ABSTRACT
Central India faced major dengue outbreaks in 2019 and 2021. In the present study, we aimed to identify the dengue virus serotypes and genotypes circulating in Central India during the COVID pre-pandemic year (2019) and ongoing-pandemic year (2021). For this purpose, the suspected cases were first tested by serological assays. Sero-positive samples were then subjected to molecular diagnosis by RT-PCR and semi-nested PCR. The serotypes obtained were confirmed by nucleotide sequencing. A phylogenetic analysis of serotypes was performed to identify the circulating genotypes. All four DENV serotypes were detected during 2019 and 2021, with the predominance of DENV2. Cases with multiple DENV serotype infections were also identified, involving DENV-2 in all the coinfections. Genotyping revealed that DENV-1 (Genotype V, American/African), DENV-2 (Genotype IV, Cosmopolitan), DENV-3 (Genotype III, Cosmopolitan), and DENV-4 (Genotype I) were involved during both outbreaks. DENV-2 detected in 2019 and 2021 has diverged from the previous strains detected in Central India (2016 and 2018), which may account for the higher transmission of DENV-2 during these outbreaks. The detection of heterologous DENV serotypes with high transmission efficiency calls for continuous viral monitoring and surveillance, which will contribute to a better understanding of changing viral dynamics and transmission patterns.
KEYWORDS: Dengue, serotype, genotype, Central India
1. Introduction
Dengue virus (DENV) infection caused by the bite of infected Aedes mosquitoes, has expanded throughout the tropical regions of the world. According to WHO, the number of dengue cases has increased 10-fold since 2000 [1]. In India also, the number of suspected dengue cases in 2021 has almost doubled since 2015 [2]. Several factors such as global warming, rapid urbanization, and increased travel have been attributed to the upsurge in cases. In 2019, a dengue outbreak was observed in Central India, while in 2020, all the healthcare resources like hospitals, laboratory testing, surveillance, etc were directed toward COVID-19, leading to under-diagnosis and under-reporting of dengue cases. Again in 2021, there was an upsurge in dengue cases during the monsoon and post-monsoon seasons.
DENV has been antigenically classified into four serotypes (DENV-1 to 4) and genetic variation between these serotypes has already been reported [3]. Each DENV serotype is further classified into different genotypes based on nucleotide variations. DENV-1 is further subdivided into five genotypes, with genotype I being the Asian genotype, while genotype II and III belonging to Thailand and Malaysia isolates respectively. Genotype IV is the South Pacific genotype and genotype V is the American/African genotype [4]. DENV-2 has been classified into six genotypes: the Asian genotypes (genotypes I and II), the American/Asian genotype (genotype III), the cosmopolitan genotype (genotype IV), and the American genotype (genotype V). The sylvatic genotype of DENV-2 strains was recovered from humans, forest mosquitoes, and sentinel monkeys in West Africa and Southeast Asia [5–7]. DENV-3 is further divided into five genotypes (I-V) in the human lineage [8,9]. The majority of DENV-3 infections are caused by genotypes I, II, and III, which are associated with DF/DHF epidemics in the regions of Southeast Asia, the South Pacific, the Indian subcontinent, East Africa, and the Americas [10]. DENV-4 genotype I (Thailand, Sri Lanka, Philippines, and Japan), genotype II (Indonesia, Malaysia, Tahiti, the Caribbean, and the Americas), genotype III (Thai isolates), and genotype IV (Sylvatic strains from Malaysia) were analyzed using complete E gene sequences [11,12].
Genotypic differences play an important role in virulence. Intra-epidemic evolution among the circulating strains has been linked to disease transmission. The emergence of DENV lineages has crucial implications for understanding dengue epidemiology and control, since it is often associated with differential disease incidence and severity. Therefore, in light of the above, the present study was conducted to elucidate the DENV serotypes and genotypes during the pre-pandemic year (2019) and the ongoing-pandemic year (2021).
2. Material and methods
2.1. Study design and ethics statement
This cross-sectional, hospital-based study was performed on Dengue-suspected patients (based on WHO case definitions) presenting with fever/joint pain to the Medicine, Pediatrics and Obstetrics & Gynaecology outpatient department of our tertiary care teaching hospital from Jan 2019 to Dec 2021. The study protocol was duly approved by the Institutional Ethics Committee (IEC) vide letter no. 629/MC/IEC/2020. The study participants were recruited after taking written informed consent.
2.2. Sample collection and Processing
Blood samples were collected at the central collection facility of our hospital (associated with Gandhi Medical College, Bhopal) and transported to our laboratory. Clinical and demographic data were collected in a predesigned case-record form. Aseptically collected blood sample was received in a vacutainer vial and then allowed to clot at room temperature (20–25°C). After which, blood was centrifuged at 3000 rpm for 3 min to separate serum. Separated serum was transferred to a microcentrifuge tube for serological diagnosis and stored at 2–8°C until used. For molecular detection, serum was stored in cryovials at −80°C until use.
2.3. Serological diagnosis
For serological diagnosis of DENV, Dengue IgM Capture ELISA kit (manufactured and supplied by National Institute of Virology, Pune, India) and Dengue NS1 Ag Microlisa kit (J. Mitra & Co. Pvt. Ltd, New Delhi, India) was used following manufacturer’s protocols.
2.4. Molecular diagnosis and serotyping
Molecular diagnosis was performed on samples positive by either the IgM ELISA and/or the NS1 antigen ELISA. Briefly, viral RNA was extracted from 140 µl of serum using HiGenoMB Viral RNA Purification Kit (Himedia, India) according to the manufacturer’s protocol. Eluted RNA was stored at − 80°C until further use. For RT-PCR, SSIII platinum one-step RT-PCR kit (Invitrogen, U.S.A.) and DENV group-specific consensus primers (D1 and D2) targeting the DENV capsid-premembrane gene were used. This cPrM-based PCR assay is routinely used in diagnostic settings as the consensus primers are broadly reactive to amplify all the circulating dengue isolates, making it suitable tool for diagnosis as well as epidemiological surveillance of dengue fever [13]. PCR cycling conditions were as per kit instructions. After amplification, the PCR products were run on 1% agarose gel and visualized on a Gel Documentation system (Aplegen, U.S.A.). A band of 511 bp was identified as a DENV positive sample. Positive and negative controls were run alongside.
For serotyping, RT-PCR positive samples were further subjected to multiplex RT- PCR/semi-nested RT-PCR using the following primers viz., forward D1 and four serotype-specific reverse primers (Ts1, Ts2, Ts3 and Ts4) [13]. Following which PCR products were electrophoresed and identified by different band sizes viz. 482 bp (DENV-1), 119 bp (DENV-2), 290 bp (DENV-3) and 392 bp (DENV-4).
2.5. Nucleotide sequencing
DENV serotypes detected by PCR were further confirmed by DNA sequencing using the Sanger’s method. For this, representative samples of each serotype (10–30%) were selected based on the prevalence from both the years (2019–2 samples of DENV-1, 8 samples of DENV-2, 2 samples of DENV-3, 1 sample of DENV-4; 2021–2 samples of DENV-1, 5 samples of DENV-2, 4 samples of DENV-3, 1 sample of DENV-4) and then the capsid-premembrane gene of DENV was sequenced using the above-mentioned primers (D1 and D2) [13] as per the following protocol. For this purpose, first, the PCR product amplified by D1 and D2 primers were agarose gel purified using the QIAquick gel extraction kit (Qiagen, Germany). Further, the sequencing reaction was carried out using a Bigdye terminator cycle sequencing kit (Applied Biosystems, U.S.A.) and the purified PCR product was used as a template. The reaction mixture was then purified through a column and loaded onto a 3130×l Genetic Analyzer (Applied Biosystems, US). After obtaining the nucleotide sequences, BLAST analysis was performed on the NCBI website (National Centre for Biotechnology Information) to confirm the serotypes.
2.6. Phylogenetic analysis
The obtained nucleotide sequences were aligned with different Indian and global DENV sequences using the clustalW module in MEGA 7.0 software [14]. After alignment, a phylogenetic tree was constructed using the Maximum Likelihood method based on the Tamura Nei model in MEGA 7.0 software [15]. The tree topologies were verified using 1000 bootstrap replicates of the dataset. The DENV-1 tree was constructed from 41 Indian and global sequences belonging to five genotypes. DENV-2 tree was constructed using 41 Indian and global sequences belonging to five genotypes. DENV-3 tree was constructed using 35 sequences belonging to three genotypes. DENV-4 tree was constructed using 35 sequences belonging to three genotypes. For the outgroup, a sequence of sylvatic viruses belonging to the same serotype or a sequence of different serotypes was used.
2.7. Statistical analysis
The chi-square test was used to compare categorical variables such as gender and clinical manifestations. An independent two-tailed t-test and ANOVA were used to compare continuous variables such as age distribution and mean disease duration. p < .05 was considered as statistically significant.
3. Results
In 2019, out of 1447 suspected samples, 606 (41.8%) (512: IgM+, 94: NS1+, 18: IgM & NS1+) were serologically positive, and in 2021, out of 1395 suspected samples, 546 (39.1%) (455: IgM+, 91: NS1+, 12: IgM & NS1+) were serologically positive. A schematic representation of the study layout has been shown in Figure 1. In both the years, males were infected more (65.6% and 60.2%) than females (34.3% and 39.7%). The age group most commonly affected by dengue infections was 16–45 years old, followed by 6–15 years. The most common clinical presentation was fever, which occurred in all the patients. Other symptoms observed in seropositive patients were chills (25%), rigor (8.3%), malaise (11.6%), myalgia (16.4%), arthralgia (4.8%), vomiting (10.2%), rash (9.7%), headache (11.0%), retro-orbital pain (5.1%), hemorrhagic manifestations (4.0%) and joint pain. Other clinical manifestations seen were sore throat, cough, abdominal pain, diarrhea, bullae, and new onset of seizures.
Figure 1.
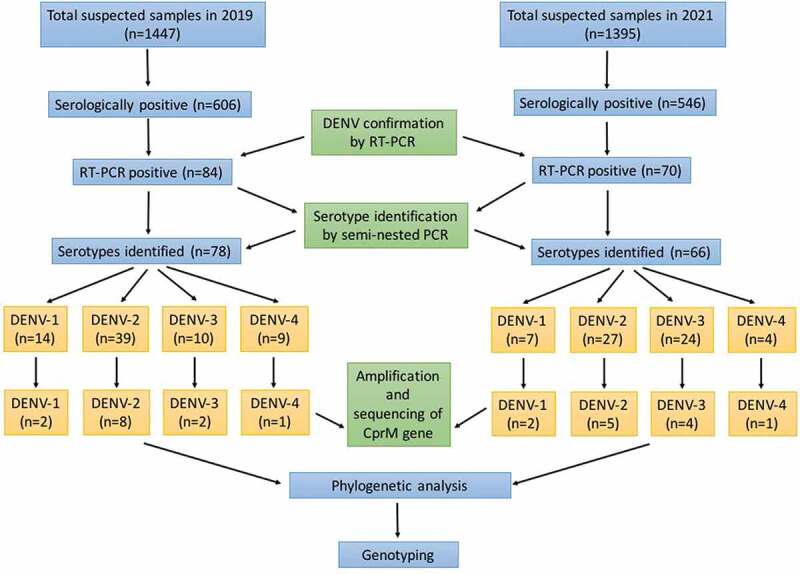
Schematic representation of the work flow.
The temporal distribution of dengue cases during the 2019 outbreak revealed seropositivity onset from June onwards, while a peak was observed during November (496) with 278 being seropositive, out of which 35 were RT- PCR positive. Due to the COVID-19 pandemic in 2020, dengue-suspected samples for the year were not included in the study as very few samples were received, low seropositivity was observed and none of the samples were positive by RT-PCR. Due to the second wave of the COVID-19 pandemic in 2021, the majority of samples were received during the monsoon and post-monsoon season, i.e. from August to October. A maximum number of samples (498) were received in the month of October with 221 being seropositive, out of which 29 were RT- PCR positive (Figure 2).
Figure 2.
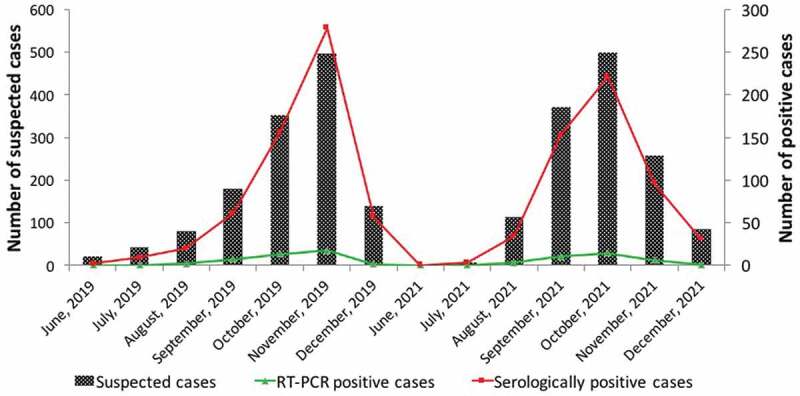
Temporal distribution of dengue cases during the 2019 and 2021 outbreak. X-axis represents the month and year of the outbreak, Y-axis represents the number of suspected cases, secondary axis represents the number of positive cases.
3.1. Identification of DENV serotypes
Among all the serologically positive samples, 84 samples (13.8%) and 70 samples (12.8%) were positive for viral RNA in 2019 and 2021 respectively. In 2019, 78 cases were found to be infected with a single serotype (DENV-1 = 14, DENV-2 = 39, DENV-3 = 10, DENV-4 = 9), and 6 cases were infected with two serotypes (DENV 1 & 2 = 3, DENV 2 & 3 = 2, DENV 2 & 4 = 1). In 2021, 66 cases were found to be infected with a single serotype (DENV-1 = 7, DENV-2 = 27, DENV-3 = 24, DENV-4 = 4), and 4 cases were infected with two serotypes (DENV 2 & 3 = 2, DENV 2 & 4 = 2). Among the coinfected cases, the patients did not show any different or unusual clinical syndromes as compared to single-infected ones. DENV-2 was the dominant serotype circulating during the study period, followed by DENV-3. DENV-4 was the least detected serotype. The serotypes identified by RT-PCR were validated by nucleotide sequencing of a panel of selected samples as shown in Figure 1. BLAST analysis of the nucleotide sequences demonstrated complete concordance with RT-PCR results. The following accession numbers of the serotypes were obtained after the submission of nucleotide sequences in GenBank (2019: OR215469, OR215470, OR215471, OR215472, OR215473, OR215474, OR215475, OR215476, OR215477, OR215478, OR215479, OR215480, OR215481; 2021: OR215482, OR215483, OR215484, OR215485, OR215486, OR215487, OR215488, OR215489, OR215490, OR215491, OR215492, OR215493).
3.2. Genotype identification of DENV-1 isolates
For genotype identification of DENV-1 isolates, 41 representative DENV-1 strains from India, as well as diverse geographical origins, were retrieved from GenBank. In this study, four DENV-1 sequences were obtained as observed by BLAST analysis. These sequences were aligned with other 41 strains and edited to a final length of 446 bp. The phylogenetic tree based on the CprM region revealed five distinct genotypic groups (Figure 3). All DENV-1 strains belong to genotype V (American/African) and are clustered into one group along with DENV-1 from Central India 2016 & 2018 strains. Indian strains from Kerala (2008–09), Gwalior (2010–11), and Delhi 2009 were forming a separate group within this genotype. Other strains that represent this genotype are Singapore (2013), Myanmar (1998), Brazil (2006), Argentina (2000), Brazil (1997), Columbia (2006), Mexico (2008), Venezuela (1997), and Indian prototype strain, Vellore (1964). Genotype I was represented by Asian isolates (Sri Lanka, Myanmar, Vietnam, Cambodia, Thailand, and Singapore). Genotypes II and III were represented by isolates from Thailand and Malaysia respectively. Genotype IV was represented by viruses from South Pacific (Seychelles, Reunion, French Polynesia, Micronesia, New Caledonia, and Nauru island). The sylvatic isolate from Brunei formed an outgroup.
Figure 3.
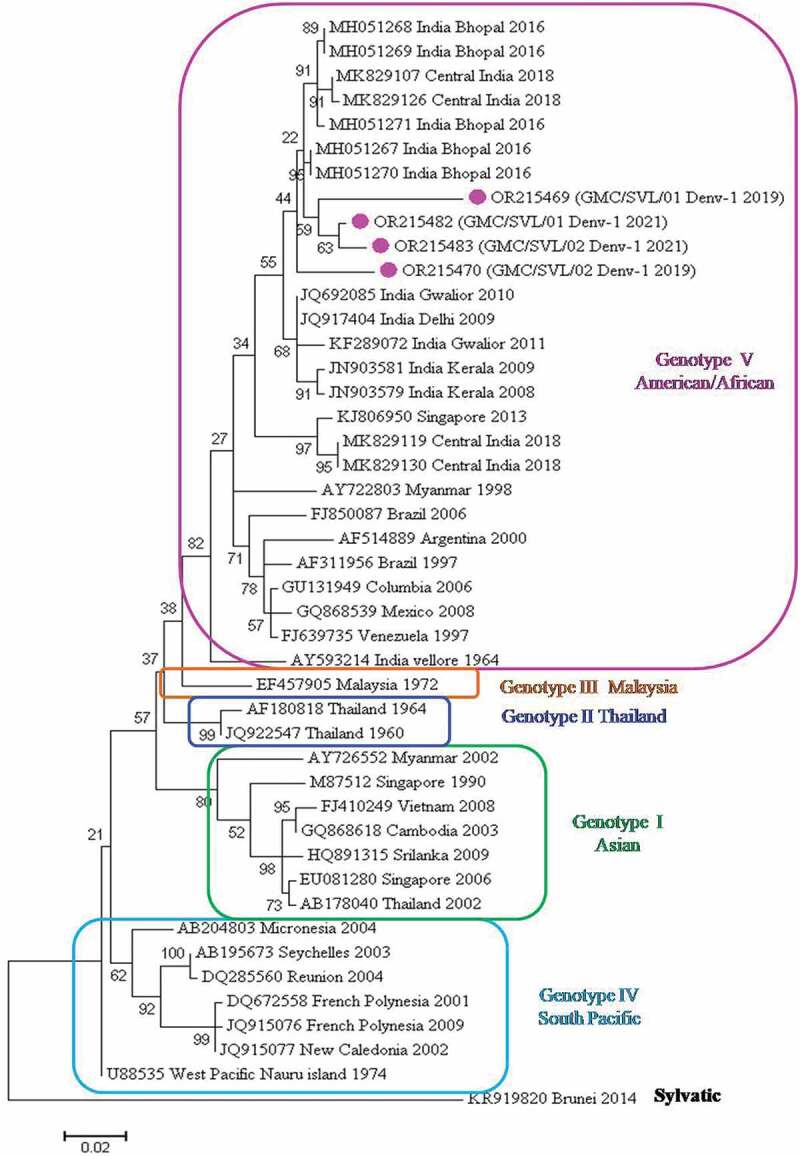
Phylogenetic tree based on CprM gene of DENV-1. Each strain is identified by its GenBank accession number, country/state/city of origin and the year of isolation. The DENV-1 viruses sequenced in this study are marked with a pink-colored circle symbol.
3.3. Genotype identification of DENV-2 isolates
For genotype identification of DENV-2 isolates, 41 representative DENV-2 strains from India, as well as diverse geographical origins, were retrieved from GenBank. In this study, 13 DENV-2 sequences were obtained as observed by BLAST analysis. These sequences were aligned along with the other 41 strains and edited to a final length of 440 bp. The phylogenetic tree based on the CprM region revealed five distinct genotypic groups (Figure 4). All the 13 strains of DENV-2 belong to genotype IV (Cosmopolitan). Only one strain from 2019 was forming a cluster along with Central Indian strains of 2016 and 2018 and recent strains from Kerala, Delhi, Kolkata, Lucknow and Karnataka. While the other 12 strains are clustered into a separate group along with Taiwan, Indonesia, and Singapore isolates indicating genetic divergence. Isolates from China belong to Genotype I (Asian-II) and isolates from Thailand and Cambodia belongs to Genotype II (Asian-I). The American/Asian genotype (III) was represented by viruses from Brazil, Puerto Rico, and U.S.A.. The American genotype (V) was represented by viruses from Colombia, Venezuela and, India. The sylvatic isolate from Malaysia formed an outgroup.
Figure 4.
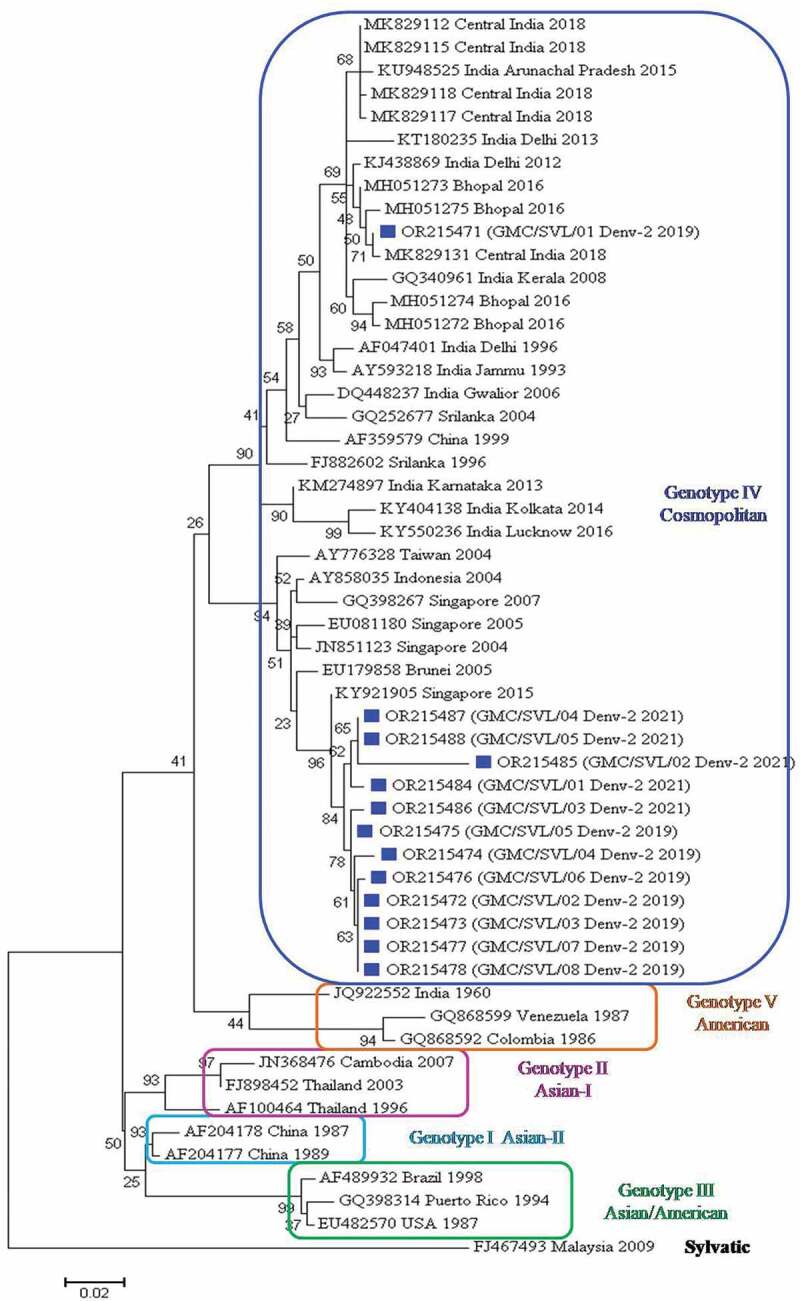
Phylogenetic tree based on CprM gene of DENV-2. Each strain is identified by its GenBank accession number, country/state/city of origin and the year of isolation. The DENV-2 viruses sequenced in this study are marked with a blue-colored square symbol.
3.4. Genotype identification of DENV-3 isolates
For genotype identification of DENV-3 isolates, 35 representative DENV-3 strains from India, as well as diverse geographical origins, were retrieved from GenBank. In this study, six DENV-3 sequences were obtained as observed by BLAST analysis. These sequences along with the other 35 strains were aligned and edited to a final length of 407 bp. The phylogenetic tree based on the CprM region revealed three distinct genotypic groups (Figure 5). All six strains of DENV-3 belong to lineage 3 within genotype III (Cosmopolitan) and are clustered together with 2016 and 2018 Central Indian strains. Other isolates grouped into genotype III belong to Asian, the Pacific Island and, South American countries. Further, genotype III is divided into five lineages. Lineage I is represented by isolates from the Americas. Lineage II & V are represented by isolates from Sri Lanka. Lineages III & IV are represented by Indian & Singapore strains respectively. Genotype I is represented by DENV-3 from Indonesia, Taiwan, Philippines, Samoa, French Polynesia, and New Caledonia. Genotype II is represented by DENV-3 from Asian countries. The DENV-2 sequence was used as an outgroup.
Figure 5.
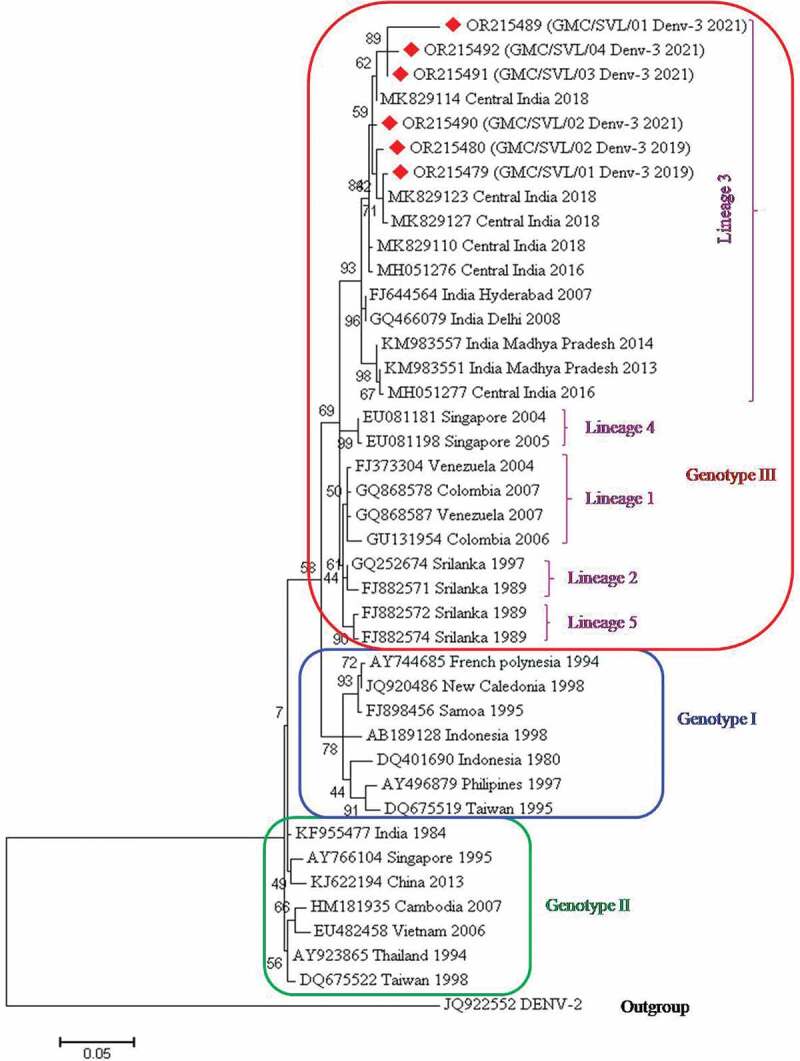
Phylogenetic tree based on CprM gene of DENV-3. Each strain is identified by its GenBank accession number, country/state/city of origin and the year of isolation. The DENV-3 viruses sequenced in this study are marked with a red-colored diamond symbol.
3.5. Genotype identification of DENV-4 isolates
For the genotype identification of DENV-4 isolates, 35 representative DENV-4 strains from India, as well as diverse geographical origins, were retrieved from GenBank. In this study, two DENV-4 sequences were obtained as observed by BLAST analysis. These sequences were aligned with the other 35 strains and edited to a final length of 427 bp. The phylogenetic tree based on the CprM region revealed three distinct genotypic groups (Figure 6). Both the strains of DENV-4 belong to genotype I and are clustered together with Central Indian strains of 2016 and 2018 and other Indian strains as described above. Other isolates grouped into this genotype I are from Asian countries (Sri Lanka, Thailand, Singapore, Cambodia, China, and the Philippines). Genotype II was represented by the isolates from South America (Venezuela, Brazil, and Colombia). Genotype III was represented by isolates from Asian countries (Thailand, Indonesia, Taiwan, China, and Singapore). The DENV-2 sequence was used as an outgroup.
Figure 6.
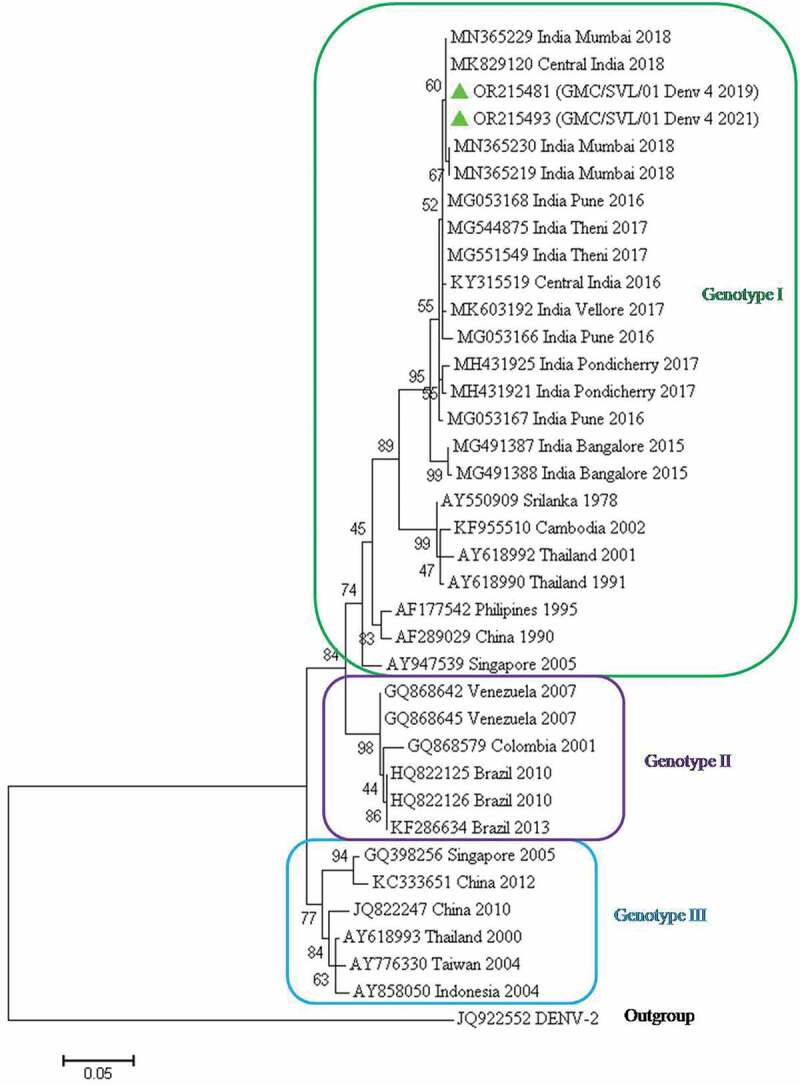
Phylogenetic tree based on CprM gene of DENV-4. Each strain is identified by its GenBank accession number, country/state/city of origin and the year of isolation. The DENV-4 virus sequenced in this study is marked with a green-colored triangle symbol.
3.6. Comparison of clinical and demographic characteristics in patients infected with different DENV serotypes
When comparing the clinical and demographic characteristics of patients infected with different DENV serotypes, there was no significant difference observed between the mean age, number of males & females, and clinical manifestations. However, the mean disease duration was significantly longer i.e. 10 days in DENV-2 infected patients as compared to 7, 8, and 6 days in DENV-1, DENV-3, and DENV-4 infected patients respectively (p < .01). In addition, hemorrhagic manifestations were found in 4 of the DENV-2 infected patients, but this symptom was not observed in patients infected with other serotypes (Table 1).
Table 1.
Comparison of clinical presentations in patients infected with different DENV serotypes.
| Clinical variables | DENV-1 (n = 21) | DENV-2 (n = 68) | DENV-3 (n = 36) | DENV-4 (n = 13) | p value |
|---|---|---|---|---|---|
| Mean Age | 27 ± 16 | 26 ± 15 | 26 ± 13 | 25 ± 16 | P=.98 |
| Males | 15 (71.4%) | 42 (61.7%) | 23 (63.8%) | 9 (69.2%) | P=.85 |
| Females | 6 (28.5%) | 26 (38.2%) | 13 (36.1%) | 4 (30.7%) | |
| Mean disease duration | 7 ± 2 days | 10 ± 3 days | 8 ± 3 days | 6 ± 2 days | P<.01 |
| Fever | 21 (100%) | 68 (100%) | 36 (100%) | 13 (100%) | cannot be calculated |
| Chills | 6 (28.5%) | 23 (33.8%) | 12 (33.3%) | 2 (15.3%) | P=.59 |
| Rigors | 3 (14.2%) | 2 (2.9%) | 4 (11.1%) | 3 (23.0%) | P=.06 |
| Malaise | 5 (23.8%) | 6 (8.8%) | 5 (13.8%) | 2 (15.3%) | P=.34 |
| Myalgia | 9 (42.8%) | 32 (47.0%) | 15 (41.6%) | 4 (30.7%) | P=.73 |
| Bone/Joint Pain | 1 (4.7%) | 1 (1.4%) | 0 (0%) | 0 (0%) | cannot be calculated |
| Joint Swelling | 1 (4.7%) | 2 (2.9%) | 1 (2.7%) | 0 (0%) | cannot be calculated |
| Headache | 4 (19.0%) | 5 (7.3%) | 3 (8.3%) | 2 (15.3%) | P=.40 |
| Rash | 1 (4.7%) | 3 (4.4%) | 2 (5.5%) | 1 (7.6%) | P=.96 |
| Retro-Orbital Pain | 3 (14.2%) | 6 (8.8%) | 4 (11.1%) | 2 (15.3%) | P=.84 |
| Jaundice | 0 (0%) | 0 (0%) | 1 (2.7%) | 0 (0%) | cannot be calculated |
| Hemorrhagic Manifestations | 0 (0%) | 4 (5.8%) | 0 (0%) | 1 (7.6%) | cannot be calculated |
4. Discussion
During 2019, there was a significant dengue outbreak in Central India and a high number of dengue-suspected samples were received at our tertiary care hospital. However, 2020 saw significant drop in dengue cases as well as positivity due to the COVID-19 pandemic. This might be due to lockdowns or movement restrictions, resulting in reduced exposure to mosquitoes. Continuous environmental sanitization and fumigation also led to a decline in mosquito populations. However, in 2021, the dengue outbreak reemerged with a large number of cases. Therefore, in this study, we attempted to elucidate the DENV serotypes and genotypes circulating in the year before the pandemic (2019) and ongoing-pandemic year (2021).
Studies on various demographic characteristics revealed that males were affected more by dengue infection than females. This may be due to the higher tendency of males to engage in more outdoor-related activities and travel more often than females making them favorable targets for Aedes mosquitoes. Further, we found that the most common age group affected by dengue infections was 16–45 years old, followed by 6–15 years. The higher prevalence of dengue fever in this age group could be due to the higher outdoor exposure to dengue mosquitoes [16]. The elderly group (55 years to 79 years old) showed a low dengue infection rate which could be due to the development of immunity from previous infections [16,17]. A comparison of clinical and demographic characteristics in patients infected with different DENV serotypes revealed no significant difference. However, the mean disease duration was significantly longer in DENV-2 infected patients as compared to patients infected with other serotypes (p < .01). Further, hemorrhagic manifestations were found only in DENV-2 infected patients. The temporal distribution of dengue cases during the 2019 and 2021 outbreaks revealed the peak in November and October respectively.
DENV has been antigenically classified into four serotypes (DENV- 1 to 4) and genetic variation between these serotypes has also been reported [3]. Exposure to one virus induces lifelong immunity against a particular serotype, but the interaction between the serotypes is less clear. Multiple hypotheses have been proposed, however, a common explanation focuses on antibody-dependent enhancement (ADE) [18]. During primary infection, neutralizing antibodies are produced against the infecting serotype, and cross-reactive antibodies produced during secondary infection (with a heterologous serotype) lead to severe manifestations. The antibodies help the virus to infect monocytes more efficiently [19]. As a result of ADE, overall replication of the virus increases, as does the chance of severe dengue. Thus, knowledge of the DENV serotype associated with a particular attack of Dengue could help identify patients with a higher likelihood of severe outcomes. Characterization of circulating DENV serotypes helps in a better understanding of genetic recombination and the evolutionary basis of their emergence.
Our molecular analysis revealed the presence of all four DENV serotypes in these 2019 and 2021 outbreaks, with the predominance of DENV-2, followed by DENV-3 and DENV-1. DENV-4 was the least detected serotype. DENV-1, DENV-2, and DENV-3 are prevalent in most of North and South India [20–22]. DENV-4 from Central India was reported in 2010, DENV-1 in 2012, and DENV-2 during 2013 [23,24]. A recent report demonstrated the predominant circulation of DENV-1, followed by DENV-2 and DENV-3 from Central India in 2016 [25]. The serotype distribution observed in our study suggests a replacement of the DENV-1 with DENV-2 in this region, along with DENV-3 and re-occurrence of DENV-4. Co-circulation of multiple DENV serotypes in a particular area is an indicator of endemicity, which is influenced by growing urbanization, deforestation, and changing weather conditions [26]. More frequent reports on the co-circulation of multiple DENV serotypes have been observed in India since 2011 [20,22,25,27,28]. In our study, DENV-2 is the commonest serotype present in all the coinfections observed during 2019 and 2021.
Genotypic differences play an important role in virulence. The emergence of genotypes/lineages has crucial implications for understanding DENV’s evolutionary process, epidemiology, and control, as it is frequently linked to variations in disease incidence and severity [29]. DENV-1 (2021) isolates from our study (Central India) belongs to genotype V (American/African) and are clustered into one group along with DENV-1 from Central India 2016 & 2018 strains. Most Indian studies have shown the prevalence of this genotype [30,31]. Another molecular study suggests that the Indian DENV-1 belonged to different lineages of this genotype till 2012; however, in a large outbreak in 2012 in Tamil Nadu, South India, infection by the Asian genotype of DENV 1 resulted in high case fatality rates [32]. DENV-2 is the predominant serotype circulating during the study period. It is regarded as the most virulent and predominant serotype associated with the outbreaks of DF and DHF cases during 1970–2000 [26]. DENV-2 sequenced in this study belongs to genotype IV (Cosmopolitan) and are clustered into one group along with Taiwan, Indonesia, and Singapore isolates but not with Central Indian 2016 & 2018 strains or other Indian strains. This demonstrates that the DENV-2 detected in 2021 has diverged from the previous strains, which might be responsible for the higher transmission of this particular serotype. Other studies from Central India, Delhi, and Tamil Nadu also found the Cosmopolitan genotype (IV) of DENV2 [31,33–35].
Phylogenetic analysis of DENV-3 strains (2021) obtained in this study revealed them to be of lineage 3 within genotype III (Cosmopolitan). These are found to be clustered into one group along with the Central Indian strains of 2016 and 2018 as well as other Indian strains. Another report from Central India showed DENV3 belonged to genotype III (lineage C) and the viruses belonging to this lineage were responsible for causing more severe diseases [36]. Similar studies from Central India (2012, 2013) and Tamil Nadu (2017) have reported the same lineage and genotype in previous years [31,35]. Phylogenetic analysis of DENV-4 (2021) sequenced in our study belongs to genotype I. These are found to be clustered into one group along with Central Indian strains of 2016 and 2018, strains from Bangalore (2015), Pune (2016), Pondicherry (2017), and Mumbai (2018). DENV-4 was previously reported from Central India in 2010 [23,24]. A study conducted by other authors also reported DENV4 virus belonging to genotype I [37,38]. Genotype I of DENV4 was also detected in Pune (Maharashtra), Jabalpur (Madhya Pradesh) and Tamil Nadu [23,32,35,37,39]. DENV4 has been rarely responsible for outbreaks; however, secondary infection by DENV4 has been associated with severe outcomes and high epidemic potential [40].
The study suffered from some limitations. Firstly, only a smaller number of seropositive samples were positive for viral RNA. This might be owing to the hospital visit during the later stage of infection or after viral clearance, leading to RNA degradation. Secondly, nested PCR was used for serotyping, while most of the laboratories are now using real-time RT-PCR. Thirdly, nucleotide sequencing and genotypic analysis of only selected samples were carried out. In addition, partial genome sequencing was done instead of complete genome due to limited resources.
5. Conclusions
In conclusion, our study reports the circulation of DENV-2 as the dominant serotype followed by DENV-3 and DENV-1, and the reappearance of DENV-4. In addition, we observed coinfection with DENV-2 & DENV-3 as well as DENV-2 & DENV-4 in two patients each. DENV-2 detected in 2019 and 2021 has diverged from the previous strains detected in Central India (2016 and 2018), which might be responsible for the higher transmission of DENV-2 during these outbreaks. Thus, continuous surveillance, vigilant monitoring, and testing are needed for the effective management of these outbreaks.
Funding Statement
The author(s) reported there is no funding associated with the work featured in this article.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
All the data related to this study is available in the manuscript.
References
- [1].WHO (World Health Organization) . 2022. [cited 21 Jun 2022]. Available from: https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue
- [2].NVBDCP (National Vector Borne Diseases Control Programme) . 2022. [cited 21 Jun 2022]. Available from: https://nvbdcp.gov.in/index4.php?lang=1&level=0&linkid=431&lid=3715
- [3].Rico-Hesse R. Microevolution and virulence of dengue viruses. Adv Virus Res. 2003;59:315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Goncalvez AP, Escalante AA, Pujol FH, et al. Diversity and evolution of the envelope gene of dengue virus type 1. Virology. 2002;303(1):110–119. doi: 10.1006/viro.2002.1686 [DOI] [PubMed] [Google Scholar]
- [5].Dash PK, Sharma S, Soni M, et al. Complete genome sequencing and evolutionary analysis of Indian isolates of dengue virus type 2. Biochem Biophys Res Commun. 2013;436(3):478–485. doi: 10.1016/j.bbrc.2013.05.130 [DOI] [PubMed] [Google Scholar]
- [6].Twiddy SS, Farrar JJ, Chau NV, et al. Phylogenetic relationships and differential selection pressures among genotypes of dengue-2 virus. Virology. 2002;298(1):63–72. doi: 10.1006/viro.2002.1447 [DOI] [PubMed] [Google Scholar]
- [7].Wang E, Ni H, Xu R, et al. Evolutionary relationships of endemic/epidemic and sylvatic dengue viruses. J Virol. 2000;74(7):3227–3234. doi: 10.1128/JVI.74.7.3227-3234.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lanciotti RS, Lewis JG, Gubler DJ, et al. Molecular evolution and epidemiology of dengue-3 viruses. J Gen Virol. 1994;75(1):65–75. doi: 10.1099/0022-1317-75-1-65 [DOI] [PubMed] [Google Scholar]
- [9].Wittke V, Robb TE, Thu HM, et al. Extinction and rapid emergence of strains of dengue 3 virus during an interepidemic period. Virology. 2002;301(1):148–156. doi: 10.1006/viro.2002.1549 [DOI] [PubMed] [Google Scholar]
- [10].Aquino VH, Amarilla AA, Alfonso HL, et al. New genotype of dengue type 3 virus circulating in Brazil and Colombia showed a close relationship to old Asian viruses. PLoS One. 2009;4(10):e7299. doi: 10.1371/journal.pone.0007299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Klungthong C, Zhang C, Mammen MP Jr, et al. The molecular epidemiology of dengue virus serotype 4 in Bangkok, Thailand. Virology. 2004;329(1):168–179. doi: 10.1016/j.virol.2004.08.003 [DOI] [PubMed] [Google Scholar]
- [12].Weaver SC, Vasilakis N. Molecular evolution of dengue viruses: contributions of phylogenetics to understanding the history and epidemiology of the preeminent arboviral disease. Infect Genet Evol. 2009;9(4):523–540. doi: 10.1016/j.meegid.2009.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lanciotti RS, Calisher CH, Gubler DJ, et al. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol. 1992;30(3):545–551. doi: 10.1128/jcm.30.3.545-551.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10(3):512–526. doi: 10.1093/oxfordjournals.molbev.a040023 [DOI] [PubMed] [Google Scholar]
- [16].Najri NI, Mazlan Z, Jaimin JJ, et al. Genotypes of the dengue virus in patients with dengue infection from Sabah, Malaysia. J Phys Conf Ser. 2019;1358(1):012019. doi: 10.1088/1742-6596/1358/1/012019 [DOI] [Google Scholar]
- [17].Dar L, Gupta E, Narang P, et al. Cocirculation of dengue serotypes, Delhi, India, 2003. Emerg Infect Dis. 2006;12(2):352. doi: 10.3201/eid1202.050767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Reich NG, Shrestha S, King AA, et al. Interactions between serotypes of dengue highlight epidemiological impact of cross-immunity. J R Soc Interface. 2013;10(86):20130414. doi: 10.1098/rsif.2013.0414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rodenhuis-Zybert IA, Wilschut J, Smit JM. Dengue virus life cycle: viral and host factors modulating infectivity. Cell Molec Life Sci. 2010;67(16):2773–2786. doi: 10.1007/s00018-010-0357-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Afreen N, Deeba F, Naqvi I, et al. Molecular investigation of 2013 dengue fever outbreak from Delhi, India. PLoS Curr. 2014;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Prakash O, Singh DD, Mishra G, et al. Observation on dengue cases from a virus diagnostic laboratory of a tertiary care hospital in North India. Indian J Med Res. 2015;142(Suppl 1):S7. doi: 10.4103/0971-5916.176596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Reddy MN, Dungdung R, Valliyott L, et al. Occurrence of concurrent infections with multiple serotypes of dengue viruses during 2013–2015 in northern Kerala, India. Peer J. 2017;5:e2970. doi: 10.7717/peerj.2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Barde PV, Godbole S, Bharti PK, et al. Detection of dengue virus 4 from central India. Indian J Med Res. 2012;136(3):491. [PMC free article] [PubMed] [Google Scholar]
- [24].Barde PV, Shukla MK, Kori BK, et al. Emergence of dengue in tribal villages of Mandla district, Madhya Pradesh, India. Indian J Med Res. 2015;141(5):584. doi: 10.4103/0971-5916.159517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Agarwal A, Gupta S, Chincholkar T, et al. Co-circulation of dengue virus serotypes in Central India: evidence of prolonged viremia in DENV-2. Infect Genet Evol. 2019;70:72–79. doi: 10.1016/j.meegid.2019.02.024 [DOI] [PubMed] [Google Scholar]
- [26].Chakravarti A, Arora R, Luxemburger C. Fifty years of dengue in India. Trans R Soc Trop Med Hyg. 2012;106(5):273–282. doi: 10.1016/j.trstmh.2011.12.007 [DOI] [PubMed] [Google Scholar]
- [27].Mishra B, Turuk J, Sahu SJ, et al. Co-circulation of all four dengue virus serotypes: first report from Odisha. Indian J Med Microbiol. 2017;35(2):293–295. doi: 10.4103/ijmm.IJMM_15_536 [DOI] [PubMed] [Google Scholar]
- [28].Shrivastava S, Tiraki D, Diwan A, et al. Co-circulation of all the four dengue virus serotypes and detection of a novel clade of DENV-4 (genotype I) virus in Pune, India during 2016 season. PLoS One. 2018;13(2):e0192672. doi: 10.1371/journal.pone.0192672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Martina BE, Koraka P, Osterhaus AD. Dengue virus pathogenesis: an integrated view. Clin Microbiol Rev. 2009;22(4):564–581. doi: 10.1128/CMR.00035-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kukreti H, Chaudhary A, Rautela RS, et al. Emergence of an independent lineage of dengue virus type 1 (DENV-1) and its co-circulation with predominant DENV-3 during the 2006 dengue fever outbreak in Delhi. Int J Infect Dis. 2008;12(5):542–549. doi: 10.1016/j.ijid.2008.02.009 [DOI] [PubMed] [Google Scholar]
- [31].Barde PV, Shukla MK, Bharti PK, et al. Co-circulation of dengue virus serotypes with chikungunya virus in Madhya Pradesh, central India. WHO South-East Asia J Public Health. 2014;3(1):36–40. doi: 10.4103/2224-3151.206881 [DOI] [PubMed] [Google Scholar]
- [32].Cecilia D, Patil JA, Kakade MB, et al. Emergence of the Asian genotype of DENV1 in South India. Virology. 2017;510:405. doi: 10.1016/j.virol.2017.07.004 [DOI] [PubMed] [Google Scholar]
- [33].Sharma P, Mittal V, Chhabra M, et al. Dominance shift of DENV-1 towards reemergence and co-dominant circulation of DENV-2 & DENV-3 during postmonsoon period of 2012 in Delhi, India. J Virol Retrovirol. 2014;1(1):1–5. [Google Scholar]
- [34].Sharma P, Mittal V, Chhabra M, et al. Continued circulation of DENV-2 (genotype IV) in Delhi, India. Br Microbiol Res J. 2016;11(3):1–8. doi: 10.9734/BMRJ/2016/21728 [DOI] [Google Scholar]
- [35].Murugesan A, Aridoss D, Senthilkumar S, et al. Molecular diversity of dengue virus serotypes 1–4 during an outbreak of acute dengue virus infection in Theni, India. Indian J Medi Microbiol. 2020;38(3–4):401–408. doi: 10.4103/ijmm.IJMM_20_89 [DOI] [PubMed] [Google Scholar]
- [36].Barde PV, Shukla MK, Joshi P, et al. Molecular studies on dengue viruses detected in patients from Central India. Indian J Med Microbiol. 2019;37(1):12–18. doi: 10.4103/ijmm.IJMM_18_377 [DOI] [PubMed] [Google Scholar]
- [37].Dash PK, Sharma S, Srivastava A, et al. Emergence of dengue virus type 4 (genotype I) in India. Epidemiol Infect. 2011;139:85761. doi: 10.1017/S0950268810001706 [DOI] [PubMed] [Google Scholar]
- [38].Neeraja M, Lakshmi V, Dash PK, et al. The clinical, serological and molecular diagnosis of emerging dengue infection at a tertiary care institute in Southern, India. J Clin Diagn Res. 2013;7:45761. doi: 10.7860/JCDR/2013/4786.2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Patil JA, Cherian S, Walimbe AM, et al. Influence of evolutionary events on the Indian subcontinent on the phylogeography of dengue type 3 and 4 viruses. Infect Genet Evol. 2012;12:175969. doi: 10.1016/j.meegid.2012.07.009 [DOI] [PubMed] [Google Scholar]
- [40].Carrington CV, Foster JE, Pybus OG, et al. Invasion and maintenance of dengue virus type 2 and type 4 in the Americas. J Virol. 2005;79:146807. doi: 10.1128/JVI.79.23.14680-14687.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data related to this study is available in the manuscript.


