover, identification methods varied greatly, as some studies used highly sensitive methods in recovering and identifying organisms while others employed relatively fewer sensitive methods. Consequently, some isolates could not be identified and reported on to the species level. Also, the varying microbiological methods across the studies could not allow for an extensive meta-analysis.
Conclusion
Many fomites that are readily used in patient care in the ICU carry nosocomial pathogens. The most common fomite appeared to be mobile phones, sphygmomanometers, and stethoscope, while the most common organism harboured was Staphylococcus. Hence, the need for rigorous disinfection and sterilization protocols on fomites in the ICU cannot be overemphasized. Additionally, heightened awareness on the subject among health professionals is crucial to mitigating the risk and burden of nosocomial infections caused by drug-resistant bacteria.
Footnotes
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This systematic review was supported by the Fogarty International Center of the National Institutes of Health, USA, through the Research and Capacity Building in Antimicrobial Resistance in West Africa (RECABAW) Training Programme hosted at the Department of Medical Microbiology, University of Ghana Medical School (Award Number: D43TW012487). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
ORCID iDs: Samuel Darkwah  https://orcid.org/0000-0003-0868-1798
https://orcid.org/0000-0003-0868-1798
Fleischer C N Kotey  https://orcid.org/0000-0003-0286-3638
https://orcid.org/0000-0003-0286-3638
Alex Odoom  https://orcid.org/0000-0001-5761-1564
https://orcid.org/0000-0001-5761-1564
Prince Hotor  https://orcid.org/0000-0001-9529-2728
https://orcid.org/0000-0001-9529-2728
Nicholas T K D Dayie  https://orcid.org/0000-0003-4491-6902
https://orcid.org/0000-0003-4491-6902
Eric S Donkor  https://orcid.org/0000-0002-5179-546X
https://orcid.org/0000-0002-5179-546X
References
- 1. Bereket W, Hemalatha K, Getenet B, et al. Update on bacterial nosocomial infections. Eur Rev Med Pharmacol Sci. 2012;16:1039-1044. [PubMed] [Google Scholar]
- 2. Edwardson S, Cairns C. Nosocomial infections in the ICU. Anaesth Intensive Care Med. 2019;20(1):14-18. [Google Scholar]
- 3. Khan HA, Ahmad A, Mehboob R. Nosocomial infections and their control strategies. Asian Pac J Trop Biomed. 2015;5:509-514. [Google Scholar]
- 4. Raoofi S, Pashazadeh Kan F, Rafiei S, et al. Global prevalence of nosocomial infection: A systematic review and meta-analysis. PLoS One. 2023;18:e0274248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kumar S, Sen P, Gaind R, et al. Prospective surveillance of device-associated health care–associated infection in an intensive care unit of a tertiary care hospital in New Delhi, India. Am J Infect Control. 2018;46:202-206. [DOI] [PubMed] [Google Scholar]
- 6. Bures S, Fishbain JT, Uyehara CF, Parker JM, Berg BW. Computer keyboards and faucet handles as reservoirs of nosocomial pathogens in the intensive care unit. Am J Infect Control. 2000;28:465-471. [DOI] [PubMed] [Google Scholar]
- 7. Wang M, Wei H, Zhao Y, et al. Analysis of multidrug-resistant bacteria in 3223 patients with hospital-acquired infections (HAI) from a tertiary general hospital in China. Bosn J Basic Med Sci. 2019;19:86-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gozel MG, Hekimoglu CH, Gozel EY, et al. National Infection Control Program in Turkey: the healthcare associated infection rate experiences over 10 years. Am J Infect Control. 2021;49:885-892. [DOI] [PubMed] [Google Scholar]
- 9. Aiesh BM, Qashou R, Shemmessian G, et al. Nosocomial infections in the surgical intensive care unit: an observational retrospective study from a large tertiary hospital in Palestine. BMC Infect Dis. 2023;23:686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mbim E, Mboto C, Agbo B. A review of nosocomial infections in Sub-Saharan Africa. Br Microbiol Res J. 2016;15:1-11. [Google Scholar]
- 11. Sikora A, Zahra F. Nosocomial infections. StatPearls. StatPearls Publishing; 2023. Accessed May 31, 2023. http://www.ncbi.nlm.nih.gov/books/NBK559312/. [PubMed] [Google Scholar]
- 12. Beggs C, Knibbs LD, Johnson GR, Morawska L. Environmental contamination and hospital-acquired infection: factors that are easily overlooked. Indoor Air. 2015;25:462-474. [DOI] [PubMed] [Google Scholar]
- 13. Yapicioglu H, Gokmen TG, Yildizdas D, et al. Pseudomonas aeruginosa infections due to electronic faucets in a neonatal intensive care unit. J Paediatr Child Health. 2012;48:430-434. [DOI] [PubMed] [Google Scholar]
- 14. Castaño N, Cordts SC, Kurosu Jalil M, et al. Fomite transmission, physicochemical origin of virus-surface interactions, and disinfection strategies for enveloped viruses with applications to sars-CoV-2. ACS Omega. 2021;6:6509-6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Donkor ES, S. Anyen NE, Akumwena A. Making a case for infection control at public places of convenience in Accra, Ghana. Environ Health Insights. 2020;14:1178630220938414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maryam A, Hadiza U, Aminu U. Characterization and determination of antibiotic susceptibility pattern of bacteria isolated from some fomites in a teaching hospital in northern Nigeria. Afr J Microbiol Res. 2014;8:814-818. [Google Scholar]
- 17. Newman MJ. Neonatal intensive care unit: reservoirs of nosocomial pathogens. West Afr J Med. 2002;21:310-312. [DOI] [PubMed] [Google Scholar]
- 18. Paul LM, Hegde A, Pai T, et al. An outbreak of Burkholderia cepacia bacteremia in a neonatal Intensive Care Unit. Indian J Pediatr. 2016;83:285-288. [DOI] [PubMed] [Google Scholar]
- 19. Sued BP, Pereira PM, Faria YV, et al. Sphygmomanometers and thermometers as potential fomites of Staphylococcus haemolyticus: biofilm formation in the presence of antibiotics. Mem Inst Oswaldo Cruz. 2017;112:188-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. O'Flaherty N, Fenelon L. The stethoscope and healthcare-associated infection: a snake in the grass or innocent bystander? J Hosp Infect. 2015;91:1-7. [DOI] [PubMed] [Google Scholar]
- 21. Cohen R, Babushkin F, Shimoni Z, et al. Water faucets as a source of Pseudomonas aeruginosa infection and colonization in neonatal and adult intensive care unit patients. Am J Infect Control. 2017;45:206-209. [DOI] [PubMed] [Google Scholar]
- 22. Voss A, Verweij PE. Faucet aerators: a source of patient colonization with Stenotrophomonas maltophilia. Am J Infect Control. 1999;27:459-460. [DOI] [PubMed] [Google Scholar]
- 23. Wang JL, Chen ML, Lin YE, Chang SC, Chen YC. Association between contaminated faucets and colonization or infection by nonfermenting Gram-negative bacteria in intensive care units in Taiwan. J Clin Microbiol. 2009;47:3226-3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xiang Q, Lv Y, Jin Y, et al. Faucet aerators as a reservoir for carbapenem-resistant Acinetobacter baumannii: a healthcare-associated infection outbreak in a neurosurgical intensive care unit. Antimicrob Resist Infect Control. 2019;8:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Braine T. Race against time to develop new antibiotics. Bull World Health Organ. 2011;89:88-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. da Costa PM, Loureiro L, Matos AJ. Transfer of multidrug-resistant bacteria between intermingled ecological niches: the interface between humans, animals and the environment. Int J Environ Res Public Health. 2013;10:278-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. El Chakhtoura NG, Saade E, Iovleva A, et al. Therapies for multidrug resistant and extensively drug-resistant non-fermenting gram-negative bacteria causing nosocomial infections: a perilous journey toward ‘molecularly targeted' therapy. Expert Rev Anti-Infect Ther. 2018;16:89-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Horcajada JP, Montero M, Oliver A, et al. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin Microbiol Rev. 2019;32:e00031-NaN19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van Duin D, Paterson DL. Multidrug-resistant bacteria in the community: trends and lessons learned. Infect Dis Clin North Am. 2016;30:377-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kotey FC, Awugah SA, Dayie NT, et al. High prevalence of methicillin-resistant Staphylococcus aureus carriage among infants at the Children’s Hospital, Accra, Ghana. J Infect Dev Ctries. 2022;16:1450-1457. [DOI] [PubMed] [Google Scholar]
- 31. Donkor ES, Kotey FC. Methicillin-resistant Staphylococcus aureus in the oral cavity: Implications for antibiotic prophylaxis and surveillance. Infect Dis Res Treat. 2020;13:1178633720976581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brown M, Willms D. Colonization of yankauer suction catheters with pathogenic organisms. Am J Infect Control. 2005;33:483-485. [DOI] [PubMed] [Google Scholar]
- 33. Opoku-Asare B, Boima V, Ganu VJ, et al. Catheter-related bloodstream infections among patients on maintenance haemodialysis: a cross-sectional study at a tertiary hospital in Ghana. BMC Infect Dis. 2023;23:664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Creamer E, Humphreys H. The contribution of beds to healthcare-associated infection: the importance of adequate decontamination. J Hosp Infect. 2008;69:8-23. [DOI] [PubMed] [Google Scholar]
- 35. Wolfe DF, Sinnett S, Vossler JL, Przepiora J, Engbretson BG. Bacterial colonization of respiratory therapists’ pens in the Intensive Care Unit. Respir Care. 2009;54:500-503. [PubMed] [Google Scholar]
- 36. Kramer A, Schwebke I, Kampf G. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect Dis. 2006;6:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Duszynska W, Rosenthal VD, Szczesny A, et al. Device associated -health care associated infections monitoring, prevention and cost assessment at intensive care unit of University Hospital in Poland (2015-2017). BMC Infect Dis. 2020;20:761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Berglund Kristiansson E, Källman U. Healthcare staff’s views on the patients’ prerequisites to be co-creator in preventing healthcare-associated infections. Scand J Caring Sci. 2020;34:314-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bjark P, Hansen E, Lingaas E. In-hospital deaths attributable to healthcare-associated infections. Tidsskr Den Nor Laegeforening Tidsskr Prakt Med Ny Raekke. 2020;140. [DOI] [PubMed] [Google Scholar]
- 40. Moher D, Liberati A, Tetzlaff J, Altman DG; PRISMA Group. Preferred Reporting Items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;151(4):264-269 [DOI] [PubMed] [Google Scholar]
- 41. Bédard E, Laferrière C, Charron D, et al. Post-outbreak investigation of Pseudomonas aeruginosa faucet contamination by quantitative polymerase chain reaction and environmental factors affecting positivity. Infect Control Hosp Epidemiol. 2015;36:1337-1343. [DOI] [PubMed] [Google Scholar]
- 42. Guyot A, Turton JF, Garner D. Outbreak of Stenotrophomonas maltophilia on an intensive care unit. J Hosp Infect. 2013;85:303-307. [DOI] [PubMed] [Google Scholar]
- 43. Macrae MB, Shannon KP, Rayner DM, et al. A simultaneous outbreak on a neonatal unit of two strains of multiply antibiotic resistant Klebsiella pneumoniae controllable only by ward closure. J Hosp Infect. 2001;49:183-192. [DOI] [PubMed] [Google Scholar]
- 44. Ganime AC, Carvalho-Costa FA, Santos M, et al. Viability of human adenovirus from hospital fomites. J Med Virol. 2014;86:2065-2069. [DOI] [PubMed] [Google Scholar]
- 45. Ganime AC, Leite JP, Figueiredo CE, et al. Dissemination of human adenoviruses and rotavirus species A on fomites of hospital pediatric units. Am J Infect Control. 2016;44:1411-1413. [DOI] [PubMed] [Google Scholar]
- 46. Veloso JO, Lamaro-Cardoso J, Neves LS, et al. Methicillin-resistant and vancomycin-intermediate Staphylococcus aureus colonizing patients and intensive care unit environment: virulence profile and genetic variability. Acta Pathol Microbiol Immunol Scand A. 2019;127:717-726. [DOI] [PubMed] [Google Scholar]
- 47. Espinoza EPS, Cortes MF, Noguera SV, et al. Are mobile phones part of the chain of transmission of SARS-CoV-2 in hospital settings? Rev Inst Med Trop Sao Paulo. 2021;63:e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Al-Beeshi NZ, Alohali RM, Torchyan AA, Somily AM. The bacterial colonization of healthcare workers’ mobile phones in a large tertiary care teaching hospital in Saudi Arabia. J Infect Dev Ctries. 2021;15:1314-1320. [DOI] [PubMed] [Google Scholar]
- 49. Anupriya A, Puhalenthi K, Keerthi S J, R P, V H. Microbial contamination of mobile phones in a teritary care hospital. Int J Community Med Public Health. 2018;5:2313. [Google Scholar]
- 50. Caldwell NW, Guymon CH, Aden JK, Akers KS, Mann-Salinas EA. Bacterial contamination of burn unit employee identity cards. J Burn Care Res. 2016;37:e470-e475. [DOI] [PubMed] [Google Scholar]
- 51. Chen KH, Chen LR, Wang YK. Contamination of medical charts: an important source of potential infection in hospitals. PLoS One. 2014;9;9(2):e78512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Coppry M, Leroyer C, Saly M, et al. Exogenous acquisition of Pseudomonas aeruginosa in intensive care units: a prospective multi-centre study (DYNAPYO study). J Hosp Infect. 2020;104:40-45. [DOI] [PubMed] [Google Scholar]
- 53. Darge A, Kahsay AG, Hailekiros H, Niguse S, Abdulkader M. Bacterial contamination and antimicrobial susceptibility patterns of intensive care units medical equipment and inanimate surfaces at Ayder comprehensive specialized hospital, Mekelle, northern Ethiopia. BMC Res Notes. 2019;12:621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. de Jonge E, de Boer MGJ, van Essen EHR, Dogterom-Ballering HCM, Veldkamp KE. Effects of a disinfection device on colonization of sink drains and patients during a prolonged outbreak of multidrug-resistant Pseudomonas aeruginosa in an intensive care unit. J Hosp Infect. 2019;102:70-74. [DOI] [PubMed] [Google Scholar]
- 55. Eiref SD, Leitman IM, Riley W. Hand sanitizer dispensers and associated hospital-acquired infections: friend or fomite? Surg Infect. 2012;13:137-140. [DOI] [PubMed] [Google Scholar]
- 56. Galazzi A, Panigada M, Broggi E. Microbiological colonization of healthcare workers’ mobile phones in a tertiary-level Italian intensive care unit. Intensive Crit Care Nurs. 2019;53:112-121. [DOI] [PubMed] [Google Scholar]
- 57. Heyba M, Ismaiel M, Alotaibi A, et al. Microbiological contamination of mobile phones of clinicians in intensive care units and neonatal care units in public hospitals in Kuwait. BMC Infect Dis. 2015;15:434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hu H, Johani K, Gosbell IB, et al. Intensive care unit environmental surfaces are contaminated by multidrug-resistant bacteria in biofilms: combined results of conventional culture, pyrosequencing, scanning electron microscopy, and confocal laser microscopy. J Hosp Infect. 2015;91:35-44. [DOI] [PubMed] [Google Scholar]
- 59. Kotris I, Drenjančević D, Talapko J, Bukovski S. Identification of microorganisms on mobile phones of intensive care unit health care workers and medical students in the tertiary hospital. Med Glas. 2017;14:85-90. [DOI] [PubMed] [Google Scholar]
- 60. Loyola S, Gutierrez LR, Horna G, et al. Extended-spectrum β-lactamase-producing Enterobacteriaceae in cell phones of health care workers from Peruvian pediatric and neonatal intensive care units. Am J Infect Control. 2016;44:910-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Loyola S, Gutierrez L, Avendaño E, Severino N, Tamariz J. Multidrug-resistant bacteria isolated from cell phones in five intensive care units: exploratory dispersion analysis. Germs. 2018;8:85-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nwankwo EO, Ekwunife N, Mofolorunsho KC. Nosocomial pathogens associated with the mobile phones of healthcare workers in a hospital in Anyigba, Kogi state, Nigeria. J Epidemiol Glob Health. 2013;4:135-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Panhotra BR, Saxena AK, Al-Mulhim AS. Contamination of patients’ files in intensive care units: an indication of strict handwashing after entering case notes. Am J Infect Control. 2005;33:398-401. [DOI] [PubMed] [Google Scholar]
- 64. Pilonetto M, Rosa EAR, Brofman PRS, et al. Hospital gowns as a vehicle for bacterial dissemination in an intensive care unit. Braz J Infect Dis. 2004;8:206-210. [DOI] [PubMed] [Google Scholar]
- 65. Whittington AM, Whitlow G, Hewson D, Thomas C, Brett SJ. Bacterial contamination of stethoscopes on the intensive care unit. Anaesthesia. 2009;64:620-624. [DOI] [PubMed] [Google Scholar]
- 66. Gonçalves CL, Mota FV, Ferreira GF, et al. Airborne fungi in an intensive care unit. Braz J Biol. 2017;78:265-270. [DOI] [PubMed] [Google Scholar]
- 67. Hartmann B, Benson M, Junger A, et al. Computer keyboard and mouse as a reservoir of pathogens in an intensive care unit. J Clin Monit Comput. 2003;18:7-12. [DOI] [PubMed] [Google Scholar]
- 68. Ulger F, Esen S, Dilek A, et al. Are we aware how contaminated our mobile phones with nosocomial pathogens? Ann Clin Microbiol Antimicrob. 2009;8:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Esteves DC, Pereira VC, Souza JM, et al. Influence of biological fluids in bacterial viability on different hospital surfaces and fomites. Am J Infect Control. 2016;44:311-314. [DOI] [PubMed] [Google Scholar]
- 70. Jabłońska-Trypuć A, Makuła M, Włodarczyk-Makuła M, et al. Inanimate surfaces as a source of hospital infections caused by fungi, bacteria and viruses with particular emphasis on SARS-CoV-2. Int J Environ Res Public Health. 2022;19:8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Arslan U, Erayman I, Kirdar S, et al. Serratia marcescens sepsis outbreak in a neonatal intensive care unit. Pediatr Int. 2010;52:208-212. [DOI] [PubMed] [Google Scholar]
- 72. Parer S, Lotthé A, Chardon P, et al. An outbreak of heterogeneous Glycopeptide-Intermediate Staphylococcus aureus related to a device source in an intensive care unit. Infect Control Hosp Epidemiol. 2012;33:167-174. [DOI] [PubMed] [Google Scholar]
- 73. Brown L, Siddiqui S, McMullen A, Waller J, Baer S. Revisiting the “leading edge” of hospital privacy curtains in the medical intensive care unit. Am J Infect Control. 2020;48:746-750. [DOI] [PubMed] [Google Scholar]
- 74. Prajapati B, Dunne M, Armstrong R. Sample size estimation and statistical power analyses. Optom Today. 2010;16:10-18. [Google Scholar]
- 75. Becker K, Heilmann C, Peters G. Coagulase-negative staphylococci. Clin Microbiol Rev. 2014;27:870-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Vink J, Edgeworth J, Bailey SL. Acquisition of MDR-GNB in hospital settings: a systematic review and meta-analysis focusing on ESBL-E. J Hosp Infect. 2020;106:419-428. [DOI] [PubMed] [Google Scholar]
- 77. Zahornacký O, Porubčin Rovňáková A, Jarčuška P. Gram-negative rods on inanimate surfaces of selected hospital facilities and their nosocomial significance. Int J Environ Res Public Health. 2022;19:6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Aedh AI, Al-Swedan AD, Mohammed AA, et al. Occurrence of multidrug-resistant strains of Acinetobacter spp.: an emerging threat for nosocomial-borne infection in Najran Region, KSA. Trop Med Infect Dis. 2023;8:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Weber DJ, Rutala WA, Miller MB, Huslage K, Sickbert-Bennett E. Role of hospital surfaces in the transmission of emerging health care-associated pathogens: norovirus, Clostridium difficile, and Acinetobacter species. Am J Infect Control. 2010;38:S25-S33. [DOI] [PubMed] [Google Scholar]
- 80. Kassim A, Pflüger V, Premji Z, Daubenberger C, Revathi G. Comparison of biomarker based matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) and conventional methods in the identification of clinically relevant bacteria and yeast. BMC Microbiol. 2017;17:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Abubakar U, Amir O, Rodríguez-Baño J. Healthcare-associated infections in Africa: a systematic review and meta-analysis of point prevalence studies. J Pharm Policy Pract. 2022;15:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Muhammad I, Ismail W, Samsuddin N, Alias N. Pathogens from fomites in clinical setting: a scoping review. IIUM J Orofac Health Sci. 2023;4:59-79. [Google Scholar]
- 83. Bhatta DR, Hamal D, Shrestha R, et al. Bacterial contamination of frequently touched objects in a tertiary care hospital of Pokhara, Nepal: how safe are our hands? Antimicrob Resist Infect Control. 2018;7:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kramer A, Assadian O. Survival of microorganisms on inanimate surfaces. Use Biocidal Surf Reduct Healthc Acquir Infect. Springer, Cham; 2014:7-26. [Google Scholar]
- 85. Koca O, Altoparlak U, Ayyildiz A, Kaynar H. Persistence of nosocomial pathogens on various fabrics. Eurasian J Med. 2012;44:28-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Gelber SE, Ratner AJ. Hospital-acquired viral pathogens in the neonatal intensive care unit. Semin Perinatol. 2002;26:346-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Estofolete CF, Banho CA, Verro AT, et al. Clinical characterization of respiratory syncytial virus infection in adults: a neglected disease? Viruses. 2023;15:1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Sheikh T, Tomcho JC, Awad MT, Zaidi SR. Candida albicans endocarditis involving a normal native aortic valve in an immunocompetent patient. BMJ Case Rep. 2020;13(11):e236902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hemaid ASS, Abdelghany MME, Abdelghany TM. Isolation and identification of Candida spp. from immunocompromised patients. Bull Natl Res Cent. 2021;45:8. [Google Scholar]
- 90. Dixit S, Varshney S, Gupta D, Sharma S. Textiles as fomites in the healthcare system. Appl Microbiol Biotechnol. 2023;107:3887-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Murray C, Ikuta K, Sharara F, et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399:629-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Datta P, Kaur M, Rawat S, Gupta V, Chander J. Stethoscope, "the friendly foe" - A study to evaluate bacterial contamination of stethoscopes and disinfection practices. J Infect Dev Ctries. 2018;12:887-893. [DOI] [PubMed] [Google Scholar]
- 93. Seah JJ, Zhao J, Wang Y, Lee HP. Review on the advancements of stethoscope types in chest auscultation. Diagnostics. 2023;13:1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Mine Y, Higuchi W, Taira K, et al. Nosocomial outbreak of multidrug-resistant USA300 methicillin-resistant Staphylococcus aureus causing severe furuncles and carbuncles in Japan. J Dermatol. 2011;38:1167-1171. [DOI] [PubMed] [Google Scholar]
- 95. Gholami-Motlagh F, Jouzi M, Soleymani B. Comparing the effects of two Swedish massage techniques on the vital signs and anxiety of healthy women. Iran J Nurs Midwifery Res. 2016;21:402-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Zargaran D, Hardwick S, Adel R, et al. Sphygmomanometer cuffs: A potential source of infection! Angiology. 2015;66:118-121. [DOI] [PubMed] [Google Scholar]
- 97. Blanc DS, Gomes Magalhaes B, Abdelbary M, et al. Hand soap contamination by Pseudomonas aeruginosa in a tertiary care hospital: no evidence of impact on patients. J Hosp Infect. 2016;93:63-67. [DOI] [PubMed] [Google Scholar]
- 98. Fanci R, Bartolozzi B, Sergi S, et al. Molecular epidemiological investigation of an outbreak of Pseudomonas aeruginosa infection in an SCT unit. Bone Marrow Transplant. 2009;43:335-338. [DOI] [PubMed] [Google Scholar]
- 99. Kobayashi T, Nakaminami H, Ohtani H, et al. An outbreak of severe infectious diseases caused by methicillin-resistant Staphylococcus aureus USA300 clone among hospitalized patients and nursing staff in a tertiary care university hospital. J Infect Chemother. 2020;26:76-81. [DOI] [PubMed] [Google Scholar]
- 100. Mahmoudi S, Pourakbari B, Rahbarimanesh A, et al. An outbreak of ESBL-producing Klebsiella pneumoniae in an Iranian referral hospital: epidemiology and molecular typing. Infect Disord Drug Targets. 2019;19:46-54. [DOI] [PubMed] [Google Scholar]
- 101. Piezzi V, Wassilew N, Atkinson A, et al. Nosocomial outbreak of vancomycin-resistant Enterococcus faecium (VRE) ST796, Switzerland, 2017 to 2020. Eurosurveillance. 2022;27(48):2200285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Huang SS, Datta R, Platt R. Risk of acquiring antibiotic-resistant bacteria from prior room occupants. Arch Intern Med. 2006;166:1945-1951. [DOI] [PubMed] [Google Scholar]
- 103. Nseir S, Blazejewski C, Lubret R, et al. Risk of acquiring multidrug-resistant Gram-negative bacilli from prior room occupants in the intensive care unit. Clin Microbiol Infect. 2011;17:1201-1208. [DOI] [PubMed] [Google Scholar]
- 104. Tekerekoğlu MS, Duman Y, Serindağ A, et al. Do mobile phones of patients, companions and visitors carry multidrug-resistant hospital pathogens? Am J Infect Control. 2011;39:379-381. [DOI] [PubMed] [Google Scholar]
- 105. Olsen M, Campos M, Lohning A, et al. Mobile phones represent a pathway for microbial transmission: a scoping review. Travel Med Infect Dis. 2020;35:101704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Rineh A, Kelso MJ, Vatansever F, Tegos GP, Hamblin MR. Clostridium difficile infection: molecular pathogenesis and novel therapeutics. Expert Rev Anti-Infect Ther. 2014;12:131-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Chen J, Chen H, Liu C, Huan H, Teng Y. Evaluation of FEAST for metagenomics-based source tracking of antibiotic resistance genes. J Hazard Mater. 2023;442:130116. [DOI] [PubMed] [Google Scholar]
- 108. De Oliveira DMP, Forde BM, Kidd TJ, et al. Antimicrobial resistance in ESKAPE pathogens. Clin Microbiol Rev. 2020;33(3):10-128. doi: 10.1128/CMR.00181-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Durso LM, Harhay GP, Bono JL, Smith TP. Virulence-associated and antibiotic resistance genes of microbial populations in cattle feces analyzed using a metagenomic approach. J Microbiol Methods. 2011;84:278-282. [DOI] [PubMed] [Google Scholar]
- 110. Hendriksen R, Munk P, Njage P, et al. Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nat Commun. 2019;10:1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Laghari AA, Liu L, Kalhoro DH, Chen H, Wang C. Mechanism for reducing the horizontal transfer risk of the airborne antibiotic-resistant genes of Escherichia coli species through microwave or UV irradiation. Int J Environ Res Public Health. 2022;19:4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Leung KY, Wang Q, Zheng X, et al. Versatile lifestyles of Edwardsiella: free-living, pathogen, and core bacterium of the aquatic resistome. Virulence. 2022;13:5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Ciacotich N, Kvich L, Sanford N, et al. Copper-silver alloy coated door handles as a potential antibacterial strategy in clinical settings. Coatings. 2020;10:790. [Google Scholar]
- 114. Colin M, Klingelschmitt F, Charpentier E, et al. Copper alloy touch surfaces in healthcare facilities: an effective solution to prevent bacterial spreading. Materials. 2018;11(12):2479. doi: 10.3390/ma11122479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Inkinen J, Mäkinen R, Keinänen-Toivola MM, Nordström K, Ahonen M. Copper as an antibacterial material in different facilities. Lett Appl Microbiol. 2017;64:19-26. [DOI] [PubMed] [Google Scholar]
- 116. Karpanen TJ, Casey AL, Lambert PA, et al. The antimicrobial efficacy of copper alloy furnishing in the clinical environment: a crossover study. Infect Control Hosp Epidemiol. 2012;33:3-9. [DOI] [PubMed] [Google Scholar]
- 117. Matthew UO, Nwanakwaugwu AC, Kazaure JS, et al. Ultra violet (UV) Light irradiation device for hospital disinfection: hospital acquired infections control. Int J Inf Commun Technol Hum Dev. 2022;14:1-24. [Google Scholar]
- 118. Osman AH, Kotey FCN, Odoom A, et al. The potential of bacteriophage-antibiotic combination therapy in treating infections with multidrug-resistant bacteria. Antibiotics. 2023;12:1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Rutala WA, Kanamori H, Gergen MF, et al.; and the CDC Prevention Epicenters Program. Enhanced disinfection leads to reduction of microbial contamination and a decrease in patient colonization and infection. Infect Control Hosp Epidemiol. 2018;39:1118-1121. [DOI] [PubMed] [Google Scholar]
- 120. Rutala WA, Weber DJ. Monitoring and improving the effectiveness of surface cleaning and disinfection. Am J Infect Control. 2016;44:e69-e76. [DOI] [PubMed] [Google Scholar]
- 121. Artasensi A, Mazzotta S, Fumagalli L. Back to basics: choosing the appropriate surface disinfectant. Antibiotics. 2021;10:613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Wilson AM, Reynolds KA, Sexton JD, Canales RA. Modeling surface disinfection needs to meet microbial risk reduction targets. Appl Environ Microbiol. 2018;84:e00709-e00718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Chiba K. Aspects of disinfectants for control of nosocomial infections. Hokkaido Igaku Zasshi. 1994;69:182-187. [PubMed] [Google Scholar]