Abstract
The cell wall of the Glycine max altered by the polygalacturonases (PGs) secreted by the fungus Sclerotinia sclerotiorum, causes disease and quality losses. In soybeans, a resistance protein called polygalacturonases-inhibiting proteins (PGIPs) binds to the PG to block fungal infection. The active site residues of PGIP3, VAL170 and GLN242 are mutated naturally by various amino acids in different types of PGIPs. Therefore, the mutation of VAL170 to GLY is ineffective but the GLN242 amino acid mutation by LYS significantly alters the structure and is crucial for interacting with the PG protein. Docking and Molecular Dynamics simulation provide a comprehensive evaluation of the interactions between gmPGIP and ssPG. By elucidating the structural basis of the interaction between gmPGIP and ssPG, this investigation lays a foundation for the development of targeted strategies in-order to enhance soybean resistance against Sclerotinia sclerotiorum. By leveraging this knowledge, researchers can potentially engineer soybean varieties with improved resistance to the fungus, thereby reducing disease incidence and improving crop yields.
Keywords: Site-directed mutagenesis, Soybean, Resistance, Pgip-pg complex
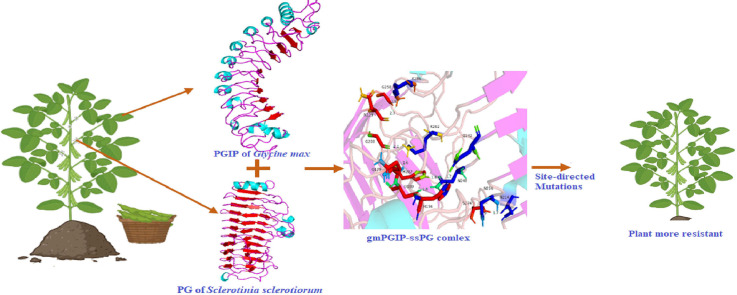
Graphical abstract
Specifications Table
| Subject | Agricultural and structural biology |
| Specific subject area | Bionformatics based host-pathogen interaction, and associated structural and functional biology |
| Type of data | Table and Figures |
| Data collection | Sequences were collected from the NCBI and PDB, modelled the desired structures which were relevant to our research. |
| Data source location | Protein sequences of the data were retrieved from the NCBI (G. max (NP_001304551.2) and S. sclerotiorum (CAF05670.1)) and the structures are from PDB (1OGQ and 2IQ7). |
| Data accessibility | Link & DOI for data deposited in Mendeley Data repository : https://data.mendeley.com/datasets/t8fnb5vmtr/1 DOI : 10.17632/t8fnb5vmtr.1 Primary source of data used https://www.ncbi.nlm.nih.gov/protein/?term=PGIP+in+glycine+max https://www.rcsb.org/structure/2IQ7 https://www.rcsb.org/structure/1OGQ |
1. Value of the Data
-
•
Fungal polygalacturonases (PG) interact with polygalacturonase-inhibiting protein (PGIP) to confer resistance in soybean.
-
•
Alteration in protein in terms of structure and function is caused via site-directed mutagenesis.
-
•
Detail data about the interactions and stability of complexes of fungal PG and G. max PGIP have been generated by docking and MD simulations studies, which will be valuable to researchers in developing resistance variety of soybean against fungal infection.
-
•
Data will serve as a standard for PG-PGIP interaction study
2. Background
Soybean crop is damaged by several biotic factors, where diseases adversely affect the production and reduces the grain yield [1]. The major losses in soybean crop are mainly caused by foliar and stem diseases, nematodes infestation, seedling blights and diseases of harvested grain. Recently, in 2022, sudden death syndrome disease (Fusarium virguliforme) reported an estimated yield loss of 21 million bushels in US and Ontario [2]. Among the diseases, the seedling diseases mainly Sclerotinia sclerotiorum led stem rot and sudden death syndromes (Fusarium virguliforme) have caused greatest yield losses in USA and Canada. The estimated yield losses from Sclerotinia stem rot disease during 1996–2009, was more than 10 million bushels (270 million kg) in seven years out of the 14 years in USA [3,4]. We have tried to look into host-pathogen interactions through the execution of molecular dynamics (MD) studies [[5], [6], [7]]. The study employed the computational approaches coupled with MD simulations to infer that how the binding of G. max PGIP proteins with S. sclerotiorum PG proteins is altered via physio-biochemical and molecular mechanisms. Additionally, study contributes to our understanding of the binding interactions between PGIP and PG, which modify the structure of the enzyme and prevent its activity. The host pathogen interaction studies of PG from S. sclerotiorum and PGIP of soybean (G. max) is essential to decipher etiology and to develop the strategies for integrated disease management and to minimize the losses.
3. Data Description
3.1. Sequence comparison and repeat identification
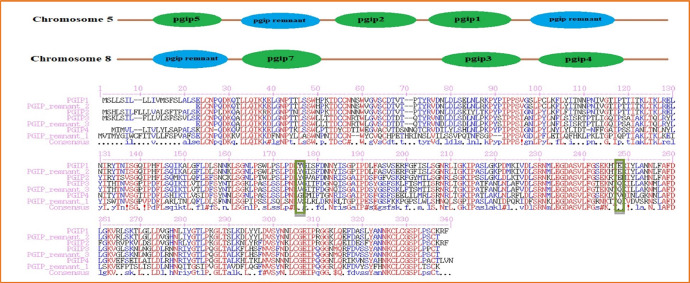
The PGIP protein is encoded by nine genes located on chromosomes 5 and 8. Pgip1, Pgip2 and Pgip5 are positioned on Chr5 whereas Pgip3, Pgip4 and Pgip7 are placed on Chr8. The pgip remnant has three copies; two of them are on chr5, while the third copy is on chr8 (Fig. 1). Recently, another PGIP has been reported in G. max and it deals with pathogens defense mechanism [8].
Fig. 1.
PGIP distribution among the chromosome 5 and 8; Sequence alignment of all PGIP protein of G. max.
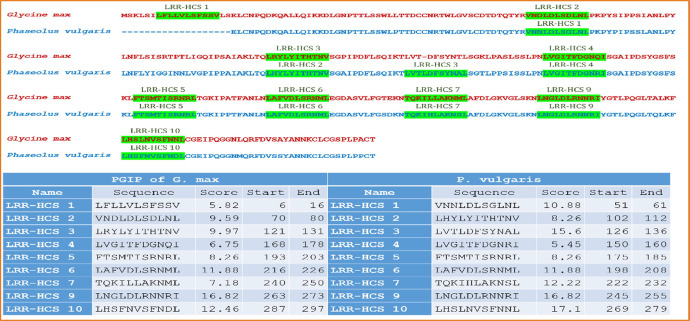
The polygalacturonase-inhibiting protein (PGIP) is a Leucine Rich Repeats (LRRs) structure composed of nine LRRs. PGIP3 of soybean is chosen for further study and it reveals homology with PGIP of P. vulgaris. The distribution of LRR in PGIP are illustrate in (Fig. 2). Only one LRR differs across the nine LRRs that are aligned in the PGIP of G. max and P. vulgaris.
Fig. 2.
Pictorial and tabular representation of LRRs in PGIP for G. max and P. vulgaris.
3.2. Quality assessment of modelled structures
Model structures serve as a visual representation of a molecule, helping to emphasize how different structures appear in living and non-living systems. A large number of PGs' crystal structures are available in the Protein Data Bank, yet just one, P. vulgaris PGIP (1OGQ), is present there. The reason for performing comparative or homology modelling is that the sequences exhibit a high enough proportion of query coverage and sequence identity with pvPGIP. gmPGIP demonstrates 87.79% sequence identity and 100% query coverage with pvPGIP (PDB: 1OGQ). The parasitic ssPG has sequence identity with C. lupini's PG (PDB: 2IQ7) was 69.73%, and its query coverage was 98%.
Finding the optimal structure requires using more than just one tool for structure modelling. By comparison of various parameters like quality score, amino acid 3D profile, Ramachandran plot, RMSD, TM-score, identity and length of modelled residues, it can be observed that the structure modelled by Modeller software is significantly better than the Swiss-model and AlphaFold. All the comparing parameters of various tools and model are mentioned in Table 1.
Table 1.
Quality of modelled gmPGIP and ssPG protein by Swiss-model, AlphaFold and Modeller.
| Tools and Parameters | gmPGIP |
ssPG |
|||||
|---|---|---|---|---|---|---|---|
| Swiss-Model | AlphaFold | Modeller | Swiss-Model | AlphaFold | Modeller | ||
| ERRAT (Overall Quality Factor) | 80.26 | 78.48 | 84.62 | 91.77 | 90.37 | 75.45 | |
| VERIFY | 96.15 % (Pass) | 91.29 % (Pass) | 86.10 % (Pass) | 87.24 % (Pass) | 88.17 % (Pass) | 85.80 % (Pass) | |
| Ramachndran plot | Most favoured regions | 79.6 % | 78.7 % | 83.7 % | 88.1 % | 87.7 % | 88.1 % |
| Additional allowed regions | 20.1 % | 21.0 % | 16.0 % | 11.6 % | 11.9 % | 11.6 % | |
| Generously allowed regions | 0.4 % | 0.4 % | 0.0 % | 0.4 % | 0.4 % | 0.4 % | |
| Disallowed regions | 0.0 % | 0.0 % | 0.3 % | 0.0 % | 0.0 % | 0.0 % | |
| Structure alignment | RMSD | 0.14 | 0.3 | 0.28 | 0.67 | 0.8 | 0.21 |
| TM-score | 1 | 0.99 | 0.94 | 0.97 | 0.97 | 0.98 | |
| Identity | 88 % | 89 % | 89 % | 71 % | 71 % | 71 % | |
| Equivalent residues | 312 | 310 | 312 | 332 | 332 | 332 | |
| Sequence length of modelled residues | 312 | 310 | 331 | 338 | 338 | 338 | |
3.3. Effect of mutation in gmPGIP
Mutations in protein at specific amino acids alter its structure as well as function. But it is not necessary that the effects of mutations get reflected in the structure and functionality of the protein. Intramolecular interactions provide the stability to the protein molecule and maintain their structure. The shape of the protein is totally dependent on the pattern of the amino acids and its properties. When any modification has been occurred, the structure is changed due to the interactions with the neighboring amino acids.
In the case of PGIP of P. vulgaris, the structure is changed by the specific site mutation and its functionality is also changed as reported by Maulik and Basu (2013) [9]. Their analysis showed that the double mutation i.e. VAL152 by GLY and GLN224 by LYS affect the concavity of the PGIP structure and help in inhibiting the binding process with PG parasitic protein. The residues of concave faces provides the interactions with the parasitic protein PG and the convex face provides the flexibility and mobility to the PGIP protein.
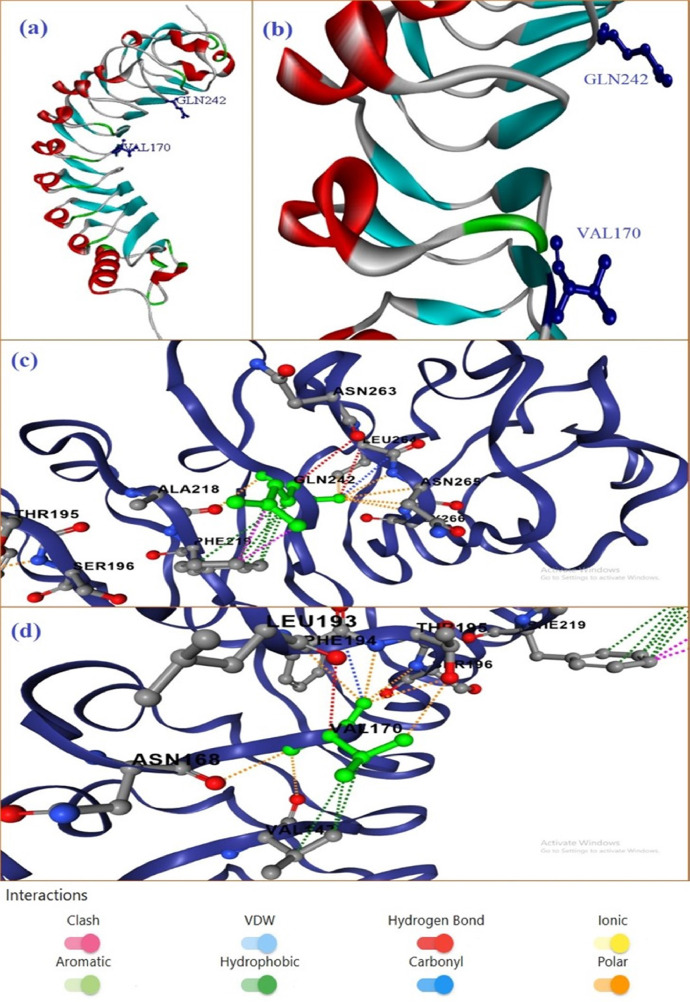
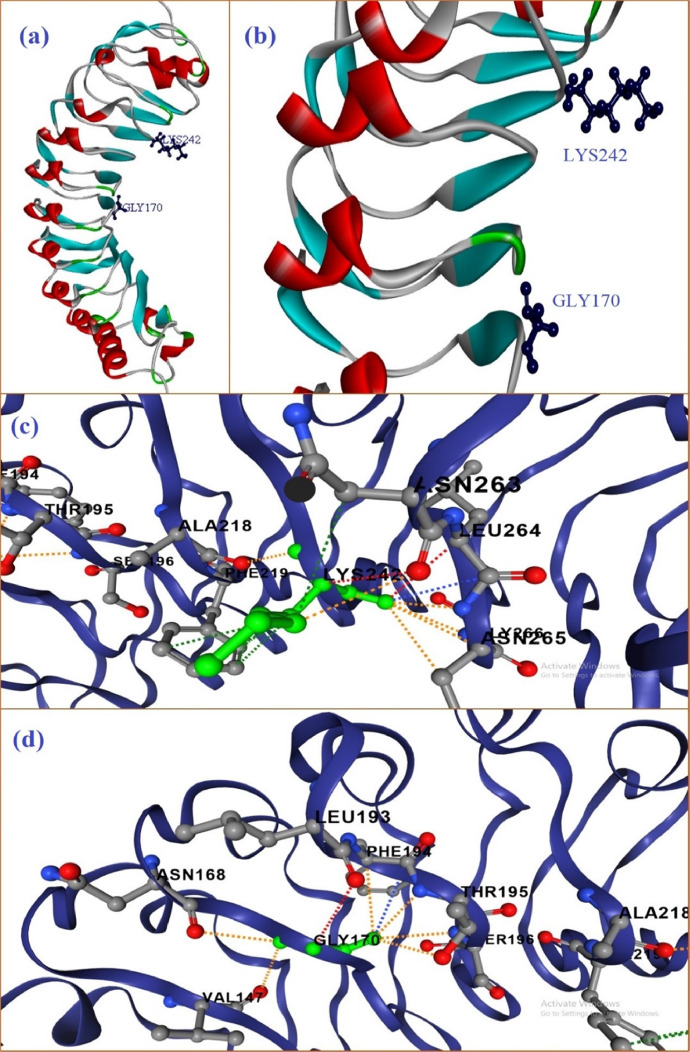
Fig 3 represents the interaction of VAL170 and GLN242 to its neighboring residues. gmPGIP looks like half solenoid structure (Fig. 3, Fig. 3), are a zoom representation of the active residues which are involved in the interactions with the parasitic PG protein. GLN242 interacted with the ALA218, PHE219, ASN263, LEU264, ASN265 and GLY266 by various types of interactions. These interactions are - two hydrogen bonds are made with the ASN263 and LEU264; one carbonyl interactions with LEU264; two clashes with PHE219; six polar interactions with ALA218, LEU264, ASN265 and GLY266; and six hydrophobic interactions with PHE219 (Fig 3(c)). Another residue VAL170 showed different interactions with ASN168, VAL147, LEU193, PHE194, THR195 and SER196. These interactions are – one hydrogen bond with LEU193; one carbonyl interaction with PHE194; three hydrophobic interactions with VAL147, seven polar interactions with ASN168, LEU193, THR195, SER196 and VAL147.
Fig. 3.
PGIP protein of G. max (gmPGIP) (a-b) active residues VAL170 and GLN242 in modelled protein; (c) interactions of amino acid GLN242 with neighboring residues; (d) various interactions of VAL170 with their neighboring residues. The interactions of selected amino acid (green) their neighboring residues are taken from DDMut (Zhou et.al, 2023).
Two residues are mutated form of gmPGIP, VAL170 mutated by GLY and GLN242 mutated by LYS by Mutation Explorer server. After the mutation of GLN242 to LYS, it interact with six amino acids, with mutated amino acids are the same but the pattern of interaction are changed. Two Hydrogen bonds, six polar interactions and one clash have been similar to WT but hydrophobic and carbonyl interactions are changed in MT. Four hydrophobic interactions are formed with PHE219 and one with ASN263. Carbonyl interactions are removed between LYS242 with neighboring residues (Fig 4(c)). GLY170 interacts with the same neighboring amino acids and similar patterns like WT gmPGIP, except hydrophobic interactions which are absent in mutated gmPGIP (Fig 4(d)). The total energy of WT gmPGIP (−187.82 kcal/mol) is little bit low as −2.41 kcal/mol than the double mutated gmPGIP (−185.41 kcal/mol).
Fig. 4.
Mutated PGIP protein of Glycine max (gmPGIP) (a-b) mutated active residues GLY170 and LYS242 in modelled protein; (c) interactions of amino acid LYS242 with neighboring residues; (d) various interactions of GLY170 with their neighboring residues.
3.4. P-P interactions of gmPGIP-ssPG and mutated-gmPGIP-ssPG
Haddock provided the 10 clusters using 158 structures that had a 79 % water-refined model. A gmPGIP-ssPG complex model with a score of −125.5 ± 8.2 HADDOCK was chosen. Other HADDOCK parameters of selected complex are Cluster size (10), RMSD from the overall lowest-energy structure (0.8 +/- 0.4), Van der Waals energy (−55.2 +/- 13.2), Electrostatic energy (−349.5 +/- 65.9), Desolvation energy (−4.5 +/- 6.9), Restraints violation energy (41.0 +/- 18.3), Buried Surface Area (2312.8 +/- 241.4) and Z-Score (−1.7).
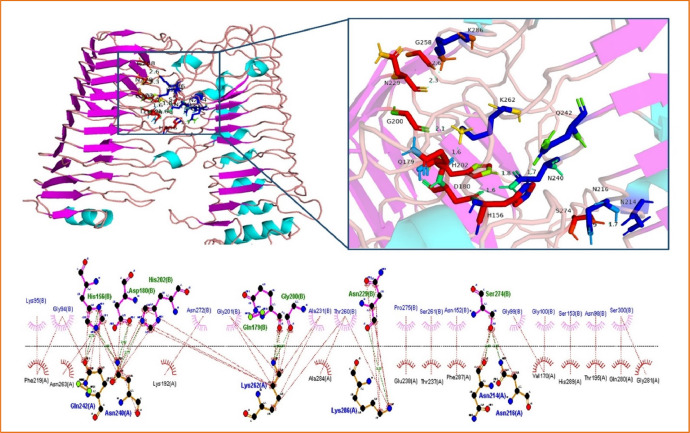
Total nine hydrogen bonds are established, interacting with seven residues of ssPG through six amino acids of gmPGIP (Fig. 5). The Gly242, one of the gmPGIP's active residues, binds with ssPG's HIS156, while VAL170, another gmPGIP residue, interacts hydrophobically with ssPG's GLY99 and GLY100. ASN240 of gmPGIP forms three hydrogen bonds with HIS156, ASP180 and HIS202 residues of ssPG. SER274 of ssPG and the amino acids ASN214 and ASN216 of gmPGIP form two hydrogen bonds. LYS286 of gmPGIP interacts with ASN229 of ssPG and two residues GLN179 and GLY200 of parasitic protein PG interacts to LYS262 of gmPGIP protein.
Fig. 5.
Docked complex of gmPGIP with ssPG and their 2D as well as 3D representation of interacted amino acids, red sticks for ssPG and blue sticks for gmPGIP. Hydrogen bonds are shown by dotted line, light blue in 3D and green in 2D; Red dotted lines are used for hydrophobic interactions.
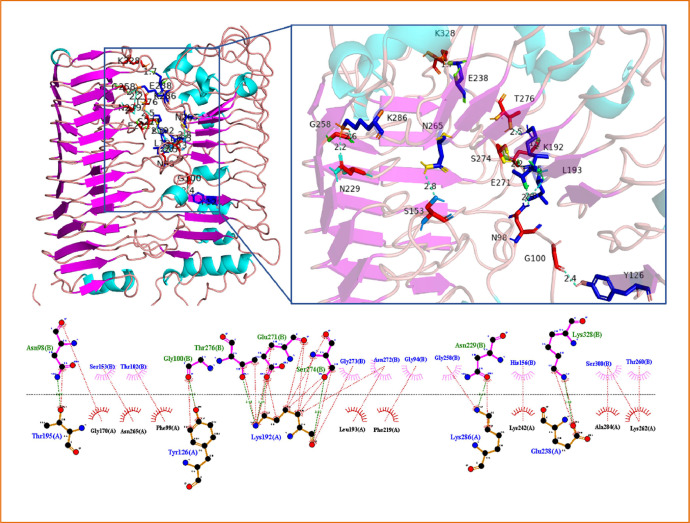
Total seven hydrogen are formed between 5 residues of gmPGIP and 7 residues of ssPG (Fig.6). In the hydrophobic interactions, 12 residues are involved in the WT of gmPGIP and this number is reduced in the MT of gmPGIP. The number of residues of ssPG are also reduced by the interaction with WT and MT. As an effect of the mutation's little alteration to the concave face, the mutant residue LYS242 of gmPGIP forms hydrophobic interactions in the MT rather than hydrogen bonds. Mutational alterations also affect the compactness of contacts between the binding residues of MT-PGIP-PG complexes.
Fig. 6.
Docked complex of mutated-gmPGIP with ssPG and their 2D as well as 3D representation of interacted amino acids, red sticks for ssPG and blue sticks for mutated-gmPGIP. Hydrogen bonds are shown by dotted line, yellow in 3D and green in 2D; Red dotted lines are used for hydrophobic interactions.
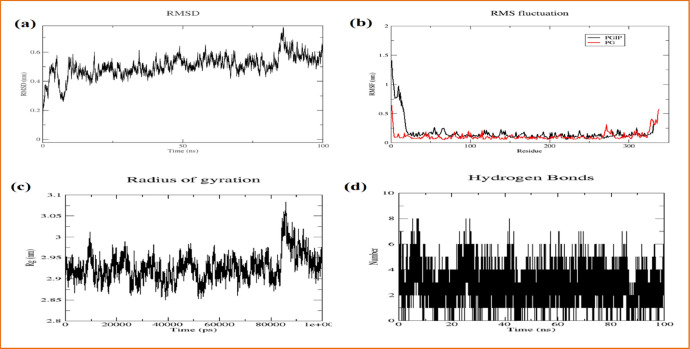
3.5. Simulation study of complexes
MD simulations were run for the WT-gmPGIP-ssPG complex at100ns. RMSD provides the structural distances between the coordinates of different confirmations over the time. It varies between 0.4 and 0.7 nm, indicating how stable the gmPGIP-ssPG complex is (Fig. 7(a)). The RMSF for the both protein gmPGIP and ssPG is extremely low (Fig. 7(b)). The rg reveals the compactness of the structure, the complex's overall compactness is maintained while the rg only fluctuates between at the 85–88 ns (Fig. 7(c)). There is also a significant conservation of hydrogen bonds across time (Fig. 7(d)). These all parameters make it clear that the complex of gmPGIP-ssPG is extremely stable.
Fig. 7.
Diagramatic representation of Molecular Simulations at 100 ns (a) RMSD, (b) RMSF, (c) Radius of gyration and (d) hydrogen bond distribution.
All above data generated in this study have been deposited in Mendeley Data repository, and Link & DOI for the deposited data are given below: https://data.mendeley.com/datasets/t8fnb5vmtr/1
DOI : 10.17632/t8fnb5vmtr.1
While links for all the primary data used in this study are available in the below given links https://www.ncbi.nlm.nih.gov/protein/?term=PGIP+in+glycine+max https://www.rcsb.org/structure/2IQ7 https://www.rcsb.org/structure/1OGQ
4. Experimental Design, Material and Methods
4.1. Protein structure prediction and validation
The accession numbers of G. max (NP_001304551.2) and S. sclerotiorum (CAF05670.1) were used to download the PGIP and PG sequences from the NCBI. To determine homologs, we ran a BLAST search against the Protein Data Bank [10,11]. Selected the template on the basis of top most hits, highest percent identity and query coverage for homology modelling. The Swiss-model [12], AlphaFold [13], and Modeller [14] programs are used very frequently to predict three-dimensional structures of the proteins. The quality of the modelled structures was confirmed using SAVES (https://saves.mbi.ucla.edu/) metaserver, and the best structure was chosen by structural alignment of modelled protein with the template structures using Pairwise Structural Alignment, a feature of the PDB system. Saves metaservers used five programs to check and validate the three-dimensional structure of proteins i.e. ERRAT, Verify3D, WHATCHECK, PROCHECK and PROVE. Comparing the modelled structures obtained by selected three programs, the Modeller exhibit the superior quality structures over the other programs. LRRsearch [15] was used to find repeats for the comparison of target PGIP protein sequence with the selected template.
4.2. Mutation in structure
It estimates the stability of mutation in the protein structure based on evaluation of torsion angle distribution and amino acid-atom potentials. Site-directed mutations were carried out using Mutation Explorer [16] utilizing the P. vulgaris as a reference.
4.3. Protein-protein docking and mutation
The PGIP and PG docking were performed using HADDOCK 2.4 at the specified active site residues whish are predicted by comparison of modelled PGIP protein with the 1OGQ protein [17]. It predict the interactions between PGIP-PG and calculates the HADDOCK score on the basis of several parameters like force constant, ambiguous restraints, radius of gyration restraint, rotation, clustering methods, Dihedral angle/hydrogen bond restraints and more. And most important are the energy and interaction parameters, that are electrostatic and intermolecular interactions as well as cross docking iterations which are most crucial.
4.4. MD simulation of complex
Molecular dynamics (MD) simulations was used to explore the stability, conformational changes and dynamic behavior of the ptotein-protein complexes i.e. gmPGIP-ssPG [18,19]. The simulations were initiated by putting the docked structures in an ion-filled water box using the GROMACS/2021 package and the CHARMm27 force field. In order to neutralize the system, the docked complexes were placed within a cubic box with the correct number of chlorine ions added. Any steric clashes were fixed using the energy minimization (EM) strategy that employed a steep descent method with a threshold of 1000 kJ/mol and 50,000 iterations or reduction steps. The solvent and ions system were stabilized by a two-phase equilibration procedure such as NVT and NPT. The system's temperature was equilibrated at 300 K using the NVT ensemble phase. Next, NPT ensemble, equilibrate and stabilized system's pressure by retaining a constant number of particles, pressure and temperature. Every phase run by 100 ps and final run was conducted for 100 ns to determine the atomic trajectories.
4.5. Trajectories analysis
The end of the Molecular Dynamics (MD) simulations got massive volumes of data by variations in confirmations of the individual atoms and molecules over the time. The stability of the complex was analysed by various intrinsic parameters that are root-mean-square deviation (RMSD), root-mean-square fluctuation (RMSF), radius of gyration (Rg), and hydrogen bond distribution. The RMSD and RMSF were calculated by the rms and rmsf modules respectively, to see the stability of the backbone of protein. Furthermore, the changes in the shape and size of the complex were determined through the Rg by gyrate modules. The obtained results were visualized by qtgrace.
Limitations
None.
Ethical Statement
We have read and followed the ethical requirements for publication in Data in Brief and confirm that the current work does not involve human subjects, animal experiments, or any data collected from social media platforms.
CRediT authorship contribution statement
Mayank Rashmi: Data curation, Writing – review & editing, Software, Writing – original draft. Sneha Murmu: Data curation, Writing – review & editing, Software, Writing – original draft. Dipak T. Nagrale: Conceptualization, Methodology, Supervision. Mahender Kumar Singh: Software. Santosh Kumar Behera: Software. Raja Shankar: Writing – review & editing. Rajiv Ranjan: Writing – review & editing. Girish Kumar Jha: Writing – review & editing. Anurag Chaurasia: Writing – review & editing. Sunil Kumar: Conceptualization, Methodology, Supervision.
Acknowledgments
Acknowledgements
We gratefully acknowledge CABin grant (F. no. Agril. Edn.4-1/2013-A & P), Indian Council of Agricultural Research, Ministry of Agriculture and Farmers’ Welfare, Govt. of India and ICAR-Indian Agricultural Statistics Research Institute, New Delhi for providing facilities and support. We acknowledge the Department of Biotechnology, Govt. of India for BIC project grant (BT/PR40161/ BTIS/137/32/2021).
Declaration of Competing Interest
The authors declare no conflicts of interest.
Contributor Information
Anurag Chaurasia, Email: govtofindia.icar@gmail.com.
Sunil Kumar, Email: Sunil.Kumar7@icar.gov.in.
Data availability
References
- 1.Bandara A.Y., Weerasooriya D.K., Bradley C.A., Allen T.W., Esker P.D. Dissecting the economic impact of soybean diseases in the United States over two decades. PLoS ONE. 2020;15(4) doi: 10.1371/journal. pone.0231141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crop protection Network, CPN Soybean disease loss estimates from the United States and Ontario. Canada-2022. 2022 CPN-1018-22. Published: 04/19/2023. [Google Scholar]
- 3.Wrather A., Koenning S. Effects of diseases on soybean yields in the United States 1996 to 2007. Plant Health Progress. 2009 doi: 10.1094/PHP-2009-0401-01-RS. [DOI] [Google Scholar]
- 4.Koenning S., Wrather J. Suppression of soybean yield potential in the continental United States by plant disease from 2006 to 2009. Plant Health Prog. 2010 doi: 10.1094/PHP-2010-1122-01-RS. [DOI] [Google Scholar]
- 5.Fazil M.H., Kumar S., Farmer R., Pandey H.P., Singh D.V. Binding efficiencies of carbohydrate ligands with different genotypes of cholera toxin B: molecular modeling, dynamics and docking simulation studies. J. Mol. Model. 2012;18(1):1–10. doi: 10.1007/s00894-010-0947-6. Epub 2011 Mar 16. PMID: 21409571. [DOI] [PubMed] [Google Scholar]
- 6.Choudhary P., Bhowmik A., Chakdar H., Khan M.A., Selvaraj C., Singh S.K., Murugan K., Kumar S., Saxena A.K. Understanding the biological role of PqqB in Pseudomonas stutzeri using molecular dynamics simulation approach. J. Biomol. Struct. Dyn. 2022;40(9):4237–4249. doi: 10.1080/07391102.2020.1854860. Epub 2020 Dec 8. PMID: 33287678. [DOI] [PubMed] [Google Scholar]
- 7.Jha Y., Dehury B., Kumar S.P.J., Chaurasia A., Singh U.B., Yadav M.K., Angadi U.B., Ranjan R., Tripathy M., Subramanian R.B., Kumar S., Simal-Gandara J. Delineation of molecular interactions of plant growth promoting bacteria induced β-1,3-glucanases and guanosine triphosphate ligand for antifungal response in rice: a molecular dynamics approach. Mol. Biol. Rep. 2021 doi: 10.1007/s11033-021-07059-5. PMID: 34914086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Acharya S., Troell H.A., Billingsley R.L., Lawrence K.S., McKirgan D.S., Alkharouf N.W., Klink V.P. Data analysis of polygalacturonase inhibiting proteins (PGIPs) from agriculturally important proteomes. Data Br. 2023;52 doi: 10.1016/j.dib.2023.109831. PMID: 38076472; PMCID: PMC10698527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maulik A., Basu S. Study of Q224K, V152G double mutation in bean PGIP2, an LRR protein for plant defense–an in silico approach. Proteins. 2013;81(5):852–862. doi: 10.1002/prot.24243. [DOI] [PubMed] [Google Scholar]
- 10.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 11.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwede T., Kopp J., Guex N., Peitsch M.C. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31(13):3381–3385. doi: 10.1093/nar/gkg520. PMID: 12824332; PMCID: PMC168927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jumper J., Evans R., Pritzel A. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Webb B., Sali A. Current Protocols in Bioinformatics. John Wiley & Sons, Inc.; 2016. Comparative protein structure modeling using modeller. 545.6.1-5.6.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bej A., Sahoo B.R., Swain B., Basu M., Jayasankar P., Samanta M. LRRsearch: an asynchronous server-based application for the prediction of leucine-rich repeat motifs and an integrative database of NOD-like receptors. Comput. Biol. Med. 2014;53:164–170. doi: 10.1016/j.compbiomed.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 16.M. Philipp, C.W. Moth, N. Ristic, J.K.S. Tiemann, F. Seufert, J. Meiler, P.W. Hildebrand, A. Stein, D. Wiegreffe, R. Staritzbichler, MutationExplorer - a webserver for mutation of proteins and 3D visualization of energetic impacts, bioRxiv 2023.03.23.533926; 10.1101/2023.03.23.533926. [DOI] [PMC free article] [PubMed]
- 17.van Zundert G.C.P., Rodrigues J.P.G.L.M, Trellet M., Schmitz C., Kastritis P.L., Karaca E., Melquiond A.S.J., van Dijk M., de Vries S.J., Bonvin A.M.J.J. The HADDOCK2.2 webserver: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016;428:720–725. doi: 10.1016/j.jmb.2015.09.014. (2015) [DOI] [PubMed] [Google Scholar]
- 18.Singh S., Sablok G., Farmer R., Singh A.K., Gautam B., Kumar S. Molecular dynamic simulation and inhibitor prediction of cysteine synthase structured model as a potential drug target for trichomoniasis. Biomed. Res. Int. 2013;2013 doi: 10.1155/2013/390920. Epub 2013 Sep 1. PMID: 24073401; PMCID: PMC3773994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varshney N., Murmu S., Baral B., Kashyap D., Singh S., Kandpal M., Bhandari V., Chaurasia A., Kumar S., Jha H.C. Unraveling the Aurora kinase A and Epstein-Barr nuclear antigen 1 axis in Epstein Barr virus associated gastric cancer. Virology. 2023;588 doi: 10.1016/j.virol.2023.109901. Epub 2023 Oct 9. PMID: 37839162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.