Abstract
Purpose
Automated machine learning (AutoML) has emerged as a novel tool for medical professionals lacking coding experience, enabling them to develop predictive models for treatment outcomes. This study evaluated the performance of AutoML tools in developing models predicting the success of pneumatic retinopexy (PR) in treatment of rhegmatogenous retinal detachment (RRD). These models were then compared with custom models created by machine learning (ML) experts.
Design
Retrospective multicenter study.
Participants
Five hundred and thirty nine consecutive patients with primary RRD that underwent PR by a vitreoretinal fellow at 6 training hospitals between 2002 and 2022.
Methods
We used 2 AutoML platforms: MATLAB Classification Learner and Google Cloud AutoML. Additional models were developed by computer scientists. We included patient demographics and baseline characteristics, including lens and macula status, RRD size, number and location of breaks, presence of vitreous hemorrhage and lattice degeneration, and physicians’ experience. The dataset was split into a training (n = 483) and test set (n = 56). The training set, with a 2:1 success-to-failure ratio, was used to train the MATLAB models. Because Google Cloud AutoML requires a minimum of 1000 samples, the training set was tripled to create a new set with 1449 datapoints. Additionally, balanced datasets with a 1:1 success-to-failure ratio were created using Python.
Main Outcome Measures
Single-procedure anatomic success rate, as predicted by the ML models. F2 scores and area under the receiver operating curve (AUROC) were used as primary metrics to compare models.
Results
The best performing AutoML model (F2 score: 0.85; AUROC: 0.90; MATLAB), showed comparable performance to the custom model (0.92, 0.86) when trained on the balanced datasets. However, training the AutoML model with imbalanced data yielded misleadingly high AUROC (0.81) despite low F2-score (0.2) and sensitivity (0.17).
Conclusions
We demonstrated the feasibility of using AutoML as an accessible tool for medical professionals to develop models from clinical data. Such models can ultimately aid in the clinical decision-making, contributing to better patient outcomes. However, outcomes can be misleading or unreliable if used naively. Limitations exist, particularly if datasets contain missing variables or are highly imbalanced. Proper model selection and data preprocessing can improve the reliability of AutoML tools.
Financial Disclosure(s)
Proprietary or commercial disclosure may be found in the Footnotes and Disclosures at the end of this article.
Keywords: Automated machine learning (AutoML), Machine learning, Medical outcome prediction, Pneumatic retinopexy, Rhegmatogenous retinal detachment
Pneumatic retinopexy (PR) is a minimally invasive procedure commonly used for the treatment of rhegmatogenous retinal detachment (RRD).1 Compared with scleral buckling or pars plana vitrectomy, PR offers several advantages, including lower costs,2 feasibility to perform in an outpatient setting, faster recovery, and reduced morbidity.3 However, the single procedure anatomic success rates for the PR have been reported to range from 60% to 91%,4 which is comparatively lower than that of pars plana vitrectomy (93%)5, scleral buckling only (82%)6, and combined pars plana vitrectomy with scleral buckling (92.2%).6
The reattachment rates after PR heavily depend on patient selection,7 surgeon experience,8 and proper postprocedure positioning.9 Pneumatic retinopexy is primarily indicated for RRDs with ≥ 1 retinal breaks within 1 clock hour located in the superior 8 clock hours of the retina.7 More favorable outcomes are observed in phakic patients10 and when procedure is performed by more experienced specialists.8 Previous studies have also indicated that the presence of vitreous hemorrhage,11 inferior retinal breaks,12 aphakic or pseudophakic lens status,13 retinal tears > 1 clock hour,14 greater number of retinal breaks,13,14 and male sex are associated with a higher rate of failure. However, no definitive predictive measures and formulas have been identified for patient selection in clinical practice or preprocedure counseling.15
Over the past decade, machine learning (ML) has made significant strides in medicine and ophthalmology.16 Machine learning has proven effectiveness in identifying pathologies and physiologic features from fundus photos and OCT,17, 18, 19 predicting treatment outcomes,20,21 and facilitating telemedicine.22,23 These algorithms can recognize intricate patterns and structures in medical datasets and images, which makes them powerful tools for classification, pattern recognition, and generation of predictive models for use in clinical practice.24 Traditionally, the development of ML models has been carried out by computer scientists and artificial intelligence (AI) experts. However, the emergence of automated machine learning (AutoML) tools has made the technology more accessible, enabling nonexperts to construct powerful ML models without requiring coding or extensive engineering expertise.25, 26, 27 This accessibility can potentially unlock numerous opportunities in the medical field, in which proficiency in ML and coding is often limited. Nevertheless, it is crucial to exercise caution in adopting these tools because their native adoption may yield unreliable outcomes.28
Given the limited data on the feasibility of using AutoML for predicting procedural success, we harnessed various AutoML platforms to predict the success rate of PR based on a combination of clinical and demographic features. Furthermore, we compared these platforms and benchmarked their discriminative performance against similar tools developed by ML experts.
Methods
Patient Population
The institutional review board approved the study protocol. The study was conducted in accordance with the tenets of the Declaration of Helsinki and the Health Insurance Portability and Accountability Act. This retrospective study included 539 eyes of 539 patients who underwent PR by vitreoretinal (VR) fellows. A total of 483 patient records were obtained from the database previously collected and described by our groups,8 which included patients who underwent PR by VR fellows at 6 training sites across the United States (Associated Retinal Consultants (Royal Oak, Michigan), Duke University Eye Center, New York Eye & Ear Infirmary, University of California (UC) Davis, UC San Diego, and Wills Eye Hospital) between 2002 and 2016. This database was used to train the ML algorithms. To test the algorithms, we retrospectively collected data on additional 56 patients who underwent PR by VR fellows at UC Davis Eye Center between 2020 and 2022. We excluded patients with prior history of ocular trauma or retinal surgery and those with < 3 months of follow-up data. A total of 10 demographic and clinical features were recorded, including age, sex, lens status (phakic, aphakic, or pseudophakic), macula status at the time of RRD diagnosis (macula involving or macula sparing), size of RRD (clock hours), number of retinal breaks, presence of inferior retinal breaks, vitreous hemorrhage, lattice degeneration, and VR fellow procedure experience (recorded as either < 16 or ≥ 16 PR cases previously performed by a VR fellow). The primary outcome of interest was single-procedure anatomic success, defined as retinal reattachment at 3 months with no additional procedures. All independent variables were binary except for the lens status (ternary), age, and the number of retinal breaks (continuous), as shown in Table 1, Tables S2 and S3.
Table 1.
Clinical and Demographic Characteristics of Dataset 1
| Training Set (n = 483) |
Test Set (n = 56) |
|||
|---|---|---|---|---|
| PR Failure |
PR Success |
PR Failure |
PR Success |
|
| n = 153 (31.7%) | n = 330 (68.3%) | n = 30 (53.6%) | n = 26 (46.4%) | |
| Mean age (SD) | 62.4 (12.1) | 64.0 (10.4) | 62.4 (10.3) | 60.5 (8.3) |
| Sex | ||||
| Female | 48 (31.4%) | 121 (36.7%) | 17 (56.7%) | 13 (50.0%) |
| Male | 104 (68.0%) | 209 (63.3%) | 13 (43.4%) | 13 (50.0%) |
| Missing | 1 (0.6%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Procedure experience | ||||
| < 16 cases | 133 (86.9%) | 231 (70.00%) | 26 (86.7%) | 10 (38.5%) |
| > 16 cases | 4 (2.6%) | 25 (7.6%) | 4 (13.3%) | 16 (61.5%) |
| Missing | 16 (10.5%) | 74 (22.4%) | 0 (0%) | 0 (0%) |
| Lens status | ||||
| Aphakic | 1 (0.6%) | 0 (0.00%) | 7 (23.3%) | 3 (11.5%) |
| Phakic | 102 (66.7%) | 244 (73.9%) | 13 (43.3%) | 7 (26.9%) |
| Pseudophakic | 50 (32.7%) | 86 (26.1%) | 10 (33.3%) | 16 (61.5%) |
| Macula status | ||||
| Detached | 76 (49.7%) | 112 (33.9%) | 20 (66.7%) | 5 (15.4%) |
| Attached | 76 (49.7%) | 218 (66.1%) | 10 (33.3%) | 22 (84.6%) |
| Missing | 1 (0.6%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Size of RRD | ||||
| < 4 clock hours | 64 (41.8%) | 194 (58.8%) | 9 (30.0%) | 23 (88.5%) |
| > 4 clock hours | 87 (56.9%) | 130 (39.4%) | 21 (70.0%) | 3 (11.5%) |
| Missing | 2 (1.3%) | 6 (1.8%) | 0 (0%) | 0 (0%) |
| Number of retinal breaks (SD) | 1.34 (0.74) | 1.35 (0.95) | 1.4 (0.6) | 1.1 (0.4) |
| Inferior break | ||||
| Absent | 142 (92.8%) | 313 (94.9%) | 25 (83.3%) | 25 (96.1%) |
| Present | 2 (1.3%) | 3 (0.9%) | 5 (16.7%) | 1 (3.9%) |
| Missing | 9 (5.9%) | 14 (4.2%) | 0 (0%) | 0 (0%) |
| Vitreous hemorrhage | ||||
| Absent | 134 (87.6%) | 293 (88.8%) | 18 (60.0%) | 26 (100%) |
| Present | 19 (12.4%) | 37 (11.2%) | 12 (40.0%) | 0 (0) |
| Lattice degeneration | ||||
| Absent | 117 (76.5%) | 273 (82.7%) | 21 (70.0%) | 22 (84.6%) |
| Present | 34 (22.2%) | 57 (17.3%) | 9 (30.0%) | 4 (15.8%) |
| Missing | 2 (1.3%) | 0 (0.00%) | 0 (0%) | 0 (0%) |
PR = pneumatic retinopexy; RRD = rhegmatogenous retinal detachment; SD = standard deviation.
Comprised of 483 patients (training set), captured from Emami-Naeini et al database8 and the test set (n = 56) comprised of patient records captured from the UC Davis electronic medical records that underwent pneumatic retinopexy (PR) by procedure outcome at 3-month follow-up.
Model Development
To generate the AutoML models, we utilized 2 commonly used platforms, Google Cloud AutoML Vertex AI (Mountain View, CA) and MATLAB Classification Learner App (Natick, MA, version R2022b [9.13.0]). These models were devised by medical professionals with no relevant coding experience. An additional custom model was generated independently by the computer scientists and compared with the AutoML models.
We used the following evaluation metrics: sensitivity (defined as the probability of correctly predicting a failed PR outcome), specificity (defined as the probability of correctly predicting a successful PR outcome), and positive predictive value (PPV, also referred to as precision: the likelihood of a patient predicted to have a failed PR outcome to actually have failed the procedure). Additionally, we used area under the receiver operating curve (AUROC) to assess the models’ overall capability to distinguish between classes and F2 scores, which is a metric that combines sensitivity and PPV, to compare the models. We developed several models, and the models were selected based on the greatest AUROC and F2 scores on validation.
Dataset Preparation
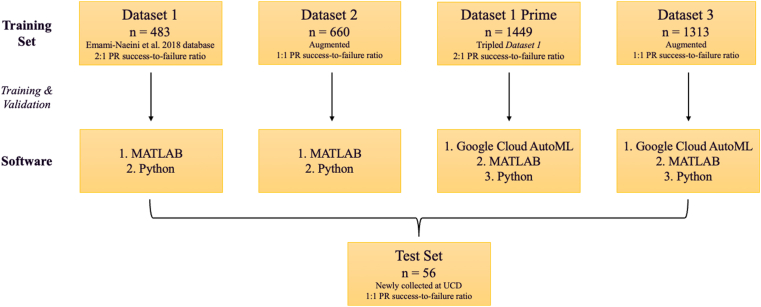
We prepared a total of 4 training datasets to compare the performance of AutoML models (Fig 1). Dataset 1 comprised of the previously collected database of 483 eyes8 (training and validation set). The training set had 5 missing data points for age, 1 for sex, 90 for procedure experience, 1 for macula status, 8 for RRD size, 23 for the inferior break, and 2 for the presence of lattice degeneration (Table 1). There were no missing values in the test set. The PR success-to-failure ratio was approximately 2-to-1 (68.3% to 31.7%) in the training set and 1-to-1 (46.4% to 53.6%) in the test set. Patients’ clinical and demographic features for the training and test sets are summarized in Table 1. In our naive adoption of the AutoML tools, we did not perform any preprocessing of the training set. Moreover, Google Cloud AutoML does not allow for analysis of a data set < 1000; therefore, we generated Dataset 1 prime by triplicating Dataset 1 (n = 1449).
Figure 1.
The flowchart illustrates the dataset preparation steps. Dataset 1 comprised of the previously collected database of 483 eyes.8 Dataset 1 prime (n = 1313) was generated by triplicating Dataset 1. The pneumatic retinopexy (PR) success-to-failure ratio was approximately 2-to-1 in Datasets 1 and 1 prime. Dataset 1 was further augmented to generate Datasets 2 (n = 660) and 3 (n = 1313) with a 1:1 PR outcome ratio. The test set was independently collected at the University of California, Davis (UCD) and was not included in the training phase.
To address missing data and the imbalance of successful versus failed PR cases, the computer scientists (A.Y. and I.S) performed data imputation and augmentation. First, we used the nearest neighbors in each class to fill in the missing data using Jaccard similarity. Between the most similar data points (per Jaccard similarity), we employed Euclidian Distance to find the most similar (closest) data points.29,30 We then replaced the mode of feature values of the missing feature to fill the candidate with similar values. Next, we performed data augmentation using the synthetic minority over-sampling technique for categorical values to generate a balanced dataset (n = 660) with a 1:1 success-to-failure ratio. The resulting dataset is referred to as Dataset 2, and the dataset breakdown is summarized in Table S2. We then followed the same methodology to generate an additional dataset (n = 1313) with a 1:1 outcome ratio to generate enough data points (> 1000) to meet Google Cloud AutoML’s minimum data threshold (as opposed to Dataset 1 prime relying on simple data replicas). This dataset was referred to as Dataset 3 (Table S3). All models were tested on the same test set (n = 56) described previously.
MATLAB Models
Two medical professionals (A.N. and P.E.N.) consulted the publicly available tutorials for the Classifications Learner App provided by MathWorks, MATLAB.31 We trained and tested all 4 training datasets using all models available in the MATLAB Classification Learner app. All datasets were evaluated using a crossvalidation scheme with 5 folds as part of the training step. On training and validation steps, we chose 2 models that yielded the best average discriminant performance as measured by the AUROC and F2 scores on validation for further evaluation in the study, namely the linear support vector machine (SVM) and Ensemble Random Undersampling Boosting (RUSBoosted) tree models. We used a linear Kernel function with an automatic kernel scale mode and a box constraint level of 1 for the SVM model. The Ensemble RUSBoosted model offered several options for hyperparameter tuning. The learning rate is a hyperparameter that determines how aggressively the model is changed in each training step and in response to classification errors. We set the learning rate to 0.1, a common setting recommended by the MATLAB tutorial.31 We further fine tuned the number of splits and learners to improve predictive power per validation results. The number of splits specifies the number of branch points of the learning tree and controls the depth of learning, and the number of learners determines the count of individual models that are combined to form an ensemble.31 We achieved optimal validation results with 11 splits and 16 learners.
Google Cloud AutoML Vertex AI Model
We constructed a binary classification model from tabular data through Google Cloud AutoML Vertex AI following the available instructions.32 This model was trained using Dataset 1 prime and Dataset 3, which met the requirement of at least 1000 data entries. We uploaded the datasets into the Google Cloud console as tabular data and chose the classification training method with a log loss optimization objective, suggested by the Google Cloud AutoML tutorial for keeping the prediction probabilities as accurate as possible. From the available options, we chose to randomly split the training sets into 80% for training and 20% for validation and used our test set to evaluate the model.
Custom Model Development
To compare and validate the automated models’ performance, custom models were generated by computer scientists. We evaluated multiple classification techniques, including RandomForestClassifier, Gaussian Naive Bayes, KNeighborsClassifier, Gaussian Processes Classifier, AdaBoostClassifier, and the support vector classification (SVC). The support vector classification method with Radial Basis Function kernel and tuned gamma yielded the best results. In Python, we implemented a 2-step method using support vector classification, which is a binary type of SVM commonly used for classification tasks. We employed the Radial Basis Function kernel with parameters such as gamma (the kernel parameter) and C (the regularization parameter). Additionally, we utilized random state for reproducibility. By specifying a fixed random state, the random process within each model runs from the same starting point. This allows to replicate the results precisely when training the model using the same dataset and hyperparameters. Furthermore, we optimized hyperparameter tuning using BayesSearchCV and GridSearchCV, 2 techniques commonly employed in machine learning. GridSearchCV conducts a brute-force search by exploring all possible combinations of hyperparameter values. BayesSearchCV utilizes a probabilistic model to predict the model's performance and intelligently selects promising hyperparameter values based on past evaluations. Hereafter, the model developed by ML experts is referred to as the custom model. This model was trained with all 4 datasets and tested using the test set.
Results
Patient Population
Our cohort included a total of 539 eyes of 539 patients (Table 1), 183 of whom had a failed single-procedure PR outcome (34.0%) among both the training and test sets. Out of these 183 patients, 159 (86.9%) underwent the procedure by VR fellows who had previously performed < 16 PR procedures, 60 (32.8%) were pseudophakic, 96 (52.5%) had a macula-involving RD, 108 (59.0%) had an RD size > 4 clock hours, 7 (3.8%) had an inferior retinal break, and 31 (16.9%) had a vitreous hemorrhage. The PR failure rate was 31.7% in the training set and 53.6% in the test set. The overall single-procedure success rate at the 3-month mark was 66.0%, with a 68.3% and 46.4% success rate in the training and test sets, respectively.
AutoML Model Performance
The performance of all models is summarized in Table 4 (validation) and 5 (test phase). Based on the performance metrics of AutoML models, the Ensemble model (MATLAB) demonstrated the best test performance when trained using Dataset 2 with the test accuracy of 82.1% (AUROC: 0.89; F2 score: 0.74; and PPV: 0.90). The sensitivity and specificity were 0.86 and 0.77, respectively. In other words, the model correctly predicted 86% of patients with a failed outcome (true-positive rate) while misclassifying the remaining 14% to have a successful outcome (false-negative rate). Comparatively, the model correctly predicted a successful outcome in 77% of the patients (true negative rate) and miscategorized the remaining 23% of cases as failure (false-positive rate). Performance metrics on test phase were lower when the model was trained on Dataset 1 (F2 score: 0.72; AUROC: 0.91), 1 prime (F2 score: 0.74, AUROC: 0.89), or 3 (F2 score: 0.72; AUROC: 0.85).
Table 4.
Performance Metrics of the Automated Machine Learning Compared to Custom Models on the Validation Phase
| ML Platform | Model Type | Dataset Characteristics |
Discriminative Model Test Performance |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Dataset # | n | Outcome Ratio | F2 Score | AUROC | Accuracy (%) | PPV | Sensitivity | Specificity | ||
| MATLAB Classification Learner App | Linear Superior Vector Machine (SVM) | Dataset 1 | 483 | 2:1 | 0.01 | 0.53 | 68.3 | 1.00 | 0.01 | 1.00 |
| Dataset 1 prime | 1449 | 2:1 | 0.02 | 0.60 | 68.5 | 0.67 | 0.01 | 1.00 | ||
| Dataset 2 | 660 | 1:1 | 0.60 | 0.66 | 61.4 | 0.62 | 0.59 | 0.63 | ||
| Dataset 3 | 1313 | 1:1 | 0.58 | 0.64 | 60.9 | 0.61 | 0.58 | 0.64 | ||
| Ensemble Random Undersampling Boosting (RUSBoosted) | Dataset 1 | 483 | 2:1 | 0.52 | 0.56 | 52.6 | 0.35 | 0.59 | 0.50 | |
| Dataset 1 prime | 1449 | 2:1 | 0.60 | 0.67 | 60.3 | 0.42 | 0.67 | 0.57 | ||
| Dataset 2 | 660 | 1:1 | 0.72 | 0.65 | 62.4 | 0.60 | 0.76 | 0.48 | ||
| Dataset 3 | 1313 | 1:1 | 0.62 | 0.66 | 61.2 | 0.61 | 0.62 | 0.61 | ||
| Python | Support Vector Classification (SVC) | Dataset 1 | 483 | 2:1 | 0.71 | 0.61 | 59.6 | 0.76 | 0.57 | 0.43 |
| Dataset 1 prime | 1449 | 2:1 | 0.71 | 0.61 | 59.6 | 0.76 | 0.57 | 0.43 | ||
| Dataset 2 | 660 | 1:1 | 0.61 | 0.62 | 61.5 | 0.64 | 0.54 | 0.60 | ||
| Dataset 3 | 1313 | 1:1 | 0.66 | 0.66 | 66.3 | 0.66 | 0.68 | 0.66 | ||
AUROC = area under the receiver operating characteristic curve; ML = machine learning; PPV = positive predictive value.
This table does not include validation results for the Google AutoML models as the software generates an output for performance metrics for the test phase only.
The linear SVM model had a low accuracy (53.6%) when trained using either Dataset 1 (AUROC = 0.87) or Dataset 1 prime (AUROC = 0.62). The use of preprocessed datasets improved the discriminative model’s performance, with an improved accuracy of 80.4% (AUROC = 0.87) when trained with Datasets 2 and 3. Additionally, a substantial improvement in sensitivity and PPV was observed, with sensitivity increasing from 0.17 (SVM, Datasets 1 and 1 prime) to 0.63 (SVM, Datasets 2 and 3) and PPV increasing from 0.83 (SVM, Datasets 1 and 1 prime) to 1.00 (SVM, Datasets 2 and 3).
The Google Cloud AutoML model showed similar test accuracy when trained using Dataset 1 prime (57.1%) and Dataset 3 (60.7%) and identical specificity (0.81). Training with a preprocessed and augmented dataset led to improvements in AUROC from 0.62 in Dataset 1 prime to 0.70 in Dataset 3, sensitivity (from 0.37 to 0.43), PPV (from 0.69 to 0.72), and F2 score (from 0.40 to 0.47).
Custom Model Performance
Training the custom model with Datasets 1 and 1 prime yielded identical results on test accuracy (83.9%), AUROC (0.85), F2 scores (0.79), as well as PPV (0.75), sensitivity (0.96), and specificity (0.96) (Table 5). Similarly, data preprocessing and augmentation improved the model performance. When trained with Dataset 2, the model’s accuracy increased to 85.7%, AUROC increased to 0.86, PPV increased to 0.78, and the resulting F2 score increased to 0.92. The custom model exhibited the best performance upon training with Dataset 3, resulting in the test accuracy of 87.5%, with an AUROC of 0.88, PPV of 0.81, sensitivity and specificity of 0.96, and F2 score of 0.93.
Table 5.
Performance Metrics of the Automated Machine Learning Compared to Custom Models on the Test Phase
| ML Platform | Model Type | Dataset Characteristics |
Discriminative Model Test Performance |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Dataset # | n | Outcome Ratio | F2 Score | AUROC | Accuracy (%) | PPV | Sensitivity | Specificity | ||
| MATLAB Classification Learner App | Linear Superior Vector Machine (SVM) | Dataset 1 | 483 | 2:1 | 0.20 | 0.81 | 53.6 | 0.83 | 0.17 | 0.96 |
| Dataset 1 prime | 1449 | 2:1 | 0.20 | 0.62 | 53.6 | 0.83 | 0.17 | 0.96 | ||
| Dataset 2 | 660 | 1:1 | 0.68 | 0.87 | 80.4 | 1.00 | 0.63 | 1.00 | ||
| Dataset 3 | 1313 | 1:1 | 0.68 | 0.87 | 80.4 | 1.00 | 0.63 | 1.00 | ||
| Ensemble Random Undersampling Boosting (RUSBoosted) | Dataset 1 | 483 | 2:1 | 0.71 | 0.91 | 80.4 | 0.95 | 0.67 | 0.96 | |
| Dataset 1 prime | 1449 | 2:1 | 0.74 | 0.89 | 82.1 | 0.95 | 0.70 | 0.96 | ||
| Dataset 2 | 660 | 1:1 | 0.85 | 0.90 | 82.1 | 0.81 | 0.86 | 0.77 | ||
| Dataset 3 | 1313 | 1:1 | 0.72 | 0.85 | 75.0 | 0.81 | 0.70 | 0.81 | ||
| Google Cloud AutoML Vertex AI | Log Loss Optimization | Dataset 1 prime | 1449 | 2:1 | 0.40 | 0.62 | 57.1 | 0.69 | 0.37 | 0.81 |
| Dataset 3 | 1313 | 1:1 | 0.47 | 0.70 | 60.7 | 0.72 | 0.43 | 0.81 | ||
| Python | Support Vector Classification (SVC) | Dataset 1 | 483 | 2:1 | 0.79 | 0.85 | 83.9 | 0.75 | 0.96 | 0.96 |
| Dataset 1 prime | 1449 | 2:1 | 0.79 | 0.85 | 83.9 | 0.75 | 0.96 | 0.96 | ||
| Dataset 2 | 660 | 1:1 | 0.92 | 0.86 | 85.7 | 0.78 | 0.96 | 0.95 | ||
| Dataset 3 | 1313 | 1:1 | 0.93 | 0.88 | 87.5 | 0.81 | 0.96 | 0.96 | ||
AUROC = area under the receiver operating characteristic curve; ML = machine learning; PPV = positive predictive value.
Discussion
Developing and implementing models to reliably predict the success of various procedures greatly enhances our ability to identify ideal candidates for delivering personalized care and optimizing the success rate of the procedures while reducing costs33 and complications.34 In the present study, we utilized electronic medical records data to generate ML algorithms for predicting the outcomes of a commonly performed retinal procedure. Moreover, we successfully used the available AutoML platforms, implemented by health care professionals without previous coding experience. Although these platforms have previously been used for the classification of images and disease outcomes24, 25, 26,35 their use in electronic medical records data and ophthalmology has been limited. Our results indicate that AutoML is a powerful tool that can be reliably used by medical professionals with no coding background, especially when some rather basic prerequisites related to data balancing, imputation, augmentation, and proper use of evaluation metrics are satisfied.
In this study, we used a multicenter database that had been previously collected from multiple practitioners to train and validate our models. It is important to note that this database was inherently different from our test data, which was independently collected at UC Davis. To ensure external validation, no part of the test set was included in the training phase.36 Although this approach introduces heterogeneity in the analysis, evaluating the model with diverse data help mitigate the risk of overfitting, addresses potential differences among data collectors, and enhances the generalizability of our ML schemes to a broader patient population.37, 38, 39, 40, 41
As evident from the results on the test phase (Table 5), our models performed well in classifying new test cases despite the heterogeneity between the training and test datasets. The improved performance further confirms higher quality, generalizability, and real-world applicability of our models. It is worth noting that when homogenous test and training data is used to develop models, the applicability of these models in real-world scenarios can be limited.42 Therefore, our approach of using diverse datasets contributes to the robustness and practicality of our ML models.
The training dataset was imbalanced, with a success rate of 68.3%. Additionally, several variables, including the vitreoretinal fellow’s procedure experience were missing. Imbalanced datasets can introduce bias, limit generalizability for minority classes,43 and compromise the replicability and validity of the findings.44 To address these issues, we employed imputation and augmentation techniques.43,45 Computer scientists chose a two-step method that incorporated a high similarity threshold based on 2 widely-used similarity measures.29,30 It is important to note that only the training data underwent augmentation and imputation preprocessing, whereas the test data maintained a higher quality with no missing values. This explains the reason cross-validation performance was lower compared to the test results. In such instances, it is beneficial to employ a crossvalidation scheme instead of a fixed training-validation split. The latter is susceptible to bias resulting from the specific partition of the data. In contrast, crossvalidation randomly divides the data into multiple train-validation splits and averages the performance across all of them. This approach helps mitigate the issue of overfitting and provides a more accurate assessment of the model’s true performance on unseen data.46
The multi-institute dataset imbalance used in the training phase prompted us to adopt 2 key metrics, namely the AUROC and F2 scores on validation, for model selection rather than relying solely on validation accuracy. According to Ling et al,47 AUROC serves as a superior measure for comparing classification models. Accuracy, on the other hand, is influenced by the distribution of samples in each class, making it unsuitable for unbalanced data and could potentially lead to inappropriate model selection. Another metric for comparing classification models is the F score.48 We specifically focused on the F2 score, because it places greater emphasis on sensitivity rather than PPV, prioritizing the reduction of false-negatives over false-positives. This prioritization aligns with our research objectives because correctly predicting an unsuccessful procedure outcome holds more significance in medical applications than predicting successful ones.
MATLAB Models
The performance of the SVM model on the test data exhibited variability based on the dataset it was trained on. Specifically, we observed that with the unbalanced Dataset 1, the SVM model had a relatively high AUROC while exhibiting low test accuracy. In this scenario, the high AUROC and near-perfect specificity resulted from the model predominantly classifying cases as successful. It misclassified most of the failed PR cases, leading to a true positive rate of only 17%. However, data preprocessing led to a significant improvement in the test accuracy, comparable to the Ensemble models, and superior specificity and PPV. In a study conducted by Antaki et al25 using a SVM model to predict the development of postoperative proliferative vitreoretinopathy, a similar distribution of specificity versus sensitivity and AUROC was observed following data preprocessing.
The Ensemble RUSboosted tree model exhibited comparable performance to the custom model when trained on Datasets 1 and 2. However, when the larger augmented dataset (Dataset 3) was used, it did not enhance the model’s performance. In fact, the test accuracy and sensitivity decreased. This outcome was somewhat expected since 63% of the Dataset 3 consisted of synthetic data. Previous research by Seiffert et al49 showed the RUSBoosted tree is highly sensitive to data distribution. Although augmentation and imputation can potentially address issues such as data scarcity and class imbalance and improve training performance, relying too heavily on data synthesis and oversampling can introduce misleading patterns and result in overfitting.50 The decrease in the performance may also be attributed to the random sampling nature of RUSBoosted, especially when working with a dataset that has been tripled without proper random shuffling. This is in contrast to the SMOTEBoost method employed here for data augmentation. Certain classification methods demonstrate greater robustness and can benefit from larger training samples, as observed in the performance improvement of our custom model trained on Dataset 3.
Overall, the Ensemble RUSBoosted model exhibited similar test performance across all 4 datasets (Table 5). This can be attributed to the model’s inherent adoption of random undersampling, where the number of samples from the least represented class in the training data is used as the basic unit for sampling. Consequently, this approach demonstrates lower sensitivity to data imbalance.49 On the other hand, the SVM model does not share this characteristic, as evidenced by its low performance when trained on the imbalanced datasets. Additionally, the Ensemble RUSboosted tree appears to be more adept at balancing sensitivity and specificity. Conversely, when properly trained, the SVM model demonstrates nearly perfect specificity, making it a reliable model51 to identify PR candidates who are most likely to experience successful outcomes.7
Google Cloud AutoML Models
We evaluated the Google AutoML model using 2 datasets. Dataset 1 prime which is a tripled version of Dataset 1. In comparison to the MATLAB models, Google AutoML was found to be inferior to the Ensemble tree model but comparable to the SVM during the test phase when trained with raw data. Training the Google AutoML model with Dataset 3 led to a slight improvement in the test accuracy, AUROC, and sensitivity. However, the overall performance metrics remained lower than those achieved by the MATLAB or custom models.
Google AutoML’s classification of tabular data has not been previously applied in ophthalmology. Although previous studies have shown high performance in the classification of retinal pathology26 and cataract surgery phases24 using platforms like Google AutoML Vision and Video Classification, our study showed that its application for tabular data may not be suitable and result in subpar performance. The model evidently lacks automated augmentation, imputation capabilities, and effective sampling strategies to handle imbalance data. Moreover, the absence of hyperparameter tuning options significantly impacts model performance. These limitations restrict medical professionals from optimizing the models, which may account for the stark differences in performance compared to the custom or MATLAB models. Additionally, the software does not provide performance metrics for the validation phase, rendering it impossible to select the optimal model from the outset. Consequently, naive applications of this software could lead to erroneous interpretation of results and should be avoided.
Custom Model
The custom model outperformed both the MATLAB and Google AutoML models. The results clearly show that training the model with preprocessed data resulted in improved performance during the test phase, achieving the highest overall AUROC and F2 scores. This underscores the importance of data preprocessing, particularly for datasets with missing values and imbalanced classes. The performance improvement attributed to data preprocessing further confirms the reliability and effectiveness of the custom model, indicating that it does not suffer from overfitting. Moreover, the superior performance of the custom model can be attributed, in part, to its flexibility and access to a broader range of additional underlying functionality.
User Experience: MATLAB versus Google Cloud AutoML
Both platforms offer user-friendly interfaces, clear instructions, and automatically generate model statistics. However, the Google Cloud platform appears to be easier to use, particularly when it comes to generating predictions for future patients or cases. Once the model is trained and tested, users can easily create an interactive model on the Google Cloud platform, allowing them to predict procedure outcomes for individual patients by inputting the necessary clinical and demographic features. In contrast, generating predictions in MATLAB would require clinicians to create a separate test dataset file and execute the algorithm in the Classification Learner application. Furthermore, the Google Cloud platform allows for the potential of making single-case prediction models publicly available on the cloud, enabling other physicians to conveniently assess patients’ procedure outcomes in real time. In terms of associated costs, MATLAB seems to be the more cost-efficient option with a 1-time finite price for software license. In contrast, Google AutoML charges per hour of model training, potentially requiring more extensive funds depending on the complexity of the models. MATLAB also provides a built-in toolbox that requires minimal programming knowledge to fine-tune models and better handle unbalanced datasets, which is not available in Google AutoML. Additionally, the MATLAB Classification Learner App does not have a minimum data threshold, allowing models to be trained with datasets of virtually any size, whereas Google Cloud AutoML does not support smaller datasets, requiring a minimum of 1000 data points.
Strengths and Limitations
In this study, we primarily focused on evaluating MATLAB and Google Cloud AutoML models; however, it is important to note that there are other AutoML tools available, such as Weka, RapidMiner, and Orange, which can also benefit non-ML experts. While our test dataset had high quality with no missing values, our training set suffered from a high rate of missing values and data imbalance, which prompted us to adopt imputation and augmentation techniques to address these issues. Although model performance trained on imbalanced Dataset 1 was comparable in some metrics to models trained on preprocessed Datasets 2 and 3 (Table 5), addition of synthetic data could have introduced bias and affected model performance. Limiting missing values in the training set and obtaining a larger, balanced dataset can potentially enhance the performance of the algorithms and limit the risk of introducing additional bias.
Our study has several strengths that contribute to its scientific value. To our knowledge, this is the first study to evaluate ML algorithms for predicting PR outcomes. We benefited from a relatively large sample size, encompassing a diverse range of patients from multiple institutions across the United States. Furthermore, to assess the reliability of AutoML tools, we conducted a comparative analysis between models developed by researchers without relevant experience using only publicly available tutorials and models designed by ML experts. We also evaluated various models using MATLAB and Google Cloud AutoML tools with both raw and preprocessed datasets, providing insights into best practices for clinicians using these tools independently or in collaboration with computer science experts.
In conclusion, our study demonstrates the feasibility of using AutoML models by clinicians for predicting procedure outcomes based on patient demographic and clinical characteristics. Our findings underscore the importance of simple data preprocessing techniques, proper data segmentation, and cross-validation techniques when using these tools. In more complex cases, consulting with ML experts may be beneficial in identifying appropriate preprocessing steps, model selection, and hyperparameter tuning strategies.
Manuscript no. XOPS-D-23-00219.
Footnotes
Supplemental material available atwww.ophthalmologyscience.org.
Disclosure(s):
All authors have completed and submitted the ICMJE disclosures form.
The authors made the following disclosures:
A.N.: Grants – J. William Kohl Summer Scholarship for Medical Students.
D.G.: Financial support – EyePoint, Regeneron, Unity Biotechnology, Iveric Bio, RegenxBio; Receipt of services – Regeneron Pharmaceuticals, Inc, Regeneron Healthcare Solutions, Inc, Bausch & Lomb Americas Inc, Heidelberg Engineering, Inc, ABBVIE INC, EyePoint Pharmaceuticals US, Inc, Carl Zeiss Meditec USA, Inc.
E.N.: Financial support – Regeneron Pharmaceuticals, Inc, Regeneron Healthcare Solutions, Inc, Bausch & Lomb Americas Inc, Heidelberg Engineering, Inc, ABBVIE INC, EyePoint Pharmaceuticals US, Inc, Carl Zeiss Meditec USA, Inc; Honoraria, Travel expenses – Genentech USA, Inc, Alcon Vision LLC; Receipt of services – Genentech USA, Inc, EyePoint Pharmaceuticals US, Inc, Oculus Surgical Inc, Alcon Vision LLC.
F.S.A.: Financial support – Genentech USA, Inc, ABBVIE INC, EyePoint; Honoraria, Travel expenses – Genentech USA, Inc; Scientific Advisory Board – Genentech, Allergan, EyePoint; Leadership – Genentech USA, Inc; Receipt of services – Bausch & Lomb Americas Inc, Genentech USA, Inc, Alimera Sciences, Inc, Alcon Vision LLC, Regeneron Pharmaceuticals, Inc, EyePoint Pharmaceuticals US, Inc, Biogen, Inc, Apellis Pharmaceuticals, Inc, Genentech USA, Inc, ABBVIE INC, EyePoint Pharmaceuticals US, Inc.
G.Y.: Financial support – Abbvie, Adverum, Alimera, Anlong, Bausch & Lomb, Cholgene, Clearside, Endogena, Genentech, Gyroscope, Intergalactic, Iridex, Janssen Research & Development, LLC, NGM Bio, Regeneron, Thea, Topcon, Zeiss; Honoraria, Travel expenses – Bausch & Lomb Americas Inc; Receipt of services – Bausch & Lomb Americas Inc, Genentech USA, Inc.
J.W.: Financial support – Alcon Vision LLC, Apellis Pharmaceuticals, Inc, ABBVIE INC, Alcon Research LLC, ABBVIE INC, F. Hoffmann-La Roche AG, Regeneron Healthcare Solutions, Inc, Genentech USA, Inc, Novartis, Allegran/Abvie; Honoraria – Regeneron Healthcare Solutions, Inc, Apellis Pharmaceuticals, Inc, Genentech USA, Inc, ABBVIE INC, Apellis Pharmaceuticals, Inc, Allergan/Abvie, Regeneron; Receipt of services – Apellis Pharmaceuticals, Inc, Genentech USA, Inc, Regeneron Healthcare Solutions, Inc, ABBVIE INC, Alcon Vision LLC, Allergan, Inc; Financial or nonfinancial interests – Allergan/Abvie, Gentech, Gyroscope, Kodiak, Regeneron, and RegenexBio, Aviceda, Caeregen, and Vortex.
J.D.: Financial support – Eyepoint Pharmaceuticals Inc, Alimera Sciences Inc; Honoraria – Alimera Sciences Inc; Receipt of services – Genentech USA, Inc, Dutch Ophthalmic, USA, EyePoint Pharmaceuticals US, Inc, Regeneron Healthcare Solutions, Inc, Alcon Vision LLC, EyePoint Pharmaceuticals US, Inc, Alimera Sciences, Inc, Apellis Pharmaceuticals, Inc.
K.C.C.: Travel – Alcon Vision LLC.
M.G.: Financial support – Regeneron, Genentech, Allergan, RegenexBio; Honoraria – Bausch & Lomb Americas Inc; Receipt of services – ABIOMED, Abbott Laboratories.
M.K.: Financial support – Biogen, Inc, Dutch Ophthalmic, USA, F. Hoffmann-La Roche AG, Allergan, RegenexBio; Honoraria – Alimera Sciences, Inc, Biogen, Inc, Genentech USA, Inc, Regeneron Healthcare Solutions, Inc; Travel – Biogen, Inc, Genentech USA, Inc, Regeneron Healthcare Solutions, Inc; Receipt of services – Alimera Sciences, Inc, Apellis Pharmaceuticals, Inc, Biogen, Inc, Dutch Ophthalmic, USA, Genentech USA, Inc, Regeneron Healthcare Solutions, Inc.
P.E.N.: Grants – Career Starter Grant, Knight Templar Eye Foundation; Financial support – EyePoint, Bausch & Lomb, Genentech; Receipt of services – EyePoint Pharmaceuticals US, Inc, Bausch & Lomb Americas Inc.
P.G.: Travel – Horizon Therapeutics plc.
Supported by training funds from the J. William Kohl Summer Scholarship for Medical Students. PEN received funding support from the Knights Templar Eye Foundation and Doris Duke Charitable Foundation COVID-19 Fund to Retain Clinical Scientists awarded to UC Davis School of Medicine by the Burroughs Wellcome Fund. The funding organizations did not play any role in the design or conduct of this study; the collection, management, analysis, or interpretation of data; or the preparation, review, approval, and submission decision of the manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
HUMAN SUBJECTS: No human subjects were included in this study. Due to the nature of the study, institutional review board approval was not required. All research adhered to the tenets of the Declaration of Helsinki.
No animal subjects were used in this study.
Author Contributions:
Conception and design: Nisanova, Yavari, Soltani, Emami
Data collection: Nisanova, Deaner, Ali, Gogte, Kaplan, Chen, Nudleman, Grewal, Gupta, Wolfe, Klufas, Yiu, Emami
Analysis and interpretation: Nisanova, Yavari, Deaner, Ali, Gogte, Kaplan, Chen, Nudleman, Grewal, Gupta, Wolfe, Klufas, Yiu, Soltani, Emami
Obtained funding: Soltani, Emami
Overall responsibility: Nisanova, Yavari, Deaner, Ali, Gogte, Kaplan, Chen, Nudleman, Grewal, Gupta, Wolfe, Klufas, Yiu, Soltani, Emami
The material in this manuscript was presented at the Annual Meeting of the Association for Research in Vision and Ophthalmology (ARVO), 2023.
Contributor Information
Iman Soltani, Email: isoltani@ucdavis.edu.
Parisa Emami-Naeini, Email: parisaemami@gmail.com.
Supplementary Data
References
- 1.Tornambe P.E. Pneumatic retinopexy. Surv Ophthalmol. 1988;32:270–281. doi: 10.1016/0039-6257(88)90175-0. [DOI] [PubMed] [Google Scholar]
- 2.Elhusseiny A.M., Yannuzzi N.A., Smiddy W.E. Cost analysis of pneumatic retinopexy versus pars plana vitrectomy for rhegmatogenous retinal detachment. Ophthalmol Retina. 2019;3:956–961. doi: 10.1016/j.oret.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Ellakwa A.F. Long term results of pneumatic retinopexy. Clin Ophthalmol. 2012;6:55. doi: 10.2147/OPTH.S22063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mandelcorn E.D., Mandelcorn M.S., Manusow J.S. Update on pneumatic retinopexy. Curr Opin Ophthalmol. 2015;26:194–199. doi: 10.1097/ICU.0000000000000148. [DOI] [PubMed] [Google Scholar]
- 5.Hillier R.J., Felfeli T., Berger A.R., et al. The pneumatic retinopexy versus vitrectomy for the management of primary rhegmatogenous retinal detachment outcomes randomized trial (PIVOT) Ophthalmology. 2019;126:531–539. doi: 10.1016/j.ophtha.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 6.Echegaray J.J., Vanner E.A., Zhang L., et al. Outcomes of pars plana vitrectomy alone versus combined scleral buckling plus pars plana vitrectomy for primary retinal detachment. Ophthalmol Retina. 2021;5:169–175. doi: 10.1016/j.oret.2020.09.013. [DOI] [PubMed] [Google Scholar]
- 7.Stewart S., Chan W. Pneumatic retinopexy: patient selection and specific factors. Clin Ophthalmol. 2018;12:493. doi: 10.2147/OPTH.S137607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emami-Naeini P., Deaner J., Ali F.S., et al. Pneumatic retinopexy experience and outcomes of vitreoretinal fellows in the United States: a multicenter study. Ophthalmol Retina. 2019;3:140. doi: 10.1016/j.oret.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hilton G.F., Kelly N.E., Salzano T.C., et al. Pneumatic retinopexy: a collaborative report of the first 100 cases. Ophthalmology. 1987;94:307–314. [PubMed] [Google Scholar]
- 10.Chan C.K., Lin S.G., Nuthi A.S.D., Salib D.M. Pneumatic retinopexy for the repair of retinal detachments: a comprehensive review (1986-2007) Surv Ophthalmol. 2008;53:443–478. doi: 10.1016/j.survophthal.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 11.Mudvari S.S., Ravage Z.B., Rezaei K.A. Retinal detachment after primary pneumatic retinopexy. Retina. 2009;29:1474–1478. doi: 10.1097/IAE.0b013e3181ae70f3. [DOI] [PubMed] [Google Scholar]
- 12.Goldman D.R., Shah C.P., Heier J.S. Expanded criteria for pneumatic retinopexy and potential cost savings. Ophthalmology. 2014;121:318–326. doi: 10.1016/j.ophtha.2013.06.037. [DOI] [PubMed] [Google Scholar]
- 13.Gorovoy I.R., Eller A.W., Friberg T.R., Coe R. Characterization of pneumatic retinopexy failures and the pneumatic pump: a new complication of pneumatic retinopexy. Retina. 2014;34:700–704. doi: 10.1097/IAE.0000000000000002. [DOI] [PubMed] [Google Scholar]
- 14.Tornambe P.E. Pneumatic retinopexy: the evolution of case selection and surgical technique. A twelve-year study of 302 eyes. Trans Am Ophthalmol Soc. 1997;95:551. [PMC free article] [PubMed] [Google Scholar]
- 15.Kiew G., Poulson A.V., Newman D.K., et al. Montgomery and informed consent during Covid-19: pneumatic retinopexy versus pars plana vitrectomy or scleral buckling for retinal detachment repair. Med Leg J. 2021;89:102–105. doi: 10.1177/0025817220984009. [DOI] [PubMed] [Google Scholar]
- 16.Ting D.S.W., Pasquale L.R., Peng L., et al. Artificial intelligence and deep learning in ophthalmology. Br J Ophthalmol. 2019;103:167. doi: 10.1136/bjophthalmol-2018-313173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee C.S., Baughman D.M., Lee A.Y. Deep learning is effective for classifying normal versus age-related macular degeneration OCT images. Ophthalmol Retina. 2017;1:322–327. doi: 10.1016/j.oret.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varadarajan A.V., Bavishi P., Ruamviboonsuk P., et al. Predicting optical coherence tomography-derived diabetic macular edema grades from fundus photographs using deep learning. Nat Commun. 2020;11:130. doi: 10.1038/s41467-019-13922-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zekavat S.M., Sekimitsu S., Ye Y., et al. Photoreceptor layer thinning is an early biomarker for age-related macular degeneration: epidemiologic and genetic evidence from UK biobank OCT data. Ophthalmology. 2022;129:694–707. doi: 10.1016/j.ophtha.2022.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bora A., Balasubramanian S., Babenko B., et al. Predicting the risk of developing diabetic retinopathy using deep learning. Lancet Digit Health. 2021;3:e10–e19. doi: 10.1016/S2589-7500(20)30250-8. [DOI] [PubMed] [Google Scholar]
- 21.Zekavat S.M., Raghu V.K., Trinder M., et al. Deep learning of the retina enables phenome- and genome-wide analyses of the microvasculature. Circulation. 2022;145:134–150. doi: 10.1161/CIRCULATIONAHA.121.057709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abràmoff M.D., Leng T., Ting D.S.W., et al. Automated and computer-assisted detection, classification, and diagnosis of diabetic retinopathy. Telemed J E Health. 2020;26:544. doi: 10.1089/tmj.2020.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.FDA permits marketing of artificial intelligence-based device to detect certain diabetes-related eye problems | FDA. https://www.fda.gov/news-events/press-announcements/fda-permits-marketing-artificial-intelligence-based-device-detect-certain-diabetes-related-eye
- 24.Touma S., Antaki F., Duval R. Development of a code-free machine learning model for the classification of cataract surgery phases. Sci Rep. 2022;12:2398. doi: 10.1038/s41598-022-06127-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antaki F., Kahwati G., Sebag J., et al. Predictive modeling of proliferative vitreoretinopathy using automated machine learning by ophthalmologists without coding experience. Sci Rep. 2020;10:1–10. doi: 10.1038/s41598-020-76665-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Antaki F., Coussa R.G., Kahwati G., et al. Accuracy of automated machine learning in classifying retinal pathologies from ultra-widefield pseudocolour fundus images. Br J Ophthalmol. 2023;107:90–95. doi: 10.1136/bjophthalmol-2021-319030. [DOI] [PubMed] [Google Scholar]
- 27.Faes L., Wagner S.K., Fu D.J., et al. Automated deep learning design for medical image classification by health-care professionals with no coding experience: a feasibility study. Lancet Digit Health. 2019;1:e232–e242. doi: 10.1016/S2589-7500(19)30108-6. [DOI] [PubMed] [Google Scholar]
- 28.Mullainathan S., Obermeyer Z. Does machine learning automate moral hazard and error? Am Econ Rev. 2017;107:476–480. doi: 10.1257/aer.p20171084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levandowsky M., Winter D. Distance between sets. Nature. 1971;234:34–35. [Google Scholar]
- 30.Moulton R., Jiang Y. Proceedings - IEEE International Conference on Data Mining, ICDM. 2018. Maximally consistent sampling and the Jaccard index of probability distributions; pp. 347–356. [Google Scholar]
- 31.Train models to classify data using supervised machine learning - MATLAB. https://www.mathworks.com/help/stats/classificationlearner-app.html
- 32.Tabular data overview | Vertex AI | Google cloud. https://cloud.google.com/vertex-ai/docs/tabular-data/overview
- 33.Waring J., Lindvall C., Umeton R. Automated machine learning: review of the state-of-the-art and opportunities for healthcare. Artif Intell Med. 2020;104 doi: 10.1016/j.artmed.2020.101822. [DOI] [PubMed] [Google Scholar]
- 34.Kumar M., Ang L.T., Png H., et al. Automated machine learning (AutoML)-derived preconception predictive risk model to guide early intervention for gestational diabetes mellitus. Int J Environ Res Public Health. 2022;19:6792. doi: 10.3390/ijerph19116792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abbas A., O'Byrne C., Fu D.J., et al. Evaluating an automated machine learning model that predicts visual acuity outcomes in patients with neovascular age-related macular degeneration. Graefes Arch Clin Exp Ophthalmol. 2022;260:2461–2473. doi: 10.1007/s00417-021-05544-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Faes L., Liu X., Wagner S.K., et al. A clinician's guide to artificial intelligence: how to critically appraise machine learning studies. Transl Vis Sci Technol. 2020;9:7. doi: 10.1167/tvst.9.2.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen J.S., Coyner A.S., Ostmo S., et al. Deep learning for the diagnosis of stage in retinopathy of prematurity: accuracy and generalizability across populations and cameras. Ophthalmol Retina. 2021;5:1027–1035. doi: 10.1016/j.oret.2020.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ting D.S.W., Cheung C.Y.L., Lim G., et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA. 2017;318:2211. doi: 10.1001/jama.2017.18152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Batra R. Accurate machine learning in materials science facilitated by using diverse data sources. Nature. 2021;589:524–525. doi: 10.1038/d41586-020-03259-4. [DOI] [PubMed] [Google Scholar]
- 40.Teo Z.L., Lee A.Y., Campbell P., et al. Developments in artificial intelligence for ophthalmology: federated learning. Asia Pac J Ophthalmol. 2022;11:500–502. doi: 10.1097/APO.0000000000000582. [DOI] [PubMed] [Google Scholar]
- 41.Lu C., Hanif A., Singh P., et al. Federated learning for multicenter collaboration in ophthalmology: improving classification performance in retinopathy of prematurity. Ophthalmol Retina. 2022;6:657–663. doi: 10.1016/j.oret.2022.02.015. [DOI] [PubMed] [Google Scholar]
- 42.Chang K., Beers A.L., Brink L., et al. Multi-institutional assessment and crowdsourcing evaluation of deep learning for automated classification of breast density. J Am Coll Radiol. 2020;17:1653–1662. doi: 10.1016/j.jacr.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dablain D.A., Bellinger C., Krawczyk B., Chawla N.V. Efficient augmentation for imbalanced deep learning. 2022. https://arxiv.org/abs/2207.06080v2
- 44.Woods A.D., Gerasimova D., Van Dusen B., et al. Best practices for addressing missing data through multiple imputation. Infant Child Dev. 2023 [Google Scholar]
- 45.Myers W.R., Statistician S. Handling missing data in clinical trials: an overview. Drug Inf J. 2000;34:525–533. [Google Scholar]
- 46.King R.D., Orhobor O.I., Taylor C.C. Cross-validation is safe to use. Nat Mach Intell. 2021;3:276. [Google Scholar]
- 47.Ling C.X., Huang J., Zhang H. Vol. 2671. 2003. AUC: a better measure than accuracy in comparing learning algorithms; pp. 329–341. (Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics)). [Google Scholar]
- 48.Hand D.J., Christen P., Kirielle N. F∗: an interpretable transformation of the F-measure. Mach Learn. 2021;110:451. doi: 10.1007/s10994-021-05964-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seiffert C., Khoshgoftaar T.M., Van Hulse J., Napolitano A. RUSBoost: a hybrid approach to alleviating class imbalance. IEEE Trans Syst Man Cybern. 2010;40:185–197. [Google Scholar]
- 50.Goodman J., Sarkani S., Mazzuchi T. Distance-based probabilistic data augmentation for synthetic minority oversampling. ACM/IMS Trans Data Sci. 2022;2:1–18. [Google Scholar]
- 51.Sackett D.L., Straus S. On some clinically useful measures of the accuracy of diagnostic tests. BMJ Evid Based Med. 1998;3:68–70. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.