Abstract
Lysine decarboxylase (LDC; EC 4.1.1.18) from Selenomonas ruminantium comprises two identical monomeric subunits of 43 kDa and has decarboxylating activities toward both l-lysine and l-ornithine with similar Km and Vmax values (Y. Takatsuka, M. Onoda, T. Sugiyama, K. Muramoto, T. Tomita, and Y. Kamio, Biosci. Biotechnol. Biochem. 62:1063–1069, 1999). Here, the LDC-encoding gene (ldc) of this bacterium was cloned and characterized. DNA sequencing analysis revealed that the amino acid sequence of S. ruminantium LDC is 35% identical to those of eukaryotic ornithine decarboxylases (ODCs; EC 4.1.1.17), including the mouse, Saccharomyces cerevisiae, Neurospora crassa, Trypanosoma brucei, and Caenorhabditis elegans enzymes. In addition, 26 amino acid residues, K69, D88, E94, D134, R154, K169, H197, D233, G235, G236, G237, F238, E274, G276, R277, Y278, K294, Y323, Y331, D332, C360, D361, D364, G387, Y389, and F397 (mouse ODC numbering), all of which are implicated in the formation of the pyridoxal phosphate-binding domain and the substrate-binding domain and in dimer stabilization with the eukaryotic ODCs, were also conserved in S. ruminantium LDC. Computer analysis of the putative secondary structure of S. ruminantium LDC showed that it is approximately 70% identical to that of mouse ODC. We identified five amino acid residues, A44, G45, V46, P54, and S322, within the LDC catalytic domain that confer decarboxylase activities toward both l-lysine and l-ornithine with a substrate specificity ratio of 0.83 (defined as the kcat/Km ratio obtained with l-ornithine relative to that obtained with l-lysine). We have succeeded in converting S. ruminantium LDC to form with a substrate specificity ratio of 58 (70 times that of wild-type LDC) by constructing a mutant protein, A44V/G45T/V46P/P54D/S322A. In this study, we also showed that G350 is a crucial residue for stabilization of the dimer in S. ruminantium LDC.
Ornithine decarboxylase (ODC) (EC 4.1.1.17) is an important enzyme for the biosynthesis of putrescine, a precursor of polyamines which are implicated in a wide variety of biological processes that include the synthesis of DNA, RNA, and protein in all living cells (26, 30, 31). Lysine decarboxylase (LDC) (EC 4.1.1.18), which exists in most bacteria, is involved in the biosynthesis of cadaverine, a molecule that participates in the closing of the porin channels in the outer membrane of Escherichia coli (5) and is also an essential component of the peptidoglycan of Selenomonas ruminantium, Veillonella alcalescens, V. parvula, and Anaerovibrio lipolytica, which are strictly anaerobic gram-negative bacteria (9, 12, 14, 15). Previously, we reported that in these bacteria, cadaverine is transferred to the d-glutamic acid residue of a lipid intermediate for the synthesis of the cadaverine-containing peptidoglycan by cadaverine transferase (11, 12, 15, 17). In S. ruminantium, cadaverine is constitutively synthesized from l-lysine (16) and its synthesis was completely prevented by dl-α-difluoromethyllysine (DFML) and dl-α-difluoromethylornithine (DFMO), which are irreversible inhibitors of LDC from Mycoplasma dispar (27) and eukaryotic ODC, respectively, resulting in growth inhibition due to the synthesis of the abortive peptidoglycan without cadaverine (11, 15). These observations suggested that S. ruminantium ODC could decarboxylate l-lysine, as well as l-ornithine. Accordingly, in our preceding study (32), we purified and characterized S. ruminantium LDC and found the following evidence. (i) S. ruminantium LDC comprises two identical monomeric subunits of 43 kDa. (ii) S. ruminantium LDC has decarboxylase activities toward both l-lysine and l-ornithine, with similar Km values. (iii) The decarboxylating activities toward l-lysine and l-ornithine are inhibited by either DFML or DFMO competitively and the catalytic domains for the enzyme activities toward both substrates are identical. We also showed in the preceding study that a drastic decrease in LDC activity occurred on entry into the stationary phase of cell growth, which was due to the degradation of LDC (32). This degradation resembled that of mouse ODC by an antizyme-mediated mechanism (8). These results indicate that S. ruminantium LDC resembles the eukaryotic ODC in both physicochemical and biochemical features except for its dual preference as a decarboxylase with l-lysine as well as l-ornithine with similar Km and Vmax values. In this study, we cloned and characterized the LDC gene (ldc) and showed that the amino acid sequence of S. ruminantium LDC is 35% identical and 53 to 60% similar to those of eukaryotic ODCs and that 26 amino acid residues, all of which are implicated in either contributing to pyridoxal phosphate (PLP)- and substrate-binding domains or in formation of the homodimeric forms of eukaryotic ODCs, are conserved in S. ruminantium LDC. From this information, we have now succeeded in converting S. ruminantium LDC to an enzyme with a preference for decarboxylating l-ornithine when five amino acid residues from the active site of S. ruminantium LDC were replaced with the corresponding residues found in mouse ODC.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The strains used in the present study included S. ruminantium subsp. lactilytica (18) and E. coli DH5α, which was used as the host strain in cloning experiments. Plasmids pUC119, Charomid 9-36 (Nippon Gene Co., Tokyo, Japan), and pTrc99A (Pharmacia) were used as the vector plasmids for subcloning and gene expression, respectively. Culture media included a tryptone-yeast extract-glucose medium (18) and a tryptone-yeast extract-lactate medium (18) and L broth for the growth of S. ruminantium and E. coli, respectively. S. ruminantium was grown under the anaerobic conditions described previously (18).
Purification of LDC from S. ruminantium.
The cells (125.7 g) derived from a 60-liter culture were suspended in 50 mM potassium phosphate buffer (pH 6.7; buffer A) containing 10 mM 2-mercaptoethanol and 0.1 mM PLP and disrupted in a French pressure cell. After centrifugation at 20,000 × g for 20 min, the supernatant was collected. Ammonium sulfate (3 M) in buffer A was added to the enzyme fraction at a final concentration of 1 M with stirring. After 2 h, the supernatant was collected by centrifugation and put on a TSKgel butyl-Toyopearl 650M column (4.0 by 24 cm). The column was washed with 3.5 liters of buffer A containing 1 M ammonium sulfate and 50 μM PLP. The enzyme was then eluted with a linear gradient created by mixing 750 ml of 1 M ammonium sulfate in buffer A with 750 ml of buffer A. The fractions which were eluted from 0.80 to 0.85 M ammonium sulfate were pooled. After 3 M ammonium sulfate in buffer A was added at a final concentration of 0.8 M, the enzyme preparation was put on a TSKgel phenyl-5PW column (21.5 by 150 mm; Tosoh, Tokyo, Japan). The column was washed with 450 ml of buffer A containing 0.8 M ammonium sulfate and then eluted with a linear gradient of ammonium sulfate in buffer A (0.8 to 0 M). The fractions eluted from 0.54 to 0.56 M ammonium sulfate were pooled. After being dialyzed against 20 mM potassium phosphate buffer containing 50 μM PLP (pH 6.5; buffer B), the enzyme preparation was put on a TSKgel DEAE-5PW column (7.5 by 75 mm; Tosoh) equilibrated with 20 mM potassium phosphate buffer (pH 6.5; buffer C). The column was washed with buffer C and then eluted with a linear gradient of NaCl in buffer C (0 to 0.4 M). The fractions eluted from 0.18 to 0.20 M NaCl were pooled and dialyzed against buffer B containing 20% glycerol.
Determination of the N-terminal and internal amino acid sequences of S. ruminantium LDC.
The purified LDC preparation (350 μg) was digested with 2.5 μg of lysyl endopeptidase (Wako Chemicals, Osaka, Japan) at 37°C for 12 h in 0.4 ml of 100 mM Tris-HCl buffer (pH 9.0). The digest was subjected to high-pressure liquid chromatography (HPLC) with a TSKgel ODS-120T column (4.6 by 250 mm; Tosoh) at 40°C using a linear gradient of acetonitrile in 0.1% trifluoroacetic acid (0 to 80%). Twenty-four fragments were obtained. Six fragments were analyzed for N-terminal amino acid sequencing by a gas-phase protein sequencer (model PSQ; Shimadzu, Kyoto, Japan) equipped with an on-line amino acid analyzer (model RF-550; Shimadzu).
Preparation of oligonucleotide primers for cloning of the ldc gene.
Oligonucleotide primers corresponding to the N-terminal (Glu8-Lys-Glu-Val-Lys-Thr-Leu-Ala15) amino acid sequence and three internal amino acid sequences of LDC (Glu-Glu-Asn-Tyr-Gln-Phe-Met, Ala-Asn-Pro-Thr-Pro-Glu-Ile, and Glu-Met-Gly-Ser-Tyr) were designed and synthesized as primers I [5′-GA(A,G) AA(A,G) GA(A,G) GTI AA(A,G) ACI (T,C)TI GC-3′], II [5′-GAG GAA AAC TAC CAG TTT ATG-3′], III [5′-AT (T,C)TC IGG IGT (G,A,T,C)GG (A,G)TT (G,A,T,C)GC-3′], and IV [5′-GT (A,G)TA (G,A,T,C)GA (G,A,T,C)CC CAT (T,C)TC-3′], respectively (in the sequences, I represents inosine). In combination with primers I and III and primers II and IV, 152- and 967-bp fragments, respectively, were amplified from the chromosomal DNA of S. ruminantium by PCR. By DNA sequencing of the amplified fragments, it was confirmed that the peptides described above correspond to the amino acid sequences of the 21-, 8-, 13-, 11-, 13-, and 9-residue segments M1-D21, V30-M37, A52-G64, G153-L163, T276-G288, and V344-Y352, respectively, of intact LDC. Therefore, we used these 152- and 967-bp fragments as DNA probes for genomic cloning of ldc from the S. ruminantium chromosomal DNA.
Cloning of ldc from chromosomal DNA of S. ruminantium.
The chromosomal DNA of S. ruminantium, which was isolated and purified by the standard method, was digested with EcoRI and resolved by 0.7% agarose gel electrophoresis. For Southern blot hybridization with the fluorescein-labeled 152- and 967-bp DNA fragments, the procedure was carried out using the ECL random prime labeling and detection systems (Amersham Life Science, Inc., Cleveland, Ohio). Only a single 4-kbp fragment was hybridized; this fragment was extracted from the gel and ligated into the EcoRI site of Charomid 9-36 vector DNA. After transduction into E. coli DH5α, a recombinant E. coli strain containing ldc was selected by colony hybridization using the labeled 152- and 967-bp DNA fragments. The plasmid containing ldc was designated pCLDC.
Nucleotide sequence determination.
The 4-kbp DNA was fragmented by digestion with various restriction enzymes; the resulting fragments were subcloned into plasmid pUC119 and sequenced. Analysis was done with a Thermo Sequenase cycle sequencing kit (Amersham) with IRD-41 dye-labeled M13 forward and reverse primers (Aloka, Ltd., Tokyo, Japan) using a model 4000 DNA sequencing system (Li-Cor, Lincoln, Nebr.).
ORF identification, homology search, and alignment of multiple nucleotide and amino acid sequences.
Protein and nucleotide sequences were compared with the sequence databases using the FASTA (version 3.0) and BLAST (version 1.49) programs implemented at the EMBL/GenBank/DDBJ nucleotide sequence databases and the SWISSPROT/NBRF-PIR protein sequence databases. Open reading frame (ORF) identification and multiple-sequence alignment were performed using the GENETYX program (Software Development Co., Tokyo, Japan).
RNA analysis.
For total cellular RNA isolation, S. ruminantium cells were grown in 50 ml of tryptone-yeast extract-glucose medium at 37°C under anaerobic conditions for various amounts of time. Total RNAs were extracted as described previously (1) and dissolved in 2 ml of distilled water and then stored at −80°C until used.
Primer extension analysis was done in accordance with the protocol of Aloka Ltd. using an IRD-41-labeled 19-mer complementary to nucleotides (nt) 1768 to 1786 (see Fig. 2B).
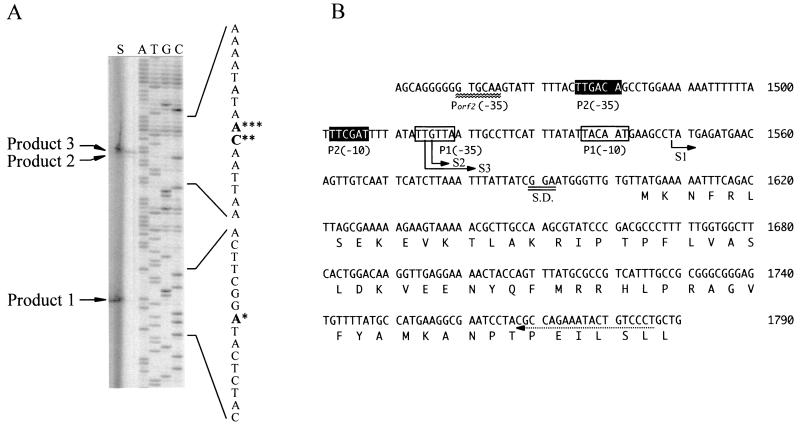
FIG. 2.
Determination of the ldc transcription start site by primer extension using a 19-mer complementary to nt 1768 to 1786, counting from the EcoRI site of the S. ruminantium chromosomal DNA sequence. The RNA preparation (30 μg) extracted from cells cultivated for 3 h was used for primer extension. Panel A (lane S) shows gel electrophoresis of primer extension products 1, 2, and 3. The sequence shown next to the sequencing gel is complementary to that displayed in panel B. The single, double, and triple asterisks indicate the bases complementary to the 5′ end of the transcript for S1, S2, and S3, respectively. Panel B represents the nucleotide sequence (nt 1451 to 1790) containing the putative promoter site (−35 region) for ORF2 and the beginning of ldc. The dashed arrow indicates the region complementary to the primer used. The transcription start sites (boldface T, G, and T) determined here (nt 1558, 1516, and 1515) are indicated by S1, S2, and S3. S.D., Shine-Dalgarno sequence.
Assay for LDC and ODC activities and kinetic analysis.
LDC and ODC activities were measured by the amount of cadaverine or putrescine formed from l-lysine or l-ornithine by HPLC using a TSK-gel Polyaminepak column (Tosoh) as described previously (13). The standard assay was done at saturating PLP (50 μM) and lysine or ornithine concentrations ranging from 0.15 to 500 mM, depending on the Km of the enzyme being tested. The incubation was done at 30°C, pH 6.0.
Construction of plasmid pTLDC for LDC expression in a trc promoter expression system.
For expression of recombinant LDC (rLDC), a DNA fragment containing only the ldc structural gene was amplified from chromosomal DNA by PCR using primers 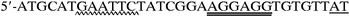
 (primer V) and
(primer V) and 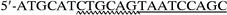
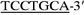 (primer VI), which are located upstream and downstream of ldc, respectively. In primer V, wavy underlining and double underlining represent the EcoRI site and the ribosomal binding site which replaced the original Shine-Dalgarno sequence of ldc (see Fig. 1), respectively. On the other hand, wavy underlining and single underlining in primer VI indicate the PstI site and the complementary sequence at positions 2892 to 2909 (see dotted underlining in Fig. 1), respectively. The amplified fragment was digested with EcoRI and PstI, and the EcoRI-PstI fragment was inserted into the EcoRI-PstI site of expression vector pTrc99A to construct plasmid pTLDC, in which ldc was under the control of the trc promoter. pTLDC was then introduced into E. coli DH5α. The ldc region of pTLDC was sequenced, and it was confirmed that there was no point mutation in the region.
(primer VI), which are located upstream and downstream of ldc, respectively. In primer V, wavy underlining and double underlining represent the EcoRI site and the ribosomal binding site which replaced the original Shine-Dalgarno sequence of ldc (see Fig. 1), respectively. On the other hand, wavy underlining and single underlining in primer VI indicate the PstI site and the complementary sequence at positions 2892 to 2909 (see dotted underlining in Fig. 1), respectively. The amplified fragment was digested with EcoRI and PstI, and the EcoRI-PstI fragment was inserted into the EcoRI-PstI site of expression vector pTrc99A to construct plasmid pTLDC, in which ldc was under the control of the trc promoter. pTLDC was then introduced into E. coli DH5α. The ldc region of pTLDC was sequenced, and it was confirmed that there was no point mutation in the region.
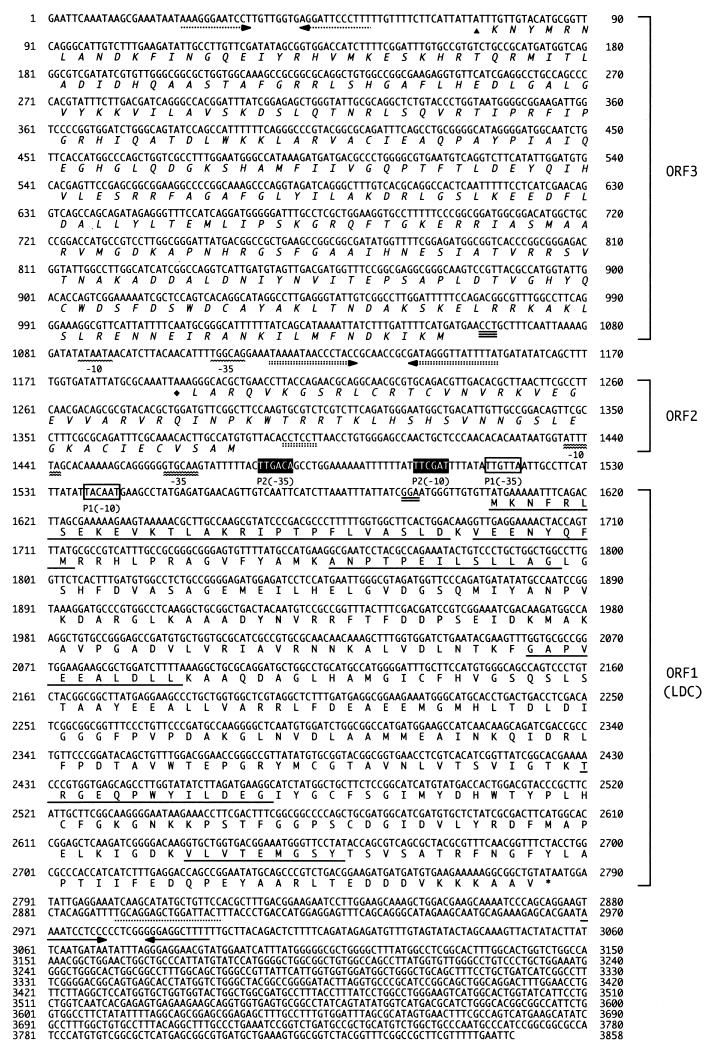
FIG. 1.
Nucleotide sequence of the ldc gene of S. ruminantium and the flanking region. The strand shown is in the 5′-to-3′ direction. The deduced amino acid sequence is indicated by the single-letter code under the nucleotide sequence. For ldc, the amino acid sequence, putative ribosomal binding site, promoter sequences, translation termination codon, and transcription termination sequences are indicated by single underlining, double underlining, boxes (white boxes, promoter 1; black-shaded boxes, promoter 2), a single asterisk, and horizontal arrows, respectively. For ORF2 and ORF3, ribosome-binding sequences, promoter sequences, translation termination codons, and transcription termination sequences are indicated by double-dotted and triple underlining, double and single wavy lines, diamonds and triangles, and double- and single-dotted inverted arrows, respectively.
Purification of rLDC.
Cells of E. coli DH5α(pTLDC) were grown in 2× TY (1.6% Bacto Tryptone–1.0% Bacto Yeast Extract–0.5% NaCl) medium containing ampicillin (100 μg/ml) at 37°C for 20 h with shaking and were collected by centrifugation at 10,000 × g for 5 min at 4°C. rLDC was purified to electrophoretic homogeneity from a sonicated extract of the cells by the method used for the purification of native LDC from S. ruminantium. This method was also applicable to the purification of mutant LDC obtained by site-directed mutagenesis of ldc.
Site-directed mutagenesis.
Site-directed mutagenesis was done by PCR using the oligonucleotide primer shown in Table 1 together with either primer VI or V, used to create plasmid pTLDC. The PCR products were digested with both SacII (Table 1) and PstI, both EcoRI and ClaI, or both EcoRI and NruI, and the mutant SacII-PstI, EcoRI-ClaI, or EcoRI-NruI fragments were used to replace the SacII-PstI, EcoRI-ClaI, or EcoRI-NruI fragments from pTLDC which had been digested with SacII and PstI, EcoRI and ClaI, or EcoRI and NruI. Mutations were confirmed by DNA sequencing.
TABLE 1.
Oligonucleotides used for site-directed mutagenesis of the ldc gene
 |
See Fig. 5.
The boxed sequences are SacII, ClaI, and NruI restriction sites. The underlined boldface sequences are those that were replaced.
Other analytical procedures.
Protein was measured by the method of Bradford (3), with bovine serum albumin as the standard. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was done as described previously (20). Circular dichroism (CD) analysis of the wild-type and mutant LDCs was done with a Jasco J-720 spectropolarimeter at room temperature in a 1-mm path length cell containing 5 μM enzyme in 5 mM potassium phosphate buffer (pH 6.5) containing 50 μM PLP. Secondary-structure computer analysis of S. ruminantium LDC was done by the new-joint method (10, 23).
Nucleotide sequence accession number.
The nucleotide sequence of ldc has been deposited in the DDBJ/EMBL/GenBank nucleotide sequence databases under accession number AB011029.
RESULTS AND DISCUSSION
Cloning and nucleotide sequencing of ldc.
We cloned ldc by obtaining N-terminal and internal amino acid sequence information from purified S. ruminantium LDC, designing appropriate oligonucleotide primers, and preparing labeled DNA fragments obtained by subsequent PCR. The screening of an EcoRI library gave a single positive clone from approximately 100,000 colonies. The positive clone contained an insert of 3,858 bp whose sequence included three ORFs. The first ORF encoded a 393-amino-acid protein of 43,213 Da whose N-terminal sequence matched that determined for purified LDC and its fragments (Fig. 1, underlined); later functional tests (described below) verified the assignment of this ORF as ldc. The ldc gene, spanning positions 1605 to 2786 within the genomic clone (pCLDC), started with an ATG codon and ended with a TAA stop codon. Thirteen base pairs upstream from the ATG codon, a ribosome-binding site consensus sequence (GGA) was found at positions 1590 to 1592. After analysis of the transcription start sites as determined by primer extension (described below), two putative promoter regions of ldc were identified, one at positions 1537 to 1542 (TACAAT) and 1514 to 1519 (TTGTTA) for the respective −10 and −35 sequences (Fig. 1, white boxes) and the second at positions 1502 to 1507 (TTCGAT) and 1476 to 1481 (TTGACA) for the −10 and −35 sequences (Fig. 1, black-shaded boxes). An inverted-repeat sequence consisting of a stem-loop with an 11-bp arm appeared at positions 2970 to 2997, as indicated by the GENETYX program. It is probably used as a transcription terminator for mRNA of ldc.
The second ORF (ORF2 in Fig. 1), consisting of 192 nt, was found at positions 1381 to 1190 in the antisense DNA strand. ORF2 encoded a 63-amino-acid protein which is tentatively called protein 2. From the primary structure of protein 2, 44% identity was observed in an alignment with the sequence of ribosomal protein L28 from E. coli (21).
The third ORF (ORF3 in Fig. 1), consisting of 987 nt, extended from position 1056 to position 70 in the antisense DNA strand. ORF3 encoded a 328-amino-acid protein which is tentatively called protein 3. The amino acid sequence of protein 3 exhibited 34% overall identity with that of E. coli recombinase XerD (2).
Functional analysis of the ldc gene product.
To prove that ldc is the gene encoding S. ruminantium LDC, the gene was expressed in E. coli; rLDC was purified, and its characteristics were compared with that of the earlier S. ruminantium LDC preparation (32). E. coli DH5α(pTLDC) expressed ldc at a final concentration of approximately 2% of its total protein. The purified rLDC preparation had characteristics identical to those of the earlier S. ruminantium LDC preparation, as judged by the molecular mass of the native enzyme (88 kDa), its subunit mass (43 kDa) determined by SDS-PAGE, and the N-terminal 25-residue and internal amino acid sequences underlined in Fig. 1. When the decarboxylase activities of rLDC toward l-lysine and l-ornithine were studied, the following became evident: (i) rLDC decarboxylated both substrates with Km values identical to those obtained with the original LDC preparation from S. ruminantium, and (ii) the decarboxylase activities toward both substrates were inhibited competitively by both DFML and DFMO with a Ki identical to that observed for the S. ruminantium LDC preparation (32). Thus, we concluded that ORF1 is the LDC-encoding gene (ldc) of S. ruminantium.
Transcriptional start site of ldc.
The transcriptional start site of ldc was determined by primer extension analysis of mRNA isolated from S. ruminantium (Fig. 2A). The ldc transcriptional start sites were identified as T at position 1549 (S1), G at position 1516 (S2), and T at position 1515 (S3) (Fig. 2B). Looking further upstream of these start sites, putative promoter structures 5′-TTGTTA(17 bp)TACAAT-3′ for S1 and 5′-TTGACA(20 bp)TTCGAT-3′ for S2 and S3 were identified with 7, 9, and 8 bp separating the −10 region of the promoter structure and the respective transcriptional start sites.
Amino acid sequence homologies of LDC with eukaryote ODCs.
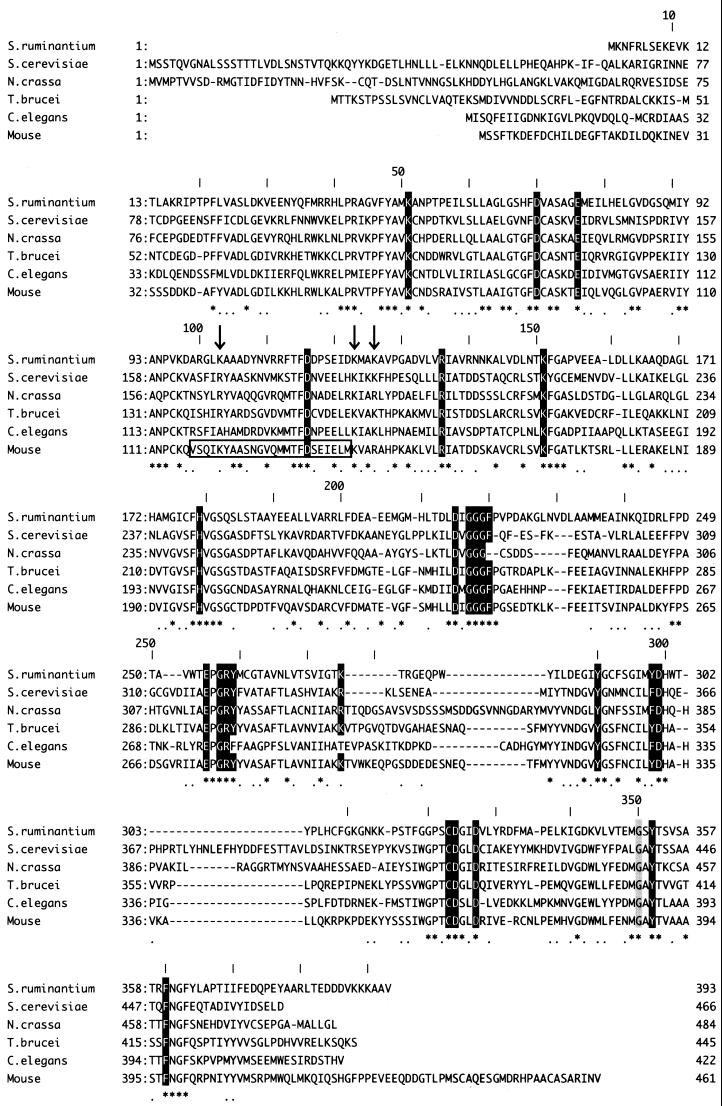
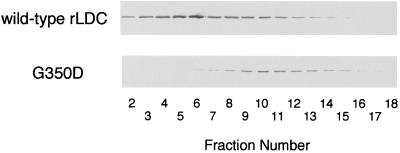
The amino acid sequence of S. ruminantium LDC had no similarity to that of the other reported bacterial LDCs and ODCs but showed 35% identity and approximately 60% similarity to eukaryotic ODCs, including the mouse, Saccharomyces cerevisiae, Neurospora crassa, Trypanosoma brucei, and Caenorhabditis elegans ODCs (Fig. 3). In addition, absolute conservation among those amino acid residues implicated in homodimer stabilization, in the PLP- and substrate-binding domains, were found for S. ruminantium LDC and mouse ODC as follows. (i) From the crystal structure (19), mouse ODC was found to consist of a symmetrical homodimer which is formed by a head-to-tail interaction between the barrel domain of one monomer and the sheet domain of the second. The two salt bridges (K169-D364′ and D134-K294′ [the primed residue belongs to the second subunit in the dimer]) and the stack of aromatic residues (F397′/Y323′/Y331), which are involved in stabilization of the dimer by formation of the primary interaction between monomers of mouse ODC, were conserved in S. ruminantium LDC (K151-D327′ and D116-K275′ and F360′/Y290′/Y298) (Fig. 3). (ii) G387 in mouse ODC, which is located between the barrel and sheet domains and is crucial for stabilization of the homodimer (33), was also conserved in S. ruminantium LDC (G350; shaded gray in Fig. 3). Mutation of G350 to D350 in S. ruminantium LDC caused the loss of both dimer formation (Fig. 4) and decarboxylase activity (data not shown). (iii) Eighteen amino acid residues, K69, D88, E94, R154, H197, D233, G235, G236, G237, F238, E274, G276, R277, Y278, D332, C360, D361, and Y389 (mouse ODC numbering) (shaded black in Fig. 3), were identified as the crucial active-site residues, especially residues K69 and E274, which interact with PLP by forming an internal Schiff base and by stabilizing the protonated pyridine nitrogen of PLP (19), respectively, in mouse ODC. Residues C360 and D361 function in the binding of ornithine as a substrate in the active center in eukaryotic ODCs (19). All of these residues were conserved in S. ruminantium LDC (Fig. 3). (iv) In S. ruminantium LDC, a unique five-residue sequence (T276-R277-G278-E279-Q280) corresponding to a five-residue sequence (S303-D304-D305-E306-D307) which had been reported to be phosphorylated [consensus sequence, S(or T)XXE(or D)X] by casein kinase II in mouse ODC (29) was also conserved. (v) Residues 117 to 140 in mouse ODC were identified as the antizyme-binding region (Fig. 3, large box), in which basic residues K121, K141, and R144 have been suggested to be pivotal in antizyme binding (19). In S. ruminantium LDC, basic residues K103, K123, and K126 correspond to these amino acids in mouse ODC (Fig. 3, residues marked with vertical arrows). In the preceding paper (32), we observed a dramatic decrease in the LDC protein level in early stationary phase due to rapid degradation. The presence of the proposed antizyme recognition site in S. ruminantium LDC suggests the presence of a similar mechanism for its degradation during entry into the stationary phase of S. ruminantium cells.
FIG. 3.
Amino acid sequence alignment of S. ruminantium LDC with ODCs of various eukaryotes. S. ruminantium LDC exhibits 60% similarity to eukaryotic ODCs, including the S. cerevisiae, N. crassa, T. brucei, C. elegans, and mouse ODCs. On the consensus line below the aligned sequences, identical amino acids are indicated by asterisks and conserved residues are indicated by dots. Amino acids required for formation of the active sites of the ODCs are boxed. The residues marked in black and gray and vertical arrows are explained in the text. A large box in the sequence of mouse ODC represents the region to which antizyme is bound.
FIG. 4.
Analysis of wild-type rLDC and G350D mutant rLDC by gel filtration. Cells of E. coli DH5α harboring plasmid pTLDC or pG350D were grown in 4 ml of 2× TY medium containing ampicillin (100 μg/ml) at 37°C for 18 h and collected by centrifugation at 10,000 × g for 5 min at 4°C. The cells were suspended in 2 ml of dimer buffer (20 mM potassium phosphate [pH 6.7] containing 0.5 mM EDTA, 1 mM dithiothreitol, 0.1 M NaCl, 50 μM PLP, 0.5 mM ornithine) (33) and sonicated. The sonicated extracts were filtrated by column chromatography with a Superdex 200 HR 10/30 column (Pharmacia) equilibrated with dimer buffer. Proteins in the fractions were analyzed by SDS-PAGE and then transferred to nitrocellulose membrane (Hybond-C pure; Amersham), and the LDC protein was visualized immunologically using anti-LDC antibodies and anti-rabbit immunoglobulin G(Fc)-alkaline phosphatase conjugate (Seikagaku Kogyo Co., Tokyo, Japan).
Comparison of the secondary-structure assignments of S. ruminantium LDC and a truncated form of mouse ODC in which the C-terminal 37 residues are missing revealed approximately 70% secondary-structure identity overall (data not shown). This suggests a strong similarity between the three-dimensional structures of S. ruminantium LDC and eukaryote ODCs. Grishin et al. (6) proposed a topographic model of the PLP-binding domain of eukaryotic ODC through the analysis of known barrel structures of ODC. They classified the PLP-utilizing enzymes into seven fold types from the predicted secondary structures, and they showed that the eukaryotic ODCs belong to fold type III while bacterial ODCs and LDCs belong to fold type I. We propose that S. ruminantium LDC belongs to fold type III.
Most recently, whole genome sequences of Aquifex aeolicus (4) and Thermotoga maritima (22), which have been placed as the deepest and most slowly evolving lineages among the Eubacteria (24), were reported. After a homology search of the amino acid sequences corresponding to putative ODCs from these bacteria, we noticed that the putative ODCs of these bacteria show 35% identity overall with S. ruminantium LDC. Interestingly, in T. maritima ODC, 22 of the 26 amino acid residues, K69, D88, E94, D134, R154, K169, H197, G235, G236, G237, F238, E274, G276, R277, Y278, K294, C360, D361, D364, G387, Y389, and F397 (mouse ODC numbering), which were described above as being essential for ODC activity, are also conserved, with D233 and Y323 of S. ruminantium LDC being replaced with N and F in T. maritima ODC. Taken together with the 35% sequence identity between S. ruminantium LDC and eukaryotic ODC, these observations indicate the occurrence of a bacterial group which includes the eukaryotic ODC in the putative bacterial ODCs. The data also suggest that the origin of the ODCs of S. ruminantium and the two species of eubacteria mentioned above is completely different from that of the ODCs of general bacteria, including E. coli, and that eukaryotic ODC and S. ruminantium LDC have the same origin.
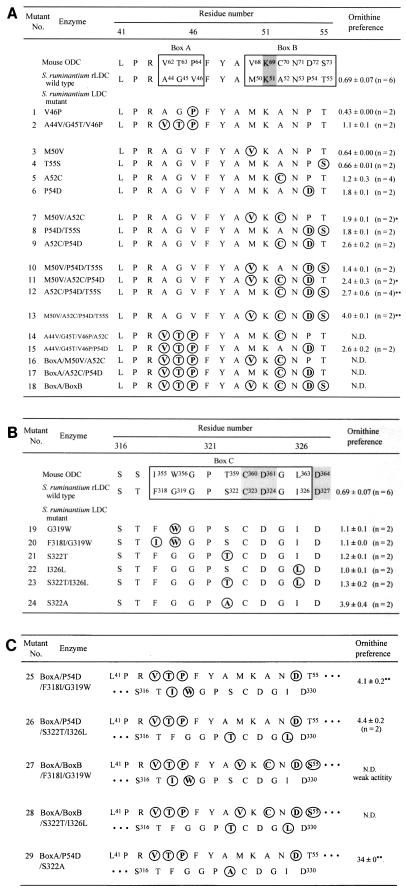
Identification of the amino acid residues conferring substrate specificity upon S. ruminantium LDC.
Eukaryotic ODC has absolute specificity as a decarboxylase for l-ornithine. In contrast, S. ruminantium LDC decarboxylates both l-lysine and l-ornithine with similar Km and Vmax values (32). Here, we identified the amino acid residue(s) in the catalytic domain of LDC that confers decarboxylase activity toward l-ornithine and/or l-lysine. To identify the amino acid residue(s) conferring substrate specificity on S. ruminantium LDC, we considered 15-residue and 12-residue segments neighboring K51 (K69 in mouse ODC) and C323 (C360 in mouse ODC), respectively, of the catalytic domain of S. ruminantium LDC (Fig. 5) as candidates for regulators of the substrate specificity of S. ruminantium LDC. In the PLP-binding domain, residue K51 (K69 in mouse ODC) could be crucial for binding to PLP by forming an internal Schiff base between the ɛ-NH2 of K69 and the aldehyde group of PLP based on the evidence from eukaryotic ODCs (19, 28). The internal Schiff base is replaced with an external Schiff base between l-ornithine and PLP when the enzyme is incubated with l-ornithine as a substrate. In the 15-residue alignment centered around K51 and K69 between S. ruminantium LDC and mouse ODC, two regions that contain different amino acid residues, i.e., (i) A44 G45 V46 and V62 T63 P64 (box A in Fig. 5A) and (ii) M50 K51 A52 N53 P54 T55 and V68 K69 C70 N71 D72 S73 (box B in Fig. 5A) (residues differing between them are underlined), were found. On the other hand, four different amino acid residues (underlined), i.e., F318 G319 G320 P321 S322 C323 D324 G325 I326 and I355 W356 G357 P358 T359 C360 D361 G362 L363 were found in the 12-residue segment including C323 (Fig. 5B). To find whether or not the residues conferring the dual-specificity decarboxylase activity on S. ruminantium LDC exist in these segments, we generated 29 mutant LDCs corresponding to the amino acid residues in boxes A, B, and C individually or all together by constructing plasmids containing a series of mutant genes by site-directed mutagenesis using the primers shown in Table 1. As a result of the manipulations, 29 different plasmids which correspond to the mutant proteins listed in Fig. 5A, B, and C were obtained. The mutant proteins were expressed in E. coli DH5α harboring the appropriate plasmid. First, we measured the decarboxylase activities of mutant LDCs 1 through 13 toward l-lysine and l-ornithine after 10 min of incubation at 37°C, using the sonicated extracts, and calculated the ODC/LDC activity ratio. With respect to the mutations in boxes A and B, mutants 1, 3, and 4, mutant 7, mutants 8 and 10, and mutants 11 and 12 showed ODC/LDC activity ratios similar to those of wild-type LDC, mutant 5, mutant 6, and mutant 9, respectively (Fig. 5A). Therefore, mutant proteins 2, 5, 6, 9, and 13 were purified to electrophoretic homogeneity from the crude extract of the recombinant E. coli cells and used in kinetic analyses. The purified recombinant wild-type and mutant protein preparations were analyzed by CD spectrometry to determine whether global structural changes were induced by the mutations. The far-UV CD spectra of the mutant LDCs were similar to that of wild-type LDC (data not shown). These results suggest that no major conformational alterations occurred in the mutant enzymes.
FIG. 5.
Comparison of amino acid sequences conferring substrate specificity upon eukaryote ODCs or S. ruminantium LDC. S. ruminantium LDC numbering is used to define residue position numbers. Residues K51 and C323 match K69 and C360 in mouse ODC, respectively. The mutated positions are in boldface and circled. Ornithine preference is expressed as the ODC/LDC activity ratio. Cells of E. coli DH5α harboring each plasmid were grown in 2 ml of LB medium containing ampicillin (100 μg/ml) at 37°C for 18 h and collected by centrifugation at 4°C. Cells suspended in 1 ml of 20 mM potassium phosphate buffer (pH 7.0) containing 0.1 mM PLP and 5 mM dithiothreitol were sonicated. The LDC or ODC activity in cell extracts was measured by the amount of cadaverine or putrescine formed from lysine or ornithine by HPLC using a TSK-gel Polyaminepak column as described in Materials and Methods. The incubation mixture contained 0.5 M sodium acetate buffer (pH 6.0), 5 mM l-lysine or l-ornithine, 50 μM PLP, and the cell extracts in a total volume of 50 μl. The reaction mixture was incubated at 37°C for 10 min. Under these conditions, the ODC and LDC activities of E. coli itself were almost negligible. Each value is the mean ± the standard deviation. In parentheses, n represents the number of experiments. Asterisks indicate mutations that caused loss of specific activities. The ODC activities of mutants 7 and 11 (∗) and mutants 12 and 13 (∗∗) were about 1/20 and 1/4, respectively, of that of wild-type LDC. N.D., not detected.
By determination of the kcat/Km ratio (catalytic efficiency), one can obtain a lower limit of the second-order rate constant for conversion of a substrate to a product (25). Combining effects due to substrate binding and transition state stabilization, this parameter is useful for assessment of altered substrate specificity. The kinetic parameters for decarboxylation of l-lysine and l-ornithine by wild-type and mutant LDCs were determined from the initial rate measurements, and the results are listed in Table 2. When the substrate specificity was defined as the kcat/Km ratio obtained with l-ornithine relative to that obtained with l-lysine as the substrate, the following findings became evident. (i) Mutant 2, in which all three of the amino acid residues of S. ruminantium LDC in box A were replaced with the corresponding ones of mouse ODC, showed a substrate specificity ratio of 2.0 (Table 2, line 3). (ii) Mutant 13, in which residues M50, A52, P54, and T55 of LDC were replaced with corresponding residues V68, C70, D72, and S73 of mouse ODC, converting the entire series of residues in box B of LDC to the corresponding ones of ODC, had a substrate specificity ratio of 1.5 (Table 2, line 7). (iii) Box B single mutants 5 and 6 showed substrate specificity ratios of 1.0 and 2.2, respectively (Table 2, lines 4 and 5), suggesting that P54 is involved in conferring substrate specificity on S. ruminantium LDC. (iv) Double mutant 9 (A52C/P54D) had a substrate specificity ratio of 1.6 (Table 2, line 6). These data suggest that the three-residue segment of box A and P54 of box B together are involved in conferring decarboxylase activities toward both l-lysine and l-ornithine on S. ruminantium LDC. Accordingly, we created mutant A44V/G45T/V46P/P54D, in which all of the amino acid residues of S. ruminantium LDC in box A and P54 in box B were replaced with the corresponding residues of mouse ODC. The mutant protein was expressed in E. coli with plasmid pA44V/G45T/V46P/P54D, purified to electrophoretic homogeneity, and then analyzed for decarboxylase activity toward both substrates. The resulting mutant, 15, showed a substrate specificity ratio of 3.8, which is 4.6 times that of wild-type S. ruminantium LDC (Table 2, line 8). Mutant 18, in which all of the residues of LDC in both boxes A and B were replaced with the corresponding residues from mouse ODC, had no enzyme activity toward either l-lysine or l-ornithine. Thus, it was concluded that in the N-terminal domain of the active site of S. ruminantium LDC, residues A44, G45, V46, and P54 together confer higher decarboxylating activity toward l-lysine than toward l-ornithine upon S. ruminantium LDC.
TABLE 2.
Kinetic parameters for the decarboxylation of lysine and ornithine by wild-type and mutant LDCs and mouse ODCa
| Line no. | Mutant no. | Enzyme |
Km (mM)
|
kcat (s−1)
|
kcat/Km ratio
|
Substrate specificity | |||
|---|---|---|---|---|---|---|---|---|---|
| Lysine | Ornithine | Lysine | Ornithine | Lysine | Ornithine | ||||
| 1 | Mouse ODCb | 5.2 | 0.090 | 1.8 | 7.8 | 350 | 87,000 | 250 | |
| 2 | Wild-type rLDC | 1.5 | 0.96 | 16 | 8.6 | 11,000 | 8,900 | 0.83 | |
| 3 | 2 | A44V/G45T/V46P | 3.0 | 0.98 | 13 | 8.4 | 4,300 | 8,600 | 2.0 |
| 4 | 5 | A52C | 1.9 | 1.3 | 10 | 7.0 | 5,500 | 5,500 | 1.0 |
| 5 | 6 | P54D | 5.7 | 2.1 | 16 | 13 | 2,900 | 6,300 | 2.2 |
| 6 | 9 | A52C/P54D | 2.4 | 1.6 | 6.8 | 7.2 | 2,900 | 4,500 | 1.6 |
| 7 | 13 | M50V/A52C/P54D/T55S | 3.6 | 4.5 | 2.6 | 4.7 | 700 | 1,000 | 1.5 |
| 8 | 15 | A44V/G45T/V46P/P54D | 8.0 | 3.1 | 9.8 | 14 | 1,200 | 4,700 | 3.8 |
| 9 | 19 | G319W | 4.0 | 1.3 | 5.6 | 7.2 | 1,400 | 5,400 | 3.9 |
| 10 | 23 | S322T/I326L | 17 | 1.5 | 21 | 24 | 1,200 | 16,000 | 13 |
| 11 | 24 | S322A | 6.7 | 1.2 | 4.8 | 20 | 720 | 17,000 | 24 |
| 12 | 26 | A44V/G45T/V46P/P54D/S322T/I326L | 22 | 3.3 | 19 | 36 | 850 | 11,000 | 13 |
| 13 | 29 | A44V/G45T/V46P/P54D/S322A | 270 | 4.0 | 5.5 | 4.8 | 21 | 1,200 | 58 |
Each reaction was carried out in 0.5 M sodium acetate buffer containing 50 μM PLP and 0.1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS; pH 6.0) at 30°C. Substrate specificity was defined as the kcat/Km ratio obtained with ornithine relative to that obtained with lysine as the substrate.
Described in reference 25.
Next, the C-terminal domain in the active center of S. ruminantium LDC was examined. Sonic extracts from the cells in which the mutant LDCs were expressed were used as enzyme preparations, the decarboxylase activities of mutant LDCs 19 through 24 toward l-lysine and l-ornithine after 10 min of incubation at 37°C were measured, and the ODC/LDC activity ratio was calculated. Mutant LDCs 19 to 23 showed the same ODC/LDC activity ratio, which was twice that of wild-type LDC (Fig. 5B). Mutant 24 (S322A) had about 5.7 times the l-ornithine decarboxylation activity of wild-type LDC. Mutant proteins 19, 23, and 24 were purified to electrophoretic homogeneity, and their decarboxylase activities toward both substrates were measured; the substrate specificity ratios were 3.9, 13, and 24, respectively. Thus, we concluded that in the C-terminal domain of the active site of S. ruminantium LDC, residue S322 also contributes to the higher decarboxylating activity toward l-lysine.
Finally, we generated mutants 26 and 29 (double mutants of mutants 15 and 23 and mutants 15 and 24, respectively). The two mutant proteins were purified, Km and kcat were measured, and the substrate specificity ratios were calculated. Surprisingly, mutant 29 showed a Km value of 270 for l-lysine, 180 times that of wild-type rLDC, resulting in a substrate specificity ratio of 58, which is approximately 70 times that of wild-type rLDC. The substrate specificity of mutant 26 was the same as that of mutant 23. Thus, it was concluded that five amino acid residues (A44, G45, V46, P54, and S322) confer the dual decarboxylation activities toward both l-lysine and l-ornithine in the case of S. ruminantium LDC and to similar extents. Consequently, we have succeeded in converting S. ruminantium LDC to an enzyme with a preference for l-ornithine with a substrate specificity ratio of approximately 58 by constructing mutant protein A44V/G45T/V46P/P54D/S322A.
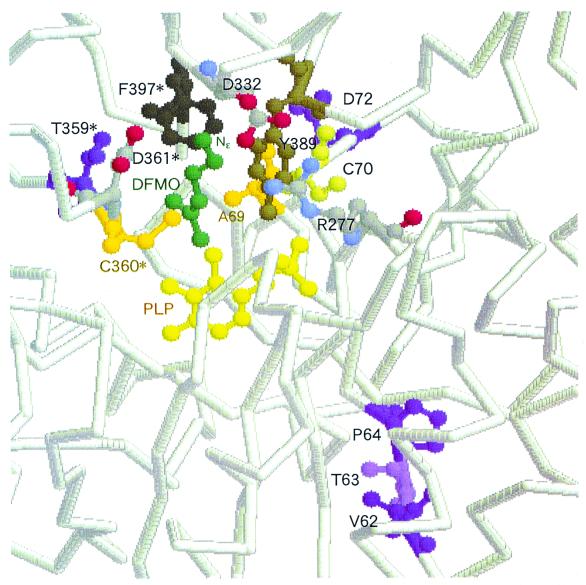
The role of each substitution may be proposed from an analysis of the X-ray crystal structure of T. brucei ODC (TbODC) complexed with a substrate analog, DFMO (Fig. 6 and reference 7). The Nɛ of DFMO is bound between two acidic residues, D361 and D332, which are contributed to the active site by opposite subunits. S322 in box C (T359 in TbODC) might influence substrate binding through second-shell interaction with the D361 loop. Since the other residues in boxes A and B, A44, G45, V46, and P54, which correspond to V62, T63, P64, and D72 in TbODC, respectively, are positioned farther away from the substrate, it is more difficult to speculate on their effects. The X-ray structure of TbODC also shows that the aliphatic portion of DFMO is cradled by the aromatic rings of F397 and Y389 and also forms a hydrogen bond with the PLP phosphate and that R277 is also involved in substrate binding as a second-shell residue. A salt bridge is formed between R277 and D332 (2.9 Å), the residue that forms a hydrogen bond with Nɛ of DFMO. A52 in box B (C70 in TbODC) may therefore influence substrate binding through second-shell interactions with Arg277 and/or Y389. However, mutation of P54, the residue positioned farther than A52 from the substrate, had a more significant effect on substrate specificity (Table 2, lines 4 and 5). Because the residues in box A are 12 Å below the PLP ring, it is impossible to speculate on the effect of the A44V/G45T/V46P substitutions. It is likely that the residues which seem not to be in a position to influence substrate binding have an effect on substrate specificity alteration. Yano et al. (34) modified the substrate specificity of aspartate aminotransferase, and they identified six amino acid residues which contributed to the substrate specificity change. Interestingly, only one of the six residues was located at a distance allowing direct interaction with the substrate and the other residues were positioned farther away from the substrate, even though one of these was located >10 Å from the substrate-binding site.
FIG. 6.
Active site of K69A T. brucei ODC in complex with DFMO. The figure was drawn based on the data of reference 7 using the RasMol 2.6 Molecular Graphics Visualisation tool (Glaxo Wellcome Research & Development Co., Stevenage, Herts., United Kingdom). In reference 7, the N terminus of recombinant T. brucei ODC corresponds to the 21st residue of intact ODC (Fig. 3). Residues (V62, T63, P64, D72, and T359) which correspond to the residues mutated in this study and were identified as functionally important for substrate specificity in S. ruminantium LDC are purple and violet. The atoms of PLP are yellow, the atoms of DFMO are green, A69 and C360 are orange, and Y389 and F397, which cradle the aliphatic portion of DFMO, are brown. R277, D332, and D361 are also shown, and nitrogen and oxygen atoms of these residues are blue and red, respectively. The asterisked residues belong to the other subunit of the dimer.
Based on the X-ray structure of TbODC, Grishin et al. (7) have suggested that the distance between the Schiff base nitrogen and the D361-D332 pair may act as a ruler to select for a side chain with a length equivalent to that of l-ornithine. Our studies are focused on the X-ray diffraction analysis of wild-type DFML-bound and uncomplexed S. ruminantium rLDC and mutant 29 in order to improve our understanding of the relationship between the protein structure and l-lysine and l-ornithine recognition of S. ruminantium LDC.
ACKNOWLEDGMENTS
We thank Al Claiborne for careful reading and helpful comments on the manuscript and Toru Nakayama of Tohoku University for his fruitful discussion.
Y.T. was a recipient of a predoctoral fellowship from the Japan Society for the Promotion of Science and is supported by a postdoctoral fellowship from the Suzuki Foundation.
REFERENCES
- 1.Aiba H, Sdhya S, de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981;256:11905–11910. [PubMed] [Google Scholar]
- 2.Blattner F R, Plunkett G, III, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olson G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 5.dela Vega A L, Delcour A H. Cadaverine induces closing of E. coli porins. EMBO J. 1995;14:6058–6065. doi: 10.1002/j.1460-2075.1995.tb00294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grishin N V, Phillips M A, Goldsmith E J. Modeling of the spatial structure of eukaryotic ornithine decarboxylase. Protein Sci. 1995;4:1291–1304. doi: 10.1002/pro.5560040705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grishin N V, Osterman A L, Brooks H B, Phillips M A, Goldsmith E J. X-ray structure of ornithine decarboxylase from Trypanosoma brucei: the native structure and the structure in complex with α-difluoromethylornithine. Biochemistry. 1999;38:15174–15184. doi: 10.1021/bi9915115. [DOI] [PubMed] [Google Scholar]
- 8.Hayashi S, Murakami Y, Matsufuji S. Ornithine decarboxylase antizyme: a novel type of regulatory protein. Trends Biochem Sci. 1996;21:27–30. [PubMed] [Google Scholar]
- 9.Hirao T, Sato M, Shirahata A, Kamio Y. Covalent linkage of polyamines to peptidoglycan in Anaerovibrio lipolytica. J Bacteriol. 2000;182:1154–1157. doi: 10.1128/jb.182.4.1154-1157.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito M, Matsuo Y, Nishikawa K. Prediction of protein secondary structure using the 3D–1D compatibility algorithm. Comput Appl Biosci. 1997;13:415–424. doi: 10.1093/bioinformatics/13.4.415. [DOI] [PubMed] [Google Scholar]
- 11.Kamio Y. Structural specificity of diamines covalently linked to peptidoglycan for cell growth of Veillonella alcalescens and Selenomonas ruminantium. J Bacteriol. 1987;169:4837–4840. doi: 10.1128/jb.169.10.4837-4840.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamio Y, Itoh Y, Terawaki Y. Chemical structure of peptidoglycan in Selenomonas ruminantium: cadaverine links covalently to the d-glutamic acid residue of peptidoglycan. J Bacteriol. 1981;146:49–53. doi: 10.1128/jb.146.1.49-53.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamio Y, Itoh Y, Terawaki Y, Kusano T. Cadaverine is covalently linked to peptidoglycan in Selenomonas ruminantium. J Bacteriol. 1981;145:122–128. doi: 10.1128/jb.145.1.122-128.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamio Y, Nakamura K. Putrescine and cadaverine are constituents of peptidoglycan in Veillonella parvula. J Bacteriol. 1987;169:2881–2884. doi: 10.1128/jb.169.6.2881-2884.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamio Y, Pösö H, Terawaki Y, Paulin L. Cadaverine covalently linked to a peptidoglycan is an essential constituent of the peptidoglycan necessary for the normal growth in Selenomonas ruminantium. J Biol Chem. 1986;261:6585–6589. [PubMed] [Google Scholar]
- 16.Kamio Y, Terawaki Y. Purification and properties of Selenomonas ruminantium lysine decarboxylase. J Bacteriol. 1983;153:658–664. doi: 10.1128/jb.153.2.658-664.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamio Y, Terawaki Y, Izaki K. Biosynthesis of cadaverine-containing peptidoglycan in Selenomonas ruminantium. J Biol Chem. 1982;257:3326–3333. [PubMed] [Google Scholar]
- 18.Kanegasaki S, Takahashi H. Function of growth factors for rumen microorganisms. I. Nutritional characteristics of Selenomonas ruminantium. J Bacteriol. 1967;93:456–463. doi: 10.1128/jb.93.1.456-463.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kern A D, Oliveira M A, Coffino P, Hackert M L. Structure of mammalian ornithine decarboxylase at 1.6 Å resolution: stereochemical implications of PLP-dependent amino acid decarboxylases. Structure. 1999;7:567–581. doi: 10.1016/s0969-2126(99)80073-2. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Lee J S, An G, Friesen J D, Isono K. Cloning and the nucleotide sequence of the genes for Escherichia coli ribosomal proteins L28 (rpmB) and L33 (rpmG) Mol Gen Genet. 1981;184:218–223. doi: 10.1007/BF00272908. [DOI] [PubMed] [Google Scholar]
- 22.Nelson K E, Clayton R A, Gill S R, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Nelson W C, Ketchum K A, McDonald L, Utterback T R, Malek J A, Linher K D, Garrett M M, Stewart A M, Cotton M D, Pratt M S, Phillips C A, Richardson D, Heidelberg J, Sutton G G, Fleischmann R D, White O, Salzberg S L, Smith H O, Venter J C, Fraser C M. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 23.Nishikawa K, Noguchi T. Predicting protein secondary structure based on amino acid sequence. Methods Enzymol. 1991;202:31–44. doi: 10.1016/0076-6879(91)02005-t. [DOI] [PubMed] [Google Scholar]
- 24.Olsen G J, Woese C R, Overbeek R. The winds of (evolutionary) change: breathing new life into microbiology. J Bacteriol. 1994;176:1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osterman A L, Kinch L N, Grishin N V, Phillips M A. Acidic residues important for substrate binding and cofactor reactivity in eukaryotic ornithine decarboxylase identified by alanine scanning mutagenesis. J Biol Chem. 1995;270:11797–11802. doi: 10.1074/jbc.270.20.11797. [DOI] [PubMed] [Google Scholar]
- 26.Pegg A E. Polyamine metabolism and its importance in neoplastic growth and a target for chemotherapy. Cancer Res. 1988;48:759–774. [PubMed] [Google Scholar]
- 27.Pösö H, McCann P P, Tanskanen R, Bey P, Sjoerdsma A. Inhibition of growth of Mycoplasma dispar by dl-α-difluoromethyllysine, a selective irreversible inhibitor of lysine decarboxylase, and reversal by cadaverine (1,5-diaminopentane) Biochem Biophys Res Commun. 1984;125:205–210. doi: 10.1016/s0006-291x(84)80355-1. [DOI] [PubMed] [Google Scholar]
- 28.Poulin R, Lu L, Ackermann B, Bey P, Pegg A E. Mechanism of the irreversible inactivation of mouse ornithine decarboxylase by α-difluoromethylornithine. Characterization of sequences at the inhibitor and coenzyme binding sites. J Biol Chem. 1992;267:150–158. [PubMed] [Google Scholar]
- 29.Rosenberg-Hasson Y, Strumpf D, Kahana C. Mouse ornithine decarboxylase is phosphorylated by casein kinase-II at a predominant single location (serine 303) Eur J Biochem. 1991;197:419–424. doi: 10.1111/j.1432-1033.1991.tb15927.x. [DOI] [PubMed] [Google Scholar]
- 30.Tabor C W, Tabor H. Polyamines. Annu Rev Biochem. 1984;53:749–790. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- 31.Tabor C W, Tabor H. Polyamines in microorganisms. Microbiol Rev. 1985;49:81–99. doi: 10.1128/mr.49.1.81-99.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takatsuka Y, Onoda M, Sugiyama T, Muramoto K, Tomita T, Kamio Y. Novel characteristics of Selenomonas ruminantium lysine decarboxylase capable of decarboxylating both l-lysine and l-ornithine. Biosci Biotechnol Biochem. 1999;63:1063–1069. doi: 10.1271/bbb.63.1063. [DOI] [PubMed] [Google Scholar]
- 33.Tobias K E, Mamroud-Kidron E, Kahana C. Gly387 of murine ornithine decarboxylase is essential for the formation of stable homodimers. Eur J Biochem. 1993;218:245–250. doi: 10.1111/j.1432-1033.1993.tb18371.x. [DOI] [PubMed] [Google Scholar]
- 34.Yano T, Oue S, Kagamiyama H. Directed evolution of an aspartate aminotransferase with new substrate specificities. Proc Natl Acad Sci USA. 1998;95:5511–5515. doi: 10.1073/pnas.95.10.5511. [DOI] [PMC free article] [PubMed] [Google Scholar]