Abstract
STAT5B has been reported as a recurrent mutation in myeloid neoplasms with eosinophilia, but its overall frequency and importance across a spectrum of myeloid neoplasms are largely unknown. We conducted a multicenter study on a series of 82 myeloid neoplasms with STAT5B mutations detected by next-generation sequencing. The estimated frequency of STAT5B mutations in myeloid neoplasms was low, <0.5%, but mutations were detected in all categories of such neoplasms, including myelodysplastic syndrome (MDS, 28%), acute myeloid leukemia (AML, 26%), myelodysplastic/myeloproliferative neoplasm (MDS/MPN, 18%), Philadelphia chromosome-negative classic MPN (12%), systemic mastocytosis (1%), and, with a notably high frequency, chronic eosinophilic leukemia, not otherwise specified (CEL-NOS, 15%). STAT5B mutations occurred preferentially in the SH2 domain (95%), involved 12 different codons, with the N642H hotspot being the most common (78%). Co-mutations were present in all cases and clonal hierarchy analysis showed that STAT5B mutations tended to be subclonal in AML, MPN, and MDS, but frequently dominant/co-dominant in CEL-NOS (83%), followed by MDS/MPN (40%). Across the group, eosinophilia and/or basophilia were common (41%), frequently observed in cases in which STAT5B mutations were detected at initial diagnosis (P<0.0001), with a high variant allele frequency (median 42.5%, P=0.0001), as a dominant/co-dominant clone (P<0.0001), involving the canonical N642H (P=0.0607), and associated with fewer co-mutations (P=0.0009). Our data show that the characteristics and importance of a STAT5B mutation differ among myeloid neoplasms, but if present as a dominant mutation and detected at initial diagnosis, it appears to be a driver mutation in a subgroup of chronic myeloid neoplasms, preferentially promoting a proliferation of eosinophils and basophils.
Introduction
STAT5 is a key component of cytokine-induced signal transduction cascades, and a critical downstream mediator of transformation by oncogenic tyrosine kinases. STAT5, encoded by STAT5A and STAT5B located at chromosome 17q21.2, is fundamental for myelopoiesis, lymphoid development, macrophage functions and megakaryopoiesis, as well as basophil, eosinophil, and mast cell functions.1 STAT5A/B activation, in most cases, is induced by upregulated function of upstream tyrosine kinases, e.g. JAK2 V617F, BCR::ABL1, FLT3-ITD, or KIT D816V.2 In humans, while STAT5A mutations have rarely been implicated in causing disease, STAT5B mutations have been linked to deregulated protein signaling and function, and hyperactivation of STAT5B3 and gain-of-function STAT5B mutations4 are associated with the development of various hematolymphoid malignancies.
STAT5B mutations are mostly reported in T/NK-cell neoplasms, including large granular lymphocytic leukemia (CD4+ type), T-prolymphocytic leukemia, hepatosplenic T-cell lymphoma and T-lymphoblastic leukemia/lymphoma.5-9 STAT5B mutations occur mainly in the SH2 domain, with N642H being the most common. N642H is close to the phosphotyrosine-binding loop of STAT5B, and this mutation stabilizes STAT5 dimers, leading to prolonged pY-STAT5 levels and increased phosphotyrosine levels upon cytokine stimulation.10 STAT5B mutations are uncommon in myeloid neoplasms. Recently, Cross et al.11 reported STAT5B N642H in 27 of 1715 (1.6%) cases of myeloid neoplasms with eosinophilia, including seven cases with a presumed diagnosis of hypereosinophilic syndrome. It has been wondered whether STAT5B N642H is a recurrent mutation in myeloid neoplasms which may represent a specific subset of chronic myeloid neoplasms with eosinophilia.
We conducted this multicenter study with three aims. First, we sought to understand the spectrum of myeloid neoplasms with STAT5B mutations. Second, we assessed the characteristics of STAT5B mutations looking for possible correlations with disease phenotype. Lastly, we examined whether STAT5B-mutated myeloid neoplasms represent a unique entity with distinct clinicopathological features.
Methods
Study group
We searched the database of eight institutions for myeloid neoplasms with STAT5B mutations tested by next-generation sequencing. Clinical and laboratory data were retrieved from the medical records. A potential concomitant T/NK-cell neoplasm carrying STAT5B mutation, including lymphocyte variant hypereosinophilic syndrome, was excluded by morphological examination, flow cytometry immunophenotyping and/or TCR gene rearrangement and, most importantly, clinical follow-up. The study was conducted according to Institutional Review Board-approved protocols of all participating institutions and in accordaancewith the Declaration of Helsinki.
Morphological evaluation
Wright-Giemsa-stained peripheral blood (PB) and bone marrow (BM) aspirate smears/touch imprints, as well as hematoxylin and eosin-stained BM clot and core biopsy specimens were reviewed and assessed for percentage of blasts, eosinophils and basophils, as well as morphological dysplasia and fibrosis. Cytochemical stains for myeloperoxidase, iron staining and histochemical stains for reticulin and collagen were performed using standard methods. The grade of myelofibrosis (MF) was assessed based on the criteria of the European Consensus on the grading of BM fibrosis.12
Immunophenotypic studies
Flow cytometry immunophenotyping was performed. The panels varied at different institutions, with the basic markers including CD2, CD3 (surface and cytoplasmic), CD4, CD5, CD7, CD13, CD14, CD15, CD19, CD25, CD33, CD34, CD36, CD38, CD45, CD56, CD64, CD117, CD123, CD133, HLA-DR, myeloperoxidase and TdT.
Cytogenetic analysis
Conventional chromosomal analysis was performed on G-banded metaphases prepared from unstimulated 24-hour and 48-hour BM cultures. Twenty metaphases were analyzed, and the results were reported using the International System for Human Cytogenetics Nomenclature, 2020.
Next-generation sequencing
Genomic DNA was amplified by polymerase chain reaction and subjected to mutation analysis by next-generation sequencing. The gene panels varied among different institutions, but all panels assessed for common mutations associated with myeloid neoplasms. STAT5B was assessed in all cases and the entire coding region was covered, with a limit of detection at a variant allele frequency (VAF) of 1%. Most cases were tested using an 81-gene panel that included ANKRD26, ASXL1, ASXL2, BCOR, BCORL1, BRAF, BRINP3, CALR, CBL, CBLB, CBLC, CEBPA, CREBBP, CRLF2, CSF3R, CUX1, DDX41, DNMT3A, EED, ELANE, ETNK1, ETV6, EZH2, FBXW7, FLT3, GATA1, GATA2, GFI1, GNAS, HNRNPK, HRAS, IDH1, IDH2, IKZF1, IL2RG, IL7R, JAK1, JAK2, JAK3, KDM6A, KIT, KMT2A, KRAS, MAP2K1, MPL, NF1, NOTCH1, NPM1, NRAS, PAX5, PHF6, PIGA, PML, PRPF40B, PTEN, PTPN11, RAD21, RARA, RUNX1, SETBP1, SF1, SF3A1, SF3B1, SH2B3, SMC1A, SMC3, SRSF2, STAG1, STAG2, STAT3, STAT5A, STAT5B, SUZ12, TERC, TERT, TET2, TP53, U2AF1, U2AF2, WT1, and ZRSR2. When a mutation had a VAF of ≥5%, and this VAF was within a 10% difference from the mutation with the highest VAF, we refer to the mutation as a dominant clone.13
Statistical analysis
Statistical analysis was performed using GraphPad Prism 9. The association between categorical variables was examined using Fisher exact and Pearson χ2 square tests. The association between continuous variables was examined using the Student t test. Overall survival was calculated from the date of initial diagnosis to the date of death or last follow-up. Survival was analyzed using the Kaplan-Meier method and compared using the log-rank test. Differences between groups were considered statistically significant if P values were <0.05 in a two-tailed test.
Results
STAT5B mutations occur in a wide spectrum of myeloid neoplasms
We identified a total of 82 cases of myeloid neoplasms with STAT5B mutations with a VAF of ≥1%. Based on the total number of myeloid neoplasms tested by the same next-generation sequencing panel at one institution (MD Anderson Cancer Center) during the study period, we estimate that the frequency of STAT5B mutations across all myeloid neoplasms is below 0.5%. STAT5B mutations were detected at the time of initial diagnosis in 45 patients and acquired later in the course of disease in 20 patients. Initial diagnostic material was not available for assessment of STAT5B mutation in 17 patients. Of the 20 patients who acquired STAT5B mutations later in the disease course, the emergence of STAT5B was often accompanied by disease progression with an increased blast count (2 myelodysplastic syndrome [MDS] and 1 chronic myelomonocytic leukemia [CMML]) or leukemic transformation (6 acute myeloid leukemia [AML] from MDS), or relapse (2 AML).
The study group included 56 men and 26 women, with a median age of 72.7 years (range, 26.2-88.0). The cases occurred across a broad spectrum of myeloid neoplasms including MDS (n=23), AML (n=21), myelodysplastic/myeloproliferative neoplasm (MDS/MPN) (n=15), chronic eosinophilic leukemia, not otherwise specified (CEL-NOS, n=12), Philadelphia chromosome (Ph)-negative classic myeloproliferative neoplasms (MPN) (n=10), and aggressive systemic mastocytosis (SM) with eosinophilia (n=1) (Table 1). The AML group was heterogeneous but remarkably enriched by AML with myelodysplasia-related gene mutations (15/21, 71%). Genes mutated in this latter category included RUNX1 (n=8), ASXL1 (n=6), SRSF2 (n=5), U2AF1 (n=5), STAG 2 (n=2) and ZRSR2 (n=1); eight cases had more than one of these mutations. Among patients with MDS/MPN, CMML was predominant (n=12, 80%). One of the two cases of MDS/MPN, NOS showed persistent eosinophilia (>10%) but slightly under 1.5x109/L (criteria for CEL-NOS). Among Ph-negative classic MPN, a case of triple-negative primary myelofibrosis (PMF) showed marked eosinophilia (>2.5x109/L) and basophilia (>1.0x109/L). The diagnoses and classifications of the patients are summarized in Table 1, and clinicopathological features are summarized in Table 2.
Table 1.
Disease categories of 82 cases of myeloid neoplasms with STAT5B mutation.
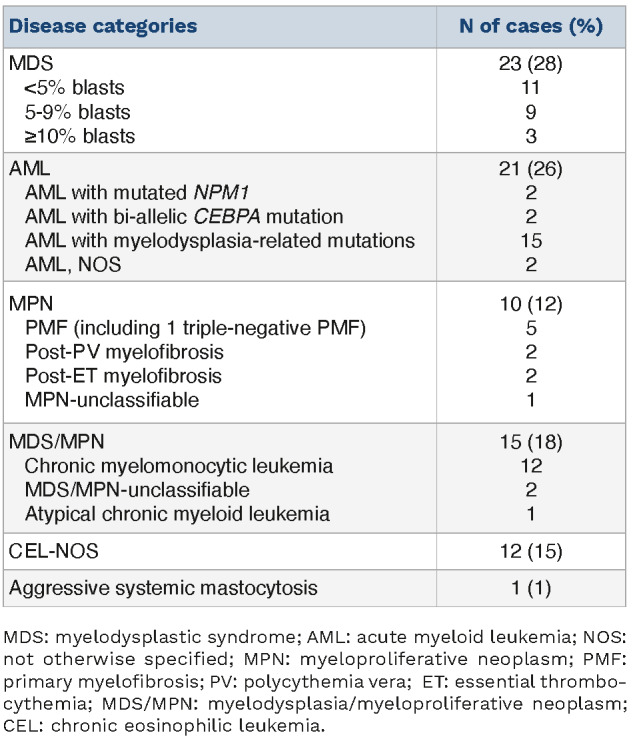
Table 2.
Clinicopathological features of cases of myeloid neoplasms with a STAT5B mutation.
Morphological findings in STAT5B-mutated myeloid neoplasms
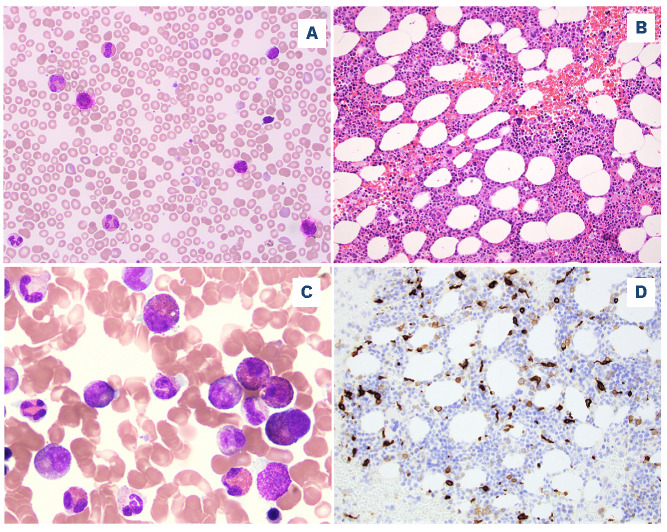
The morphological findings reported here were from the first available BM samples in which a STAT5B mutation was detected. The BM biopsy specimens were overall hypercellular (median, 80%; range, 10-100%). Dysplasia was common in all disease categories, with multilineage dysplasia (involving ≥2 lineages) observed in 12/15 (80%) patients with MDS/MPN, 18/23 (78%) with MDS, 8/13 (62%) with AML and 6/12 (50%) of those with CEL-NOS. Increased ring sideroblasts (>5%) were uncommon, being found in three cases of MDS, two cases each of AML and CEL-NOS, and one case of polycythemia vera. In addition to AML, increased blasts (≥5%) were present in 9/15 (60%) MDS/MPN, 12/23 (52%) MDS, 4/10 (40%) MPN, and 4/12 (33%) CEL-NOS patients. Significant myelofibrosis (≥ MF-2) was observed in 16/71 (23%) cases, most frequently in MPN (6/9, 67%), and less often in CEL-NOS (2/10, 20%), MDS (4/22, 18%), MDS/MPN (2/13, 15%), and AML (2/17, 12%). The morphological findings from representative CEL-NOS cases are illustrated in Figure 1.
Figure 1.
A representative case of chronic eosinophilic leukemia, not otherwise specified. (A) A peripheral blood smear shows eosinophilia and basophilia (Wright-Giemsa, 500x). (B) The bone marrow biopsy is hypercellular with small hypolobated dysplastic megakaryocytes (hematoxylin and eosin, 200x). (C) A bone marrow aspirate smear shows increased eosinophils and precursors, some intermediate-stage eosinophils with eo-basophilic granules (Wright-Giemsa, 500x). (D) CD117 highlights increased scattered spindle mast cells (200x). The mast cells were positive for CD25 by flow cytometry (not shown).
Eosinophilia, defined as an absolute eosinophil count of >0.5x109/L and ≥6% eosinophils in PB, was present in 27 (33%) patients (12 CEL-NOS, 6 MDS/MPN, 4 MDS, 3 MPN, 1 AML and 1 aggressive SM). Basophilia, defined as having an absolute basophil count of ≥0.2 x109/L and ≥2% basophils in PB,14 was identified in 20 (24%) patients (7 CEL-NOS, 5 MPN, 5 MDS/MPN and 3 MDS). Thirteen (16%) patients had both eosinophilia and basophilia. Increased BM eosinophils (≥6%) were observed in 17 (21%) patients, and increased BM basophils (≥2%) in 10 (12%) patients.
Other than cases classified as CMML, absolute and relative monocytosis were only observed in two cases of AML with monocytic differentiation. Relative monocytosis (≥10%) but not absolute monocytosis (≥1.0 x109/L) was present in four MDS, one PMF and none of the CEL-NOS.
Mast cells were not systemically evaluated in this study. The case of aggressive SM had large aggregates of mast cells meeting the major criteria for SM, with aberrant CD25 expression, and associated with eosinophilia. One case of CEL-NOS had increased scattered spindle mast cells with aberrant CD25 co-expression (Figure 1). Mast cells in two other cases of CEL-NOS were evaluated by flow cytometry and were negative for CD2, CD25 and CD30.
Cytogenetic findings of STAT5B-mutated myeloid neoplasms
Forty-six of 80 (58%) patients had a normal karyotype, 27 (34%) had a simple abnormal karyotype defined as one or two chromosomal aberrations, and seven (9%) had a complex karyotype with three or more chromosomal aberrations (Table 3). Karyotype was not available for two patients (1 AML and 1 MPN). The most common karyotypic aberrations included trisomy 8 (n=9, 11%), del7/del(7q) (n=7, 9%), del(20q) (n=6, 8%), del(11q) (n=4, 5%), del5/del(5q) (n=2, 5%), and del17/ del(17p) (n=2, 3%). There was no rearrangement of PDGFRA, PDGFRB, FGFR1, or JAK2 in cases which were assessed for these rearrangements.
Table 3.
Cytogenetic and molecular features of cases of myeloid neoplasms with STAT5B mutation.
Molecular characterization
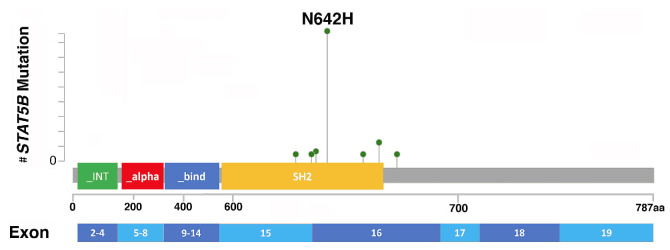
STAT5B mutations were clustered in the SH2 domain in 78 (95%) cases and were missense mutations in all cases. In four (5%) remaining patients, mutations occurred in the transactivation domain (n=3) and the coiled coil domain (n=1). A total of 12 different mutations were detected, with N642H being the most common hotspot, in 64 (78%) cases, followed by Y665F (n=6, 7%), T628S (n=3, 4%) and E637K (n=2, 2%). Other mutation codons (n=8) were observed in single cases (Online Supplementary Table S1). One patient with CEL-NOS had two STAT5B mutations, one in the SH2 domain, Y665F, and the other in the DNA binding domain, D428N. The location of STAT5B mutations is shown in Figure 2. Interpretation of non-canonical (non-N642H) STAT5B mutations, including their distribution in different disease entities, if they have been reported in the Catalogue of Somatic Mutations in Cancer (COSMIC) database, confirmed as somatic or activating mutations, and disease types in which they were reported previously, are provided in Online Supplementary Table S2. The VAF of STAT5B mutations ranged from 1.1-81.2% (median, 22.5%), with 62 (76%) cases having a VAF ≥5%.
Figure 2.
The location and number of STAT5B mutations. In total, 83 mutations were identified in 82 patients. One patient with chronic eosinophilic leukemia, not otherwise specified had two concurrent mutations, Y665F and D428N. STAT5B mutations clustered in the SH2 domain in 78 (95%) cases and were missense mutations in all cases, with N642H being the most common, seen in 64 (78%) cases. aa: amino acids. (Note: the number of amino acids on the X-axis is not proportionally scaled).
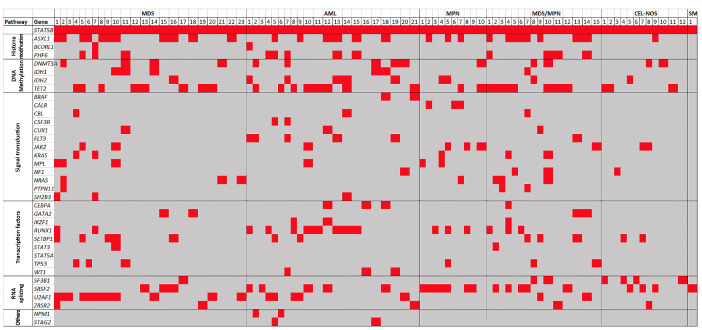
We also evaluated the mutational profile of other genes included in the next-generation sequencing panels (Table 3). The presence of a STAT5B mutation was accompanied by other gene mutations (referred to as “co-mutations”) in all cases, with a median of four co-existing mutations per case (range, 2-10). A heatmap illustrating the co-existing mutations is shown in Figure 3. Overall, the most frequent concurrent mutations were ASXL1 and TET2, each in 29 (35%) cases, followed by SRSF2 (n=23, 28%), U2AF1 (n=19, 23%), RUNX1 (n=16, 20%) and DNMT3A (n=14, 17%). In MDS, the most frequent co-existing mutation, present in 12/23 (52%) cases, was U2AF1, followed by ASXL1 (n=11) and TET2 (n=8). In AML, RUNX1 and TET2 were each mutated in 8/21 (38%) cases, followed by ASXL1 (n=6). In MPN, SRSF2 was mutated in 7/10 (70%) cases, followed by ASXL1 (n=3), CALR (n=3), JAK2 (n=3) and RUNX1 (n=3). In MDS/MPN, ASXL1 and TET2 mutations each occurred in 8/15 (53%) cases, followed by SRSF2 (n=5) and PHF6 (n=4) mutations. Cases of CELNOS had the lowest number of concurrent mutated genes (median 2, range 2-4), with SF3B1 mutation being most common in 4/12 (33%) cases, followed by SRSF2 (n=3) and TET2 (n=3). STAT5B mutations represented the dominant or co-dominant clone in 30 (37%) cases and a subclone in the remaining 52 (63%) cases (Table 3). Of note, in 18/20 (90%) cases in which STAT5B mutations were acquired during the course of disease, the STAT5B mutations were subclonal. Among 18 cases with mutations in codons other than N642H, 15 (83%) were non-dominant and three were co-dominant; eight (44%) were detected at initial diagnosis, four were acquired and six were unknown.
Figure 3.
The STAT5B mutation profile heatmap. The cases were divided into six categories: myelodysplastic syndrome, acute myeloid Leukemia, myeloproliferative neoplasm, myelodysplasia/myeloproliferative neoplasm, chronic eosinophilic leukemia, not otherwise specified, and systemic mastocytosis. Each column represents a single case. Only mutations seen in two or more cases are listed. Overall, the most frequent concurrent mutations were ASXL1 and TET2, followed by SRSF2, U2AF1, RUNX1, and DNMT3A. MDS: myelodysplastic syndrome; AML: acute myeloid leukemia; MPN: myeloproliferative neoplasm; MDS/MPN: myelodysplasia/myeloproliferative neoplasm; CEL-NOS: chronic eosinophilic leukemia, not otherwise specified; SM: systemic mastocytosis.
Comparison of STAT5B-mutated myeloid neoplasms with and without eosinophilia and/or basophilia
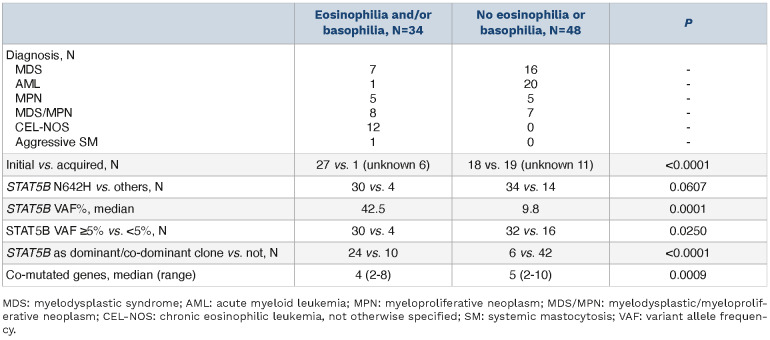
Twenty-seven (33%) patients had PB eosinophilia, 20 (24%) PB basophilia and 13 (16%) had both. Cases with eosinophilia and/or basophilia (n=34) included 12 CEL-NOS, eight MDS/MPN, seven MDS, five MPN, one AML and the case of SM. Compared to the remaining cases, cases with eosinophilia and/or basophilia tended to have STAT5B mutations detected at initial diagnosis (27/28 vs. 18/37; P<0.0001), more commonly had the canonical N642H mutation (30/34 vs. 34/48; P=0.0607), and had a higher VAF (42.5% vs. 9.8%; P=0.0001). In addition, cases with eosinophilia and/ or basophilia more often had a VAF ≥5% (30/34 vs. 32/48; P=0.0250), the mutation was a dominant or co-dominant clone (24/34 vs. 6/48; P<0.0001), and were associated with fewer concurrently mutated genes (4 vs. 5, P=0.0009) (Table 4).
Table 4.
Comparison between STAT5B-mutated myeloid neoplasms with and without eosinophilia/basophilia.
By definition, all 12 cases of CEL-NOS had hypereosinophilia, both relative (≥10%) and absolute (≥1.5x109/L).15,16 Of note, 7/12 (58%) CEL-NOS cases also had PB basophilia. The STAT5B mutations were detected at initial diagnosis in all CEL-NOS patients, with a median VAF of 44.2% (range, 7-78.5%), being a dominant clone in 10 (83%) cases. The mutation involved the N642H hotspot in 11 (92%) CELNOS cases, while one case had two mutations, Y665F and D428N.
Clinical outcome of patients with STAT5B-mutated myeloid neoplasms
Among 76 patients with treatment information available, all received disease- and risk-adapted therapies with the exception of three patients with MDS, one with CMML and one with CEL-NOS who were observed. Thirteen patients underwent allogeneic stem cell transplantation, of whom four had MDS, three had AML, three had CMML, and one each had PMF, CEL-NOS, and MDS/MPN-unclassified. With a median follow-up of 18 months (range, 1-101.5 months), AML transformation occurred in 4/12 (33%) CEL-NOS, 3/15 (20%) MDS/MPN, and 4/23 (17%) MDS patients. At the end of follow-up, 41 patients had died of disease, 17 had persistent disease, and eight had achieved complete remission, including four with MDS, two with AML, and two CMML. Fourteen patients were alive with unknown disease status. Two patients were lost to follow-up (Figure 4, Table 2).
Figure 4.
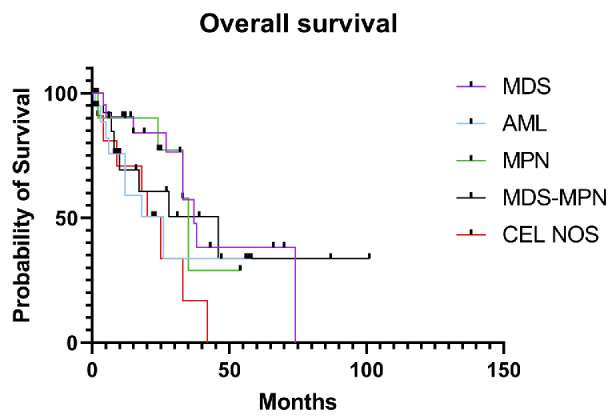
Overall survival of cases of STAT5B-mutated myeloid neoplasms based on disease categories. MDS: myelodysplastic syndrome; AML: acute myeloid leukemia; MPN: myeloproliferative neoplasm; MDS-MPN: myelodysplastic/myeloproliferative neoplasm; CEL-NOS: chronic eosinophilic leukemia, not otherwise specified.
Discussion
In this study, we show that the overall frequency of STAT5B mutations in myeloid neoplasms, estimated from one of the participating institutions, is low, being below 0.5%. This low frequency was also reported in a study by Umrau and colleagues who detected STAT5B mutations in five of 2,266 (0.22%) myeloid neoplasms including three AML, one MDS and one CMML.17 With this large cohort of cases, we show that STAT5B mutations are not confined to a specific myeloid disease category, occur most frequently at codon N642H, and are frequently associated with eosinophilia and/ or basophilia.
Unlike the study by Cross and colleagues,11 which was focused on the STAT5B N642H hotspot mutation in eosinophilic disorders, we included all myeloid neoplasms tested by next-generation sequencing panels that covered the entire coding region of STAT5B. We showed that all STAT5B mutations were missense, predominantly occurring in the SH2 domain (95%), with occasional cases involving the transactivation domain and the coiled coil domain. A total of 12 different mutations were detected, with N642H being the most common, occurring in nearly 80% of cases, followed by Y665F (7%), T628S (4%), and one each of the others. STAT5B Y665F, the second most frequent mutation, was not detected in MDS or AML, but only seen in chronic myeloid neoplasms with proliferative features including Ph-negative classic MPN, MDS/MPN and CEL-NOS. On the other hand, except for D428N as a second STAT5B mutation in a case of CEL-NOS, other rare mutations, including T628S, were only found in MDS and AML cases. The Y665F, a somatic and activating mutation, has been reported in an aggressive variant of CD8+ T-cell large granular lymphocytic leukemia,6 and T628S, a somatic mutation but of unknown status for activating, was enriched in CD4+ large granular lymphocytic leukemia5 and T-prolymphocytic leukemia.18 T628S and V712E were previously reported in one case of MDS with ring-sideroblasts.17 Notably, these non-N642H STAT5B mutations mostly occurred as a non-dominant clone (83%) and, except for Y665F and I704L, the activating status is largely unknown (Online Supplementary Table S2); their pathogenic significance needs further studies. These findings indicate likely genotype-phenotype correlations between specific STAT5B mutation sites and affected codons and disease subtypes, but the phenomenon is probably confounded by dominant versus subclonal mutations of STAT5B.
STAT5B mutations can present at the time of diagnosis or be acquired during the course of disease. The proportion of STAT5B mutations detected at initial diagnosis was highest in CEL-NOS (100%), followed by MDS/MPN in about 80% and less than 50% in AML. The median VAF of STAT5B mutations varied among different disease subtypes, being highest in CEL-NOS, followed by MDS and MDS/MPN, and lowest in Ph-negative classic MPN and AML. Concurrent gene mutations were found in all cases, but the genes mutated differed by disease categories, as expected. A similar high frequency of co-mutations with STAT5B was described in cases of myeloid neoplasms with eosinophilia,11,19 and five cases of myeloid neoplasms reported by Umrau and colleagues.17 STAT5B was a dominant or co-dominant clone in slightly over one third of the cases and was very common in CEL-NOS (>80%), followed by MDS/MPN (~40%) and lowest in AML and Ph-negative classic MPN (≤20%). Furthermore, CEL-NOS had the lowest number of concurrent gene mutations. These findings suggest that STAT5B is likely a driver gene in CEL-NOS.
One of the prominent clinical features of myeloid neoplasms with STAT5B mutations was associated eosinophilia and/or basophilia, observed in nearly half of the cases, resulting in a diagnosis of CEL-NOS in 15% cases. Of note, CEL-NOS is extremely rare. According to the Surveillance, Epidemiology, and End Results (SEER) data between 2004 and 2015, CEL-NOS together with myeloid/lymphoid neoplasms with eosinophilia and tyrosine kinase gene fusion (M/LN-eo-TK) and some idiopathic hypereosinophilic syndromes occurred in only 0.4 persons per 1,000,000 population20 while the incidence of AML, MDS and MPN each was 30-40 cases per million, and CMML 4 cases per million.21 In our previous study, only 21 cases of CEL-NOS were identified in a period of 13 years, an incidence significantly below 1% of all myeloid neoplasms.22 In addition to eosinophilia, basophilia was also common, co-existing with eosinophilia in more than half of the cases of CEL-NOS. Notably, basophilia was extremely uncommon in previously published series of CEL-NOS, although the STAT5B mutation status was mostly unknown in those studies.23-25 Interestingly, basophilia was mentioned in the series of cases of eosinophilia with STAT5B mutations reported by Cross et al.11 but the authors did not provide a frequency. Other than CEL-NOS, in this case series, eosinophilia and/or basophilia was observed in around half of MDS/MPN and Ph-negative classic MPN cases but was uncommon in MDS and AML. Eosinophilia/basophilia was significantly associated with STAT5B N642H hotspot mutations, mutations detected at initial diagnosis, with a high VAF, as a dominant or co-dominant clone and having fewer co-mutations. In addition to its function in eosinophils, STAT5 is known to be critical for basophil and mast cell differentiation and maintenance through the STAT5-GATA2 pathway.26 We only had one case presenting as SM in which STAT5B N642H was detected as a dominant mutation at initial diagnosis. This case was associated with eosinophilia and lacked a KIT mutation, raising the differential diagnosis of an eosinophilic myeloid neoplasm associated with a mast cell proliferation. One case of CEL-NOS showed increased scattered spindle mast cells with aberrant CD25 expression. The question of whether mast cell proliferation is also a common feature of STAT5B mutation, as that seen in M/LN-eo-TK,27 will require a systemic evaluation of mast cells in these cases. Aside from this case, there were several cases blurring the boundaries of classifications. A patient with triple-negative PMF showed marked eosinophilia (>2.5x109/L) and basophilia (>1.0x109/L), and a case of MDS/MPN, NOS showed persistent eosinophilia (>10%) although the count was slightly under 1.5x109/L. These cases all had a STAT5B N642H mutation detected at initial diagnosis, with a high VAF (>40%) and as a dominant clone. We question whether these cases are better considered within the spectrum of CEL-NOS given the context of STAT5B mutations.
In summary, STAT5B mutations occur across a wide spectrum of myeloid neoplasms, but show different mutational characteristics among different subtypes. In CEL-NOS, STAT5B mutations were frequently detected at initial diagnosis, with a high VAF, as a dominant clone, involving the canonical N642H hotspot, and associated with fewer co-mutations. In contrast, in MDS and AML, STAT5B mutations were more frequently present at a low VAF and as a subclone, were more likely acquired in the course of disease, often involved non-canonical mutations (i.e., other than N642H), and were usually not associated with significant eosinophilia or basophilia; a minority of MDS cases may demonstrate or develop relative eosinophilia and/or basophilia during the disease course. In Ph-negative classic MPN in which the disease phenotype is dictated by MPN-driver CALR/MPL/JAK2 mutations, STAT5B mutations preferentially occurred in PMF or fibrotic stages of polycythemia vera and essential thrombocythemia, often as a subclone. Eosinophilia and/or basophilia was seen in about half of these MPN patients, but it is difficult to attribute this entirely to STAT5B mutations because of the inherent association of increased eosinophils and/ or basophils in the fibrotic stage of MPN. These data suggest that STAT5B mutation is unlikely to be a driver in MDS, AML, and MPN with canonical JAK2/CALR/MPL mutations. STAT5B mutation features in cases classified as MDS/MPN were closer to CEL-NOS than to MDS, AML and MPN, although the median VAF was lower and STAT5B was less frequently a dominant or co-dominant clone than in CEL-NOS. It is known that STAT5B is a strong oncogenic driver in T-cell malignancies through its effect of enhancing phospho-Tyr:SH2 domain interactions and escaping negative regulatory phosphatase attack.1 We believe that STAT5B mutation in myeloid neoplasms, if occurring as a dominant clone at the time of diagnosis, is likely a driver mutation in a subgroup of chronic myeloid neoplasms lacking other genetic drivers, preferentially promoting the proliferation of eosinophils, basophils and possibly mast cells. It will be of interest for future studies to examine whether STAT5B mutations may identify a unique subset of CEL-NOS cases with distinctive clinicopathological features. Further research is warranted to determine whether STAT5B-mutated cases currently classified as MDS/MPN or MPN (lacking canonical JAK2/CALR/MPL driver mutations) may be biologically related to STAT5B-mutated CEL-NOS and may be more appropriately classified together as a novel molecularly defined entity. As tyrosine kinase inhibitors28 or STAT5 inhibitors are being evaluated in pre-clinical models,1 with potential future development of novel therapeutic strategies, STAT5B mutations may help to genetically define those chronic myeloid neoplasms that may benefit from targeted therapy.
Supplementary Material
Data-sharing statement
Data presented in this study are available upon request.
References
- 1.de Araujo ED, Erdogan F, Neubauer HA, et al. Structural and functional consequences of the STAT5B(N642H) driver mutation. Nat Commun. 2019;10(1):2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Halim CE, Deng S, Ong MS, Yap CT. Involvement of STAT5 in oncogenesis. Biomedicines. 2020;8(9):316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith MR, Satter LRF, Vargas-Hernandez A. STAT5b: a master regulator of key biological pathways. Front Immunol. 2022;13:1025373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pham HTT, Maurer B, Prchal-Murphy M, et al. STAT5BN642H is a driver mutation for T cell neoplasia. J Clin Invest. 2018;128(1):387-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhattacharya D, Teramo A, Gasparini VR, et al. Identification of novel STAT5B mutations and characterization of TCRbeta signatures in CD4+ T-cell large granular lymphocyte leukemia. Blood Cancer J. 2022;12(2):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rajala HL, Eldfors S, Kuusanmaki H, et al. Discovery of somatic STAT5b mutations in large granular lymphocytic leukemia. Blood. 2013;121(22):4541-4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wahnschaffe L, Braun T, Timonen S, et al. JAK/STAT-activating genomic alterations are a hallmark of T-PLL. Cancers (Basel). 2019;11(12):1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kucuk C, Jiang B, Hu X, et al. Activating mutations of STAT5B and STAT3 in lymphomas derived from gammadelta-T or NK cells. Nat Commun. 2015;6:6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bandapalli OR, Schuessele S, Kunz JB, et al. The activating STAT5B N642H mutation is a common abnormality in pediatric T-cell acute lymphoblastic leukemia and confers a higher risk of relapse. Haematologica. 2014;99(10):e188-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maurer B, Kollmann S, Pickem J, Hoelbl-Kovacic A, Sexl V. STAT5A and STAT5B - twins with different personalities in hematopoiesis and leukemia. Cancers (Basel). 2019;11(11):1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cross NCP, Hoade Y, Tapper WJ, et al. Recurrent activating STAT5B N642H mutation in myeloid neoplasms with eosinophilia. Leukemia. 2019;33(2):415-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thiele J, Kvasnicka HM, Facchetti F, Franco V, van der Walt J, Orazi A. European consensus on grading bone marrow fibrosis and assessment of cellularity. Haematologica. 2005;90(8):1128-1132. [PubMed] [Google Scholar]
- 13.Palomo L, Meggendorfer M, Hutter S, et al. Molecular landscape and clonal architecture of adult myelodysplastic/ myeloproliferative neoplasms. Blood. 2020;136(16):1851-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valent P, Sotlar K, Blatt K, et al. Proposed diagnostic criteria and classification of basophilic leukemias and related disorders. Leukemia. 2017;31(4):788-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arber DA, Orazi A, Hasserjian RP, et al. International consensus classification of myeloid neoplasms and acute leukemia: integrating morphological, clinical, and genomic data. Blood. 2022;140(11):1200-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khoury JD, Solary E, Abla O, et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: myeloid and histiocytic/dendritic neoplasms. Leukemia. 2022;36(7):1703-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Umrau K, Naganuma K, Gao Q, et al. Activating STAT5B mutations can cause both primary hypereosinophilia and lymphocyte-variant hypereosinophilia. Leuk Lymphoma. 2023;64(1):238-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andersson EI, Putzer S, Yadav B, et al. Discovery of novel drug sensitivities in T-PLL by high-throughput ex vivo drug testing and mutation profiling. Leukemia. 2018;32(3):774-787. [DOI] [PubMed] [Google Scholar]
- 19.Sreedharanunni S, Jamwal M, Balakrishnan A, et al. Chronic eosinophilic leukemia with recurrent STAT5B N642H mutation-an entity with features of myelodysplastic syndrome/ myeloproliferative neoplasm overlap. Leuk Res. 2022;112:106753. [DOI] [PubMed] [Google Scholar]
- 20.Ruan GJ, Smith CJ, Day C, et al. A population-based study of chronic eosinophilic leukemia-not otherwise specified in the United States. Am J Hematol. 2020;95(10):E257-E260. [DOI] [PubMed] [Google Scholar]
- 21.Srour SA, Devesa SS, Morton LM, et al. Incidence and patient survival of myeloproliferative neoplasms and myelodysplastic/ myeloproliferative neoplasms in the United States, 2001-12. Br J Haematol. 2016;174(3):382-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu Z, Boddu PC, Loghavi S, et al. A multimodality work-up of patients with hypereosinophilia. Am J Hematol. 2018;93(11):1337-1346. [DOI] [PubMed] [Google Scholar]
- 23.Wang SA, Hasserjian RP, Tam W, et al. Bone marrow morphology is a strong discriminator between chronic eosinophilic leukemia, not otherwise specified and reactive idiopathic hypereosinophilic syndrome. Haematologica. 2017;102(8):1352-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang SA, Tam W, Tsai AG, et al. Targeted next-generation sequencing identifies a subset of idiopathic hypereosinophilic syndrome with features similar to chronic eosinophilic leukemia, not otherwise specified. Mod Pathol. 2016;29(8):854-864. [DOI] [PubMed] [Google Scholar]
- 25.Kelemen K, Saft L, Craig FE, et al. Eosinophilia/ hypereosinophilia in the setting of reactive and idiopathic causes, well-defined myeloid or lymphoid leukemias, or germline disorders. Am J Clin Pathol. 2021;155(2):179-210. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Qi X, Liu B, Huang H. The STAT5-GATA2 pathway is critical in basophil and mast cell differentiation and maintenance. J Immunol. 2015;194(9):4328-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pozdnyakova O, Orazi A, Kelemen K, et al. Myeloid/lymphoid neoplasms associated with eosinophilia and rearrangements of PDGFRA, PDGFRB, or FGFR1 or w.ith PCM1-JAK2. Am J Clin Pathol. 2021;155(2):160-178. [DOI] [PubMed] [Google Scholar]
- 28.Eisenberg R, Gans MD, Leahy TR, et al. JAK inhibition in early-onset somatic, nonclonal STAT5B gain-of-function disease. J Allergy Clin Immunol Pract. 2021;9(2):1008-1010.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data presented in this study are available upon request.