Abstract
CASZ1 is a conserved transcription factor involved in neural development, blood vessel assembly and heart morphogenesis. CASZ1 has been implicated in cancer, either suppressing or promoting tumor development depending on the tissue. However, the impact of CASZ1 on hematological tumors remains unknown. Here, we show that the T-cell oncogenic transcription factor TAL1 is a direct positive regulator of CASZ1, that T-cell acute lymphoblastic leukemia (T-ALL) samples at diagnosis overexpress CASZ1b isoform, and that CASZ1b expression in patient samples correlates with PI3K-AKT-mTOR signaling pathway activation. In agreement, overexpression of CASZ1b in both Ba/F3 and T-ALL cells leads to the activation of PI3K signaling pathway, which is required for CASZ1b-mediated transformation of Ba/F3 cells in vitro and malignant expansion in vivo. We further demonstrate that CASZ1b cooperates with activated NOTCH1 to promote T-ALL development in zebrafish, and that CASZ1b protects human T-ALL cells from serum deprivation and treatment with chemotherapeutic drugs. Taken together, our studies indicate that CASZ1b is a TAL1-regulated gene that promotes T-ALL development and resistance to chemotherapy.
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive hematological malignancy that results from transformation and clonal expansion of developmentally-arrested T-cell progenitors.1,2 Conventional risk-adjusted multi-agent chemotherapy allows for high 5-year event-free survival rates in children. However, a significant number of patients still relapse or do not respond to therapy (features that are striking in adults), and the intensive-treatment regimens are often associated with severe complications. Consequently, there have been considerable efforts to better understand the cell-intrinsic lesions and microenvironmental underpinnings of the disease, resulting in the identification of numerous key genetic,3-7 epigenetic or posttranscriptional,8-12 and cell-extrinsic13-15 alterations involved in the development and resistance to treatment of T-ALL. Amongst these, several have revealed potential to translate into novel, hopefully less toxic and more efficient therapies,2,16-19 which may contribute to circumventing resistance to chemotherapy. Thus, identification of new molecular regulators of T-ALL should contribute to a better understanding of the disease and, consequently, to the improvement of therapeutic approaches. CASZ1 is a highly conserved20 zinc finger transcription factor essential for blood vessel assembly,21 cardiomyocyte differentiation and proliferation22,23 and heart morphogenesis.24,25 CASZ1 is also critical during neural development.26,27 In accordance, loss of CASZ1 expression associates with high risk and poor prognosis in neuroblastoma.28,29 In agreement with a tumor suppressor role for CASZ1, restoration of CASZ1 expression in neuroblastoma cell lines resulted in cell differentiation, increased adhesion and reduced migration, and inhibition of cell growth in vitro and in vivo.28,29 CASZ1 displays an anti-oncogenic role also in hepatocellular carcinoma30 and embryonal rhabdomyosarcoma.31 Nevertheless, the involvement of CASZ1 in cancer may vary depending on the tissue. For instance, CASZ1 is upregulated in epithelial ovarian cancer cells and promotes epithelial-mesenchymal transition, cell migration, invasion and metastasis.32 There are two known CASZ1 isoforms in humans: CASZ1a (also known as CASZ11 or CASZ1 transcript variant 1), which has 21 exons, is roughly 8 Kb-long and encodes for a nuclear protein containing 11 zinc fingers; and CASZ1b (CASZ5 or CASZ1 transcript variant 2), which has 16 exons, is 4.4 kb-long and encodes for a mainly nuclear but also cytoplasmic protein with five zinc fingers. The sequence of CASZ1b is identical to that of the longer CASZ1 isoform until the end of exon 16, where a 5’ splice donor located 1 bp before the stop codon of CASZ1b can be used with a 3’ splice acceptor in exon 17 of CASZ1a to produce the latter by alternative splicing.20 Interestingly, CASZ1a and CASZ1b are often co-expressed (although not always regulated in a similar fashion) and appear to exert similar functions in particular tissues.29 For example, both isoforms were shown to have anti-tumoral activity in neuroblastoma,28,29 while exerting protumoral effects in ovarian cancer.32 In the current study, we aimed to define the role of CASZ1 in T-ALL. We demonstrate that CASZ1b, which is upregulated in T-ALL cells in part via transcriptional activity of TAL1, has an oncogenic role, transforming Ba/F3 cells and cooperating with NOTCH1 in promoting T-ALL in zebrafish. In human T-ALL cells, CASZ1b expression correlates with poor prognosis and activation of the PI3K-AKT-mTOR signaling pathway. Consistent with these observations, CASZ1b overexpression leads to PI3K-AKT-mTOR signaling activation, which is required for T-ALL cell viability. Overall, our findings establish CASZ1 as a putative novel T-cell oncogene and suggest that PI3K targeting drugs may offer clinical benefit for relapsed T-ALL cases exhibiting high CASZ1 levels.
Methods
Primary T-cell acute lymphoblastic leukemia samples, cell lines and culture conditions
Primary T-ALL cells collected from pediatric patients at diagnosis were isolated as described.14 Informed consent was obtained in accordance with the Declaration of Helsinki after institutional ethical review board approval (authorization #13-105-1 obtained from the Institutional Review Board [IRB00003888] of the French Institute of Medical Research and Health). Upon isolation, primary samples were cultured in RPMI 1640 medium (Life Technologies) supplemented with 10% fetal bovine serum (FBS, Biowest), hereafter named RP-MI-10. The human T-ALL cell lines CEM, DND-41, HPB-ALL, Jurkat, Loucy, MOLT4 and P12 were obtained commercially and cultured in RPMI-10. The IL-3-dependent murine pro-B cell line Ba/F3 was maintained in RPMI-10 with IL-3. Mouse IL-3 was produced by WEHI3B cells, whose supernatant was used to maintain Ba/F3 cells. The human embryonic fibroblast 293T cell line was maintained in DMEM medium (Life Technologies) supplemented with 10% FBS (DMEM-10). T-ALL primary cells, cell lines and Ba/F3 cells were cultured at 37°C with a 5% CO2 atmosphere in RPMI-10 alone (with the appropriate vehicle), RPMI (when indicated) or in RPMI-10 plus the following pharmacological agents: 4-hydroxy-tamoxifen (4OHT; Sigma); NVP-BEZ235 (Selleckchem); LY294002 (Cayman Chemical), daunorubicin (Selleckchem); dexamethasone (Sigma) and L-asparaginase (Sigma). At defined time points, cells were harvested and processed as indicated below for assessment of cell viability, RNA extraction and immunobloting.
Assessment of cell viability, activation and proliferation
Determination of cell viability was performed by flow cytometry analysis of forward scatter versus side scatter (FSC vs. SSC) distribution using a LSRFortessa cell analyzer (BD Biosciences). We have confirmed previously that this strategy measures lymphocyte viability as accurately as when using annexin V and propidium iodide staining.8 Cell size, as a measure of cell activation, was assessed by FSC versus SSC gated on the live cell population. The samples were acquired using a LSRFortessa cell analyzer with a high-throughput-sampler (HTS) with a fixed volume of sample to be analyzed, which allowed us to extrapolate the number of cells that were analyzed and thus calculate proliferation.
Immunobloting
Cell lysates were prepared as described16 and equal amounts of protein were analyzed by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis, transferred onto nitrocellulose membranes, and immunoblotted with antibodies against p-mTOR (S2448), p-AKT (S473), AKT, p-S6 (S235/236) and S6 (all from Cell Signaling Technology), CASZ1 (Rockland), Lamin B (Santa Cruz Biotechnology), TAL1 (EMD Millipore), and p27Kip1 (BD Transduction Labs). Immunodetection was performed by incubation with horseradish peroxidase-conjugated appropriate secondary antibodies and developed by chemiluminescence. Where indicated, densitometry analysis was performed using Adobe Photoshop CS3 software (version 10.0, Adobe Systems Incorporated, San Jose, CA, USA). Each band was analyzed with a constant frame and normalized to the respective loading control.
Statistical analysis
The GraphPad Prism software was used for statistical analysis. Differences between mean values were calculated using two-tailed Student’s t test and one-way analysis of variance, as appropriate. Correlation in gene expression levels was determined using the Pearson correlation coefficient. Differences in survival curves were analyzed using the log-rank (Mantel-Cox) test. Differences were considered significant at P<0.05.
Information on plasmid generation, electroporation, viral transduction, mouse and zebrafish in vivo models, quantitative PCR (including primer sequences: Online Supplementary Table S1), chromatin immunoprecipitation, RNA sequencing and bioinformatics analyses are available in the Online Supplementary Appendix.
Results
CASZ1b is upregulated in T-cell acute lymphoblastic leukemia, especially in TAL1-positive cases
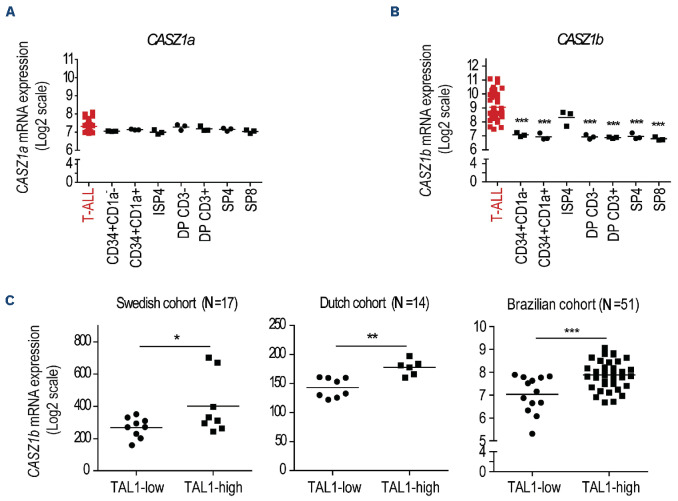
We first analyzed the pattern of Casz1 expression during normal mouse hematopoiesis using the BloodSpot database.33 Casz1 was expressed at low levels in long-term hematopoietic stem cells (HSC) and upregulated in short-term HSC (Online Supplementary Figure S1). Casz1 expression appeared to be highest in common lymphoid progenitors and it was downregulated upon commitment to either T or B lymphoid lineage, with both B- and T-cell precursors displaying low Casz1 levels (Online Supplementary Figure S1). Because T-ALL arises from clonal expansion of T-cell precursors arrested at different stages of differentiation, we next used a publicly available dataset to assess CASZ1 expression in leukemia samples collected from T-ALL patients at presentation, as compared to normal thymocyte subsets representative of the main T-cell developmental stages.34 We found that, in contrast to CASZ1a (Figure 1A), the short isoform of CASZ1 (CASZ1b) was significantly upregulated in T-ALL cells (Figure 1B). These observations, together with the fact that CASZ1b is the most conserved of the two CASZ1 isoforms,20 drove us to focus on CASZ1b (hereafter referred to simply as CASZ1) for the remainder of our studies.
Figure 1.
CASZ1b is overexpressed in T-cell acute lymphoblastic leukemia. (A, B) CASZ1a (A) and CASZ1b (B) transcript levels in T-cell acute lymphoblastic leukemia (T-ALL) patients versus normal thymocyte subpopulations. Gene expression data set GSE33469-33470. ISP4 - immature single positive CD4; DPCD3- - CD4 CD8 double positive, without CD3 expression; DPCD3+ -CD4 CD8 double positive, with CD3 expression; SP4 - single positive CD4; SP8 - single positive CD8. (C) CASZ1b transcript levels in 3 different T-ALL patients cohorts (Swedish - GSE41621, Dutch - GSE18497 and Brazilian - GSE51001 and GSE66638) analyzed according to the expression levels of the TAL1 transcript (TAL1-high vs. TAL1-low). *P<0.05, **P<0.01; ***P<0.001, student’s t test.
Although CASZ1 levels were generally increased in the T-ALL patients (Figure 1A, B) and T-ALL cell lines (Online Supplementary Figure S2) as compared to normal T-cell precursors, we noticed that TAL1 high cases expressed higher levels of CASZ1 than TAL1 low samples in three different T-ALL patient cohorts (Figure 1C; Online Supplementary Figure S3).35,36
TAL1 upregulates CASZ1 in T-cell acute lymphoblastic leukemia cells
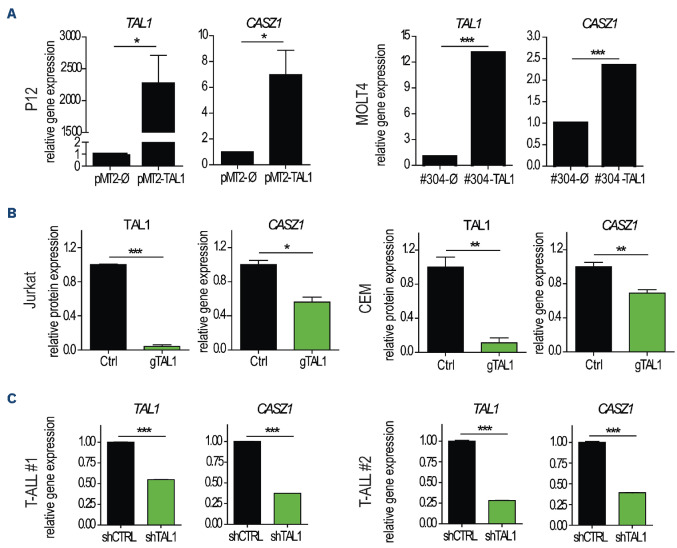
The association between TAL1 and CASZ1 expression in T-ALL patient samples prompted us to evaluate whether TAL1 might transcriptionally regulate CASZ1. Forced expression of TAL1 in two TAL1-negative T-ALL cell lines, P12 (data not shown) and MOLT4 (Online Supplementary Figure S4A), led to the upregulation of CASZ1 expression (Figure 2A), whereas TAL1 inactivation in TAL1-positive Jurkat and CEM cells, using CRISPR/cas9 technology (Figure 2B; Online Supplementary Figure S4B), downregulated CASZ1 (Figure 2B). Importantly, TAL1 silencing in two primary T-ALL patient samples also resulted in CASZ1 downregulation (Figure 2C), further reinforcing the positive link between TAL1 and CASZ1 in T-ALL.
Figure 2.
CASZ1b is regulated by TAL1 in T-cell acute lymphoblastic leukemia cells. (A) P12 and MOLT4 cells ectopically expressing the TAL1 protein were evaluated for TAL1 and CASZ1 expression by quantitative polymerase chain reaction (qPCR). Values were normalized to the respective control condition (empty vector). (B) TAL1 expression was inactivated in Jurkat and CEM cells by CRISPR/cas9. Three control clones (Ctrl) and 3 clones with inactivated TAL1 (gTAL1) from each of the cell lines were analyzed for TAL1 protein expression and CASZ1 mRNA levels. Values were normalized to the average of the control clones. (C) Two primary T-cell acute lymphoblastic leukemia (T-ALL) patient samples collected at diagnosis were transduced with short hairpin RNA (shRNA) against TAL1. Cells were expanded for 72 hours, and TAL1 (left) and CASZ1 (right) transcript levels were detected by qP-CR. Values were normalized to the control condition (shCTRL). Values represent the mean ± standard deviation of experimental triplicates of a representative experiment (N=3). *P<0.05, **P<0.01; ***P<0.001, student’s t test.
CASZ1 transforms Ba/F3 cells through the activation of PI3K-AKT-mTOR signaling pathway
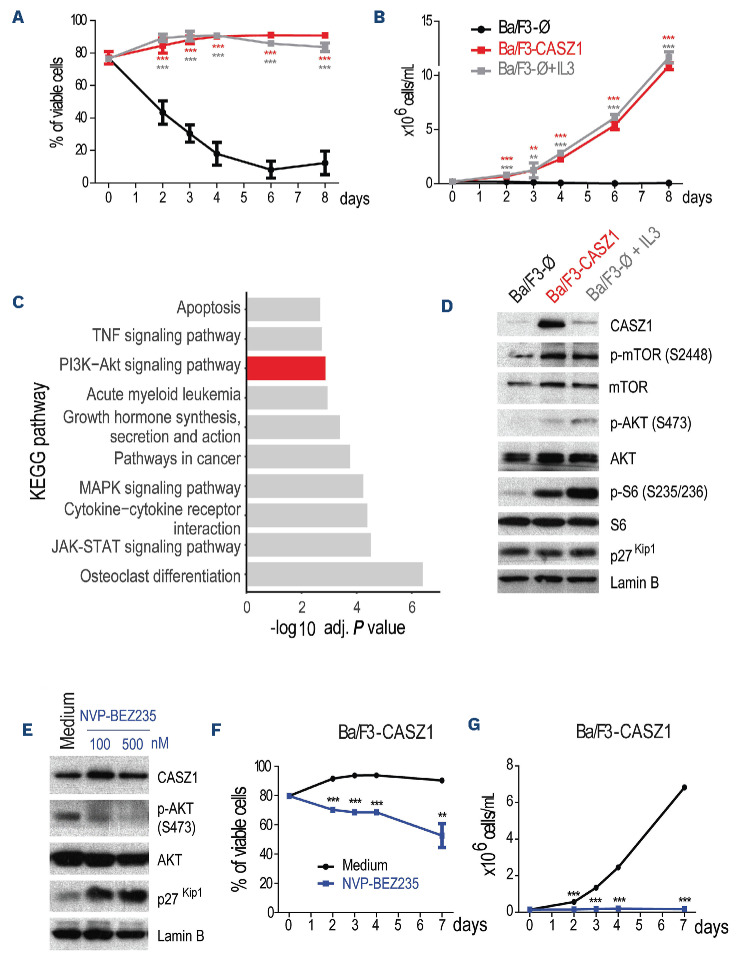
Given that CASZ1 is overexpressed in T-ALL cells, including downstream of the major T-cell oncogene TAL1, we postulated that CASZ1 should have an oncogenic role in lymphoid cells. To test this possibility, we ectopically expressed CASZ1 in a stable manner in IL-3-dependent Ba/ F3 cells7 (Online Supplementary Figure S5A). CASZ1 rescued viability, cell size and proliferation of Ba/F3 cells under IL-3 deprivation (Online Supplementary Figure S5B, C), which allowed Ba/F3 cells expressing CASZ1 in the absence of IL-3 to maintain their viability (Figure 3A) and proliferate (Figure 3B) throughout time to similar levels as mock-transduced control cells cultured with the growth factor. These findings indicate that CASZ1 has the capacity to transform Ba/F3 cells, rendering them growth factor-independent. To identify potential mechanisms by which CASZ1 exerted its oncogenic effects, we analyzed the transcriptional program engaged by CASZ1 in Ba/F3 cells. RNA-sequencing analysis showed that CASZ1 affected the expression of 1,207 genes (Online Supplementary Figure S6; Online Supplementary Table S2), and subsequent KEGG pathway overrepresentation analysis indicated that PI3K-AKT signaling was highly enriched in CASZ1 overexpressing cells (Figure 3C). In agreement, immunoblot analysis showed that CASZ1 overexpression in Ba/F3 cells (Online Supplementary Figure S7A) upregulated the phosphorylation levels of members of the PI3K-AKT-mTOR signaling pathway, such as AKT, mTOR and S6 (Figure 3D). In addition, CASZ1 promoted the downregulation of p27Kip1, a readout of cell cycle progression consistent with PI3K-AKT activation.37 Notably, CASZ1 upregulated Pik3cd (the gene that encodes PI3Kδ) transcript levels (Online Supplementary Figure S6B) and PI3Kδ protein expression (Online Supplementary Figure S8A), suggesting that CASZ1 may promote PI3K-AKT-mTOR pathway activation, at least in part, via upregulation of PI3KCD/PI3Kδ.
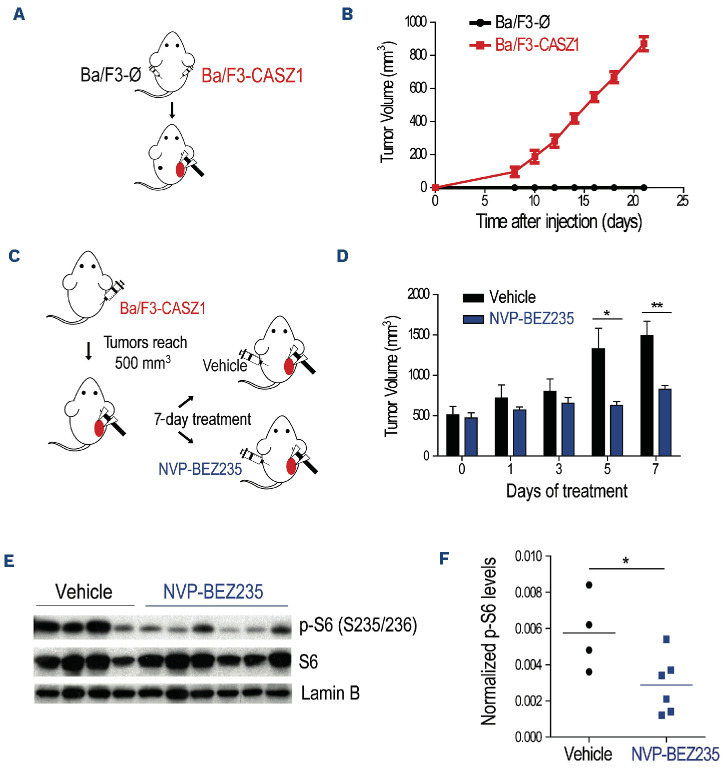
Figure 3.
CASZ1 overexpression transforms Ba/F3 cells by activating the PI3K-AKT-mTOR signaling pathway. (A-B) Ba/F3 cells stably transduced with empty vector (Ba/F3-Ø) or CASZ1 (Ba/F3-CASZ1) were cultured in medium without growth factors. As a positive control, Ba/F3-Ø cells were also cultured in the presence of IL-3 (Ba/F3-Ø + IL-3). Viability (A) and proliferation (B) were determined at the indicated time points. Values represent the mean ± standard deviation of at least 3 independent experiments. **P<0.01; ***P<0.001 (C) Top 10 most significantly enriched KEGG pathways (adj. P<0.05) from upregulated genes (adj. P<0.05, log2 fold change>1) in the RNA-sequencing analysis of Ba/F3-CASZ1 versus Ba/F3-Ø cells. (D) Ba/F3-Ø and Ba/F3-CASZ1 cells were cultured in the indicated conditions for 24 hours. PI3K signaling activation was evaluated by immunoblot detection of the phosphorylation levels of AKT and S6. Total levels of CASZ1 and p27Kip1 were also analyzed. Lamin B was used as loading control. Data are representative of 2 independent experiments. (E) Ba/F3-CASZ1 cells were cultured for 24 hours in the presence of 100 or 500 nM of NVP-BEZ235 or vehicle (medium). PI3K signaling activation was evaluated by immunoblot detection of the phosphorylation levels of AKT. Total levels of CASZ1 and p27Kip1 were also analyzed. Lamin B was used as loading control. Data are representative of 2 independent experiments. (F, G) Ba/F3-CASZ1 cells were cultured in the presence or absence of 500 nM of NVP-BEZ235. Viability (F) and proliferation (G) were determined at the indicated time points. Values represent the mean ± standard deviation of experimental triplicates of a representative experiment (N=3). **P<0.01; ***P<0.001, one-way analysis of variance.
In order to test the involvement of PI3K-AKT-mTOR signaling in CASZ1-mediated transformation, we treated CASZ1-expressing Ba/F3 cells with two distinct specific small molecule PI3K/mTOR inhibitors. As expected, NVP-BEZ235 (dactolisib)38 and LY294002 dampened PI3K-AKT-mTOR signaling (Figure 3E; Online Supplementary Figure S9A), and abrogated the effects of CASZ1 on viability (Figure 3F; Online Supplementary Figure S9B) and proliferation (Figure 3G; Online Supplementary Figure S9C) of Ba/F3 cells. These results demonstrate the importance of maintaining the integrity of the PI3K-AKT-mTOR signaling cascade for CASZ1-mediated transformation.
CASZ1 expression activates the PI3K-AKT-mTOR signaling pathway in T-cell acute lymphoblastic leukemia
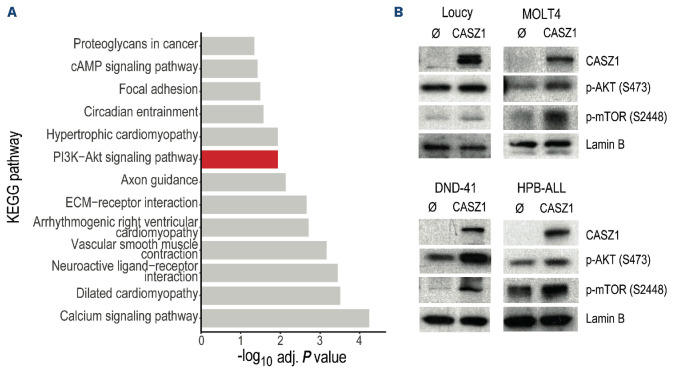
We next sought to evaluate whether, similar to Ba/F3 cells, CASZ1-mediated effects in T-ALL cells involved upregulation of PI3K-AKT-mTOR signaling. We compared expression levels of all the transcripts to those of CASZ1 in the T-ALL GSE18497 dataset36 and calculated their Pearson correlation coefficient. Genes whose correlation coefficient was ≥0.7 (indicating a strong positive correlation with CASZ1) were subsequently employed to perform KEGG pathway overrepresentation analysis. Our analysis shows that genes whose expression correlates with that of CASZ1 were enriched in different KEGG pathways, including PI3K-AKT signaling pathway (Figure 4A). Immunoblot analysis confirmed that T-ALL cell lines overexpressing CASZ1 (Online Supplementary Figure S7B) displayed higher levels of PI3K-AKT-mTOR signaling activation than mock-transduced controls (Figure 4B). In agreement with our transcriptomics data indicating elevated levels of Pik3cd (the gene that encodes PI3Kδ) in CASZ1-overexpressing Ba/F3 cells (Online Supplementary Figure S6B), we found that CASZ1 upregulates PI3Kδ protein levels in DND-41 cells (Online Supplementary Figure S8B). Interestingly, PI3Kβ was the isoform that was upregulated by CASZ1 in MOLT4 cells (Online Supplementary Figure S8C).
Figure 4.
CASZ1 overexpression activates PI3K-AKT-mTOR signaling in T-cell acute lymphoblastic leukemia. (A) KEGG pathway analysis of the genes whose expression levels display a correlation coefficient of at least 0.7 with CASZ1 expression in a cohort of 14 T-cell acute lymphoblastic leukemia (T-ALL) patient samples (GEO database, accession number GSE18497). (B) T-ALL cell lines without (Ø) or with CASZ1 overexpression were assessed for PI3K signaling activation by immunoblot detection of the phosphorylation levels of AKT and mTOR. Total CASZ1 levels were also examined. Lamin B was used as loading control. Data are representative of 2 independent experiments.
CASZ1 promotes tumorigenesis in vivo
Next, we evaluated whether CASZ1 was able to promote tumorigenesis in vivo. We subcutaneously transplanted empty vector- and CASZ1-transduced Ba/F3 cells into opposite flanks of each recipient NOD/SCID mouse (Figure 5A). Whereas none of the control transplants originated any detectable tumors, ten of ten CASZ1-expressing transplants originated large tumor masses within 20 days (Figure 5B). Then, to ascertain whether PI3K-AKT-mTOR signaling pathway is essential for the maintenance of CASZ1-triggered tumors in vivo, we let tumors from subcutaneously transplanted Ba/F3 cells overexpressing CASZ1 grow until 500 mm3, at which point the mice were randomly divided into two groups that either received vehicle or NVP-BEZ235/ dactolisib (Figure 5C). Treatment with NVP-BEZ235/dactolisib clearly delayed tumor growth (Figure 5D) and the effect of the PI3K-AKT-mTOR pathway inhibitor was specific, as shown by the downregulation of S6 phosphorylation levels (Figure 5E, F). Taken together, these results clearly indicate that CASZ1 promotes in vivo tumorigenesis by activating PI3K-AKT-mTOR signaling.
Figure 5.
CASZ1 overexpression in Ba/F3 cells drives in vivo tumorigenesis by activating the PI3K-AKT-mTOR signaling pathway. (A, B) Eight-week old NOD/SCID mice (N=10) were injected subcutaneously with 107 Ba/F3-Ø or Ba/F3-CASZ1 cells and tumor growth was measured as described in the methods. (A) Schematic representation of the experimental layout. (B) Longitudinal analysis of tumor growth. (C-F) Eight-week-old NSG mice were injected subcutaneously with 107 Ba/F3-CASZ1 cells. Once the tumors reached 500 mm3, mice were randomized to receive either NVP-BEZ235 (N=6) or vehicle (N=5) for 7 consecutive days. (C) Schematic representation of the experimental layout. (D) Longitudinal analysis of tumor growth upon treatment initiation. *P<0.05; **P<0.01, one-way analysis of variance. (E) Vehicle- or NVP-BEZ235-treated tumors were collected and phosphorylation and total levels of S6 assessed by immunoblot analysis. Lamin B was used as loading control. (F) S6 phosphorylation levels in (E) were measured by densitometry analysis, and then normalized to total S6 and Lamin B levels. *P<0.05; **P<0.01, student’s t test.
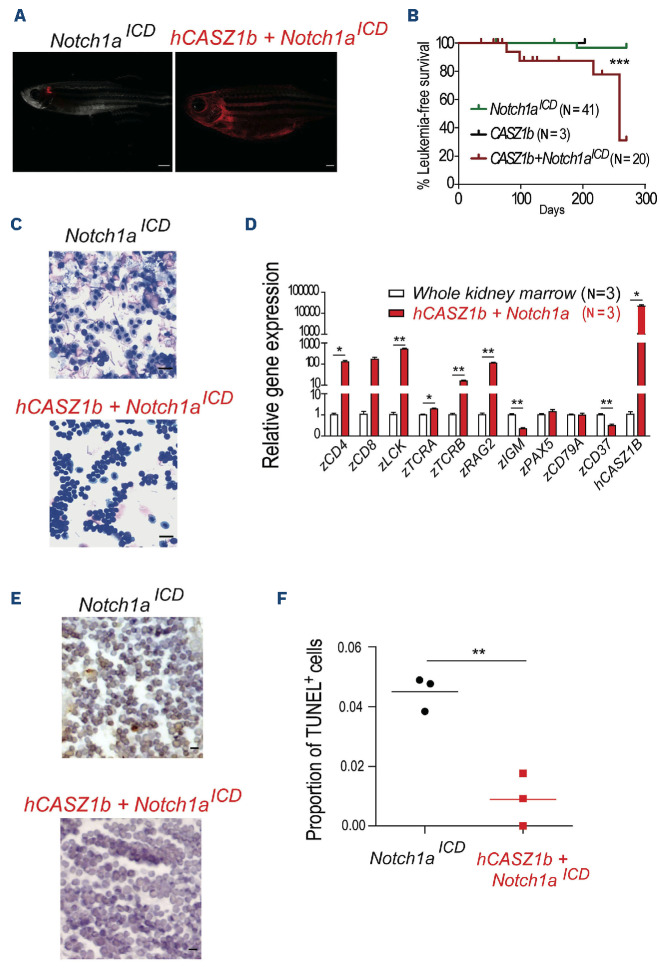
To determine whether CASZ1 promotes tumorigenesis in the context of T-ALL, we next tested whether CASZ1 accelerates NOTCH-induced T-cell leukemia in zebrafish.39,40 W e generated mosaic zebrafish lines expressing intracellular NOTCH1 (ICN1) and/or CASZ1. Although with long latency, we found that CASZ1 accelerated the development of thymic hyperplasia with subsequent invasion of adjacent tissues (Figure 6A) leading to leukemia/lymphoma development in six of 20 animals expressing CASZ1 and ICN1 (starting at day 77, median survival: 259 days), as opposed to only one of 41 animals expressing ICN1 alone that developed leukemia by day 190 (median survival not reached during the duration of the experiment; Figure 6B). The lymphoid neoplastic cells from CASZ1+ICN1 zebrafish displayed a typical blast morphology, which contrasted to the cells collected from the vast majority of the ICN1 animals (Figure 6C). As expected, leukemia/lymphoma cells were of T-cell origin, as determined by the expression of CD4, CD8 and LCK, and the absence of B-cell markers such as PAX5, IGM or CD79a (Figure 6D). Finally, malignant cells in CASZ1+ICN1 zebrafish displayed decreased apoptosis
Figure 6.
CASZ1 cooperates with NOTCH1 to drive T-cell leukemogenesis in transgenic zebrafish. Tu/AB strain embryos were injected at one cell stage with rag2:notch1aICD, rag2:h-CASZ1b or a mixture of both. (A) Images representative of mosaic transgenic zebrafish at 77 days post injection. (B) Kaplan-Meier analysis of leukemic fish comparing Notch1aICD (N=15), hCASZ1b (N=3, sacrificed at 203 days without evidence of disease) and hCASZ1b + Notch1aICD (N=20). ***P<0.001, log-rank (Mantel-Cox) test. (C) May-Grünwald and Wright-Giemsa stained cytospins showing lymphoblast morphology; 400x magnification. Images are representative of at least 3 independent fish analyzed of each genotype up to 100 days post injection. Scale bar equals 10 µm. (D) Transcript levels of the indicated genes were determined by quantitative polymerase chain reaction (qPCR) and normalized to β-actin. The values were further normalized to the control condition (whole kidney marrow) and represent the mean ± standard deviation of at least 2 independent replicates. (E) Representative hematoxylin- and eosin-stained histological sections juxtaposed to immunohistochemistry for TUNEL. Scale bar equals 10 μm. (F) Values are the quantification of the immunohistochemistry data represented in (E). * P<0.05; **P<0.01, student’s t test.
(Figure 6E, F) compared to control cells in ICN1 zebrafish. Altogether, our results indicate that CASZ1 can have a tumorigenic effect in lymphoid cells and cooperate with ICN1 to promote T-cell leukemia in vivo.
CASZ1 protects human T-cell acute lymphoblastic leukemia cells from death triggered by serum starvation and chemotherapy
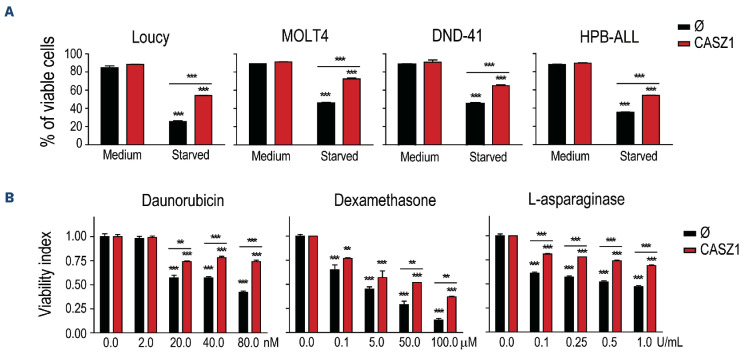
To investigate the biological impact of CASZ1 in human T-ALL, we tried to silence CASZ1 in T-ALL cell lines. However, we were able to do so only mildly in Jurkat cells, after transfection with small interfering RNA (siRNA) for CASZ1. Although CASZ1 knock down was only around 25-30% (Online Supplementary Figure S10A), there was a tendency for decreased viability in cells with lower CASZ1 levels (Online Supplementary Figure S10B) and a statistically significant negative impact on proliferation (Online Supplementary Figure S10C). We then ectopically expressed CASZ1 in several T-ALL cell lines using lentiviral transduction (Online Supplementary Figure S11A). Overexpression of CASZ1 had no impact on T-ALL cell viability (Online Supplementary Figure S11B) or proliferation (Online Supplementary Figure S11C). Basal viability and growth rate of T-ALL cell lines in regular culture conditions are high. This may mask positive effects of some oncogenes. For instance, in vitro prosurvival effects of TAL1 in T-ALL cells were exposed only upon demonstration that TAL1 protected leukemia cells from stress-induced apoptosis.41 Similar to TAL1, CASZ1 overexpression protected T-ALL cells from serum starvation-induced apoptosis (Figure 7A). Moreover, CASZ1 conferred resistance to treatment with daunorubicin, dexamethasone and L-asparaginase (Figure 7B), chemotherapeutic drugs currently used to treat patients with T-ALL.
Figure 7.
CASZ1 overexpression promotes T-cell acute lymphoblastic leukemia resistance to chemotherapy. (A) T-cell acute lymphoblastic leukemia (T-ALL) cells without (Ø) or with CASZ1 overexpression were cultured in regular culture conditions (medium) or under serum starvation (starved) for 48 hours [h] (Loucy), 72 h (MOLT4) or 144 h (DND-41 and HPB-ALL). (B) Loucy cells were cultured with the indicated concentrations of daunorubicin (72h), dexamethasone (72 h) and L-asparaginase (24 h). (A, B) Viability was determined, and the indicated values represent the mean ± standard deviation of at least 3 independent experiments *P<0.05; **P<0.01; ***P<0.001, student’s t test.
Based on our findings that CASZ1 overexpression could rescue T-ALL cell viability under stress conditions and contribute to in vitro resistance to chemotherapeutic drugs commonly used in the treatment of T-ALL patients, we hypothesized that high expression of CASZ1 might correlate with unfavorable outcome. However, we did not observe any significant association between CASZ1 expression levels and relapse-free or overall survival in children with newly diagnosed T-ALL enrolled in the St. Jude Children’s Research Hospital Total Therapy 15 and 16 studies (Online Supplementary Figure S12A, B), even when specifically examining relapse cases (Online Supplementary Figure S12C).
Nevertheless, in another cohort comprising exclusively of relapse T-ALL patients (GSE18497), for which gene expression and clinical parameters are publicly available,36 high CASZ1 levels associated with particularly poor prognosis, as the CASZ1-high group displayed faster progression to disease relapse and decreased overall survival (Online Supplementary Figure S12D-F). Interestingly, in agreement with our findings that TAL1 regulates CASZ1 in T-ALL, TAL1 was also associated with poor prognosis in the GSE18497 dataset (Online Supplementary Figure S12G-I). Overall, these analyses suggest that CASZ1 is not an independent prognostic factor in T-ALL, although it may associate with poor prognosis in relapse T-ALL depending on the specific treatment protocol.
Discussion
CASZ1 is a conserved zinc finger transcription factor that is essential for heart and neural development.20,22-25,28,29 Notwithstanding its pivotal role in embryogenesis, CASZ1 has been also implicated in cancer, either by acting as tumor suppressor in neuroblastoma,28,29 embryonal rhabdomyosarcoma31 and hepatocellular carcinoma30 or by promoting tumor metastasis in an ovarian cancer model.32 Here, we demonstrate that CASZ1 is overexpressed and acts in an oncogenic manner in T-ALL.
The transcription factor TAL1 is a major oncogene in the context of T-ALL,42 being overexpressed in a large portion of the patients and defining one of the subgroups of the disease.43 TAL1 transgenic mice develop aggressive T-cell leukemia albeit with long latency.44,45 Previous studies unveiled the transcriptional program associated with TAL1 in T-ALL.46-48 Here, we demonstrated that TAL1 can regulate CASZ1. However, analysis of publicly-available ChIP-seq data revealed that TAL1 binding to the CASZ1 locus in T-ALL cells is limited, with small and non-conserved peaks (Online Supplementary Figure S13), suggesting that TAL1 may have, at best, a mild direct regulatory effect on CASZ1. In addition, the negative impact of TAL1 genetic inactivation on CASZ1 expression varied in degree amongst the T-ALL cell lines and primary leukemia cells we analyzed. This, together with the fact that T-ALL samples in general (including TAL1-negative cases) displayed upregulation of CASZ1 as compared to normal thymocytes, suggests that additional factors should contribute to CASZ1 upregulation in T-ALL. Preliminary analyses of publicly-available data and treatment of T-ALL cell lines with small molecule inhibitors did not find evidence for NOTCH1, MYC or EZH2 being consistent upstream regulators of CASZ1 (data not shown). Thus, the identification of CASZ1 upstream regulators in T-ALL, including transcription factors directly activating CASZ1, warrants further investigation. The role of CASZ1 in hematopoietic differentiation remains unaddressed. However, our analyses of a mouse dataset from the BloodSpot database33 allowed us to trace CASZ1 expression throughout T-cell ontogeny. CASZ1 expression is low in developing and terminally differentiated T cells, paralleling that of TAL1 expression.49 Compatible with an oncogenic role for CASZ1 overexpression, CASZ1 levels are normally low in healthy T-cell progenitors, which are the targets for malignant transformation in T-ALL.1 In agreement, we showed that CASZ1 is able to cooperate with ICN1 to drive T-cell leukemia in zebrafish, by promoting the survival of T-ALL blasts (effects that are recapitulated upon CASZ1-mediated transformation of Ba/F3 cells). Interestingly, in established human leukemia, the prosurvival role of CASZ1 was exposed (reminiscent of what happens with TAL141) when T-ALL cells were cultured under stress conditions, either via serum starvation or treatment with chemotherapeutic drugs. This suggests that CASZ1 may protect, to some extent, against chemotherapy. In agreement with this assumption, we found that, among patients with relapsed T-ALL, high CASZ1 expression can associate with accelerated time to relapse and decreased overall survival. The PI3K-AKT-mTOR signaling pathway regulates multiple cellular processes, including those that are associated with the tumorigenic process, such as viability, proliferation, migration and metabolism. The pivotal contribution of this signaling pathway to the malignant process in T-ALL has been established.5,8,50 In the present study, we demonstrated that CASZ1 expression activates PI3K-AKT-mTOR signaling in both Ba/F3 cells and T-ALL cells, which is required for CASZ1-mediated cell transformation. These results demonstrate that one of the major molecular events elicited by CASZ1 expression in T-ALL is the activation of the PI3K-AKT-mTOR pathway. The mechanisms by which CASZ1 activates PI3K signaling in lymphoid cells warrant investigation. Our bioinformatics analyses of RNA-sequencing data also indicated that CASZ1 can activate JAK-STAT5 and MAPK pathways (Figure 5C). We confirmed these findings at the protein level (Online Supplementary Figures S14 and S15). Whereas we did not test the functional impact of JAK-STAT signaling downstream from CASZ1, we found that MAPK is required for in vitro transformation of Ba/F3 cells but dispensable for CASZ1-mediated expansion of Ba/ F3 cells in vivo (Online Supplementary Figure S16). Overall, our studies demonstrate that CASZ1 is upregulated in T-ALL (at least in part via TAL1 activity), protecting T-ALL cells from stress induced apoptosis and inducing the activation of the PI3K-AKT-mTOR signaling pathway, which is required for transformation in vitro and tumor growth in vivo. Our findings identify CASZ1 as a putative novel oncogene in T-ALL.
Supplementary Material
Acknowledgments
We thank Dr Thiele for kindly providing the CASZ1 plasmids and Francisco Alexandrino, Hannah Taylor, Ana Sofia Moreira, Danyl Shatalov and Veronika Waas for their technical support. We also thank the mouse, zebrafish and flow cytometry core facilities of Instituto de Medicina Molecular João Lobo Antunes for their technical support.
Funding Statement
Funding: This work was supported by the ERC-CoG-648455 and ERC-POC-101069429 grants from the European Research Council, under the European Union’s Horizon 2020 research and innovation program; PTDC/SAU-OBD/69974 grant from Fundação para a Ciência e a Tecnologia (FCT), Portugal; and a grant from Children with Leukemia (now Children with Cancer) Charity, UK (to JTB). The work was also supported by EXPL/ MEC-HEM/0571/2021 grant from FCT (to BAC), and grants R01CA211734 and MGH Scholars Award (to DML); and US NCI CA21765 grant to the Cancer center of St. Jude Children’s Research Hospital. ARG and RF were the recipients of FCT Investigator Grants (CEECIND/02699/2017 and CEECIND/03459/2018, respectively). JRA is the recipient of a Howard Hughes Medical Institute Hanna H. Gray Fellowship. ABS and JAY (301596/2017-4) received a fellowship from the Brazilian National Counsel of Technological and Scientific Development (CNPq).
Data-sharing statement
For original data that are not publicly deposited, please contact the corresponding author.
References
- 1.Girardi T, Vicente C, Cools J, De Keersmaecker K. The genetics and molecular biology of T-ALL. Blood. 2017;129(9):1113-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belver L, Ferrando A. The genetics and mechanisms of T cell acute lymphoblastic leukaemia. Nat Rev Cancer. 2016;16(8):494-507. [DOI] [PubMed] [Google Scholar]
- 3.Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269-271. [DOI] [PubMed] [Google Scholar]
- 4.Clappier E, Cuccuini W, Kalota A, et al. The C-MYB locus is involved in chromosomal translocation and genomic duplications in human T-cell acute leukemia (T-ALL), the translocation defining a new T-ALL subtype in very young children. Blood. 2007;110(4):1251-1261. [DOI] [PubMed] [Google Scholar]
- 5.Gutierrez A, Sanda T, Grebliunaite R, et al. High frequency of PTEN, PI3K, and AKT abnormalities in T-cell acute lymphoblastic leukemia. Blood. 2009;114(3):647-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Homminga I, Pieters R, Langerak AW, et al. Integrated transcript and genome analyses reveal NKX2-1 and MEF2C as potential oncogenes in T cell acute lymphoblastic leukemia. Cancer Cell. 2011;19(4):484-497. [DOI] [PubMed] [Google Scholar]
- 7.Zenatti PP, Ribeiro D, Li W, et al. Oncogenic IL7R gain-of-function mutations in childhood T-cell acute lymphoblastic leukemia. Nat Genet. 2011;43(10):932-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silva A, Yunes JA, Cardoso BA, et al. PTEN posttranslational inactivation and hyperactivation of the PI3K/Akt pathway sustain primary T cell leukemia viability. J Clin Invest. 2008;118(11):3762-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mansour MR, Sanda T, Lawton LN, et al. The TAL1 complex targets the FBXW7 tumor suppressor by activating miR-223 in human T cell acute lymphoblastic leukemia. J Exp Med. 2013;210(8):1545-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durinck K, Wallaert A, Van de Walle I, et al. The Notch driven long non-coding RNA repertoire in T-cell acute lymphoblastic leukemia. Haematologica. 2014;99(12):1808-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Correia NC, Fragoso R, Carvalho T, Enguita FJ, Barata JT. MiR-146b negatively regulates migration and delays progression of T-cell acute lymphoblastic leukemia. Sci Rep. 2016;6:31894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Y, Han C, Wang E, et al. Posttranslational regulation of the exon skipping machinery controls aberrant splicing in leukemia. Cancer Discov. 2020;10(9):1388-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silva A, Laranjeira AB, Martins LR, et al. IL-7 contributes to the progression of human T-cell acute lymphoblastic leukemias. Cancer Res. 2011;71(14):4780-4789. [DOI] [PubMed] [Google Scholar]
- 14.Uzan B, Poglio S, Gerby B, et al. Interleukin-18 produced by bone marrow-derived stromal cells supports T-cell acute leukaemia progression. EMBO Mol Med. 2014;6(6):821-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pitt LA, Tikhonova AN, Hu H, et al. CXCL12-producing vascular endothelial niches control acute T cell leukemia maintenance. Cancer Cell. 2015;27(6):755-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cardoso BA, de Almeida SF, Laranjeira AB, et al. TAL1/SCL is downregulated upon histone deacetylase inhibition in T-cell acute lymphoblastic leukemia cells. Leukemia. 2011;25(10):1578-1586. [DOI] [PubMed] [Google Scholar]
- 17.Peirs S, Matthijssens F, Goossens S, et al. ABT-199 mediated inhibition of BCL-2 as a novel therapeutic strategy in T-cell acute lymphoblastic leukemia. Blood. 2014;124(25):3738-3747. [DOI] [PubMed] [Google Scholar]
- 18.Maude SL, Dolai S, Delgado-Martin C, et al. Efficacy of JAK/STAT pathway inhibition in murine xenograft models of early T-cell precursor (ETP) acute lymphoblastic leukemia. Blood. 2015;125(11):1759-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Smedt R, Morscio J, Reunes L, et al. Targeting cytokine- and therapy-induced PIM1 activation in preclinical models of T-cell acute lymphoblastic leukemia and lymphoma. Blood. 2020;135(19):1685-1695. [DOI] [PubMed] [Google Scholar]
- 20.Liu Z, Yang X, Tan F, Cullion K, Thiele CJ. Molecular cloning and characterization of human Castor, a novel human gene upregulated during cell differentiation. Biochem Biophys Res Commun. 2006;344(3):834-844. [DOI] [PubMed] [Google Scholar]
- 21.Charpentier MS, Christine KS, Amin NM, et al. CASZ1 promotes vascular assembly and morphogenesis through the direct regulation of an EGFL7/RhoA-mediated pathway. Dev Cell. 2013;25(2):132-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christine KS, Conlon FL. Vertebrate CASTOR is required for differentiation of cardiac precursor cells at the ventral midline. Dev Cell. 2008;14(4):616-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorr KM, Amin NM, Kuchenbrod LM, et al. Casz1 is required for cardiomyocyte G1-to-S phase progression during mammalian cardiac development. Development. 2015;142(11):2037-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang RT, Xue S, Wang J, et al. CASZ1 loss-of-function mutation associated with congenital heart disease. Gene. 2016;595(1):62-68. [DOI] [PubMed] [Google Scholar]
- 25.Sojka S, Amin NM, Gibbs D, Christine KS, Charpentier MS, Conlon FL. Congenital heart disease protein 5 associates with CASZ1 to maintain myocardial tissue integrity. Development. 2014;141(15):3040-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mellerick DM, Kassis JA, Zhang SD, Odenwald WF. castor encodes a novel zinc finger protein required for the development of a subset of CNS neurons in Drosophila. Neuron. 1992;9(5):789-803. [DOI] [PubMed] [Google Scholar]
- 27.Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106(4):511-521. [DOI] [PubMed] [Google Scholar]
- 28.Liu Z, Yang X, Li Z, et al. CASZ1, a candidate tumor-suppressor gene, suppresses neuroblastoma tumor growth through reprogramming gene expression. Cell Death Differ. 2011;18(7):1174-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Z, Naranjo A, Thiele CJ. CASZ1b, the short isoform of CASZ1 gene, coexpresses with CASZ1a during neurogenesis and suppresses neuroblastoma cell growth. PLoS One. 2011;6(4):e18557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang JL, Yang MY, Xiao S, Sun B, Li YM, Yang LY. Downregulation of castor zinc finger 1 predicts poor prognosis and facilitates hepatocellular carcinoma progression via MAPK/ERK signaling. J Exp Clin Cancer Res. 2018;37(1):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Z, Zhang X, Lei H, et al. CASZ1 induces skeletal muscle and rhabdomyosarcoma differentiation through a feed-forward loop with MYOD and MYOG. Nat Commun. 2020;11(1):911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu YY, Chang CL, Chuang YJ, et al. CASZ1 is a novel promoter of metastasis in ovarian cancer. Am J Cancer Res. 2016;6(6):1253-1270. [PMC free article] [PubMed] [Google Scholar]
- 33.Bagger FO, Sasivarevic D, Sohi SH, et al. BloodSpot: a database of gene expression profiles and transcriptional programs for healthy and malignant haematopoiesis. Nucleic Acids Res. 2016;44(D1):D917-D924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Vlierberghe P, Ambesi-Impiombato A, Perez-Garcia A, et al. ETV6 mutations in early immature human T cell leukemias. J Exp Med. 2011;208(13):2571-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borssen M, Palmqvist L, Karrman K, et al. Promoter DNA methylation pattern identifies prognostic subgroups in childhood T-cell acute lymphoblastic leukemia. PLoS One. 2013;8(6):e65373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staal FJ, de Ridder D, Szczepanski T, et al. Genome-wide expression analysis of paired diagnosis-relapse samples in ALL indicates involvement of pathways related to DNA replication, cell cycle and DNA repair, independent of immune phenotype. Leukemia. 2010;24(3):491-499. [DOI] [PubMed] [Google Scholar]
- 37.Barata JT, Silva A, Brandao JG, Nadler LM, Cardoso AA, Boussiotis VA. Activation of PI3K is indispensable for interleukin 7-mediated viability, proliferation, glucose use, and growth of T cell acute lymphoblastic leukemia cells. J Exp Med. 2004;200(5):659-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seront E, Rottey S, Filleul B, et al. Phase II study of dual phosphoinositol-3-kinase (PI3K) and mammalian target of rapamycin (mTOR) inhibitor BEZ235 in patients with locally advanced or metastatic transitional cell carcinoma. BJU Int. 2016;118(3):408-415. [DOI] [PubMed] [Google Scholar]
- 39.Blackburn JS, Liu S, Raiser DM, et al. Notch signaling expands a pre-malignant pool of T-cell acute lymphoblastic leukemia clones without affecting leukemia-propagating cell frequency. Leukemia. 2012;26(9):2069-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J, Jette C, Kanki JP, Aster JC, Look AT, Griffin JD. NOTCH1-induced T-cell leukemia in transgenic zebrafish. Leukemia. 2007;21(3):462-471. [DOI] [PubMed] [Google Scholar]
- 41.Bernard M, Delabesse E, Novault S, Hermine O, Macintyre EA. Antiapoptotic effect of ectopic TAL1/SCL expression in a human leukemic T-cell line. Cancer Res. 1998;58(12):2680-2687. [PubMed] [Google Scholar]
- 42.Correia NC, Arcangeli ML, Pflumio F, Barata JT. Stem cell leukemia: how a TALented actor can go awry on the hematopoietic stage. Leukemia. 2016;30(10):1968-1978. [DOI] [PubMed] [Google Scholar]
- 43.Ferrando AA, Neuberg DS, Staunton J, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1(1):75-87. [DOI] [PubMed] [Google Scholar]
- 44.O’Neil J, Shank J, Cusson N, Murre C, Kelliher M. TAL1/SCL induces leukemia by inhibiting the transcriptional activity of E47/HEB. Cancer Cell. 2004;5(6):587-596. [DOI] [PubMed] [Google Scholar]
- 45.Kelliher MA, Seldin DC, Leder P. Tal-1 induces T cell acute lymphoblastic leukemia accelerated by casein kinase IIalpha. Embo J. 1996;15(19):5160-5166. [PMC free article] [PubMed] [Google Scholar]
- 46.Sanda T, Lawton LN, Barrasa MI, et al. Core transcriptional regulatory circuit controlled by the TAL1 complex in human T cell acute lymphoblastic leukemia. Cancer Cell. 2012;22(2):209-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palomero T, Odom DT, O’Neil J, et al. Transcriptional regulatory networks downstream of TAL1/SCL in T-cell acute lymphoblastic leukemia. Blood. 2006;108(3):986-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kusy S, Gerby B, Goardon N, et al. NKX3.1 is a direct TAL1 target gene that mediates proliferation of TAL1-expressing human T cell acute lymphoblastic leukemia. J Exp Med. 2010;207(10):2141-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Herblot S, Steff AM, Hugo P, Aplan PD, Hoang T. SCL and LMO1 alter thymocyte differentiation: inhibition of E2A-HEB function and pre-T alpha chain expression. Nat Immunol. 2000;1(2):138-144. [DOI] [PubMed] [Google Scholar]
- 50.Palomero T, Sulis ML, Cortina M, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13(10):1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
For original data that are not publicly deposited, please contact the corresponding author.