Abstract
Amyotrophic lateral sclerosis (ALS) is characterized by the progressive loss of somatic motor neurons. A major focus has been directed to motor neuron intrinsic properties as a cause for degeneration, while less attention has been given to the contribution of spinal interneurons. In the present work, we applied multiplexing detection of transcripts and machine learning–based image analysis to investigate the fate of multiple spinal interneuron populations during ALS progression in the SOD1G93A mouse model. The analysis showed that spinal inhibitory interneurons are affected early in the disease, before motor neuron death, and are characterized by a slow progressive degeneration, while excitatory interneurons are affected later with a steep progression. Moreover, we report differential vulnerability within inhibitory and excitatory subpopulations. Our study reveals a strong interneuron involvement in ALS development with interneuron specific degeneration. These observations point to differential involvement of diverse spinal neuronal circuits that eventually may be determining motor neuron degeneration.
A combined transcriptomics and computational approach reveals early and differential dysregulation of spinal interneurons in ALS.
INTRODUCTION
In amyotrophic lateral sclerosis (ALS), somatic motor neurons degenerate leading to muscle denervation and wasting (1). Hence, people affected by the disease progressively lose the ability to control muscles and perform movements. Execution of movements requires coordinated activation of motor neuron pools for appropriate muscle activation (2). The activation is mediated by a complex network of inhibitory and excitatory intraspinal neurons decoding the input from the descending motor pathways and the afferent sensory input (3). Within the ventral horn of the spinal cord, inhibitory and excitatory neurons can be divided into four cardinal classes: V0, V1, V2, and V3, identified by the expression of specific molecular markers defining cell fate during development (4). Research in a number of vertebrate species has shown that specific subpopulations of inhibitory spinal neurons [expressing glycine or γ-aminobutyric acid (GABA)] and excitatory spinal neurons (expressing glutamate or acetylcholine) form central pattern generators (CPGs), controlling rhythmic movements, and regulating the coordination of locomotion (2, 3, 5, 6).
These cardinal classes are further diversified. Thus, V0 interneurons are, based on their axonal projections and excitatory or inhibitory neurotransmitters, divided into several classes of neurons. A small group of V0 neurons expressing the Pitx2 transcription factor, the V0C/G subpopulations, are ipsilaterally projecting, contain either acetylcholine or glutamate, and are localized around the central canal (7). The cholinergic V0C neurons are the main source of C boutons (7, 8). The largest population of V0 interneurons are contralaterally projecting, inhibitory (V0D) or excitatory (V0V), and are characterized by expression or lack thereof of the Dbx1 and Evx1 transcription factors. They control left-right alternation at slow (V0D) and high speed (V0V) of locomotion (9–11). V1 interneurons are a large ipsilaterally projecting inhibitory population. In the adult mice, 80% of them are glycinergic, although often coexpressing GABA (12). Previous studies have shown that this population is highly heterogeneous, and, depending on marker coexpression, up to 50 subpopulations can be identified (13, 14). In mammals, V1 interneurons are known to control the speed of locomotion (15), and Renshaw cells (RCs) as well as Ia interneurons belong to the V1 class (12). V2 interneurons are also ipsilaterally projecting and divided into V2a excitatory interneurons and V2b inhibitory interneurons. V2a neurons are characterized by expression of the transcription factor Chx10 (16) and are rhythmically active during locomotion (17). V2b neurons are identified by Gata2/3 expression (18) and, together with V1 interneurons, are required for the coordination of flexor-extensor alternation (19, 20). The V3 interneurons project both contralaterally and ipsilaterally (21, 22) and are excitatory; ablation or silencing of those neurons leads to changes in left-right coordination (21). Two other molecularly defined excitatory populations are the Shox2 (17) and the Hb9-positive (also called Mnx1) (23) neurons. Shox2 neurons are found in the intermediate area of the spinal cord, 75% of them coexpress Chx10 while the remaining 25%, which are Shox2-positive and Chx10-negative, are known to be involved in rhythm generation (17). Hb9 is expressed by motor neurons as well as many interneurons in the intermediate area, some of which also contribute to locomotor rhythm generation (23).
Growing evidence has reported spinal circuit dysregulations in ALS [for review (24)]. The cholinergic C boutons on motor neurons undergo changes during ALS progression (25). Glycinergic interneurons change their electrophysiological properties at early postnatal (P) stages, with the most ventrally located neurons being less excitable (26). Our recent findings showed that already at P45, before motor neuron degeneration and muscle denervation, there is a loss of glycinergic synapses on fast motor neurons (27), including the V1 interneuron synapses (27). Other studies have also reported a loss of V1 and V2a interneurons at different disease stages (28, 29). However, the temporal dynamics of potential loss of excitatory and inhibitory interneurons in the ventral spinal cord motor–related circuits throughout disease development have not been revealed. Since these interneuron populations are essential for movement, we set out to perform such studies.
A clear challenge to this type of investigation is the inherent difficulties in being able to study the fate of multiple interneuron subpopulations during disease progression. To succeed, it requires the simultaneous visualization of several molecular markers in the spinal cord as the disease develops. Previous studies have used multiplexed antibody staining with up to four different markers to identify neuronal subpopulations within the spinal cord (13). However, recent advancement in the multiplexing detection of transcripts now allows for the visualization of tens and up to hundreds of markers at the same time (30, 31). While multiplexing detection and visualization are now routinely performed in several laboratories (32–34), reliable quantification of multiple transcripts in the same cells has been proven difficult and often requires manual annotation. Hence, in the present study, we developed machine learning–based tools to automize cell segmentation and subsequent transcript registration, as well as a bioinformatic analysis pipeline to perform spatial analysis of differentially expressed transcripts. The combination of the multiplexing detection and the computational analysis allowed us to investigate the changes in expression of molecular markers identifying multiple classes of interneurons in the spinal cord as well as motor neuron populations in the SOD1G93A mouse at three time points of disease progression: (i) presymptomatic disease stage, P30; (ii) onset of locomotor disease stage, P63, and (iii) end disease stage with motor neuron death, P112. We find that inhibitory subpopulations of interneurons start losing their specific markers [e.g., Engrailed-1 (En1) and Calb1] early in the disease. In contrast, excitatory interneurons (e.g., Chx10-positive) lose their specific markers at late stages of the disease, although these changes in expression appear quickly at symptomatic stages. Together, these results point toward a large contribution of spinal interneuron circuits in disease, with distinct degenerative temporal dynamics between inhibitory and excitatory interneurons.
RESULTS
Developmentally defining neuronal markers are maintained in the adult spinal cord
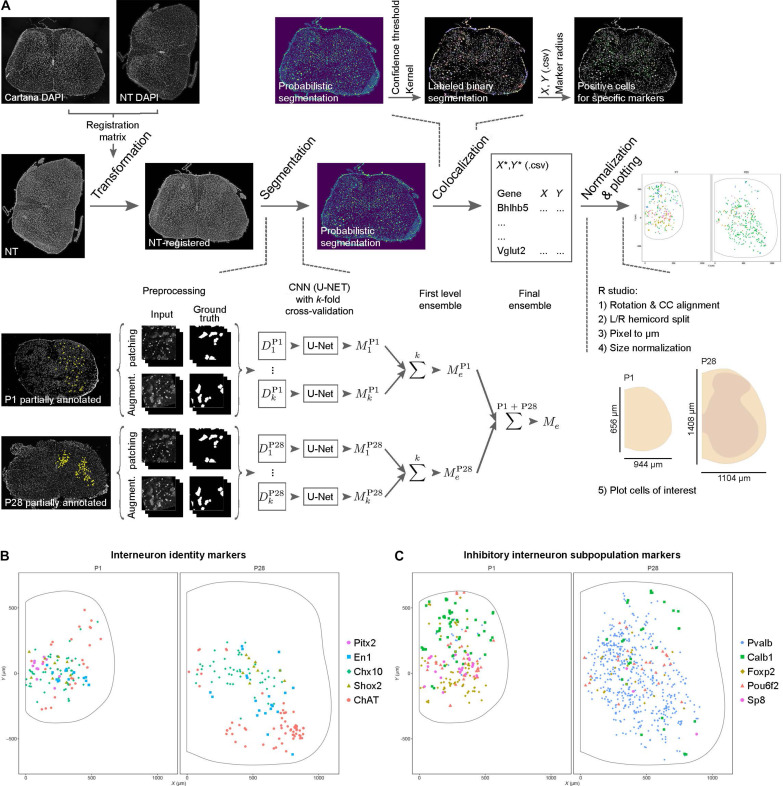
The expression of molecularly defining interneuron markers has been mainly characterized during mouse development at embryonic and postnatal stages (4, 13). Among these markers are transcription factors, whose expression appears to be down-regulated in the adult tissue making antibody detection less sensitive. We previously showed that the En1 transcript, identifying V1 interneurons, can be reliably detected in the mouse adult spinal cord by in situ hybridization (27, 35). In the present study, we compared the expression of 24 markers (Fig. 1, A and B) in P1 and P28 mouse spinal cord tissue to validate their presence and spatial location in the adult, by using an in situ sequencing technique (CARTANA) (Fig. 2A), now replaced by Xenium (10x Genomics) (31). This technique allows for the detection of more than 250 markers at the same time, by using barcoded probes detecting the specific transcripts (31). Upon sequencing, information about the spatial localization of the barcodes is provided as X-Y coordinates within the acquired tile images (31) which allow for exact localization of transcript within the spinal cord (Fig. 2A). However, for exact subpopulation identification, coexpression of multiple markers is often required. Therefore, outlines of neuronal soma or cell bodies are needed to assign potential expression of multiple markers within the same cell. Previous computational methods have been developed to identify cell body boundaries based on the segmentation of nuclear [4′,6-diamidino-2-phenylindole (DAPI)] staining (32, 36). However, neurons found within the ventral and dorsal areas of the spinal cord differ substantially in shape and size, with ventral neurons being larger and dorsal neurons being characterized by smaller and rounder somata. Therefore, segmentation based on nuclear staining might not capture the diverse cell body size in the spinal cord. To segment spinal neurons, we therefore implemented a machine learning deep ensemble approach as shown in Fig. 2A. To get an outline of cell bodies we used NeuroTrace, which provides a fluorescent Nissl staining of the cytoplasm of neurons and nuclei. NeuroTrace images of the analyzed spinal cord tissue were obtained after in situ sequencing and registered to the DAPI images obtained by CARTANA for X-Y coordinate registration (Fig. 2A). Probabilistic segmentation of the cell bodies was then obtained on the basis of the NeuroTrace staining, using an updated version of a deep ensemble method previously developed for limited label training data (37). Neuron annotations for both P1 (n = 52 annotated patches, from N = 2 sections from two independent mice) and P28 tissue (n = 20 annotated patches, from N = 2 sections from two independent mice) were generated and used to train parallel models using U-Net convolutional neural networks (CNN) with k-fold cross-validation (Fig. 2A). The final ensemble model obtained from the combination of P1 and P28 models was used for probabilistic segmentation of the tile images. A threshold of 0.4 confidence and a structural kernel of 5 were applied to define the final segmentation used for analysis (neurons visualized in yellow and green; Fig. 2A). Transcript X-Y coordinates were then registered to segmented neurons, and spatial analysis was performed per hemicord. A total of N = 6 P1 and N = 29 P28 spinal cord sections (from 3 and 11 mice, respectively) were analyzed to validate transcript expression and spatial localization. Cross sections obtained from lumbar (L) segments 1 to 3 were included in the study. Figure 2B shows an example of spatial analysis for the cardinal neuronal identity markers Pitx2, En1, Chx10, Shox2, and choline acetyltransferase (ChAT) in P1 versus P28 tissue (the graph includes data from two spinal cord sections from one mouse at P1, and two sections from two mice at P28; markers are shown in different colors and shapes). An example of spatial segregation of markers for inhibitory interneuron subpopulations including parvalbumin (Pvalb) (12), Calbindin (Calb1), Foxp2, Pou6f2, and Sp8 (13), which can be used in combination with En1 to represent V1 interneurons, is shown in Fig. 2C. Examples of combinatorial expression of interneuron markers are shown in fig. S1 (A and B), including colocalization of inhibitory markers with En1. The lack of representation of some markers at P28 can be explained by the fact that the in situ sequencing technique detects only 30% of all transcript, thus especially underrepresenting the more lowly expressed markers (such as Pitx2 or combinatorial En1/Sp8) in the portrayed dataset (2 to 6 sections out of 29). However, when taking into account the full dataset (not shown here to facilitate visualization), all markers were detected.
Fig. 1. Interneuronal markers investigated in the present study and their circuitry.
(A) Schematic of the locomotor circuits in the mammalian lumbar spinal cord. Shown are the cardinal ventral interneuron subpopulations (V0D, V0V, V0C, V1, V1 RCs, V1 Ia interneurons, V2a, V2b, and V3) as well as Shox2 and Hb9 interneurons, and their main ipsilateral (solid) or contralateral (dashed) projections. Inhibitory inputs are shown as triangles, and excitatory are shown as circles. (B) Panel of markers detected throughout the study representing the different interneuron populations of interest. Transcript expression of underlined markers was quantified for characterization of interneuron dysregulation in ALS progression.
Fig. 2. Fate-defining markers are present in adult mouse tissue.
(A) Pipeline used for processing and analysis of in situ sequencing data. NeuroTrace (NT) images were acquired and registered to in situ sequencing data based on 4′,6-diamidino-2-phenylindole (DAPI) staining. These were then used for segmentation of neuronal cells using a machine learning deep ensemble model based on the use of U-Net convolutional neural network with k-fold cross-validation, which was applied separately to patches (512 × 512 pixels) of annotated data from P1 (52 annotated patches) and P28 (20 annotated patches) and merged through two ensemble steps to obtain probabilistic segmentation. A confidence threshold and structural kernel were applied to obtain final segmentation, to which in situ sequencing coordinates were registered. Only transcripts that localized to segmented cells were used for final processing in R, where coordinates were normalized and plotted for visualization and qualitative analysis. (B) Validation of identity markers by in situ sequencing technique in wild-type (wt) young adult mouse lumbar spinal cord (P28, right) compared to early postnatal (P1, left), for the interneuron populations included in the study: Pitx2 (pink circle) for V0C/G, En1 (blue square) for V1, Chx10 (green diamond) for V2a, Shox2 (khaki triangle), and choline acetyltransferase (ChAT; salmon circle) for V0C and motor neurons. (C) Validation of markers for inhibitory interneuron subpopulations in adult mouse tissue including parvalbumin (Pvalb; blue circle), Calb1 (green square), Foxp2 (khaki diamond), Pou6f2 (salmon triangle), and Sp8 (pink circle). Data in (B) and (C) are pooled from n = 2 sections from N = 1 mouse for P1, and N = 2 mice for P28.
This analysis demonstrates that the interneuron markers that have been used embryonically and early postnatally to characterize spinal interneuron populations (4, 38, 39) are also found in the adult spinal cord. On the basis of spatial localization, it appears that the marker-identified interneuron populations maintain their identity in the adult spinal cord.
Interneuron markers in the SOD1G93A mouse
For single mRNA molecule quantifications in healthy and SOD1G93A tissue, we used the RNAscope HiPlexUp technique (ACD, BioTechne) (Fig. 3 and fig. S2A). RNAscope is known to detect ~95% of mRNA molecules of the targeted transcripts, and the HiPlexUp technique allows for simultaneous detection of up to 48 transcripts in the same tissue. We used the 24 transcripts originally investigated by in situ sequencing (Fig. 1B). The probes were designed and validated by ACD BioTechne and the procedure was performed as shown in fig. S2A. Upon two series of hybridization-amplification and four labeling and cleaving rounds per series, images were acquired in a total of eight rounds (three probes imaged per round). DAPI and NeuroTrace staining were included for image registration and cell segmentation, respectively. Using ImageJ, registration matrixes for the eight HiPlexUp rounds plus NeuroTrace were generated on the basis of DAPI staining, and these were used to transform and merge 26 superimposed tile images of each spinal cord section (one per transcript + DAPI + NeuroTrace) which were used for further processing in the ZEN software (Zeiss). The machine learning–based Intellesis tool (Zeiss) was used for cell segmentation and neuron enrichment (fig. S2, B and C; in orange is the positive fraction = neurons included in the study, in blue is the negative fraction = background and other cells). X-Y coordinates and area were obtained for each segmented neuron, while the intensity means and SD were obtained for each transcript expressed within each neuron (fig. S2B).
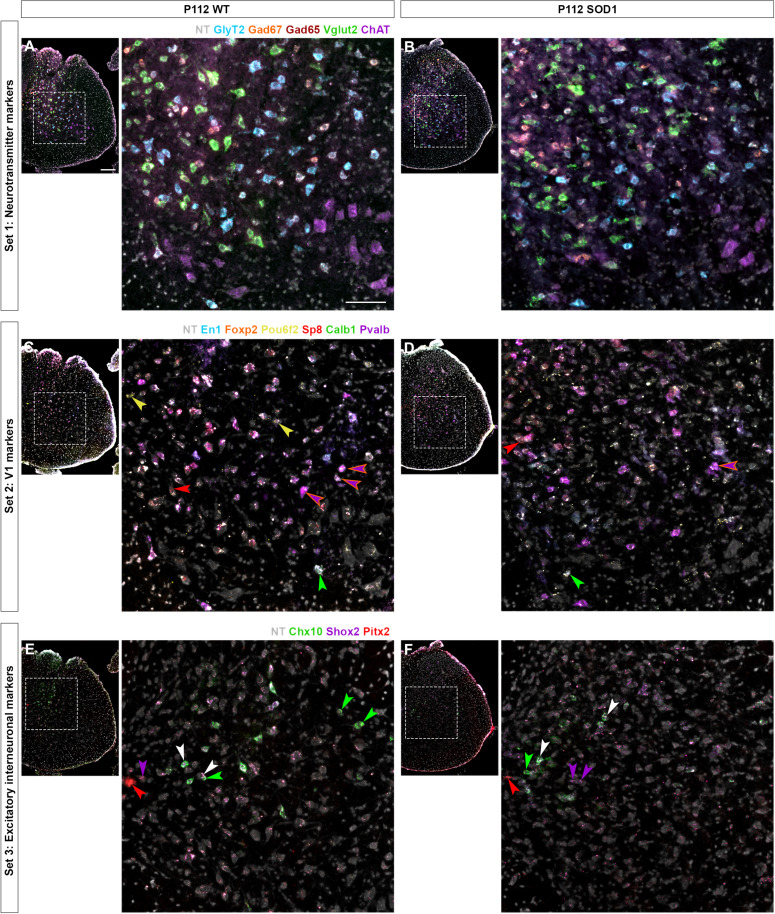
Fig. 3. Interneuron marker transcript detected by RNAscope HiPlexUp.
Microscopy images obtained by RNAscope HiPlexUp multiplexed in situ hybridization in wt (left) and SOD1G93A mice (right) at P112, including all markers used for characterization of interneuron dysregulation. To facilitate visualization, markers have been divided into three subsets. For each subset and condition, an overview of a full hemisection is shown, together with an inset with higher magnification. (A and B) Subset 1 includes neurotransmitter markers: GlyT2 (blue), Gad67 (orange), Gad65 (deep red), Vglut2 (green), and ChAT (purple). (C and D) Subset 2 includes markers for inhibitory and V1 population: En1 (blue), Foxp2 (orange), Pou6f2 (yellow), Sp8 (red), Calb1 (green), and Pvalb (purple). Arrows point to neurons that are En1+/Pou6f2+ (yellow), En1+/Sp8+ (red), En1+/Calb1+ (green, putative for Renshaw cells), En1+/Foxp2+/Pvalb+ (purple with orange stroke, putative for V1 Ia interneurons). (E and F) Subset 3 includes markers for some excitatory populations: Chx10 (green, for V2a), Shox2 (purple), and Pitx2 (red, for V0C/G). Arrows point to neurons that are Chx10+ (green), Shox2+ (purple), Chx10+/Shox2+ (white), and Pitx2+ (red).
Three different time points were included in the study: P30 (presymptomatic disease stage), P63 (onset-of-locomotor-phenotype disease stage), and P112 (symptomatic disease stage with motor neuron death). Three wild-type (wt) and three SOD1G93A mice were included per time point. Analysis was performed per hemicord since SOD1G93A shows asymmetrical loss of neurons within the spinal cord similar to patients with ALS with spinal cord onset (40–43). Both left and right hemicords were analyzed for each mouse, with two to five hemisections included per hemicord. With this approach, 98.943 neurons were segmented, of which 84.791 were included in the study after applying a soma size threshold of 70 μm2. When comparing the number of segmented neurons per condition and time point, we observed that a comparable number of neurons were included in the study, with an average of 625 segmented cells per hemisection (multiple unpaired t tests with Welch correction; P30: t = 0.5782, df = 38, P > 0.9999; P63: t = 1.311, df = 41, P = 0.5918; P112: t = 0.09803, df = 46, P > 0.9999; n = 20 to 26 hemisections, N = 3 mice per condition) (fig. S2D; the spatial distribution of the segmented neurons is shown for individual mice in fig. S2E). At P112, changes in expression levels are visually notable for some of the markers (e.g., En1 and Chx10), due to the clear decrease in intensity when compared to control conditions (Fig. 3, C to F). All 24 markers were investigated in disease progression in the SOD1G93A mouse. However, in the present work, only the following markers related to major classes of ventral interneurons will be discussed (underlined in Fig. 1B): En1, FoxP2, Pou6f2, and Sp8 for V1 interneurons; Calb1 for V1 putative RCs; Pvalb and FoxP2 for V1 putative Ia interneurons; Chx10 for V2a interneurons; Shox2 for excitatory interneurons; Pitx2, ChAT, and Vglut2 for V0C/G interneurons; ChAT for motor neurons; GlyT2, Gad67, Gad65, and Vglut2 for neurotransmitter-defined neurons.
V1 inhibitory interneurons show early dysregulation and differential vulnerability among subpopulations
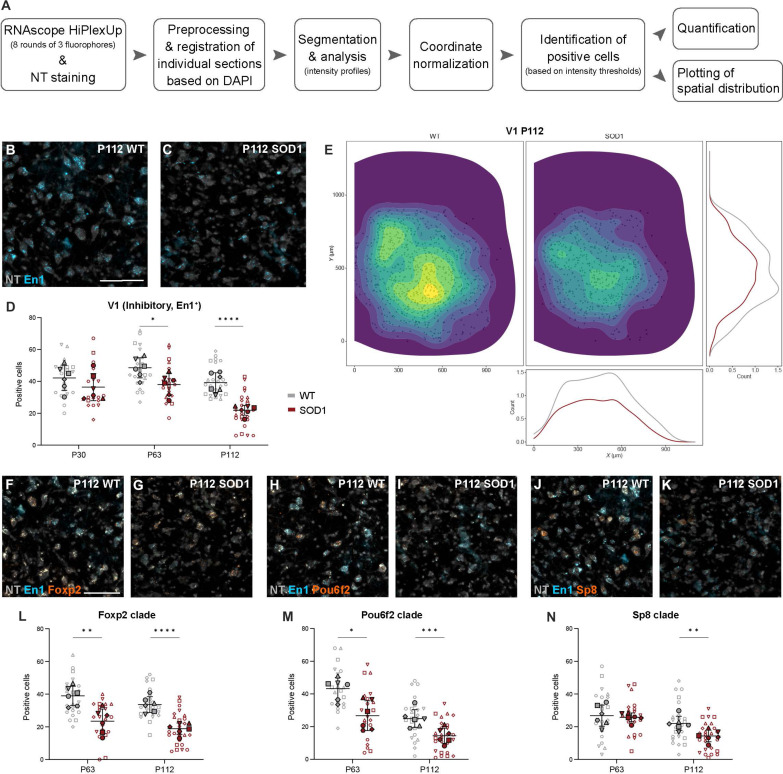
Upon image postprocessing (Fig. 4A and fig. S2, A and B), V1 interneurons were defined as inhibitory neurons (GlyT2-, Gad65-, and/or Gad67-positive), positive for En1 (Fig. 4, B and C). Analysis performed at the presymptomatic disease stage did not show alterations in En1 expression (P30: nested unpaired two-tailed t test, t = 1.305, df = 10, P = 0.2210, n = 20 to 22 hemisections, N = 6 hemicords from three mice per condition) (Fig. 4D). Our previous data with manual detection showed ~25% decrease in the En1 transcript expression already at P63 in the SOD1G93A mice (27). Here, we found a similar decrease in expression at P63 using the automized approach (P63: 19.2% down-regulation, nested unpaired two-tailed t test, t = 2.493, df = 10, P = 0.0318, n = 21 to 22 hemisections, N = 6 hemicords from three mice per condition) (Fig. 4D). The En1 loss was further exacerbated in symptomatic mice with a total decrease of 45.4% compared to controls (P112: nested unpaired two-tailed t test, t = 6.379, df = 47, P < 0.0001, n = 23 to 26 hemisections, N = 6 hemicords from three mice per condition), which was also evident upon visualization of spatial distribution and count-based kernel density estimation (Fig. 4, B and E).
Fig. 4. The V1 interneuron populations are dysregulated in the SOD1G93A mouse.
(A) Analysis workflow. (B and C) Images of HiPlexUp assay showing detection of En1 (blue) and NT (gray) in control (B) and SOD1G93A (C) lumbar spinal cord at P112. (D) Significant reduction in inhibitory (GlyT2+, Gad67+, and/or Gad65+) En1+ neurons in SOD1G93A mice (red) compared to controls (gray) at P63 and P112 (nested unpaired two-tailed t tests; P30 P = 0.2210, P63 P = 0.0318, P112 P < 0.0001). (E) Loss of En1+ neurons in the SOD1G93A mouse (right/red) compared to healthy controls (left/gray), especially in the ventral-most region, at P112. Data were pooled from all P112 sections and shown as individual cells (black dots) with count-based kernel density estimations in two dimensions (main panel), and in X (bottom) and Y dimensions (right). (F to K) HiPlexUp images of En1 (blue) colocalized with V1 subpopulation markers (orange) Foxp2 (E and F), Pou6f2 [(G) and (H)], and Sp8 [(I) and (J)] with NT (gray), in controls and SOD1G93A mice at P112. (L to N) Quantification in SOD1G93A mice versus controls shows significant decrease in Foxp2+ neurons at P63 and P112 (L) (nested unpaired two-tailed t tests; P63 P = 0.0036, P112 P < 0.0001), Pou6f2+ neurons at P63 and P112 (M) (nested unpaired two-tailed t tests; P63 P = 0.0100, P112 P = 0.0007), and Sp8+ neurons at P112 (N) (nested unpaired two-tailed t tests; P63 P = 0.7444, P112 P = 0.0083). Scale bars, 100 μm. 2D kernel densities in (E) were plotted with 10 bins using viridis scale. N = 6 hemicords from three mice (filled shapes, representing biological replicates). Number of hemisections (empty shapes, technical replicates for biological replicate): P30 n(wt) = 22, n(SOD1) = 20; P63 n(wt) = 22, n(SOD1) = 21; P112 n(wt) = 23, n(SOD1) = 26. Data are shown as means ± SD.
We next investigated potential differential vulnerability within V1 subpopulations by analyzing FoxP2-, Pou6f2-, and Sp8-positive neurons (13) at P63 and P112 time points (Fig. 4, F to N). Quantified neurons were positive for GlyT2, Gad65, and/or Gad67, and En1 markers as well as FoxP2, Pou6f2, or Sp8 (Fig. 4, F to K). The analysis revealed a differential vulnerability among neurons, with FoxP2 (P63: 37.4% down-regulation, nested unpaired two-tailed t test, t = 3.777, df = 10, P = 0.0036, n = 22 to 21 hemisections, N = 6 hemicords from three mice per condition) (Fig. 4L) and Pou6f2-positive neurons (P63: 35.1% down-regulation, nested unpaired two-tailed t test, t = 3.168, df = 10, P = 0.0100, n = 22 to 21 hemisections, N = 6 hemicords from three mice per condition) (Fig. 4M) decreasing in number at earlier time-points compared to the Sp8-positive neurons (P63: ns, nested unpaired two-tailed t test, t = 0.328, df = 41, P = 0.7444, n = 22 to 21 hemisections, N = 6 hemicords from three mice per condition) (Fig. 4N). Despite appearing initially more resistant, Sp8-positive neurons were reduced in numbers at P112 (P112: 36.4% down-regulation, nested unpaired two-tailed t test, t = 2.754, df = 47, P = 0.0083, n = 23 to 26 hemisections, N = 6 hemicords from three mice per condition) (Fig. 4N). At the same time point, FoxP2- and Pou6f2-expressing neurons were further decreased in numbers (P112 FoxP2: 44.3% down-regulation, nested unpaired two-tailed t test, t = 5.502, df = 47, P < 0.0001; P112 Pou6f2: 43.9% downregulation, nested unpaired two-tailed t test, t = 3.637, df = 47, P = 0.0007; n = 23 to 26 hemisections, N = 6 hemicords from three mice per condition) (Fig. 4, L and M). Spatial information regarding changes in expression at P63 of En1 transcripts, as well as Foxp2, Pou6f2, and Sp8, is shown in fig. S3 (A to D). All positive neurons were included in the analysis, independently of whether they were low or high expression for the transcript. Moreover, the shown contour plots also display all neurons without any cutoff in spatial density.
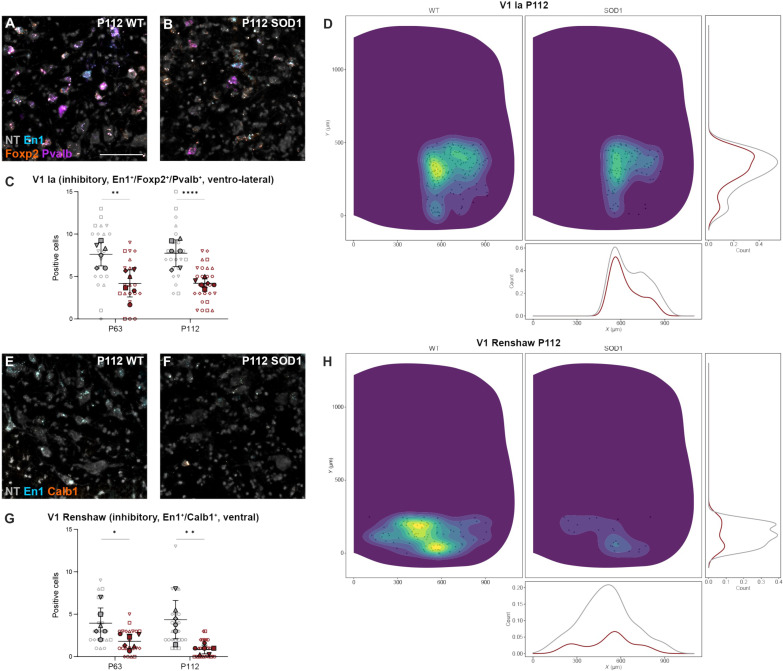
Putative RCs and Ia interneurons, members of the V1 subpopulation, were also analyzed at P63 and P112. These functionally defined interneuron populations are found in specific areas of the ventral spinal cord (12). In this study, putative Ia interneurons were identified as inhibitory interneurons (GlyT2+, Gad65+, and/or Gad67+), positive for En1, FoxP2, and Pvalb (Fig. 5, A and B) (12). Moreover, we restricted their location to the ventrolateral area of the spinal cord, where Ia interneurons are found (Fig. 5D). Decreased numbers of inhibitory En1+/FoxP2+/Pvalb+ neurons were found at P63 (42.0% down-regulation) as well as at P112 (45.8% down-regulation) (Fig. 5, C and D) (P63: nested unpaired two-tailed t test, t = 3.279, df = 41, P = 0.0021, n = 22 to 21 hemisections, N = 6 hemicords from three mice per condition; P112: nested unpaired two-tailed t test, t = 4.760, df = 47, P < 0.0001, n = 23 to 26 hemisections, N = 6 hemicords from three mice per condition). Putative RCs were identified by their expression of the inhibitory markers GlyT2, Gad65, and/or Gad67, in combination with En1 and Calb1 (Fig. 5, E and F) and their ventral location in the spinal cord (Fig. 5H). The number of ventral inhibitory En1+/Calb1+ neurons was reduced by 52.8% at P63 (P63: nested unpaired two-tailed t test, t = 2.587, df = 10, P = 0.0271, n = 22 to 21 hemisections, N = 6 hemicords from three mice per condition) (Fig. 5G), and this reduction was exacerbated at P112 to a 79.5% decrease (P112: nested unpaired two-tailed t test, t = 3.630, df = 10, P = 0.0046, n = 23 to 26 hemisections, N = 6 hemicords from three mice per condition) (Fig. 5, G and H). The spatial distributions of putative V1 Ia and RCs at P63 are shown in fig. S4 (A and B).
Fig. 5. Putative V1 Ia interneurons and Renshaw cells show dysregulation in the SOD1G93A mouse.
(A and B) Colocalization of En1 (blue), Foxp2 (orange), and Pvalb (purple) transcript detected by RNAscope HiPlexUp in situ hybridization in wt (A) and SOD1G93A mice (B) at P112. (C) Significant reduction in inhibitory En1+/Foxp2+/Pvalb+ neurons within the ventrolateral region of the spinal cord, where V1 Ia interneurons are located, in SOD1G93A mice (gray) versus wt (red) at P63 and P112 (nested unpaired two-tailed t tests; P63 P = 0.0498, P112 P = 0.0016). (D) Spatial distribution with count-based kernel density estimations of putative Ia interneurons from pooled P112 spinal cord sections. Data show a decrease in En1+/ Foxp2+/Pvalb+ neurons in the SOD1G93A mouse (right/red) compared to healthy controls (left/gray). (E and F) Colocalization of En1 (blue) and Calb1 (orange) transcript in wt (E) and SOD1G93A spinal cord (F) at P112. (G) Marked loss of inhibitory En1+/Calb1+ neurons located within the ventral spinal cord, where RCs are found, in the SOD1G93A mouse at P63, with exacerbation at P112 (nested unpaired two-tailed t tests; P63 P = 0.0271, P112 P = 0.0046). (H) Spatial distribution of putative RCs (En1+/Calb1+) at P112 with very few cells in the ventral spinal cord of SOD1G93A mice compared to healthy control mice, evidencing the marked reduction in En1+/Calb1+ neurons. Scale bars, 100 μm. NT counterstaining in gray. 2D kernel densities in (D) and (H) were plotted with 10 bins using viridis scale. N = 6 hemicords from three mice (filled). Number of hemisections (empty): P30 n(wt) = 22, n(SOD1) = 20; P63 n(wt) = 22, n(SOD1) = 21; P112 n(wt) = 23, n(SOD1) = 26. Data are shown as means ± SD.
Together, these data show that all the analyzed V1 subpopulations are affected at a symptomatic stage in the SOD1G93A mice when compared to the wt control. However, earlier in the disease, V1 subpopulations show differential vulnerability, with Sp8+ neurons being more resistant to loss of transcripts, and putative RCs being most strongly affected.
Inhibitory neurotransmitter expression and V1 interneuron survival decrease only at symptomatic stages, while the En1 transcript is lost earlier on
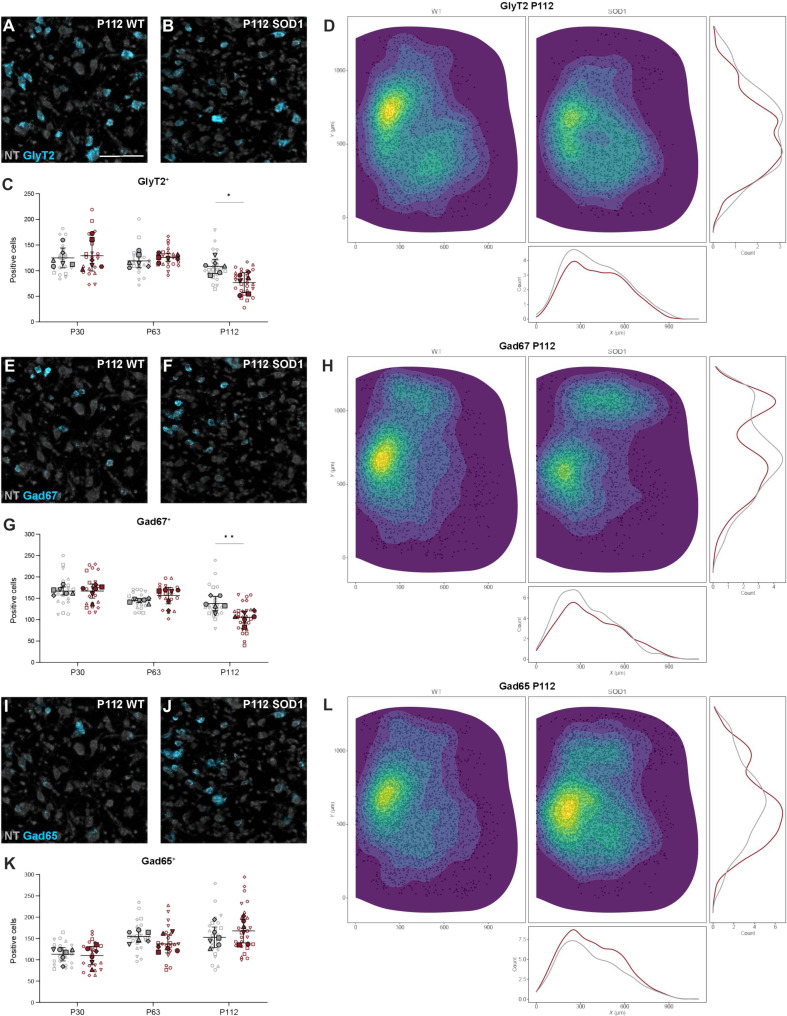
The expression of the glycine transporter 2 (Glyt2) and the enzymes catalyzing the GABA neurotransmitter Gad67 (also called Gad1) and Gad65 (also called Gad2) was investigated at the three time points (Fig. 6 and figs. S5 and S6). These markers are used to visualize glycinergic and GABA-releasing (GABAergic) neurons, respectively. GlyT2 transcript (Fig. 6, A and B) was down-regulated only at P112 (P30: nested unpaired two-tailed t test, t = 0.332, df = 10, P = 0.7469, n = 22 to 20 hemisections, N = 6 hemicords from three mice per condition; P63: nested unpaired two-tailed t test, t = 0.881, df = 41, P = 0.3836, n = 22 to 21 hemisections, N = 6 hemicords from three mice per condition; P112: nested unpaired two-tailed t test, t = 3.058, df = 10, P = 0.0121, n = 23 to 26 hemisections, N = 6 hemicords from three mice per condition) (Fig. 6, C and D). Gad67 expression (Fig. 6, E and F) was also decreased only at P112 (P30: nested unpaired two-tailed t test, t = 0.105, df = 40, P = 0.9172, n = 22 to 20 hemisections, N = 6 hemicords from three mice per condition; P63: nested unpaired-two tailed t test, t = 1.604, df = 10, P = 0.1398, n = 22 to 21 hemisections, N = 6 hemicords from three mice per condition; P112: nested unpaired two-tailed t test, t = 3.399, df = 47, P = 0.0014, n = 23 to 26 hemisections, N = 6 hemicords from three mice per condition) (Fig. 6, G and H), while no changes in Gad65 transcript levels (Fig. 6, I and J) were observed at any of the investigated time points (P30: nested unpaired two-tailed t test, t = 0.414, df = 10, P = 0.6879, n = 22 to 20 hemisections, N = 6 hemicords from three mice per condition; P63: nested unpaired two-tailed t test, t = 1.273, df = 41, P = 0.2101, n = 22 to 21 hemisections, N = 6 hemicords from three mice per condition; P112: nested unpaired two-tailed t test, t = 0.995, df = 10, P = 0.3431, n = 23 to 26 hemisections, N = 6 hemicords from three mice per condition) (Fig. 6, K and L).
Fig. 6. Inhibitory neurotransmitter markers show late affection in the SOD1G93A mouse.
(A, B, E, F, I, and J) Microscopy images showing detection of transcript (blue) by RNAscope HiPlexUp in situ hybridization in healthy control mice and SOD1G93A mice at P112 for GlyT2 [(A) and (B)], Gad67 [(E) and (F)], and Gad65 [(I) and (J)], with NT counterstaining (gray). (C, G, and K) Quantification of positive cells for the different neurotransmitter markers in SOD1G93A mice (red) versus healthy controls (gray). Data show significant decrease in GlyT2+ neurons at P112 (C) (nested unpaired two-tailed t tests; P30 P = 0.7469, P63 P = 0.3836, P112 P = 0.0121), as well as Gad67+ cells at P112 (G) (nested unpaired two-tailed t tests; P30 P = 0.9172, P63 P = 0.1398, P112 P = 0.0014), but no changes in Gad65+ cells (K) (nested unpaired two-tailed t tests; P30 P = 0.6879, P63 P = 0.2101, P112 P = 0.3431). (D, H, and L) Spatial distribution showing the location and count-based kernel density estimations for GlyT2+ (D), Gad67+ (H), and Gad65+ (L) cells within the spinal cord of healthy control (left/red) and SOD1G93A (right/gray) mice at P112. Scale bars, 100 μm. 2D kernel densities in (D), (H), and (L) were plotted with 10 bins using viridis scale. N = 6 hemicords from three mice (filled). Number of hemisections (empty): P30 n(wt) = 22, n(SOD1) = 20; P63 n(wt) = 22, n(SOD1) = 21; P112 n(wt) = 23, n(SOD1) = 26. Data are shown as means ± SD.
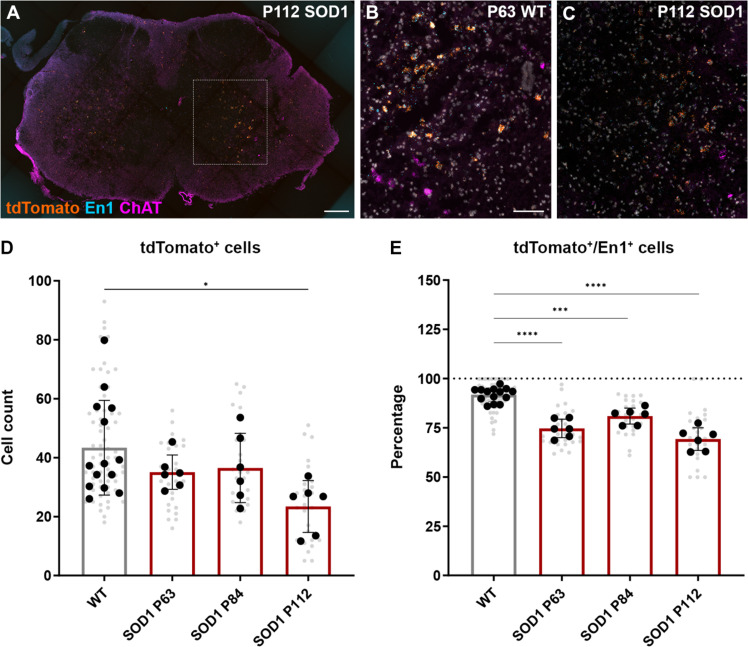
To further investigate inhibitory interneuron survival, we performed lineage tracing for the V1 subpopulation in the SOD1G93A mouse. For this, we generated a triple transgenic mouse carrying the SOD1G93A mutation and expressing tdTomato (44) under the En1 promoter (SOD1G93A;En1Cre;R26RtdTomato mouse) (Fig. 7A). TdTomato-positive neurons were quantified in wt mice at different time points and compared with P63, P84, and P112 SOD1G93A mice (Fig. 7, B and C). Quantifications of tdTomato-positive neurons (Fig. 7D) were used to evaluate V1 interneuron fate, while quantifications of double-positive tdTomato and En1 neurons (Fig. 7E) were used to evaluate transcript fate in the V1 population. These quantifications revealed a significant decrease in number of tdTomato+ cells in the symptomatic mice at P112 compared to wt mice [nested one-way analysis of variance (ANOVA) with Dunnett’s post hoc, F3,28 = 3.332; wt as control group, n = 65 hemisections, N = 14 hemicords from 7 mice; SOD1 P63: P = 0.4608, n = 26 hemisections, N = 6 hemicords from three mice; SOD1 P84: P = 0.6398, n = 25 hemisections, N = 6 hemicords from three mice; SOD1 P112: P = 0.0115, n = 27 hemisections, N = 6 hemicords from three mice] (Fig. 7D). The analysis of transcript fate in the V1 population showed a decreased expression of En1 already at P63, which was maintained at P84 and then further decreased at P112 (nested one-way ANOVA with Dunnett’s post hoc, F3,28 = 38.24; wt as control group, n = 65 hemisections, N = 14 hemicords from 7 mice; SOD1 P63: P < 0.0001, n = 26 hemisections, N = 6 hemicords from three mice; SOD1 P84: P = 0.0001, n = 25 hemisections, N = 6 hemicords from three mice; SOD1 P112; P < 0.0001, n = 27 hemisections, N = 6 hemicords from three mice) (Fig. 7E). These results show that V1 inhibitory interneurons die at a late disease stage as shown by the significant loss of tdTomato-positive neurons at P112, while the En1 transcript is lost at earlier stages of the disease.
Fig. 7. V1 interneuron fate in the SOD1G93A mouse.
(A) Microscopy image of a P112 SOD1G93A;En1Cre;R26RtdTomato mouse lumbar spinal cord after RNAscope in situ hybridization, showing tdTomato (orange), En1 (blue), and ChAT (violet) transcript. (B and C) Magnification of tdTomato, En1, and ChAT transcript, with DAPI counterstaining (gray), in the intermediate area of the spinal cord of P63 En1Cre;R26RtdTomato healthy control mice (wt) (B) and P112 SOD1G93A;En1Cre;R26RtdTomato (SOD1) (C). (D) Quantification of tdTomato+ cells in healthy controls En1Cre;R26RtdTomato (gray) and SOD1G93A;En1Cre;R26RtdTomato at different time points (red). A significant reduction of tdTomato+ cells is observed in the SOD1G93A group at P112 (nested one-way ANOVA with Dunnett’s post hoc; wt control group, SOD1 P63 P = 0.4608, SOD1 P84 P = 0.6398, SOD1 P112 P = 0.0115). (E) Percentage of tdTomato+ cells that are also positive for the En1 transcript in healthy controls versus the SOD1G93A groups. Expression of En1 transcript is reduced in neurons in the SOD1G93A mice at all time points compared to wt control (nested one-way ANOVA with Dunnett’s post hoc; wt control group, SOD1 P63 P < 0.0001, SOD1 P83 P = 0.0001, SOD1 P112 P < 0.0001). Scale bars, 200 μm (A) and 100 μm [(B) and (C)]. Biological replicates (hemicords) are shown in black, and technical replicates (hemisections) are shown in gray. Sample sizes: wt N = 14 hemicords (from 7 mice), n = 65 hemisections; SOD1 P63 N = 6 hemicords (from three mice), n = 26 hemisections; SOD1 P84 N = 6 hemicords (from three mice), n = 25 hemisections; SOD1 P112 N = 6 hemicords (from three mice), n = 27 hemisections. Data are shown as means ± SD.
Glutamatergic V2a neurons lose Chx10 transcript late in disease, while Shox2+/Chx10− and V0G interneurons remain unaffected, as does Vglut2 expression
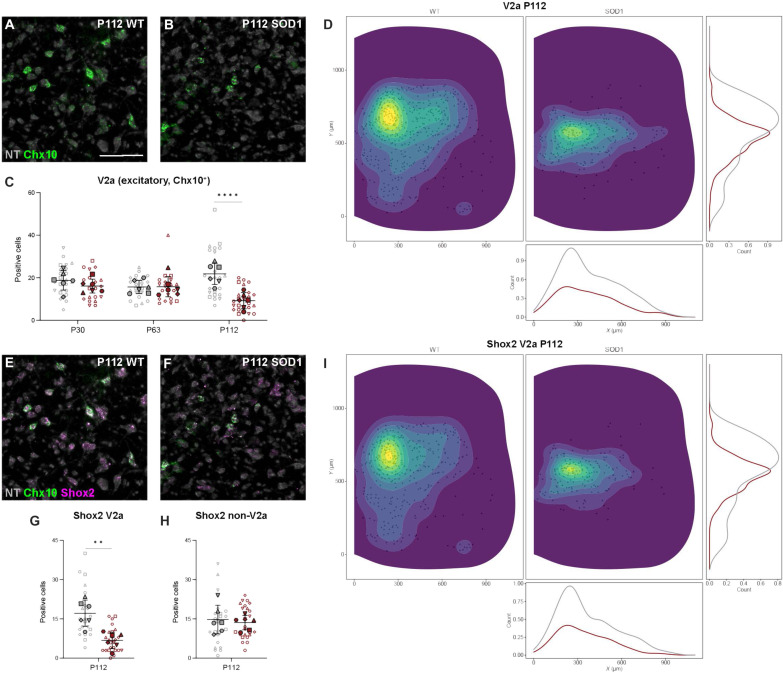
Excitatory glutamatergic (Vglut2+) interneurons were also analyzed at the three different time points. We investigated three populations: the V2a (Chx10+), the Shox2+ (further divided into Chx10+ or Chx10−), and the V0G (Pitx2+). V2a interneurons were defined as Chx10+ neurons found in the intermediate area of the spinal cord. When comparing transcript expression between wt and SOD1G93A mice (Fig. 8, A and B), a steep down-regulation (57.6%) was observed only at P112 (P30: nested unpaired two-tailed t test, t = 1.392, df = 10, P = 0.1941, n = 22 to 20 hemisections, N = 6 hemicords from three mice per condition; P63: nested unpaired two-tailed t test, t = 0.110, df = 10, P = 0.9150, n = 22 to 21 hemisections, N = 6 hemicords from three mice per condition; P112: nested unpaired two-tailed t test, t = 4.894, df = 47, P < 0.0001, n = 23 to 26 hemisections, N = 6 hemicords from three mice per condition) (Fig. 8C). Analysis of the spatial distribution at P112 revealed changes in both the ventral and dorsal areas (Fig. 8D), while no changes were observed at P30 and P63 (fig. S7, A and B).
Fig. 8. V2a and Shox2+-V2a interneurons are dysregulated at later stages of disease in the SOD1G93A mouse.
(A and B) RNAscope HiPlexUp in situ hybridization microscopy images showing detection of Chx10 transcript (green) in the lumbar spinal cord of healthy control (wt) (A) and SOD1G93A mice (B) at P112. (C) Significant reduction in the number of excitatory (Vglut2+) Chx10+ neurons in SOD1G93A mice (red) compared to healthy control mice (gray) only at P112 (nested unpaired two-tailed t tests; P30 P = 0.1941, P63 P = 0.9150, P112 P < 0.0001). (D) Spatial distribution of excitatory Chx10+ neurons with count-based kernel density estimations in SOD1G93A mice (right/gray) versus healthy control mice (left/red) from pooled P112 sections show the loss of positive cells in the intermediate region of the spinal cord. (E and F) Colocalization of Chx10 (green) and Shox2 (violet) transcript by RNAscope HiPlexUp in wt (E) and SOD1G93A spinal cord (F) at P112. (G and H) Quantification of excitatory Shox2+ neurons in combination with Chx10+ in SOD1G93A mice versus healthy control mice at P112 reveals a steep decrease in Shox2+/Chx10+ neurons (G) but no changes in Shox2+/Chx10− (H) (nested unpaired two-tailed t tests; Shox2+ P = 0.0137, Shox2+/Chx10− P = 0.8914, Shox2+/Chx10+ P = 0013). (I) Spatial distribution of Shox2+/Chx10+ neurons within the spinal cord at P112 in SOD1G93A mice versus healthy control mice shows a similar pattern to that observed for Chx10+ neurons. Scale bars, 100 μm. NT counterstaining in gray. 2D kernel densities in (D) and (I) were plotted with 10 bins using viridis scale. N = 6 hemicords from three mice (filled). Number of hemisections (empty): P30 n(wt) = 22, n(SOD1) = 20; P63 n(wt) = 22, n(SOD1) = 21; P112 n(wt) = 23, n(SOD1) = 26. Data are shown as means ± SD.
Shox2+ neurons can be divided into Shox2+/Chx10+ and Shox2+/Chx10− (17) (Fig. 8, E to H). These two subpopulations are differentially affected in the SOD1G93A mice. In the case of Shox2+/Chx10+ neurons, there was a significant 59.6% reduction at P112 (P112: nested unpaired two-tailed t test, t = 4.410, df = 10, P = 0.0013, n = 23 to 26 hemisections, N = 6 hemicords from three mice per condition) (Fig. 8G). In contrast, there was no change in the expression of Shox2+/Chx10− neurons at this same time point (P112: nested unpaired two-tailed t test, t = 0.137, df = 47, P = 0.8914, n = 23 to 26 hemisections, N = 6 hemicords from three mice per condition) (Fig. 8H). Changes in spatial distribution are shown in Fig. 8I for Shox2+/Chx10+ neurons and in fig. S7 (C and D) for overall Shox2+ and Shox2+/Chx10− neurons.
The glutamatergic Pitx2-positive V0G neurons are located close to the central canal (7). This population remained unchanged as the number of Pitx2-positive neurons was constant at all time points in the SOD1G93A mice (P30: nested unpaired two-tailed t test, t = 1.765, df = 40, P = 0.0852, n = 22 to 20 hemisections, N = 6 hemicords from three mice per condition; P63: nested unpaired two-tailed t test, t = 0.578, df = 10, P = 0.5762, n = 22 to 21 hemisections, N = 6 hemicords from three mice per condition; P112: nested unpaired two-tailed t test, t = 0.294, df = 10, P = 0.7746, n = 23 to 26 hemisections, N = 6 hemicords from three mice per condition) (figs. S8, A to D, and S9, B, D, F, and H).
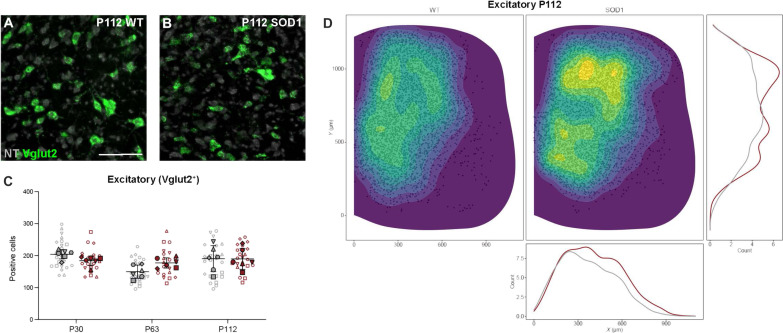
Chx10+, Shox2+, and V0G (Pitx2+) neurons are all positive for the vesicular glutamate transporter 2 (Vglut2) (7, 17). We therefore also investigated Vglut2 expression at the three time points (Fig. 9). No changes were detected in Vglut2 expression at these stages (P30: nested unpaired two-tailed t test, t = 1.557, df = 40, P = 0.1273, n = 22 to 20 hemisections, N = 6 hemicords from three mice per condition; P63: nested unpaired two-tailed t test, t = 1.832, df = 30, P = 0.0742, n = 22 to 21 hemisections, N = 6 hemicords from three mice per condition; P112: nested unpaired two-tailed t test, t = 0.099, df = 10, P = 0.9225, n = 23 to 26 hemisections, N = 6 hemicords from three mice per condition) (Fig. 9C) nor was there a difference in the spatial distribution (Fig. 9D and fig. S10).
Fig. 9. Expression of Vglut2 excitatory transcript shows no changes in the SOD1G93A mouse.
(A and B) Microscopy images after RNAscope HiPlexUp in situ hybridization showing Vglut2 transcript (green) within the lumbar spinal cord of wt (A) and SOD1G93A mice (B) at P112, NT in gray. (C) Quantification of Vglut2+ neurons in SOD1G93A mice versus healthy control mice reveals no changes at any of the tested time points (nested unpaired two-tailed t tests; P30 P = 0.1273, P63 P = 0.0742, P112 P = 0.9225). (D) Spatial distribution of Vglut2+ neurons within the spinal cord in healthy control mice (right/gray) and SOD1G93A mice (left/red) at P112, including count-based kernel density estimations. The apparent higher density observed in the SOD1G93A mouse is an effect of the higher number of sections used in this group. Scale bar, 100 μm. 2D kernel densities in (D) were plotted with 10 bins using viridis scale. N = 6 hemicords from three mice (filled). Number of hemisections (empty): P30 n(wt) = 22, n(SOD1) = 20; P63 n(wt) = 22, n(SOD1) = 21; P112 n(wt) = 23, n(SOD1) = 26. Data are shown as means ± SD.
Together, these data suggest that some excitatory populations are affected late in disease and that, also in this case, specific markers are lost before the neurotransmitter itself. Moreover, the Chx10 and Shox2 populations show differential vulnerability with regard to the transcription factor, with Chx10+ and Shox2+/Chx10+ neurons being affected at P112, while the Shox2+/Chx10− and Pitx2+ neurons appear resistant.
Cholinergic populations are affected at P112 except for V0C interneurons
The ChAT marker is known to be down-regulated upon muscle denervation (45), and somatic motor neuron loss is observed at P112 in the SOD1G93A mouse model (27, 46, 47). Here, we also found down-regulation of ChAT+ cells located in the most ventral part of the spinal cord, corresponding to putative somatic motor neurons, at P112 (P112: nested unpaired two-tailed t test, t = 2.348, df = 47, P = 0.0231, n = 23 to 26 hemisections, N = 6 hemicords from three mice per condition), with 28.4% loss (fig. S11). The V0C interneurons, positive for Pitx2 and ChAT (7), were also analyzed (figs. S8 and S9, A, C, E, and G). This population, which is known to give rise to C boutons (7, 8), was not affected at any of the investigated time points (P30: nested unpaired two-tailed t test, t = 0.257, df = 40, P = 0.7989, n = 22 to 20 hemisections, N = 6 hemicords from three mice per condition; P63: nested unpaired two-tailed t test, t = 0.578, df = 10, P = 0.6215, n = 22 to 21 hemisections, N = 6 hemicords from three mice per condition; P112: nested unpaired two-tailed t test, t = 0.123, df = 10, P = 0.6579, n = 23 to 26 hemisections, N = 6 hemicords from three mice per condition) (fig. S9, A, C, E, and G). Thus, as expected, motor neurons are affected in the SOD1G93A mice at the symptomatic stage, while at least some cholinergic interneuron populations remain unaffected in terms of transcript expression.
DISCUSSION
This study shows that spinal interneurons exhibit differential susceptibility to disease progression in a mouse model of ALS. Inhibitory interneurons exhibit early onset, starting at P63, characterized by slow loss of neuronal identity followed by cell death at later stages, while excitatory interneurons display later onset, starting at P112, but faster loss of transcript.
A condition for the success of this study was the development of methods to reliably detect the coexpression of multiple markers in spinal cell populations. We developed two analysis pipelines that enable the tracing of cell populations with high fidelity based on the detection and the quantification of multiple markers. With this approach, we obtained a quantitative dataset on the relative changes of transcript expression in subpopulations of spinal interneurons throughout disease progression in the SOD1G93A mouse line. We demonstrate that molecular markers that hitherto have been considered only expressed during development are present into adulthood in spinal cord tissue and that these markers can be used to delineate specific cell populations derived from developmental progenitors. This finding allowed us to use these transcripts as cellular markers also in the adult mouse spinal cord.
Of particular interest are the V1 interneurons, given their early dysregulation and involvement in the SOD1G93A locomotor phenotype (27). Our study reveals distinct patterns of dysregulation in transcription factor expression within different V1 interneuron populations. We find that putative RCs show the highest susceptibility to the disease, followed by putative Ia interneurons, Foxp2- and Pou6f2-positive neurons, and lastly Sp8-positive neurons. While our analysis of Renshaw cell (RC) and Ia interneuron degeneration relies on putative definitions for these populations of interest, we assert that these neuronal populations are involved, as the markers we used are widely accepted within the field for identifying RCs and Ia interneurons (12, 48, 49).
Our study reveals that putative RCs lose their markers (En1 and Calb1) early in the disease course, corroborating earlier research indicating a loss of Calb1-positive neurons in the SOD1G93A mouse model. This loss was previously documented at 10 weeks (P70) using immunohistochemistry (50), with subsequent losses of Calb1 mRNA and protein observed at P80 and P100, respectively (51). In addition, calbindin- and gephyrin-positive neurons were reported to diminish in this model (52). However, our findings contrast with those of Salamatina et al. (29), who also investigated V1 interneurons in the SOD1G93A mouse but did not observe changes in antibody labeling of putative RCs. The discrepancies between our results and those of Salamatina et al. (29) may stem from methodological and experimental design differences. Notably, Salamatina et al. (29) reported a low average number of RCs, between 0.5 and 1.5 cells per hemicord in control mice, compared to the four to six cells per hemicord found in our study. This lower detection level in their research might have affected their ability to observe changes at the protein level, contributing to the differing outcomes. The lack of observed changes in Pou6f2 in Salamatina et al. (29) might also be explained by such differences. As previously noted in studies like Wootz et al. (51), alterations at the mRNA level can precede protein level changes, further explaining the observed differences between studies. Nonetheless, our study identified similar patterns of loss in Foxp2 and Sp8 neurons. In the case of Chx10-positive neurons, our findings also showed a similar pattern of loss to that reported by Salamatina et al. (29), albeit more pronounced, beginning around P110. This onset is slightly later than the loss reported by Romer et al. (28).
Another point of divergence in our study from previous research is the spatial mapping of V1 neurons. We observed less segregated maps for Pou6f2, Sp8, and Foxp2 than previously reported in antibody staining studies (13, 14, 29). This discrepancy might arise from differences in method sensitivity or in the presentation of spatial maps. In Bikoff et al. (13), contour plots represent 30th to 90th percentile or 60th to 90th percentile densities, a more restricted range than the densities we used, where no cutoff was applied. Therefore, our contour plots, which include all positive neurons found across multiple technical replicates, show more overlap than those in (13) and (29), where a similar contour cutoff was presumably used.
In regards to cell fate, our results also show that V1 inhibitory interneurons lose their En1 transcript at early stages of the disease before they die at a later stage. While transcription factors are essential during cell differentiation, they may also play a crucial role in maintaining the neuronal fate in adult postmitotic neurons (53). Such research has demonstrated that depleting specific neuron-type genes in adulthood can lead to disturbances in neural identity and neurotransmitter levels in various neuron types, including dopaminergic, serotonergic, and cholinergic neurons (54–56). Disruption of these postmitotic maintenance mechanisms may initiate neurodegenerative conditions (53). In the context of our study, a similar mechanism may be at play. The early loss of En1 in the disease process could be a contributing factor to initiating V1 neuron degeneration, which is eventually followed by the down-regulation of neurotransmitter markers such as GlyT2 at later stages of the disease, and by cell death. Taking into account our previous findings, these events occur after inhibitory and V1 interneurons first lose connectivity with motor neurons (27).
Moreover, we also observed changes in excitatory markers which coincide with motor neuron death. These findings clearly demonstrate that inhibitory interneuron dysregulation precedes motor neuron death and thereby changes the balance of the inhibitory/excitatory synaptic input onto motor neurons. Therefore, pronounced motor neuron degeneration reflects a larger and consecutive impairment of spinal inhibitory and excitatory circuits in disease. Inhibitory neurotransmitter deficits emerge at later stages of the disease compared to transcription factors. This implies the presence of a reservoir of transmitters earlier in the disease, suggesting the maintenance of the inhibitory/excitatory phenotype of the neurons despite the ongoing degeneration and down-regulation of transcription factors, especially in the inhibitory populations. This aspect could be targeted as a potential ALS treatment aiming to maintain circuit connectivity (35) and restore the excitatory/inhibitory balance on motor neurons.
The observed degeneration of specific spinal interneuron subpopulations raises the key question of which mechanisms underlie their selective vulnerability. Although a definitive answer remains elusive, one possible framework, as suggested by Salamatina et al. (29), proposes that interneurons with direct motor neuron connectivity are more prone to degeneration. This explanation aligns with some of the available data, such as the vulnerability of RCs, Ia interneurons, and Shox2+ V2a neurons. However, the locomotor phenotype previously described in the SOD1G93A mice (27) cannot be explained by the unique loss of last-order V1 interneuron projections. The loss of speed and changes in cadence suggest changes in rhythm modulation, a process that does not occur at the motor neuron level. Consequently, it is plausible that the loss extends beyond V1 neurons projecting onto motor neurons, thus encompassing V1 interneurons and other interneurons that do not have such projections. Moreover, also V0C interneurons, which project onto motor neurons via C boutons, do not appear affected in the SOD1G93A mice at the investigated time points (7).
Thus, further gene discovery studies investigating molecular changes in interneurons and motor neurons over time are required to clarify whether interneurons are the initial targets of degeneration, subsequently spreading the disease anterogradely to motor neurons, or if they degenerate due to a retrograde signal from motor neurons or because of connectivity loss. A strong indication of the role of inhibitory interneurons in disease initiation is the loss of V1 synaptic inputs onto motor neurons, which precedes transcript down-regulation and neuronal death (27). This type of morphological evidence points toward an anterograde cascade starting from inhibitory interneurons; this idea is also supported by a recent study reporting motor deficits and motor neuron loss upon En1 down-regulation (57). However, to gain further insights into the potential role of the synaptic spread of the disease, it would be crucial to conduct a comprehensive analysis of the connectivity patterns between different interneuron populations and fast/slow motor neurons in both healthy and diseased conditions. In addition, investigating the molecular mechanisms underlying the described synaptic loss could provide valuable information. These endeavors have the potential to shed light on the intricate mechanisms driving the selective vulnerability observed in spinal interneurons.
MATERIALS AND METHODS
Experimental animals
All animal experiments were performed in accordance with the EU Directive 20110/63/EU and approved by the Danish Animal Inspectorate (Dyreforsøgstilsynet, ethical permit 2018-15-0201-01426) and the Department of Experimental Medicine at the Faculty of Health and Medical Sciences. wt C57BL6/J mice (Jackson Laboratory, strain #000664) were used for in situ sequencing experiments. Heterozygous SOD1G93A mice [Jackson Laboratory, strain #004435 - B6.Cg-Tg(SOD1*G93A)1Gur/J] were used as the ALS model. For V1 interneuron fate experiments, SOD1G93A mice were crossed with heterozygous En1Cre mice, a gift from J. Bikoff (St. Jude Children’s Hospital, St Louis, TX, USA), and homozygous R26RtdTomato [Jackson Laboratory, strain #007914 - B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J]. All strains and multiple transgenics were kept on a C57BL6/J genetic background. For experiments with SOD1G93A mice, wt littermates were used as controls. Genotyping of SOD1G93A was performed following supplier instructions. For quantification of human mutated SOD1 copy number by quantitative polymerase chain reaction, Jackson’s SOD1G93A founder breeder carrying 25 copies of the human mutated SOD1 gene was used as positive control, while a SOD1127X carrying 19 copies was used as negative control. Mice were group-housed on a 12:12-hour light:dark cycle with food and water ad libitum, 23° to 24°C temperature, and 45 to 65% humidity.
Preparation of fresh-frozen tissue
Samples were collected at P1 and P28 for in situ sequencing experiments, P30, P63, and P112 for RNAscope HiPlexUp experiments, and P63, P84, and P112 for V1 fate experiments. Both males and females were used. Mice were anesthetized with an overdose of pentobarbital solution (250 mg/kg) and euthanized by decapitation. The lumbar region of the spinal cord was then quickly dissected free, removed from the vertebrate canal, and fresh-frozen by immersion in dry ice-cold isopentane for cryoprotection. For better preservation, the samples were stored at −80°C and sectioned closely before use. Spinal cord coronal sections were embedded in Tissue-Tek OCT Compound (Sakura, ref. 4583), cut on a cryostat at 12 μm (Thermo Fisher Scientific, Cryostar NX70 or Microm HM550), collected on SuperFrost Plus glass slides (Thermo Fisher Scientific), and further stored at −80°C. To avoid RNA degradation, all equipment and tools involved in RNA work were cleaned with RNaseZap (Thermo Fisher Scientific, AM9780 or AM9786) before use.
In situ sequencing
Assay
In situ sequencing was performed using CARTANA’s Library Prep Kit (CARTANA/10x Genomics, art. no. 1010-01/02; later replaced by Xenium, 10x Genomics). Sample preparation was performed as described in the user manual (doc. no. D023). Briefly, samples were first fixated in the provided fixation solution, followed by incubation in permeabilization solution, and dehydration in ethanol series. After rehydration, samples were incubated for reverse transcription, postfixation, padlock probe ligation, rolling circle amplification, and fluorescent labeling in the provided mixes and dehydrated and mounted following the supplier’s instructions. All incubations were performed in SecureSeal Hybridization Chambers (Grace Bio-Labs, ref. 621502). The used chimeric padlock probes were custom made by CARTANA and included the 24 neuron markers in Fig. 1B: Pitx2, Evx1, ChAT, En1, Foxp2, Pou6f2, Sp8, Lmo3, Bhlhb5, Nr3b3, Nr4a2, Prdm8, Calb1, Calb2, Pvalb, Chx10, Shox2, Mnx1, Vglut2, GlyT2, Gad67, Gad65, Lhx1, and Lhx5. Quality control was performed using anchor probes labeled with Cy3 fluorophore to detect all amplification products. Quality control images were taken in-house with a 20× air objective using a Zeiss Axio Imager.Z1 or a Zeiss Axio Scan.Z1 slide scanner (EC Plan-Neofluar 20×/0.50 M27 or Plan-Apochromat 20×/0.8 M27, respectively; 0.227 μm/pixel) and approved by CARTANA/10x Genomics. Processed samples were then sent to CARTANA/10x Genomics for in situ barcode sequencing, imaging, and data processing. Briefly, adapter probes and sequencing pools (containing AF488, Cy3, Cy5, and AF750 labels) were hybridized to the ligated padlock probes to detect the first nucleotide in the gene-specific barcodes through a sequence-specific signal. This was followed by imaging and performed four times in a row to allow for decoding of full barcoded probes. Raw data was obtained using a Nikon microscope system with 40× objective (0.165 μm/pixel) or CFI Plan-Apochromat λ 20×/0.75 (0.32 μm/pixel) and images included DAPI plus the four sequencing labels, taken as Z-stack and flattened using maximum intensity projection. After sequencing, the samples were sent back for further staining with NeuroTrace 530/615 or 640/660 (Invitrogen, N21482 or N21483) for segmentation purposes, which was performed by a 2-hour incubation at room temperature [1:200 in phosphate-buffered saline (PBS)].
Image and data processing
After image sequencing and processing by CARTANA/10x Genomics, the data received included DAPI images of the sequenced spinal cord sections, and csv files with the gene identity and coordinates in pixel units (0.32 μm/pixel) of the detected transcripts. Transcript data were presented in the format of low and high threshold, of which we used the low threshold. NeuroTrace imaging was then performed using a Zeiss Axio Imager.Z1 or a Zeiss Axio Scan.Z1 slide scanner with EC Plan-Neofluar 20×/0.50 M27 or Plan-Apochromat 20×/0.8 M27, respectively (0.227 μm/pixel). DAPI counterstaining was also reimaged together with the NeuroTrace for later image registration. Cell segmentation based on NeuroTrace staining was performed using a custom-made deep learning model (Fig. 2A). The segmentation model was based on an ensemble of CNNs trained and cross-validated on different partitions of the partial, manually annotated image data. Specifically, U-Net type architecture was used as the CNN configuration for each ensemble member. Predictions from the ensemble network yielded a probabilistic segmentation map for the presence of neurons. The cell boundary delineation was then obtained using a 0.4 confidence threshold and a structural kernel of 5 on these probabilistic segmentation masks. These cell segmentation results were registered to the sequencing data based on the NeuroTrace DAPI and the DAPI obtained during the in situ sequencing assay. Keypoint-based registration using the random sample consensus (RANSAC) (58) algorithm was performed to obtain the transformation. The transformation obtained by registering the DAPIs was then applied to the NeuroTrace cell segmentation. These registered cells were then colocalized with the transcripts from CARTANA. Only transcripts that localized within segmented cells were considered. A total of N = 6 spinal cord sections (from three mice) for P1 and N = 29 sections (from 11 mice) for P28 were processed and analyzed. Further analysis was performed in RStudio (see the “Spatial profiling and quantifications” section, code at https://doi.org/10.5281/zenodo.10600983).
Quantitative multiplexed in situ hybridization (RNAscope HiPlexUp)
Assay
Quantitative multiplexed in situ hybridization was performed using the RNAscope HiPlex12 Reagents (488, 550, 647) Kit Assay (Advanced Cell Diagnostics/Bio-Techne, catalog no. 324194; since replaced by HiPlex v2 Assay) in combination with the RNAscope HiPlexUp Reagent (Advanced Cell Diagnostics/Bio-Techne, catalog no. 324190). Sample pretreatment and HiPlexUp assay were performed as described in the user manual (324100-UM). Briefly, samples were first pretreated through fixation with 4% paraformaldehyde (extended to 90 min compared to the supplier), ethanol dehydration, and incubation with the provided protease. The first HiPlex12 assay was then performed through hybridization of the first 12 probes (followed by overnight storage in 5× SSC at room temperature), then amplification and sequential probe labeling, imaging, and fluorophore cleaving (fig. S2A). Notably, the concentration of Tween in the PBST was reduced to 0.05%, and the assay was adapted to the use of three fluorescent channels (four rounds of fluorophores instead of three). Once the first 12 probes had been imaged, they were cleaved using the HiPlexUp reagent, and the second round of HiPlex12 was processed in the same manner (fig. S2A). The combination of probes and fluorophores used, including catalog numbers, is listed in Table 1. After the assay, samples were stained with NeuroTrace 530/615 (Invitrogen, N21482) by a 2-hour incubation at room temperature (1:200 in PBS) and reimaged for segmentation purposes. Counterstaining with DAPI (provided in the kit) was performed as described in the manual by a 30-s incubation before mounting for every other imaging round, and imaged in every round (including the NeuroTrace one) for image registration.
Table 1. RNAscope HiPlexUp probes. All probes were specific for mouse.
| HiPlexUp round | HiPlex12 round | Marker | Catalog no. | Fluorophore |
|---|---|---|---|---|
| 1 | 1 | Lhx5 | 885621-T1 | AF488 |
| Bhlhb5 | 467641-T2 | ATTO550 | ||
| Pitx2 | 412841-T3 | ATTO647N | ||
| 2 | Nr4a2 | 423351-T4 | AF488 | |
| Shox2 | 579051-T5 | ATTO550 | ||
| Pou6f2 | 839401-T6 | ATTO647N | ||
| 3 | Foxp2 | 428791-T7 | AF488 | |
| Nr3b3 (Errg) | 495121-T8 | ATTO550 | ||
| Slc17a6 (Vglut2) | 319171-T9 | ATTO647N | ||
| 4 | Calb1 | 428431-T10 | AF488 | |
| ChAT | 1029981-T11 | ATTO550 | ||
| Gad1 (Gad67) | 400951-T12 | ATTO647N | ||
| 2 | 5 | Lhx1 | 488581-T1 | AF488 |
| Lmo3 | 497631-T2 | ATTO550 | ||
| Evx1 | 509161-T3 | ATTO647N | ||
| 6 | En1 | 442651-T4 | AF488 | |
| Vsx2 (Chx10) | 438341-T5 | ATTO550 | ||
| Prdm8 | 467501-T6 | ATTO647N | ||
| 7 | Mnx1 (Hb9) | 1063561-T7 | AF488 | |
| Sp8 | 547521-T8 | ATTO550 | ||
| Slc6a5 (GlyT2) | 409741-T9 | ATTO647N | ||
| 8 | Pvalb | 421931-T10 | AF488 | |
| Calb2 (Calretinin) | 485811-T11 | ATTO550 | ||
| Gad2 (Gad65) | 439371-T12 | ATTO647N |
Image acquisition and processing
All imaging rounds were performed using a Zeiss Axio Scan.Z1 slide scanner. SOD1 samples and their corresponding age-matching controls were processed and imaged together. The same microscopy settings were used for all samples and imaging rounds. Full coronal spinal cord sections were scanned using a Plan-Apochromat 20×/0.8 M27 air objective (0.227 μm/pixel), with three Z-stack slices in increments of 2.5 μm and autofocused based on DAPI staining. A minimum of five sections, spaced 120 μm apart, were imaged per mouse—with three mice per condition and time point. Using the ZEN software (Zeiss, ZEN desk version 3.5), the resulting tiled images were (i) split by scene (each scene corresponding to a different section and imaging round—728 HiPlexUp images plus 91 NeuroTrace images), (ii) stitched based on DAPI staining, (iii) Z-stacks were flattened by maximum intensity projection, and (iv) single-channel images for each marker were exported to TIF format keeping the original quality (3094 images) (fig. S2A). Sections that were too broken or folded were discarded at this point (at least three sections per mouse were kept). Images from different rounds but originating from the same sections were then registered on the basis of DAPI staining using the “Register Virtual Stack Slices” and “Transform Virtual Stack Slices” plugins in Fiji (ImageJ 2.3.0/1.53f51) and merged into single images (68 images) with 26 channels each (NeuroTrace, DAPI, and the 24 interneuron markers) (fig. S2A). The merged images were converted back to CZI format for further analysis. To improve segmentation and reduce file sizes, images were cropped to include only the gray matter region of the spinal cord using the ZEN software “Extract polygons” macro (fig. S2A). These were then segmented on the basis of NeuroTrace staining using the ZEN software Intellesis tool (80% confidence, split cells by watershed, 50-μm minimum size) and analyzed to obtain the coordinates and marker intensity profiles (including intensity means and SD) for each detected cell (fig. S2B). To detect positive cells, intensity thresholds were selected for each image and interneuron marker based on signal and background. Intensity means were used for transcripts with abundant and widespread signal, while intensity SDs were used for transcripts with sparse signal. For each group, six hemicords from three mice were included, with two to five hemisections being analyzed per hemicord (table S1). Further analysis was performed in RStudio (see the “Spatial profiling and quantifications” section, code at https://doi.org/10.5281/zenodo.10600965).
Spatial profiling and quantifications
All coordinate normalizations, spatial profiling, and positive cell quantifications for in situ sequencing and quantitative RNAscope HiPlexUp in situ hybridization were performed using a custom analysis pipeline scripted in RStudio (2022.02.1 Build 461) (codes at https://doi.org/10.5281/zenodo.10600983 and https://doi.org/10.5281/zenodo.10600965). The orientation angle of the individual images, central canal coordinates, and maximum width and height of each hemisection were measured for coordinate normalization to a standardized mouse spinal cord hemisection. Spinal cord orientation was corrected using the ‘rearrr::rotate_2d’ function. Left and right hemisections were then split on the basis of the coordinates of the central canal using the ‘dplyr::mutate’ function. Last, coordinates were corrected for hemisection size and position using basic R functions. For in situ sequencing normalization, the used standardized dimensions for a full spinal cord are as follows: 2050 pixels (656 μm) mediolateral, 2950 pixels (944 μm) dorsoventral in early postnatal (P1); 3450 pixels (1104 μm) mediolateral, 4400 pixels (1408 μm) dorsoventral in young adult (P28) (Fig. 2A). For RNAscope HiPlexUp in adult, standardized dimensions used for the gray matter are: 930 μm mediolateral, 1280 μm dorsoventral (fig. S2B). The x axis was defined as parallel to the mediolateral axis with a medial origin, while the y axis was defined as parallel to the dorsoventral axis with a ventral origin. For in situ sequencing experiments, only transcripts that localized within segmented cells were visualized and considered for qualitative analysis. Colocalization of En1 transcript with other V1 markers was also visualized to represent V1 subpopulations. For RNAscope HiPlexUp experiments, positive cells for each marker were identified on the basis of the selected intensity thresholds as described above using basic R functions, and combinations of markers with cell coordinates were used to describe and quantify putative interneuron populations. Spatial distributions show estimated counts and were calculated using a two-dimensional kernel density estimation with the ‘ggplot2::geom_density_2d_filled’ function and ‘count_var = “count”’ argument, and displayed using 10 bins and the viridis scale with no density cutoffs.
In situ hybridization (RNAscope)
Assay
For assessment of V1 interneuron fate, in situ hybridization was performed using the RNAscope Multiplex Fluorescent V2 Assay (Advanced Cell Diagnostics/Bio-Techne, catalog no. 323100). Sample pretreatment and assay were performed as described for fresh frozen samples in the user manual (323100-USM) and in (27), with overnight storage of samples in 5× SSC at room temperature after probe hybridization. The following probes (Advanced Cell Diagnostics/Bio-Techne) were used: Mm-En1-C1 (catalog no. 442651), tdTomato-C3 (catalog no. 317041-C3), and Mm-Chat-C2 as counterstaining (catalog no. 408731-C2). Opal 690, 570, and 520 dyes (Akoya Biosciences, FP1497001KT, FP1488001KT, and FP147001KT) were used as fluorophores, respectively (1:1500 in trichostatin A buffer, TSA, provided in the kit). DAPI was used as counterstaining (provided in the kit).
Image acquisition, processing, and analysis
Images were acquired using a Zeiss Axio Scan.Z1 slide scanner with Plan-Apochromat 20×/0.8 M27 air objective (0.227 μm/pixel), with five Z-stack slices in increments of 1.5 μm and autofocused based on DAPI staining. A minimum of five sections (120 μm apart) were imaged per mouse—with three mice per condition and time point. Using the ZEN software (Zeiss, ZEN desk version 3.5) images were split by scene, stitched based on DAPI staining, Z-stacks were flattened by maximum intensity projection, and the ventral region of each hemisection was cropped (760 μm × 650 μm). These were then segmented on the basis of tdTomato transcript signal using the ZEN software Intellesis tool (80% confidence, split cells by watershed, 50-μm minimum size) and analyzed to obtain the cell coordinates and intensity profiles. The number of segmented cells was used to quantify tdTomato+ cells. Further tdTomato+/En1+ quantification was obtained on the basis of En1 intensity SD thresholds for the segmented cells, and normalized to tdTomato+ numbers to calculate percentages.
Statistical analysis
All statistical analyses were performed using the GraphPad Prism software [version 10.1.2(324)]. Sample sizes were not predetermined by any statistical methods and were instead similar to previous publications, such as Allodi et al. (27). For all experiments, mice were randomly allocated to different groups using a block design. Data collection was not blind to experimental group allocation but was semiautomated, and analysis of data was performed blind. All mice used were included in the datasets. All RNAscope HiPlexUp data were analyzed using a nested unpaired two-tailed t test. Effect sizes were calculated as Cohen’s ds based on statistical data following the formula (where t is t test value and n is the number of sections), and classified as trivial (<0.2), small (0.2 to 0.49), medium (0.5 to 0.79) or large (≥0.8) as suggested by Cohen (59). Reported means were calculated from the nested data. In the figures, data are presented as individual values of replicates (empty shapes), mean values for each hemicord (filled shapes), and means ± SD of hemicord values. The corresponding statistical values including n (and N), t, df, and P values are specified in the text and figure legends, while extra information including mean difference ± SEM, 95% confidence interval of differences, and effect sizes can be found in tables S1 to S3. V1 interneuron fate data were analyzed using nested one-way ANOVA with Dunnett’s post hoc, with wt as the control group. Here, data are presented as individual replicates (gray), mean values for each hemicord (black), and means ± SD of hemicords. The corresponding statistical values including n (and N), FDFn,DFd, and P values are specified in the text and figure legends. Statistical significance is reported as follows: *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Acknowledgments
We acknowledge the Department of Experimental Medicine at University of Copenhagen, especially A. S. Laugesen, and the Core Facility for Integrated Microscopy, Faculty of Health and Medical Sciences, University of Copenhagen. We thank I. Vesth-Hansen and M. Bjerre for technical assistance and N. Meola and S. Wrobel at ACD Bio (BioTechne) for technical support and discussion.
Funding: This work was supported by the Lundbeck Foundation (R346-2020-2025 and R250-2017-586, I.A.), Louis-Hansen Foundation (21-2B-9477/L102, to I.A.), the Læge Sofus Carl Emil Friis og hustru Olga Doris Friis Foundation (1218471001, to I.A.), the Novo Nordisk Laureate Program (NNF15OC0014186, to O.K.), The Lundbeck Foundation (R345-2020-1769, O.K.), the Louis-Hansen foundation (18-2B-3570, to R.M.R.), the Faculty of Health and Medical Sciences at University of Copenhagen (to O.K. and I.A.), and the School of Psychology and Neuroscience at University of St Andrews (to I.A.).
Author contributions: Conceptualization: I.A., O.K., and R.M.-R. Methodology: I.A., R.M.-R., R.S., P.H.-V., and J.M.K. Investigation: I.A., R.M.-R., R.S., D.B.A., and S.K.S. Supervision: I.A. and O.K. Writing—original draft: I.A., O.K., and R.M.-R.
Competing interests: ACD Bio and CARTANA supported this study with in-kind contributions. The authors declare that they have no other competing interests.
Data and material availability: The data that support the findings of this study are available at the University of St Andrews Research Portal using this link https://doi.org/10.17630/6985fab0-c649-4e71-9592-dbda527040ec. The code used to analyze data is available on Zenodo at https://zenodo.org/doi/10.5281/zenodo.10600982 and https://zenodo.org/doi/10.5281/zenodo.10600964. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Details about the in situ sequencing and HiplexUP probes can be provided by 10x Genomics and ACD Bio pending scientific review and a completed material transfer agreement. Requests for the probe sequences and details should be submitted to: 10x Genomics Inc., a Delaware corporation having its principal place of business at 6230 Stoneridge Mall Road, Pleasanton, CA, 94588, USA, or Advanced Cell Diagnostics Inc., located at 7707 Gateway Blvd., Newark, CA 94560, USA.
Supplementary Materials
This PDF file includes:
Figs. S1 to S11
Tables S1 to S3
REFERENCES AND NOTES
- 1.Nijssen J., Comley L. H., Hedlund E., Motor neuron vulnerability and resistance in amyotrophic lateral sclerosis. Acta Neuropathol. 133, 863–885 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kiehn O., Locomotor circuits in the mammalian spinal cord. Annu. Rev. Neurosci. 29, 279–306 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Kiehn O., Decoding the organization of spinal circuits that control locomotion. Nat. Rev. Neurosci. 17, 224–238 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Briscoe J., Pierani A., Jessell T. M., Ericson J., A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell 101, 435–445 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Grillner S., El Manira A., The intrinsic operation of the networks that make us locomote. Curr. Opin. Neurobiol. 31, 244–249 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Brownstone R. M., Bui T. V., Spinal interneurons providing input to the final common path during locomotion. Prog. Brain Res. 187, 81–95 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zagoraiou L., Akay T., Martin J. F., Brownstone R. M., Jessell T. M., Miles G. B., A cluster of cholinergic premotor interneurons modulates mouse locomotor activity. Neuron 64, 645–662 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miles G. B., Hartley R., Todd A. J., Brownstone R. M., Spinal cholinergic interneurons regulate the excitability of motoneurons during locomotion. Proc. Natl. Acad. Sci. U.S.A. 104, 2448–2453 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lanuza G. M., Gosgnach S., Pierani A., Jessell T. M., Goulding M., Genetic identification of spinal interneurons that coordinate left-right locomotor activity necessary for walking movements. Neuron 42, 375–386 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Talpalar A. E., Bouvier J., Borgius L., Fortin G., Pierani A., Kiehn O., Dual-mode operation of neuronal networks involved in left-right alternation. Nature 500, 85–88 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Bellardita C., Kiehn O., Phenotypic characterization of speed-associated gait changes in mice reveals modular organization of locomotor networks. Curr. Biol. 25, 1426–1436 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alvarez F. J., Jonas P. C., Sapir T., Hartley R., Berrocal M. C., Geiman E. J., Todd A. J., Goulding M., Postnatal phenotype and localization of spinal cord V1 derived interneurons. J. Comp. Neurol. 493, 177–192 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bikoff J. B., Gabitto M. I., Rivard A. F., Drobac E., Machado T. A., Miri A., Brenner-Morton S., Famojure E., Diaz C., Alvarez F. J., Mentis G. Z., Jessell T. M., Spinal inhibitory interneuron diversity delineates variant motor microcircuits. Cell 165, 207–219 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gabitto M. I., Pakman A., Bikoff J. B., Abbott L. F., Jessell T. M., Paninski L., Bayesian sparse regression analysis documents the diversity of spinal inhibitory interneurons. Cell 165, 220–233 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gosgnach S., Lanuza G. M., Butt S. J., Saueressig H., Zhang Y., Velasquez T., Riethmacher D., Callaway E. M., Kiehn O., Goulding M., V1 spinal neurons regulate the speed of vertebrate locomotor outputs. Nature 440, 215–219 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Crone S. A., Zhong G., Harris-Warrick R., Sharma K., In mice lacking V2a interneurons, gait depends on speed of locomotion. J. Neurosci. 29, 7098–7109 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dougherty K. J., Zagoraiou L., Satoh D., Rozani I., Doobar S., Arber S., Jessell T. M., Kiehn O., Locomotor rhythm generation linked to the output of spinal shox2 excitatory interneurons. Neuron 80, 920–933 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Lundfald L., Restrepo C. E., Butt S. J., Peng C. Y., Droho S., Endo T., Zeilhofer H. U., Sharma K., Kiehn O., Phenotype of V2-derived interneurons and their relationship to the axon guidance molecule EphA4 in the developing mouse spinal cord. Eur. J. Neurosci. 26, 2989–3002 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Zhang J., Lanuza G. M., Britz O., Wang Z., Siembab V. C., Zhang Y., Velasquez T., Alvarez F. J., Frank E., Goulding M., V1 and v2b interneurons secure the alternating flexor-extensor motor activity mice require for limbed locomotion. Neuron 82, 138–150 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Britz O., Zhang J., Grossmann K. S., Dyck J., Kim J. C., Dymecki S., Gosgnach S., Goulding M., A genetically defined asymmetry underlies the inhibitory control of flexor-extensor locomotor movements. eLife 4, e04718 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y., Narayan S., Geiman E., Lanuza G. M., Velasquez T., Shanks B., Akay T., Dyck J., Pearson K., Gosgnach S., Fan C. M., Goulding M., V3 spinal neurons establish a robust and balanced locomotor rhythm during walking. Neuron 60, 84–96 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chopek J. W., Nascimento F., Beato M., Brownstone R. M., Zhang Y., Sub-populations of spinal V3 interneurons form focal modules of layered pre-motor microcircuits. Cell Rep. 25, 146–156.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caldeira V., Dougherty K. J., Borgius L., Kiehn O., Spinal Hb9:: Cre-derived excitatory interneurons contribute to rhythm generation in the mouse. Sci. Rep. 7, 41369 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mora S., Allodi I., Neural circuit and synaptic dysfunctions in ALS-FTD pathology. Front. Neural. Circuits 17, 1208876 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herron L. R., Miles G. B., Gender-specific perturbations in modulatory inputs to motoneurons in a mouse model of amyotrophic lateral sclerosis. Neuroscience 226, 313–323 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Cavarsan C. F., Steele P. R., Genry L. T., Reedich E. J., McCane L. M., LaPre K. J., Puritz A. C., Manuel M., Katenka N., Quinlan K. A., Inhibitory interneurons show early dysfunction in a SOD1 mouse model of amyotrophic lateral sclerosis. J. Physiol. 601, 647–667 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allodi I., Montanana-Rosell R., Selvan R., Low P., Kiehn O., Locomotor deficits in a mouse model of ALS are paralleled by loss of V1-interneuron connections onto fast motor neurons. Nat. Commun. 12, 3251 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romer S. H., Seedle K., Turner S. M., Li J., Baccei M. L., Crone S. A., Accessory respiratory muscles enhance ventilation in ALS model mice and are activated by excitatory V2a neurons. Exp. Neurol. 287, 192–204 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Salamatina A., Yang J. H., Brenner-Morton S., Bikoff J. B., Fang L., Kintner C. R., Jessell T. M., Sweeney L. B., Differential loss of spinal interneurons in a mouse model of ALS. Neuroscience 450, 81–95 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Wang F., Flanagan J., Su N., Wang L. C., Bui S., Nielson A., Wu X., Vo H. T., Ma X. J., Luo Y., RNAscope: A novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 14, 22–29 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ke R., Mignardi M., Pacureanu A., Svedlund J., Botling J., Wahlby C., Nilsson M., In situ sequencing for RNA analysis in preserved tissue and cells. Nat. Methods 10, 857–860 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Qian X., Harris K. D., Hauling T., Nicoloutsopoulos D., Munoz-Manchado A. B., Skene N., Hjerling-Leffler J., Nilsson M., Probabilistic cell typing enables fine mapping of closely related cell types in situ. Nat. Methods 17, 101–106 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y., Wang W., Liu D., Hou W., Zhou T., Ji Z., GeneSegNet: A deep learning framework for cell segmentation by integrating gene expression and imaging. Genome Biol. 24, 235 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X., Andrusivova Z., Czarnewski P., Langseth C. M., Andersson A., Liu Y., Gyllborg D., Braun E., Larsson L., Hu L., Alekseenko Z., Lee H., Avenel C., Kallner H. K., Akesson E., Adameyko I., Nilsson M., Linnarsson S., Lundeberg J., Sundstrom E., Profiling spatiotemporal gene expression of the developing human spinal cord and implications for ependymoma origin. Nat. Neurosci. 26, 891–901 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mora S., von Huth Friis R., Stuckert A., Noes-Holt G., Montañana-Rosell R., Sørensen A. T., Selvan R., Allodi I., Stabilization of V1 interneuron-motor neuron connectivity ameliorates motor phenotype in a mouse model of ALS. bioRxiv 2022.12.15.520568 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bankhead P., Loughrey M. B., Fernandez J. A., Dombrowski Y., McArt D. G., Dunne P. D., McQuaid S., Gray R. T., Murray L. J., Coleman H. G., James J. A., Salto-Tellez M., Hamilton P. W., QuPath: Open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.J. M. Kaminski, I. Allodi, R. Montañana-Rosell, R. Selvan, O. Kiehn, Deep ensemble model for segmenting microscopy images in the presence of limited labeled data. Paper presented at the MIDL 2021 Conference, Lübeck, Germany, 7 to 9 July 2021. [Google Scholar]
- 38.Goulding M., Circuits controlling vertebrate locomotion: Moving in a new direction. Nat. Rev. Neurosci. 10, 507–518 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kiehn O., Development and functional organization of spinal locomotor circuits. Curr. Opin. Neurobiol. 21, 100–109 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Lubben N., Ensink E., Coetzee G. A., Labrie V., The enigma and implications of brain hemispheric asymmetry in neurodegenerative diseases. Brain Commun. 3, fcab211 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ravits J., Paul P., Jorg C., Focality of upper and lower motor neuron degeneration at the clinical onset of ALS. Neurology 68, 1571–1575 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Mochizuki Y., Mizutani T., Takasu T., Amyotrophic lateral sclerosis with marked neurological asymmetry: Clinicopathological study. Acta Neuropathol. 90, 44–50 (1995). [DOI] [PubMed] [Google Scholar]
- 43.Acevedo-Arozena A., Kalmar B., Essa S., Ricketts T., Joyce P., Kent R., Rowe C., Parker A., Gray A., Hafezparast M., Thorpe J. R., Greensmith L., Fisher E. M., A comprehensive assessment of the SOD1G93A low-copy transgenic mouse, which models human amyotrophic lateral sclerosis. Dis. Model. Mech. 4, 686–700 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Madisen L., Zwingman T. A., Sunkin S. M., Oh S. W., Zariwala H. A., Gu H., Ng L. L., Palmiter R. D., Hawrylycz M. J., Jones A. R., Lein E. S., Zeng H., A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fischer L. R., Culver D. G., Tennant P., Davis A. A., Wang M., Castellano-Sanchez A., Khan J., Polak M. A., Glass J. D., Amyotrophic lateral sclerosis is a distal axonopathy: Evidence in mice and man. Exp. Neurol. 185, 232–240 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Schaefer A. M., Sanes J. R., Lichtman J. W., A compensatory subpopulation of motor neurons in a mouse model of amyotrophic lateral sclerosis. J. Comp. Neurol. 490, 209–219 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Allodi I., Comley L., Nichterwitz S., Nizzardo M., Simone C., Benitez J. A., Cao M., Corti S., Hedlund E., Differential neuronal vulnerability identifies IGF-2 as a protective factor in ALS. Sci. Rep. 6, 25960 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alvarez F. J., Dewey D. E., McMillin P., Fyffe R. E., Distribution of cholinergic contacts on Renshaw cells in the rat spinal cord: A light microscopic study. J. Physiol. 515, 787–797 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Geiman E. J., Knox M. C., Alvarez F. J., Postnatal maturation of gephyrin/glycine receptor clusters on developing Renshaw cells. J. Comp. Neurol. 426, 130–142 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Chang Q., Martin L. J., Glycinergic innervation of motoneurons is deficient in amyotrophic lateral sclerosis mice: A quantitative confocal analysis. Am. J. Pathol. 174, 574–585 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wootz H., Fitzsimons-Kantamneni E., Larhammar M., Rotterman T. M., Enjin A., Patra K., Andre E., Van Zundert B., Kullander K., Alvarez F. J., Alterations in the motor neuron-renshaw cell circuit in the Sod1(G93A) mouse model. J. Comp. Neurol. 521, 1449–1469 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fornai F., Longone P., Cafaro L., Kastsiuchenka O., Ferrucci M., Manca M. L., Lazzeri G., Spalloni A., Bellio N., Lenzi P., Modugno N., Siciliano G., Isidoro C., Murri L., Ruggieri S., Paparelli A., Lithium delays progression of amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. U.S.A. 105, 2052–2057 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deneris E. S., Hobert O., Maintenance of postmitotic neuronal cell identity. Nat. Neurosci. 17, 899–907 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kadkhodaei B., Alvarsson A., Schintu N., Ramskold D., Volakakis N., Joodmardi E., Yoshitake T., Kehr J., Decressac M., Bjorklund A., Sandberg R., Svenningsson P., Perlmann T., Transcription factor Nurr1 maintains fiber integrity and nuclear-encoded mitochondrial gene expression in dopamine neurons. Proc. Natl. Acad. Sci. U.S.A. 110, 2360–2365 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song N. N., Xiu J. B., Huang Y., Chen J. Y., Zhang L., Gutknecht L., Lesch K. P., Li H., Ding Y. Q., Adult raphe-specific deletion of Lmx1b leads to central serotonin deficiency. PLOS ONE 6, e15998 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu C., Maejima T., Wyler S. C., Casadesus G., Herlitze S., Deneris E. S., Pet-1 is required across different stages of life to regulate serotonergic function. Nat. Neurosci. 13, 1190–1198 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leboeuf M., Vargas-Abonce S. E., Peze-Hedsieck E., Dupont E., Jimenez-Alonso L., Moya K. L., Prochiantz A., ENGRAILED-1 transcription factor has a paracrine neurotrophic activity on adult spinal α-motoneurons. EMBO Rep. 24, e56525 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fischler M. A., R. C. Bolles, Random sample consensus. Commun. ACM 24, 381–395 (1981). [Google Scholar]
- 59.J. Cohen, Statistical Power Analysis for the Behavioral Sciences (Lawrence Erlbaum Associates, ed. 2, 1988). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S11
Tables S1 to S3