Abstract
The Cox protein of bacteriophage P2 is a multifunctional protein of 91 amino acids. It is directly involved in the site-specific recombination event leading to excision of P2 DNA out of the host chromosome. In this context, it functions as an architectural protein in the formation of the excisome. Cox is also a transcriptional repressor of the P2 Pc promoter, thereby ensuring lytic growth. Finally it promotes derepression of prophage P4, a nonrelated defective satellite phage, by activating the P4 PLL promoter that controls P4 DNA replication. In this case it binds upstream of the PLL promoter, which normally is activated by the P4 Delta protein. In this work we have analyzed the native form of the Cox protein in vivo, using a bacteriophage λ cI-based oligomerization assay system, and in vitro, using gel filtration, cross-linking agents, and gel retardation assays. We found that P2 Cox has a strong oligomerization function in vivo as well as in vitro. The in vitro analysis indicates that its native form is a tetramer that can self-associate to octamers. Furthermore we show that oligomerization is necessary for the biological activity by characterizing different cox mutants and that oligomerization is mediated by the C-terminal region.
Bacteriophage P2 belongs to a group of serologically related, nonlambdoid phages that can infect several enteric bacterial species (4). It is a temperate DNA phage; i.e., after infection it can either grow lytically, leading to cell lysis and release of progeny phage particles, or form lysogeny. In the latter case, the infected cell survives, and the P2 DNA becomes integrated into the host chromosome through a site-specific recombination event. P2 cox (control of excision) mutants were originally isolated as P2 phages that were unable to liberate phages spontaneously during growth of a lysogenic strain (16). In addition, the cox mutants showed an increased frequency of lysogenization and integrase-mediated site-specific recombination between infecting phages (2, 16).
The cox gene is the first gene of the P2 early operon, and it encodes a 91-amino-acid-long, slightly basic polypeptide of 10.3 kDa (12). P2 Cox is a multifunctional protein with at least three distinct functions: (i) P2 prophage excisionase, (ii) transcriptional repressor of P2 Pc promoter, and (iii) transcriptional activator of the unrelated phage P4 PLL promoter.
In the site-specific recombination event, P2 Cox protein is directly involved as an architectural protein, where it has been shown to be required for excisive recombination and inhibitory for integrative recombination (32, 33). In this case Cox is analogous to λ Xis. As a transcriptional repressor it represses the P2 Pc promoter that controls the expression of the P2 integrase and the immunity repressor C, thereby ensuring lytic growth (21). At high concentrations it autoregulates its own expression, since it reduces the activity of the early promoter Pe (21). Thus, the P2 Cox protein has in this case a function analogous to that of the well-studied λ Cro protein.
The Cox protein has also been shown to be involved in the derepression of satellite phage P4 (27). P4 is a defective phage that needs a helper, like P2, for lytic growth, since P4 lacks all structural genes, DNA packaging, and lysis functions (17). P2 and P4 are unrelated phages with a mutual capacity to derepress each other. The derepression of prophage P4 by P2 is mediated by the Cox protein, which functions as an activator of the P4 late promoter PLL (20). The PLL promoter controls the P4 α gene required for P4 DNA replication. During P4 lytic growth, the P4 PLL promoter is activated by the P4 Delta protein, which is under immunity control (8, 15). Thus the P2 Cox protein bypasses the normal P4 immunity and mimics the activity of the P4 Delta protein.
The P2-related phages HP1 and 186 have proteins analogous to P2 Cox, named Cox and Apl, respectively; i.e., they act like repressors and excisionases (9, 10, 19). The Cox protein of HP1 has been shown to form tetramers, which can self-associate into octamers (9), while Apl of phage 186 is a monomer in solution (26). The biological significance of the multimeric forms of HP1 Cox has not yet been determined.
To gain further insights into the action of the multifunctional P2 Cox protein, we have characterized the native form of the protein and have shown that oligomerization is required for biological activity.
MATERIALS AND METHODS
Chemicals and enzymes.
Media were from Difco and acrylamide was from Saveen. Enzymes were purchased from Pharmacia Biotech, except for Vent DNA polymerase (New England Biolabs). Radioactive isotopes and the automatic sequencing kit were from Amersham. All other fine chemicals were purchased from Sigma, unless otherwise stated.
Oligonucleotides.
Oligonucleotides used are listed in Table 1 and were purchased from DNA-Technology, KEBO, or Pharmacia.
TABLE 1.
Synthetic oligonucleotides used
| Oligonucleotide | Sequence (5′ to 3′) |
|---|---|
| 7R | GTT GTC TGC TCA AAT ACT CTG A |
| 72.0R | TGG TCA ATG TGT GGA CGT GA |
| 72.5L | GGC CGA GAG TGT CCA CAC AG |
| 77.4R | TGA GCA AGC AAG TAA CAC TC |
| 77.4R-2 | AGC AAG CAA GTA ACA CTC ATG |
| 77.6L | CAG CTT TCC TTT ATC GAT CAT C |
| 79.0L | GGG TGA TGG TGA AGT CAA TC |
| cox2-R | GCT GCC TGT AAT TGA TAC GAC CGA TCC ACA ATC AGC |
| cox2-L | GCT GAT TGT GGA TCG GTC GTA TCA ATT ACA GGC AGC |
| cox3-R | CGC AAA ACT AAT AGA AAA ATC GAC AGG AGC G |
| cox3-L | CGC TCC TGT CGA TTT TTC TAT TAG TTT TGC G |
| cox4-R | GCT TCA GGT CGT GCA GGT TAA TAT TGG GTA TAC C |
| cox4-L | GGT ATA CCC AAT ATT AAC CTG CAC GAC CTG AAG C |
| cox4-R2 | GCT TCA GGT CGT GCA GGT AAA TAT TGG GTA TAC C |
| cox4-L2 | GGT ATA CCC AAT ATT TAC CTG CAC GAC CTG AAG C |
| cox107-R | CGA CAG GAG CGG TTC ATC GGA TGA TCG ATA AAG G |
| cox107-L | CCT TTA TCG ATC ATC CGA TGA ACC GCT CCT GTC G |
| cox129-R | CGG ACT AAA ACT GAC TTA TGA AAG CCG C |
| cox129-L | GCG GCT TTC ATA AGT CAG TTT TAG TCC G |
| cox130-R | GCG ATT CCT TAT CAG AAG TTC GCA AAA CTA ATA GG |
| cox130-L | CCT ATT AGT TTT GCG AAC TTC TGA TAA GGA ATC GC |
| T7 forward | TAA TAC GAC TCA CTA TAG GG |
| T7 reverse | GCT AGT TAT TGC TCA GCG G |
Bacterial strains and plasmids.
The bacteria and plasmids used are listed in Table 2. All plasmids were made according to standard procedures (23) and sequenced using a Thermo Sequenase fluorescence-labeled primer cycle sequencing kit (Amersham Pharmacia Biotech) and run on an ALFexpressII (Pharmacia Biotech), to confirm the orientation of the inserts and to exclude possible PCR-induced mutations. Escherichia coli C-1a was used as a host for all cloning procedures, except for pEE727 to pEE734, when AG1688 was used as a recipient.
TABLE 2.
Bacterial strains and plasmids
| Bacterial strain or plasmid | Pertinent feature(s)a | Origin or reference |
|---|---|---|
| Strains | ||
| AG1688 | E. coli MC1061 F′128 lacIqlacZ::Tn5 | 13 |
| AG1688 (λ112OSPS) | AG1688 containing λ112 OSPS with lacZ driven by λ OSPS and a strong upstream lambda operator | 35 |
| AG1688 (λXZ970) | AG1688 containing λ112 OSPS with lacZ driven by λ OSPS and a 434 operator | 35 |
| BL21(DE3) | ompT hsdR hsdM lon E. coli B strain, T7 RNA polymerase under control of lacUV5 promoter | 28 |
| BL21(DE3) pLysE | Strain BL21(DE3) containing a pACYC184 derivative expressing T7 lysozyme | 28 |
| C-1a | F− prototrophic E. coli C strain | 24 |
| C-1757 | Polyauxotrophic E. coli C strain str supD | 29 |
| C-6005 | P2 cox3 lysogenic C-1a | 21 |
| JH372 | AG1688 containing λ202 with lacZ driven by λ PROR | 13 |
| Plasmids | ||
| pEE720 | P2 cox gene cloned in pET-8c under control of the T7 promoter φ10 | This paper |
| pEE721 | pET8-c derivative containing cox2 gene (M42T); primers cox2-R and cox2-L were used | This paper |
| pEE722 | pET8-c derivative containing cox3 gene (G22E); primers cox3-R and cox3-L were used | This paper |
| pEE723 | pET8-c derivative containing cox4 gene (E54K); primers cox4-R2 and cox4-L2 were used | This paper |
| pEE724 | pET8-c derivative containing cox107 gene (R29H); primers cox107-R and cox107-L were used | This paper |
| pEE725 | pET8-c derivative containing cox129 gene (A69T); primers cox 129-R and cox129-L were used | This paper |
| pEE726 | pET8-c derivative containing cox130 gene (E16K); primers cox130-R and cox130-L were used | This paper |
| pEE727 | P2 cox gene cloned in frame downstream of λ cI indl in pJH391 | This paper |
| pEE728 | pJH391 derivative containing cox2 gene (M42T); primers cox2-R and cox2-L were used | This paper |
| pEE729 | pJH391 derivative containing cox3 gene (G22E); primers cox3-R and cox3-L were used | This paper |
| pEE730 | pJH391 derivative containing cox4 gene (E54K); primers cox4-R2 and cox4-L2 were used | This paper |
| pEE731 | pJH391 derivative containing cox107 gene (R29H); primers cox107-R and cox107-L were used | This paper |
| pEE732 | pJH391 derivative containing cox129 gene (A69T); primers cox129-R and cox129-L were used | This paper |
| pEE733 | pJH391 derivative containing cox130 gene (E16K); primers cox130-R and cox130-L were used | This paper |
| pEE734 | pJH391 derivative containing truncated cox gene, cox38c (amino acids 1–53); primers cox4-R and cox4-L were used | This paper |
| pEE735 | pACYC177 derivative containing wt cox gene | This paper |
| pEE736 | pACYC177 derivative containing cox2 gene (M42T) | This paper |
| pEE737 | pACYC177 derivative containing cox4 gene (E54K) | This paper |
| pEE738 | pACYC177 derivative containing cox107 gene (R29H) | This paper |
| pEE739 | pACYC177 derivative containing cox129 gene (A69T) | This paper |
| pEE740 | pACYC177 derivative containing cox130 gene (E16K) | This paper |
| pEE741 | pKK232-8 derivative containing the cat gene under control of the P4 P11 promoter | Xiushan Wu, unpublished data |
| pET-8c | pBR322 derivative containing T7 promoter φ10 | 28 |
| pFG157 | λ cI indl driven by lacUV5 promoter | 13 |
| pJH370 | Fusion between λ cI indl and the leucine zipper of GCN4 driven by the lacUV5 promoter | 13 |
| pJH391 | First 132 nt of λ cI indl fused with lacZ driven by lacUV5 promoter | 13 |
| pJH622 | Fusion between λ cI indl and the reengineered leucine zipper of GCN4 driven by the lacUV5 promoter | 35 |
| pKH101 | N-terminal DNA-binding domain of λ cI indl | 13 |
| pSS27-5 | Ampicillin-sensitive derivative of pACYC177 | 21 |
| pSS39-6 | pKK232-8 derivative containing the cat gene under the control of the P2 Pc promoter, and the early Pe promoter is inactivated | 21 |
| pZ150 | pBR322 derivative with an M13 single-strand origin | 34 |
wt, wild type; nt, nucleotide(s)
(i) pEE720.
Plasmid pEE720 was formed by cloning the cox gene into expression vector pET-8c. The cox gene was amplified by PCR using primers 77.4R-2 and 79.0L and inserted into the filled-in NcoI site of pET-8c.
(ii) pEE721 to pEE726.
To form plasmids pEE721 through pEE726, the cox mutations were introduced into pEE720 by site-directed mutagenesis using the QuickChange site-directed mutagenesis kit (Stratagene) with primers as indicated in Table 2.
(iii) pEE727.
For plasmid pEE727, the cox gene was cloned into pJH391. pJH391 was cleaved with SalI and BamHI, and the ends were filled in using Klenow fragment. Thereafter the PCR amplified cox gene (using primers 77.4R and 79.0L) was ligated in frame with the part of the λ cI indl gene that codes for amino acids 1 to 132 (the DNA binding part of the λ CI repressor). Since the indl mutation, which generates the E117K amino acid substitution, does not affect the dimerization or DNA-binding activity of the CI protein, its presence is not further indicated. Protein expression from the clones was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
(iv) pEE728 to pEE734.
For plasmids pEE728 through pEE734, the cox mutations were made by site-directed mutagenesis using the QuickChange site-directed mutagenesis kit (Stratagene) with pEE727 as a template and primers as indicated in Table 2.
(v) pEE735 to pEE740.
To produce plasmids pEE735 through pEE740, the cox genes were cloned in pACYC177. The cox genes were amplified by PCR with primers T7 forward and T7 reverse from plasmids pEE721 to pEE726, and these amplified genes were inserted into the HincII site of pACYC177.
Protein purification.
P2 Cox protein was purified from E. coli BL21(DE3)pLysE containing plasmid pEE720, pEE721, pEE722, pEE723, pEE724, pEE725, or pEE726. The bacteria were grown with shaking at 37°C until mid-log phase, when isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mM, and the incubation was continued for about 4 h. The cells were harvested by centrifugation, and the pellet was resuspended in Buffer A+ (0.3 M potassium phosphate buffer [pH 7.5], 3 mM EDTA, 0.5 M KCl) and lysed by sonication on ice in three 30-s bursts, at 8 to 14 μm with an MSE Soniprep150. The extract was clarified by centrifugation in a Sorvall RC5C at 23,000 × g for 1 h, and ammonium sulfate was added to 25% saturation in the supernatant. After being stirred at 4°C for 30 min, the mixture was centrifuged at 17,000 × g for 30 min, and the pellet was resuspended in 4 ml of Buffer A+. The extract was loaded on a Sephacryl S-200 HR (Pharmacia) column. The Cox-containing fractions were analyzed by SDS-PAGE and concentrated using Centricon 3 or 10 (Amicon). The Cox protein was at least 95% pure as judged by SDS-PAGE. Glycerol was added to 40%, and the purified protein was stored at −20°C. The protein concentration was determined by the method of Bradford (5), with bovine serum albumin as a standard.
SDS-PAGE.
Samples were precipitated with trichloroacetic acid (TCA) (18) and separated by electrophoresis using conditions developed to separate low-molecular-weight proteins (25) and stained with Coomassie brilliant blue R-250.
Cox activity.
The activity was determined by gel retardation assay, and one unit was defined as the amount of protein extract needed for shifting 50% of the Cox box containing DNA substrate amplified by PCR from the P2 PePc region, using primers 7R and 77.6L. The intensities of the bands were measured with a PhosphorImager (Molecular Dynamics) and analyzed with the ImageQuant 3.3 software.
Gel retardation assay.
The gel retardation assays were performed as described previously (33). DNA fragments were 5′ end labeled with [γ-32P]ATP and were purified using MicroSpin S-200 HR columns (Pharmacia Biotech). The binding reactions were run on a 5% nondenaturing polyacrylamide gel (29:1) and run on a water-cooled Protean II xi Cell (Bio-Rad) at 20 mA. The gels were dried prior to autoradiography and PhosphorImager analysis.
In vivo dimerization/oligomerization assay.
Dimerization/oligomerization of the CI-Cox fusion protein was analyzed by cross-streaking bacteriophage λ KH54 and λ KH54h80 (13) against bacteria containing plasmid pEE727. Dimerization or oligomerization of the CI-Cox fusion should lead to λ immunity. Repression of the reporter gene in the λORPR-lacZ (strain JH372), λ112OSPS-lacZ (strain JH607), and λXZ970-lacZ (strain XZ980) constructs (13) by the CI-Cox fusion proteins was determined by measuring β-galactosidase activity in cells grown in A medium supplemented with 0.4% glucose–1 mM MgSO4–1% Casamino Acids–1 μg of vitamin B1 per ml as previously described (23). The OR operator contains an OR2− mutation that eliminates cooperative binding by the wild-type CI repressor to the operator (13).
Gel filtration.
The size of native Cox was determined by using a Sephacryl S-300 HR column (Pharmacia) connected to the GradiFrac system (Pharmacia). All runs were performed at 4°C, with a flow rate of 0.4 ml/min. The column was equilibrated with Buffer A+ and calibrated with blue dextran, ribonuclease A (13.7 kDa), chymotrypsinogen A (25 kDa), ovalbumin (43 kDa), albumin (67 kDa), aldolase (158 kDa), catalase (232 kDa), ferritin (440 kDa), and thyroglobulin (669 kDa). The eluate was monitored at an optical density at 280 nm (OD280). Two milliliters of Cox protein (5 mg/ml) was loaded. Two-milliliter fractions were collected, and the Cox-containing fractions were detected by SDS-PAGE. The Kav values were calculated according to the formula Kav = (Ve − V0)/(Vt − V0), where Ve is the elution volume for the protein, V0 is the column void volume, and Vt is the total bed volume.
Cross-linking.
The assays were performed using the 6.4 Å long Sulfo-disulfosuccinimidyl tartrate (DST) (Pierce). Cox was incubated with different amounts of Sulfo-DST cross-linker in Buffer A+ at room temperature for 1 h. The Sulfo-DST reactions were quenched with 50 mM Tris (pH 7.5) and TCA precipitated. The precipitated protein was dissolved in 1× sample buffer without mercaptoethanol (18) and thereafter analyzed by SDS-PAGE. The protein–cross-linking agent ratio was chosen where monomeric ribonuclease A still migrated as a monomer.
Determination of CAT activity.
BL21(DE3) cells harboring plasmids pSS27-5, pEE735, pEE736, pEE737, pEE738, pEE739, or pEE740 together with pSS39-6 or pEE741 were grown to an OD600 of 0.8, and the extracts for chloramphenicol acetyltransferase (CAT) activity were prepared as described before (22). Total protein concentrations were determined by the method of Bradford (5). The extracts from cells harboring plasmid pSS39-6 were diluted to 0.1 μg, and the extracts from cells harboring plasmid pEE741 were diluted to 2 μg in the CAT reactions, which were performed as described in reference 11, with 14C-labeled d-threo-dichloroacetyl-1-1-chloramphenicol (Amersham). CAT activities, determined by PhosphorImager analysis, were calculated as the ratio of acetylated chloramphenicol to total chloramphenicol.
In vivo excision assay for Cox activity.
The P2 cox3 lysogenic strain C-6005 was transformed with plasmid pSS27-5, pEE735, pEE736, pEE737, pEE738, pEE739, or pEE740. The level of spontaneous phage production was assayed in overnight cultures grown without aeration at 30°C in Luria-Bertani broth (LB) supplemented with kanamycin and 10 mM potassium phosphate (pH 6.8), which was used to prevent readsorption of released phages (3). The phage titer was determined as described earlier (3) using C-1757 as an indicator strain, after removing the bacteria by centrifugation and adding two drops of chloroform to the supernatants. The bacterial titer was determined by analysis of colony-forming ability, and the number of PFUs per CFU was calculated.
RESULTS
Alignment and secondary structure prediction of the P2 Cox, HP1 Cox and 186 Apl proteins.
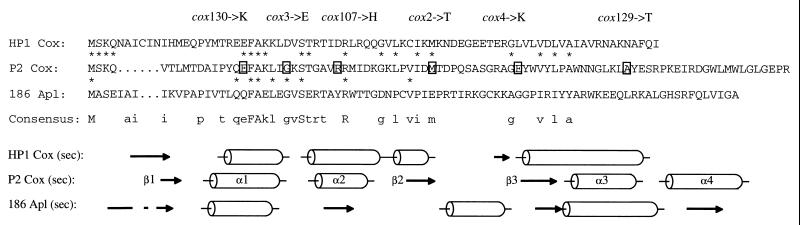
Alignment of the three analogous proteins was performed using the Pile Up program (Genetics Computer Group, Madison, Wis.), and the results (Fig. 1) show higher amino acid identity between P2 Cox and HP1 Cox (19 identical residues) than between P2 Cox and 186 Apl (9 identical residues). Further, the alignment revealed that the N-terminal parts of the proteins show higher identity than the C-terminal parts. The region with the highest resemblance between all three proteins is the first helix, α1, which together with helix α2 is believed to be the DNA-binding helix-turn-helix motif. It should be noted that the Apl protein does not contain the second helix. Cox mutants analyzed in this study affect amino acid residues which are evenly dispersed throughout the protein (boxed residues).
FIG. 1.
Alignment and secondary structure prediction of P2 Cox, HP1 Cox, and 186 Apl proteins. Asterisks symbolize amino acids that are identical between the proteins. Uppercase letters in the consensus sequence signify that the amino acid is identical in all three proteins, while lowercase letters mean identity between two of the proteins. The boxed amino acids refer to the exchanged amino acid in the different cox mutants listed above. The consensus secondary structures are shown below. Arrows symbolize β-strands, and barrels symbolize α-helices.
The secondary structure predictions were done on the Jpred server (http://jura.ebi.ac.uk:8888/) (7). The secondary structures of the three P2 Cox analogs in Fig. 1 make up the consensus structure of the following prediction programs: dsc, nnssp, pblock, phd, predator, and zpred (7).
Purification of P2 Cox protein.
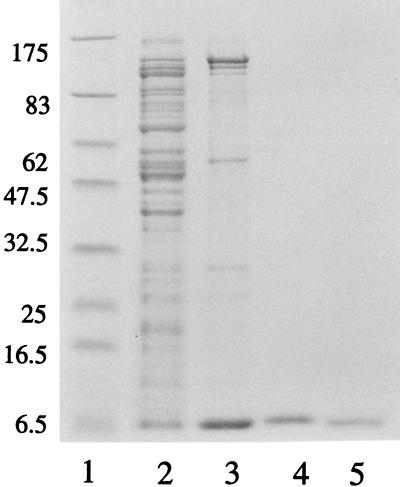
The P2 cox gene was inserted into the high-copy-number plasmid pET-8c, under the control of the T7 promoter φ10, generating plasmid pEE720. This plasmid was transformed into E. coli strain BL21(DE3)pLysE, containing the T7 RNA polymerase under the control of the lacUV5 promoter. In a strain without the pLysE plasmid, the induced Cox protein formed inclusion bodies. Induction by IPTG produced extracts containing the P2 Cox protein as judged by the size on SDS-PAGE and by gel retardation assays with the P2 PePc region. P2 Cox was purified as described in Materials and Methods. The results of the protein purification are summarized in Table 3, and the protein composition after the different purification steps is visualized by SDS-PAGE, shown in Fig. 2.
TABLE 3.
Example of P2 Cox purificationa
| Fraction | Volume (ml) | Activity (U/ml) | Protein concn (mg/ml) | Sp act (U/μg) | Yield (%) | Relative purity |
|---|---|---|---|---|---|---|
| Extract | 14.5 | 100 × 103 | 7.2 | 14 | 100 | 1.0 |
| Ammonium sulfate | 3.0 | 333 × 103 | 7.5 | 44 | 69 | 3.0 |
| Sephacryl | 1.5 | 5 × 103 | 0.06 | 83 | 0.5 | 5.9 |
| Concentrated | 0.35 | 50 × 103 | 0.25 | 200 | 1.2 | 14.3 |
One unit of Cox activity is defined as the amount needed for shifting 50% of a 0.05-pmol Cox box containing DNA (300 nt long) in a gel retardation assay. The purification and assays are described in Materials and Methods.
FIG. 2.
SDS–15% polyacrylamide gel of the fractions obtained during purification of Cox. Approximately 2 μg of protein was loaded. Lane 1, marker proteins (sizes in kilodaltons); lane 2, crude cell extract; lane 3, 25% ammonium sulfate precipitate; lane 4, Sephacryl S-200 Cox-containing fraction; lane 5, concentrated S-200 fraction.
Cox forms multimers in vivo.
The ability of Cox to form dimers or higher multimers was investigated by using the repressor fusion system, which is based on the interaction between the bacteriophage λ CI repressor and the λ OR (13). The C-terminal domain of the λ CI repressor mediates the dimerization, whereas the N terminus is involved in protein-DNA interaction. The N-terminal domain alone cannot function as a repressor, since it binds DNA very poorly, due to inefficient dimerization (30). It has been shown that, on replacing the C terminus of the λ CI repressor with other proteins, their ability to form dimers or higher oligomers in vivo can be analyzed by comparing the biological activity of the fusion protein with that of the wild-type λ CI repressor (13).
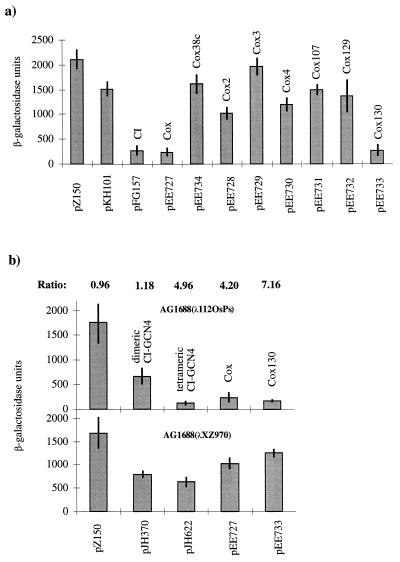
P2 Cox was fused to the N-terminal domain of the λ CI repressor, and the construct, pEE727, was transformed into reporter strain JH372, which is an AG1688 derivative containing a λ202 prophage carrying the lacZ gene driven by λORPR. In JH372, the ability of the CI-Cox fusion protein to form dimers (or oligomers) was first investigated by cross-streaking against bacteriophage λ. Bacteria expressing the CI-Cox fusion protein were found to be immune to λ infection, which indicates that the CI-Cox fusion protein can form dimers (or oligomers) like the wild-type λ CI repressor (results not shown). The ability of the CI-Cox fusion protein to bind to the λORPR was then quantified by measuring the β-galactosidase activity, with the wild-type CI-expressing clone as a positive control (pFG157) and no repressor (pZ150) and just the N-terminal part of the CI repressor (pKH101) as negative controls. As can be seen in Fig. 3a the lacZ expression in the presence of the CI-Cox fusion protein (pEE727) is about sixfold lower than its expression with pKH101. Furthermore, the CI-Cox fusion protein represses the lacZ expression to the same extent as the intact CI repressor (pFG157). These results show that the fusion protein represses transcription from the PR promoter to the same extent as wild-type λ CI repressor, which implies that the P2 Cox protein has a strong dimerization (or oligomerization) function.
FIG. 3.
In vivo assays of dimerization and multimerization/cooperative binding of Cox. Each panel shows the results from three independent β-galactosidase measurements with pJH391 derivatives transformed in three different E. coli strains. (a) The dimerization in assay strain JH372. pZ150 (no CI repressor) and pKH101 (N terminus of the CI repressor) are negative controls. pFG157 (wild-type CI repressor) is the positive control. Wild-type Cox (pEE727), the truncated Cox (pEE734), and the different Cox mutants (pEE728 to pEE733) were analyzed and plotted against β-galactosidase units. (b) The multimerization/cooperative binding assay in strain AG1688(λ112OSPS) and in the control strain AG1688(λXZ970). β-Galactosidase assays were performed as in strain JH372. pJH370 (dimeric CI-GCN repressor) was used as the negative control. pJH622 (tetrameric CI-GCN modified repressor) is a positive control. pEE727 is the wild-type cox gene cloned in pJH391, and pEE733 contains the cox130 mutation. AG1688(λXZ970) is the strain used for detecting binding to the weak promoter-proximal operator only. The ratio between the values obtained in assay strain AG1688(λ112OSPS) and the control strain AG1688(λXZ970) is indicated above their respective columns.
To distinguish between dimerization and oligomerization/cooperative binding, the CI-Cox protein was analyzed in strain AG1688 lysogenized with λ112OSPS or λXZ970 (35). AG1688(λ112OSPS) contains a synthetic promoter that controls the expression of lacZ. A weak λ operator (OS2) overlaps the promoter, and a strong operator (OS1) is situated upstream of the weak operator. If a repressor fusion protein binds cooperatively or as an oligomer to the strong upstream OS1 operator, then the lacZ expression from the PS promoter will be repressed. In AG1688(λXZ970), the OS1 operator is replaced by a 434 operator. This strain is used to analyze how much of the repression is caused by binding of the fusion protein to the weak promoter-proximal OS2 operator alone. Fig. 3b shows the β-galactosidase activity of the different extracts of AG1688(λ112OSPS)- and AG1688(λXZ970)-transformed cells. pZ150 (no repressor) and pJH370 (CI-GCN4 repressor fusion that only forms dimers) were used as negative controls. pJH622 (modified CI-GCN4 repressor fusion that forms tetramers) was used as a positive control. As can be seen in Fig. 3b, the CI-Cox fusion protein represses transcription almost as well as the tetrameric GCN4-repressor (pJH622) in strain AG1688(λ112OSPS). The values obtained for the CI-Cox fusion protein in the control strain AG1688(λXZ970) exclude the possibility that the low β-galactosidase activity in AG1688(λ112OSPS) was due to binding to the weak OS2 operator alone. In fact, the tetrameric CI-GCN4 protein has a higher affinity for the weak OS2 operator than does CI-Cox. The activities obtained with the positive control (pJH622), and the CI-Cox fusion protein decreased the lacZ expression four- to fivefold in strain AG1688(λ112OSPS) compared to the level obtained in strain AG1688(λXZ970). This indicates that the CI-Cox protein has the ability either to bind cooperatively to the strong OS1 upstream operator in λ112OSPS or to form multimers in vivo.
Cox forms tetramers and octamers in vitro.
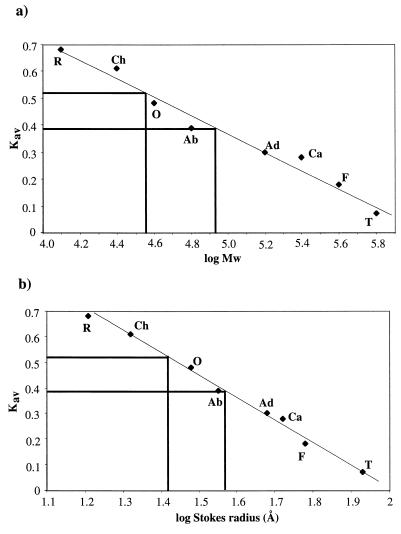
To determine the multimeric forms of Cox in vitro, a gel filtration experiment was performed loading Cox protein onto a calibrated Sephacryl S-300 HR (Pharmacia) column. Two Cox-containing peaks were obtained (Fig. 4), one corresponding to a molecular mass of about 36 kDa and a Stokes radius of about 26 Å. The other, more rapidly eluting, Cox-containing peak corresponds to a molecular mass of approximately 81 kDa and a Stokes radius of 37 Å. The latter contained the majority of the Cox protein, which suggests that the Cox protein has the capacity to form tetramers and octamers, under the conditions used.
FIG. 4.
Gel filtration analysis of native Cox protein (5 mg/ml). Gel filtration experiments were done on a Sephacryl S-300 HR column, calibrated with ribonuclease A (R), chymotrypsinogen A (Ch), ovalbumin (O), albumin (Ab), aldolase (Ad), catalase (Ca), ferritin (F), and thyroglobulin (T). (a) Kav values plotted against log molecular weights. The first Cox peak eluted at a Kav value of 0.39, which corresponds to a log molecular weight value of about 4.91 and a molecular mass of 81 kDa. The second peak eluted at a Kav value of 0.52, which corresponds to a molecular mass of about 36 kDa. (b) Kav values plotted against log Stokes radius (Å). A Kav value of 0.39 corresponds to a Stokes radius of about 37 Å, and a Kav value of 0.52 corresponds to a Stokes radius of 26 Å.
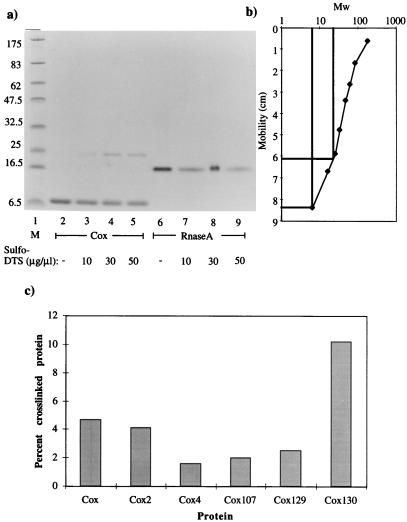
The native size of the Cox protein was further investigated at a lower protein concentration by incubating Cox protein with the lysine-specific, 6.4 Å, cross-linking agent, Sulfo-DST, as described in Materials and Methods. The SDS-PAGE gel in Fig. 5a and the plot in Fig. 5b show the result, where monomeric as well as tetrameric forms of Cox were clearly obtained, under conditions where only the monomeric form of ribonuclease A was apparent. The monomeric Cox protein migrates in SDS-PAGE as a 6.5-kDa protein rather than the expected 10.3 kDa. The apparent lack of dimeric Cox suggests that the protein tetramerizes very efficiently. Using glutaraldehyde as a cross-linking agent gave the same result (results not shown). Under the conditions used, the amount of cross-linked protein appeared to be independent of the concentration of Cox (0.8 to 12.5 μM) (results not shown).
FIG. 5.
Cross-linking of Cox. (a) SDS-PAGE of Cox preincubated with Sulfo-DST. Cox (12.5 μM, lanes 2 to 5) and 12.5 μM RNase A (lanes 6 to 9) were incubated with increasing amounts of Sulfo-DST. The Sulfo-DST concentration was as follows: no cross-linker (lanes 2 and 6), 10 μg/μl (lanes 3 and 7), 30 μg/μl (lanes 4 and 8), and 50 μg/μl (lanes 5 and 9). Lane 1 contains marker proteins (sizes in kilodaltons). (b) Plot from the results from the cross-linking assay. The curve is linear in the range where monomeric and tetrameric Cox migrated. Monomeric Cox migrates like the 6.5-kDa marker, and the size of cross-linked Cox is approximately 23 kDa. (c) Cross-linking of mutant Cox proteins. Cox (12.5 μM) was incubated with Sulfo-DST (50 μg/μl). The Cox proteins were cross-linked with Sulfo-DST as described in Materials and Methods, the SDS-PAGE gel was scanned, and the bands were quantified. The Cox proteins were plotted against the ratio between cross-linked Cox and the total amount of Cox.
Cooperative binding of Cox to DNA.
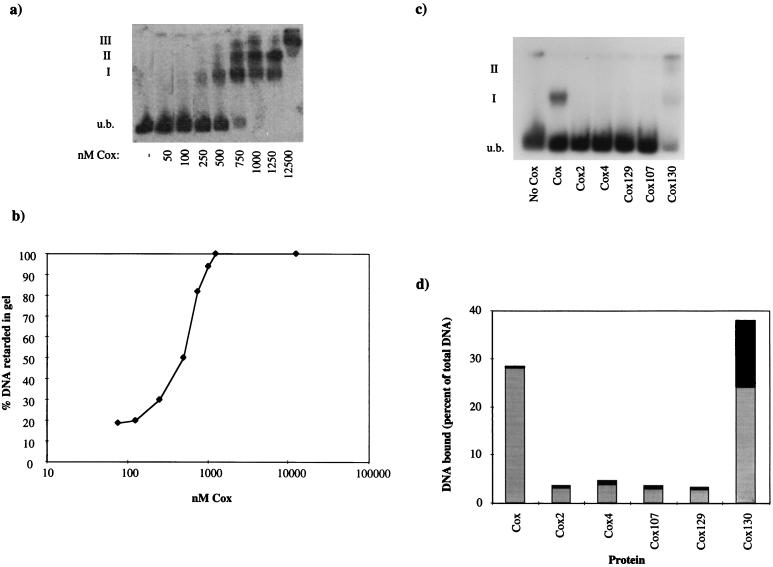
To analyze how Cox binds to its cognate DNA site, a gel retardation assay was used, incubating Cox with PCR-amplified DNA from the attP region (primers 72.OR and 72.5L). Three bands with retarded electrophoretic mobilities were formed (Fig. 6a). Complex I was preferentially formed at low protein concentrations, whereas at high protein concentrations, complex II and complex III were detected. The dependence of DNA retardation on concentration of the protein followed a sigmoidal curve (Fig. 6b). The sigmoid nature of the curve suggests that tight binding of the Cox protein to DNA requires the interaction of two or more Cox subunits with the DNA and that Cox binds cooperatively to its cognate sites.
FIG. 6.
Gel retardation assays. (a) Different amounts of Cox were incubated with 0.05 pmol of 5′ end-labeled attP DNA substrate. Three shifts were detected as indicated. (b) Fraction of DNA bound by increasing concentrations of Cox obtained from quantification of the autoradiography shown in panel a. (c) Autoradiograph showing the shifts with the different Cox proteins. Of each respective Cox protein, 21 ng was incubated with 0.05 pmol of 5′ end-labeled attP DNA substrate. The unbound (u.b.) DNA is indicated, as are the first (I) and the second (II) protein-DNA complexes. (d) Quantitated results from the gel retardation assay in panel c. The proteins were plotted against the percent DNA shifted. The total height of the bar is the total shifted DNA; the lower part indicates the first shift (I), and the upper part symbolizes the values for the second shift (II).
cox mutations can affect the ability to form oligomers.
Several mutations have been located in the cox gene (12; E. Haggård-Ljungquist, unpublished results). Three mutations, cox130, cox3, and cox107 are located in the N-terminal part containing the helix-turn-helix motif believed to be involved in DNA binding (Fig. 1), while three other mutations, cox2, cox4, and cox129, are located outside this region. The cox2, cox3, and cox4 mutants were originally isolated as excision-defective mutants of P2 (16), and they were later shown to be deficient also in derepression of satellite phage P4 (27). On the other hand, cox107, cox129, and cox130 were originally isolated as P2 mutants defective in the induction of a P4 lysogen after infection while maintaining the capacity to form phage spontaneously in growing lysogenic cultures, although at a reduced frequency (E. W. Six and M. G. Sunshine, personal communication).
To analyze the oligomerization capacity of the mutated Cox proteins and a truncated version, Cox38c, containing only the first 53 amino acids, these proteins were fused to the DNA-binding N-terminal part of the λ CI protein, and their respective capacity to form at least dimers was analyzed in the repression assay system described above. As can be seen in Fig. 3a, the fusion protein containing the cox130 mutation (pEE733), which is located in the α-helix believed to be the structural helix of the DNA-binding domain, maintains the capacity to dimerize (or oligomerize) like the wild-type Cox fusion protein. Fusion proteins containing cox3 (pEE729), cox107 (pEE731), and cox129 (pEE732) mutations, as well as the truncated Cox protein (pEE734), seem to have lost the capacity to dimerize (or oligomerize), since the β-galactosidase activities overlap with those of the negative controls (pZ150 and pKH101), while Cox2 (pEE728) and Cox4 (pEE730) seem to have some residual dimerization (or oligomerization) function. The fact that cox130, located at the N-terminal end, is able to form at least dimers made it interesting to analyze Cox130 for multimerization/cooperative binding in AG1688(λ112OSPS) and the negative control strain AG1688(λXZ970), where cox130 showed a greater ability to repress the expression of the lacZ gene than the wild-type Cox (Fig. 3b).
The mutated Cox proteins were also analyzed in vitro in cross-linking assays where the SDS-PAGE gel was scanned with an AGFA Snapscan and analyzed using NIH Image 1.61. The mutated Cox proteins were plotted against the ratio between cross-linked Cox protein and total amount of Cox protein (Fig. 5c). These data confirmed the in vivo data that the mutated Cox proteins are defective in oligomerization (except Cox130). It also showed that Cox2 is not completely defective in oligomerization, since the percentage of cross-linked Cox tetramers is almost as high as that of wild-type Cox. Cox3 was not analyzed in the cross-linking assays, since it formed inclusion bodies upon induction.
The mutated Cox proteins are defective in excision, transcriptional repression, and activation.
The respective mutated Cox proteins were characterized in an in vivo excision assay. The plasmids pEE735 (wild-type cox), pEE736 (cox2), pEE737 (cox4), pEE738 (cox107), pEE739 (cox129), pEE740 (cox130), and pSS27-5 (no cox) were transformed into a cox3-defective P2 lysogenic strain (C-6005). The capacity of the mutated Cox proteins to complement the cox-defective prophage was determined by scoring the level of free phage per bacterium in a bacterial overnight culture (Table 4). The results show that the cox2 mutation makes the Cox protein unable to complement the defective prophage, while Cox130 is almost as efficient as wild-type Cox. Cox4, Cox107, and Cox129 proteins have intermediate complementation capacities.
TABLE 4.
Effects of defective Cox proteins on the spontaneous phage production of the P2 cox3 lysogen C-6005
| Plasmid | Protein produced by the plasmid | Amt (titers) of phage produced per 1,000 bacteriaa |
|---|---|---|
| pSS27-5 | None | <0.00001 |
| pEE735 | Cox | 61 |
| pEE736 | Cox2 | <0.00007 |
| pEE737 | Cox4 | 0.0017 |
| pEE738 | Cox107 | 0.07 |
| pEE739 | Cox129 | 0.008 |
| pEE740 | Cox130 | 10.4 |
The titers of free phages and bacteria were determined with overnight cultures as described in Materials and Methods.
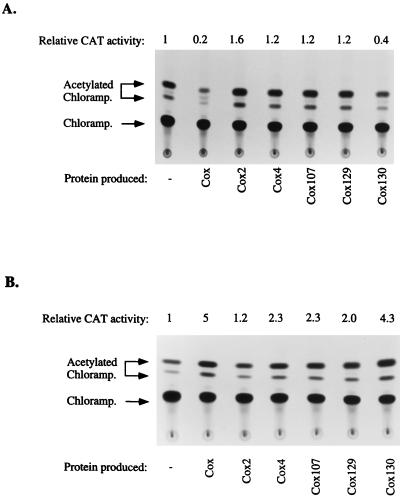
The capacity of the respective mutated Cox proteins to act as transcriptional repressors and activators was investigated using a reporter system containing vectors with the P2 Pc promoter (pSS39-6) or the P4 PLL promoter (pEE741) directing a cat reporter gene. The cox mutants were cloned into a compatible plasmid, pACYC177, and their activities as transcriptional repressors (Fig. 7a) and transcriptional activators (Fig. 7b) were analyzed. The results show that all cox mutants are defective in both repression and activation. Cox2 is the most defective protein, while Cox130 shows half the repressing capacity and almost normal activating capacity when compared to wild-type Cox.
FIG. 7.
Cox repression and activation assays. CAT activities were measured as described in Materials and Methods. The locations of chloramphenicol, 1-acetate chloramphenicol, and 3-acetate chloramphenicol are shown. CAT activities are related to the promoter in the absence of Cox proteins. (a) The ability of wild-type or mutated Cox to repress transcription was analyzed using cox and cox mutants cloned in pACYC177, which were tested for their ability to transcriptionally repress the P2 Pc promoter (pSS39-6). (b) In this Cox activation assay, the ability of wild-type or mutated Cox to activate the P4 PLL promoter (pEE741) was analyzed.
The mutated Cox proteins are defective in DNA binding.
The ability of the mutated Cox protein to bind to its cognate DNA site was examined by gel retardation assay (Fig. 6c). Each Cox protein was incubated with 5′-end-labeled DNA from the attP region (amplified by PCR, using primers 72.0R and 72.5L), and the amount of shifted DNA was quantified (Fig. 6d). The results show that the mutated proteins, which were defective in oligomerization, were also unable to bind DNA. Cox130 on the other hand seemed to be able to bind to DNA. In fact, under the same conditions, the Cox130 protein bound better to DNA than the wild-type protein. Cox130 gave the second shift under conditions in which wild-type Cox gave only the first shift.
DISCUSSION
The P2 Cox protein is a small, multifunctional protein, which is vitally important for excision of the phage genome, lytic development, and derepression of the unrelated bacteriophage P4. Functionally it resembles λ Xis in site-specific recombination and λ Cro as a transcriptional repressor.
To facilitate the purification of Cox, the cox gene was cloned under the control of the strong T7 promoter φ10. Under the conditions used, induction leads to overexpression of soluble Cox protein, when adding the T7 lysozyme on a plasmid (pLysE). A simple purification procedure was developed that gives a >95% pure Cox preparation, and this preparation specifically binds its DNA target.
Cox native size.
The analogous Cox proteins of bacteriophage HP1 and Apl of bacteriophage 186 have been characterized, and their native sizes have been determined. HP1 Cox consists of four protomers, which can self-associate to octamers (9), while Apl is monomeric in solution (26). Both proteins have a dual role in prophage induction, as does P2 Cox. The native size of the P2 Cox protein was determined in this work both in vivo and in vitro.
In the in vivo repressor fusion system (13), the cox gene replaced the part of the cI gene encoding the dimerization domain of the CI repressor of bacteriophage λ, and the capacity of the fusion protein was analyzed using a reporter gene controlled by the λ operator. In this system, the CI-Cox fusion protein repressed transcription of the reporter gene as strongly as wild-type λ CI protein, which implies that Cox forms at least dimers as efficiently as λ CI. To further analyze the capacity of P2 Cox proteins to form higher-order structures in vivo, the same CI-Cox fusion protein was assayed in a system discriminating between dimerization and cooperative binding or multimeric formation. In this system, the lacZ reporter gene is under the control of a synthetic promoter which is overlapped by a weak λ operator but preceded by a strong λ operator. The CI-Cox fusion protein repressed the reporter gene almost as well as a positive control known to form tetramers. This indicates either that (i) binding of CI-Cox to the strong upstream operator increases the affinity of the fusion protein to the weak operator, that is, cooperative binding or (ii) the fusion protein has the capacity to form oligomers.
The native size was further characterized by gel filtration experiments on a Sephacryl S-300 HR column. Cox eluted at two distinct peaks, corresponding to tetrameric and octameric P2 Cox, as does HP1 Cox. The native size of Cox was further determined at lower protein concentration using the lysine-specific 6.4 Å, cross-linker Sulfo-DST, which readily cross-linked Cox monomers, forming tetramers (Fig. 5a). The cross-linking experiment showed that Cox can form tetramers at a broad concentration range which indicates that the cross-links are formed within the Cox tetramers and not between tetramers. In a gel retardation assay, the cooperative binding of Cox to its DNA-binding site at attP was analyzed, resulting in three shifts. The sigmoidal nature of the binding curve indicates that tight binding of the Cox protein to its cognate site requires the action of two or more subunits. The results presented in this study, together with the results in the extensive study of the Cox protein from bacteriophage HP1 (9), make us believe that P2 Cox acts like HP1 Cox; that is, the first shift contains tetrameric Cox while the second shift includes octameric Cox bound to the DNA fragment. We also propose that the third shift contains Cox binding nonspecifically to the DNA, analogous to HP1 Cox.
Cox oligomerization is essential for Cox activity.
The results obtained with the cox mutants are summarized in Table 5. The biological relevance of Cox functioning as an oligomer was analyzed using cox mutants in repressor fusion assays and cross-linking assays. Three classes of cox mutants were distinguished regarding their ability to oligomerize in vivo and in vitro. The cox130 mutation seemed not to influence oligomerization negatively, while cox2 and probably cox4 affected oligomerization to a certain degree, whereas the C terminally deleted cox, cox38c, and the point mutants cox3, cox107, and cox129 completely blocked oligomerization. The fact that the mutations located in the presumed DNA-binding motif also affected oligomerization may be explained by the small size of the Cox protein. Cox probably lacks independently folded domains, and a mutation in one part of the protein affects other epitopes. This hypothesis is further supported by the fact that the Cox3 protein forms inclusion bodies when overexpressed in the same strain in which the wild-type protein is soluble (results not shown). The cox3 mutation leads to the replacement of a glycine with a glutamate, which may distort the overall three-dimensional structure of the protein, leading to an inactive protein. On the other hand, the cox2 and cox130 mutations give defective Cox proteins, which formed tetramers almost as well as wild-type Cox. Therefore, we suggest that some amino acid substitutions, like cox3, affect the folding of the entire protein, while other substitutions, like cox130, do not change the overall structure and thus do not affect other epitopes.
TABLE 5.
Summary of results for the cox mutants
| Protein | Amino acid substition | Tetramerization | Repression | Activation | Excision | DNA binding |
|---|---|---|---|---|---|---|
| Cox | − | +++ | +++ | +++ | +++ | +++ |
| Cox2 | Met42Thr | + | − | − | − | − |
| Cox3 | Gly22Glu | − | n.d. | n.d. | n.d. | n.d. |
| Cox4 | Glu54Lys | − | − | + | + | − |
| Cox107 | Arg29His | − | − | + | + | − |
| Cox129 | Ala69Thr | − | − | + | + | − |
| Cox130 | Glu16Lys | +++ | ++ | ++ | ++ | ++++ |
−, no activity; +++, wild-type activity; n.d., not determined due to the formation of inclusion bodies.
Since three mutations, cox2, cox4, and cox129, located downstream of the presumed DNA-binding epitope, are affecting the ability to form oligomers, we propose that the C-terminal half of the protein is involved in oligomerization and that oligomerization is required for the biological activity of the Cox protein.
What is the function of the presumed secondary structures?
When aligning the P2 Cox protein with the multimeric HP1 Cox protein and the monomeric Apl of phage 186 (Fig. 1), the C-terminal part of the P2 Cox protein is more similar to HP1 Cox. Around the cox4 mutation, four amino acids out of nine are identical to those of the HP1 Cox, and they could be a part of the protein-protein interacting interface. The β-strand next to the cox4 mutation, β3, is made up of 100% hydrophobic amino acid residues, which could be a potential protein-interacting epitope. Another mutation, cox129, gives a protein that is unable to oligomerize, which further supports the idea that the interacting interface is located at the C-terminal part of the Cox protein. The cross-linking experiments indicate that lysines are situated near or in the interacting interface. The P2 Cox protein contains seven lysines, where five are situated in the N terminus around the helix-turn-helix motif and two are situated around the cox129 mutation, Lys67 and Lys75. Therefore, it is possible that one of these lysines or both, together with Ala69 (cox129), are situated in the interacting interface. This was further supported by the fact that the presumed helix α3, which contains the cox129 mutation, is an amphipatic helix with Ala-69 as a part of the hydrophobic side of the helix. This kind of helix is easily visualized as being involved in protein-protein interfaces, where the hydrophobic side interacts with the hydrophobic side of the helix of another protomer. The cox129 mutation could also affect the length of the α3 helix, since when a secondary prediction of the cox129-mutated protein was run, the helix became shorter compared to the wild-type Cox protein (results not shown).
When comparing the different cox-mutated proteins in regard to their capacities as transcriptional repressors, activators, and excisionases, they are divided into three different classes: (i) no Cox activity, cox2; (ii) intermediate Cox activity, cox4, cox107, and cox129; and (iii) almost wild-type phenotype, cox130. These results together with those of the oligomerization analysis suggest that the effect of the cox2 mutation on oligomerization is indirect, since Cox2 can oligomerize to a rather large extent, but still it is the most severe cox mutation. The β-strand next to the cox2 mutation could be a hydrophobic core for the DNA-binding domain, since there are several (seven out of nine) hydrophobic amino acid residues. The cox130 mutation on the other hand does not affect tetramerization negatively; it rather enhances oligomerization of the Cox protein. Gel retardation assays showed that it even binds better to DNA than wild-type Cox. The appearance of the second shift could indicate a tendency to more easily form oligomers compared to wild-type Cox, which would correlate with the results from the in vivo cooperative-binding/multimerization assay.
In summary, we propose that the P2 Cox protein is tetrameric in solution and that it binds cooperatively to DNA, as a tetramer at low protein concentration and as an octamer at high protein concentrations. Our results further show that oligomerization is essential for Cox activity. A coupling between oligomerization and gene regulation has been observed in other systems (31), like the λ cI repressor (1), the tryptophan repressor (14), and the lac repressor (6). The results show that the protein-protein interacting interface is situated in the C terminus. We also suggest that β-strand β3 and α-helix α3 could together make up the oligomerization epitope. Further, the Cox protein seems to lack independent domain structures.
ACKNOWLEDGMENTS
We thank J. Hu for providing strains, phages, and plasmids for the repressor fusion assay and Erich W. Six for providing unpublished cox mutants and helpful discussions.
This work was supported by grant nb 72 from the Swedish Medical Research Council.
REFERENCES
- 1.Bain D L, Ackers G K. Self-association and DNA binding of λ cI repressor N-terminal domains reveal linkage between sequence-specific binding and the C-terminal cooperativity domain. Biochemistry. 1994;33:14679–14689. doi: 10.1021/bi00253a005. [DOI] [PubMed] [Google Scholar]
- 2.Bertani L E. Genetic interaction between the nip1 mutation and genes affecting integration and excision in phage P2. Mol Gen Genet. 1980;178:91–99. doi: 10.1007/BF00267217. [DOI] [PubMed] [Google Scholar]
- 3.Bertani L E, Bertani G. Preparation and characterization of temperate, non-inducible bacteriophage P2 (host: Escherichia coli) J Gen Virol. 1979;6:201–212. doi: 10.1099/0022-1317-6-2-201. [DOI] [PubMed] [Google Scholar]
- 4.Bertani L E, Six E W. The P2-like phages and their parasite, P4. Vol. 2. New York, N.Y: Plenum Publishing Corp.; 1988. [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Chakerian A E, Matthews K S. Effect of lac repressor oligomerization on regulatory outcome. Mol Microbiol. 1992;6:963–968. doi: 10.1111/j.1365-2958.1992.tb02162.x. [DOI] [PubMed] [Google Scholar]
- 7.Cuff J A, Clamp M E, Siddiqui A S, Finlay M, Barton G J. Jpred: a consensus secondary structure prediction server. Bioinformatics. 1998;14:892–893. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
- 8.Deho G, Zangrossi S, Ghisotti D, Sironi G. Alternative promoters in the development of bacteriophage plasmid P4. J Virol. 1988;62:1697–1704. doi: 10.1128/jvi.62.5.1697-1704.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esposito D, Scocca J J. Purification and characterization of HP1 Cox and definition of its role in controlling the direction of site-specific recombination. J Biol Chem. 1997;272:8660–8670. doi: 10.1074/jbc.272.13.8660. [DOI] [PubMed] [Google Scholar]
- 10.Esposito D, Wilson J C E, Scocca J J. Reciprocal regulation of the early promoter region of bacteriophage HP1 by the Cox and CI proteins. Virology. 1997;234:267–276. doi: 10.1006/viro.1997.8646. [DOI] [PubMed] [Google Scholar]
- 11.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haggård-Ljungquist E, Kockum K, Bertani L E. DNA sequences of bacteriophage P2 early genes cox and B and their regulatory sites. Mol Gen Genet. 1987;208:52–56. doi: 10.1007/BF00330421. [DOI] [PubMed] [Google Scholar]
- 13.Hu J C, O'Shea E K, Kim P S, Sauer R T. Sequence requirements for coiled-coils: analysis with repressor-GCN4 leucine zipper fusions. Science. 1990;250:1400–1403. doi: 10.1126/science.2147779. [DOI] [PubMed] [Google Scholar]
- 14.LeTilly V, Royer C A. Fluorescence anisotropy assays implicate protein-protein interactions in regulating trp repressor DNA binding. Biochemistry. 1993;32:7753–7758. doi: 10.1021/bi00081a021. [DOI] [PubMed] [Google Scholar]
- 15.Lin C-S. Nucleotide sequence of the essential region of bacteriophage P4. Nucleic Acids Res. 1984;12:8667–8684. doi: 10.1093/nar/12.22.8667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindahl G, Sunshine M. Excision-deficient mutants of bacteriophage P2. Virology. 1972;49:180–187. doi: 10.1016/s0042-6822(72)80019-9. [DOI] [PubMed] [Google Scholar]
- 17.Lindqvist B H, Dehò G, Calendar R. Mechanisms of genome propagation and helper exploitation by satellite phage P4. Microbiol Rev. 1993;57:683–702. doi: 10.1128/mr.57.3.683-702.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marshak D R, Kadanoga J T, Burgess R R, Knuth M W, Brennan W A, Lin S-H. Strategies for protein purification and characterization. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 19.Reed M R, Shearwin K E, Pell L M, Egan J B. The dual role of Apl in prophage induction of coliphage 186. Mol Microbiol. 1997;23:669–681. doi: 10.1046/j.1365-2958.1997.2521620.x. [DOI] [PubMed] [Google Scholar]
- 20.Saha S, Haggård-Ljungquist E, Nordström K. Activation of prophage P4 by the Cox protein and the sites of action of the Cox protein on the two phage genomes. Proc Natl Acad Sci USA. 1989;86:3973–3977. doi: 10.1073/pnas.86.11.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saha S, Haggård-Ljungquist E, Nordström K. The Cox protein of bacteriophage P2 inhibits the formation of the repressor protein and autoregulates the early operon. EMBO J. 1987;6:3191–3199. doi: 10.1002/j.1460-2075.1987.tb02631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saha S, Lundqvist B, Haggård-Ljungquist E. Autoregulation of bacteriophage P2 repressor. EMBO J. 1987;6:809–814. doi: 10.1002/j.1460-2075.1987.tb04823.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Sasaki I, Bertani G. Growth abnormalities in Hfr derivatives of Escherichia coli strain C. J Gen Virol. 1965;40:365–376. doi: 10.1099/00221287-40-3-365. [DOI] [PubMed] [Google Scholar]
- 25.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 26.Shearwin K E, Egan J B. Purification and self-association equilibria of the lysis-lysogeny switch proteins of coliphage 186. J Biol Chem. 1996;271:11525–11531. doi: 10.1074/jbc.271.19.11525. [DOI] [PubMed] [Google Scholar]
- 27.Six E W, Lindqvist B H. Mutual derepression in the P2–P4 bacteriophage system. Virology. 1978;87:217–230. doi: 10.1016/0042-6822(78)90127-7. [DOI] [PubMed] [Google Scholar]
- 28.Studier E W, Moffat B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. Methods Enzymol. 1986;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 29.Sunshine M G, Thorn M, Gibbs W, Calendar R, Kelly B. P2 phage amber mutants: characterization by use of a polarity supressor. Virology. 1971;46:691–702. doi: 10.1016/0042-6822(71)90071-7. [DOI] [PubMed] [Google Scholar]
- 30.Weiss M A, Pabo C O, Karplus M, Sauer R T. Dimerization of the operator binding domain of phage lambda repressor. Biochemistry. 1987;26:897–904. doi: 10.1021/bi00377a034. [DOI] [PubMed] [Google Scholar]
- 31.Wong I, Lohman T M. Linkage of protein assembly to protein-DNA binding. Methods Enzymol. 1995;259:95–127. doi: 10.1016/0076-6879(95)59040-4. [DOI] [PubMed] [Google Scholar]
- 32.Yu A, Haggård-Ljungquist E. Characterization of the binding sites of two proteins involved in the bacteriophage P2 site-specific recombination system. J Bacteriol. 1993;175:1239–1249. doi: 10.1128/jb.175.5.1239-1249.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu A, Haggård-Ljungquist E. The Cox protein is a modulator of directionality in bacteriophage P2 site-specific recombination. J Bacteriol. 1993;175:7848–7855. doi: 10.1128/jb.175.24.7848-7855.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zagursky R J, Berman M L. Cloning vectors that yield high levels of single-stranded DNA for rapid DNA sequencing. Gene. 1984;27:183–191. doi: 10.1016/0378-1119(84)90139-2. [DOI] [PubMed] [Google Scholar]
- 35.Zeng X, Hu J C. Detection of tetramerization domains in vivo by cooperative DNA binding to tandem lambda operator sites. Gene. 1997;185:245–249. doi: 10.1016/s0378-1119(96)00652-x. [DOI] [PubMed] [Google Scholar]