Abstract
Immunohistochemistry (IHC) is a well-established and commonly used staining method for clinical diagnosis and biomedical research. In most IHC images, the target protein is conjugated with a specific antibody and stained by Diaminobenzidine (DAB), resulting in a brown coloration, while hematoxylin serves as a blue counterstain for cell nuclei. The protein expression level is quantified through the H-score, calculated from DAB staining intensity within the target cell region. Traditionally, this process requires evaluation by two expert pathologists, which is both time-consuming and subjective.
To enhance the efficiency and accuracy of this process, we have developed an automatic algorithm for quantifying the H-score of IHC images. To characterize protein expression in specific cell regions, a deep learning model for region recognition was trained based on hematoxylin staining only, achieving pixel accuracy for each class ranging from 0.92 to 0.99. Within the desired area, the algorithm categorizes DAB intensity of each pixel as negative, weak, moderate, or strong staining and calculates the final H-score based on the percentage of each intensity category.
Overall, this algorithm takes an IHC image as input and directly outputs the H-score within a few seconds, significantly enhancing the speed of IHC image analysis. This automated tool provides H-score quantification with precision and consistency comparable to experienced pathologists but at a significantly reduced cost during IHC diagnostic workups. It holds significant potential to advance biomedical research reliant on IHC staining for protein expression quantification.
Keywords: Deep learning, IHC image, H-score, Protein expression quantification, Pathology image analysis
INTRODUCTION
Immunohistochemistry (IHC)-stained tissue imaging is the most common approach to characterize the expression of a specific protein across tissues. Initially, the target protein fixed on the IHC slide is recognized by a specific antibody, forming an antigen-antibody complex and is then visualized through a colored histochemical reaction with reporter molecules such as Diaminobenzidine (DAB)1,2. Additionally, a counterstain, typically hematoxylin, is applied to contrast the staining of the target protein. Due to its essential role in demonstrating the distribution and expression of protein biomarkers within tissue sections, IHC staining has become a routine method in basic research and diagnostic pathological examination3–5.
In IHC images, DAB intensity serves as an indicator of protein expression and can be effectively quantified using the H-score. The H-score is a reliable metric calculated as follows: (1 × percentage of weak staining) + (2 × percentage of moderate staining) + (3 × percentage of strong staining) within the target region, ranging from 0 to 3006. IHC staining and H-score provide superior protein detection capabilities compared to other methods (e.g., western blot) because they can measure protein expression levels within specific cell regions, which is especially valuable for quantifying proteins within specific types of cells of interest. The H-score calculation has been widely employed to establish links between proteins and tumors, playing crucial roles in diagnosis, prognosis, and therapeutic decision-making7–13. For example, over 60% of non-small cell lung cancer (NSCLC) patients exhibit overexpression of epidermal growth factor receptor (EGFR)14, and a higher EGFR H-score is strongly associated with shorter overall survival15. Similarly, breast cancer patients can be stratified into different treatment groups based on the expression levels of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) measured by the H-scores16.
Traditionally, the H-score is manually determined by proficient pathologists, involving the differentiation of diverse cell regions based on morphological features, categorization of DAB intensity within the tumor region, and calculation of the final H-score. This process demands a significant amount of time, attention, and specialized pathology expertise. Furthermore, the results obtained are not consistently reproducible even among experienced pathologists, mainly due to subjectivity in several aspects: 1) Human eyes tend to focus on tumor cells with intense DAB staining while disregarding other tumor cells; 2) The classification of DAB intensity into four categories, including negative, weak, moderate, and strong staining, lacks definitive criteria; 3) The percentage of each DAB intensity category is roughly estimated rather than accurately measured; 4) Heavy workload may also interfere with pathologists’ attention to details. All of these factors hinder the adoption of the H-score as a quantitative prognostic indicator in clinical practice17,18. Therefore, an automated analysis tool is highly desired to assist pathologists in evaluating the H-score efficiently and objectively.
Over the past decade, several studies19–27 have developed automated algorithms for the quantitative assessment of IHC images. However, significant efforts are still needed to improve quantification accuracy and efficiency. The algorithms developed before 201019–21 required hand annotations or fluorescent tags to locate the tumor region, demanding extra time and effort from pathologists. With the advancements in artificial intelligence, it has become feasible to automatically separate different regions, allowing the H-score analysis to focus on the identified tumor region. However, previous models22–26 were trained using original IHC images containing both hematoxylin staining and DAB staining. If the majority of tumor cells are strongly stained by DAB in the prepared training and validation sets, the trained model may ignore tumor cells with weak staining, leading to a higher H-score than the actual value, and vice versa. Additionally, as for the categorization of regions, former models segmented the whole image into either two categories: tumor and non-tumor22,23,26, or three categories: tumor, stroma, and background24,25. This ambiguous classification diminishes the recognition capability of the developed models. Consequently, other common cell regions whose morphological characteristics were not learned by the model during the training process, such as necrosis or lymphocyte regions, may be predicted as tumor regions and mistakenly involved in H-score calculation. Furthermore, these algorithms are challenging to generalize due to their restrictions to the subcellular localization of the protein. Some of these algorithms22,23,25,26 are exclusively designed for the H-score analysis of nuclear proteins (e.g., ER/PR), while others22,24,25 are designed for cytoplasmic or membrane proteins (e.g., EGFR/HER2). Moreover, most papers22–25,28 heavily rely on existing software from vendors (e.g., Leica, QuPath29) for region recognition, color deconvolution, and intensity measurement, necessitating pathologists to navigate multiple software programs, resulting in a time-consuming process that generates numerous intermediate files. Most importantly, many of these algorithms are not publicly accessible, significantly limiting the application of automatic H-score evaluation.
In this paper, we present an automatic algorithm for H-score quantification of IHC images. The main advantages of this developed method are fast processing, unbiased prediction, and consistent classification. The workflow is summarized in Figure 1. Initially, color deconvolution [30] of the original IHC images was performed to separate hematoxylin staining from DAB staining. Based solely on hematoxylin staining, a UNet-MobileNet model30,31 was trained to recognize the most common cell regions, including tumor regions, stroma regions, necrosis regions, lymphocyte regions, and background. DAB staining usually only appears in part of the cell, as the target protein is expressed on a specific cell structure such as the nucleus, cytoplasm, or cell membrane. Including unwanted structures in the calculation can lead to erroneous H-scores, reducing model accuracy. Therefore, it is imperative to separate different structures inside the cell and exclusively quantify DAB intensity within the target cell structure. In our method, the cell nuclei were segmented based on hematoxylin staining, enabling the separation of the nuclei and the cytoplasm of the target cells, with one of them considered the target area for a specific protein. Each pixel within the target area was classified as negatively, weakly, moderately, or strongly stained by comparing its DAB intensity to predefined standards. The final H-score was calculated as a weighted average of the percentages of weak staining (weight = 1), moderate staining (weight = 2), and strong staining (weight = 3) within the target area. Finally, a user-friendly website for automatic H-score quantification of IHC images was developed to make the algorithm publicly accessible (Supplementary Figure 1).
Figure 1.
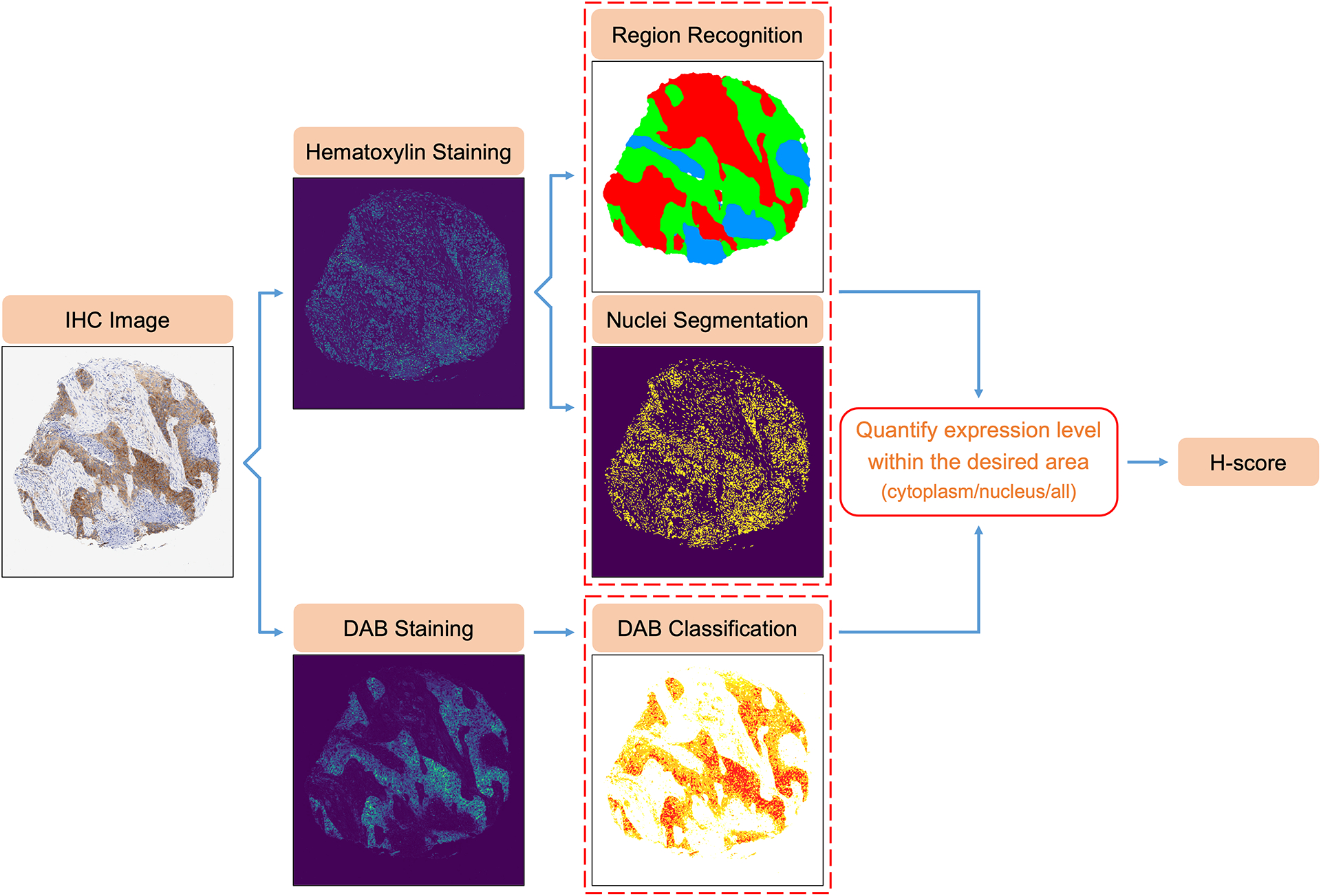
Flowchart of Automatic H-score Quantification for IHC Images.
The main contributions of this paper are: First, we developed an innovative automated algorithm that can be easily adapted for the H-score quantification of nuclear proteins, cytoplasmic proteins, and membrane proteins. Second, we trained a deep learning model to identify the most common cell regions on IHC images based solely on hematoxylin staining, thereby eliminating any interference from DAB staining. Third, we incorporated a step to generate the nuclei mask based on hematoxylin staining, allowing subsequent H-score quantification to exclusively focus on the desired cell structure. Fourth, we designed a calibration approach that significantly enhances the agreement between pathologist H-scores and machine H-scores for membrane proteins. Fifth, we built a user-friendly website to promote the widespread utilization of this algorithm.
METHODS
Dataset
Our H-score quantification algorithm was developed using one development dataset and validated through two independent validation datasets.
Development dataset
For the development set, three tissue microarray (TMA) samples from each of 498 non-small cell lung cancer (NSCLC) patients were collected and underwent IHC staining with an antibody specific for eukaryotic translation initiation factor 5B (eIF5B). IHC analysis was performed on a Dako Autostainer Link 48 system. Briefly, the slides were baked for 20 minutes at 60°C, then de-paraffinized and hydrated before the antigen retrieval step. Heat-induced antigen retrieval was performed at pH9 for 20 minutes in a Dako PT Link. The tissue was incubated with a peroxidase block and then an antibody incubation (1:200 dilution) for 20 minutes. The staining was visualized using the EnVision FLEX visualization system. The IHC images of these samples were captured at 10× magnification by a whole slide scanner and cropped into small image patches, with each containing one whole TMA core. Only the IHC image patches, where the tumor region occupied more than 15% of the tissue section, were selected for algorithm development, similar to the H-score analysis by pathologists. A total of 927 IHC images were included in this study.
Independent validation dataset 1
The first validation set comprised tissue samples from 69 pancreatic cancer patients. Paraffin-embedded human pancreatic ductal adenocarcinoma (PDAC) samples were provided by the Tissue Management Shared Resource within the Simmons Comprehensive Cancer Center at the University of Texas Southwestern Medical Center. An AKT serine/threonine kinase 3 (AKT3) antibody was first optimized by the core using the Dako autostainer, and the dilution of 1:200 was selected based on limited background staining. Then, all of these samples underwent automated IHC staining using the AKT3 antibody at the optimized concentration. Images of these slides were captured at 10× magnification using a Hamamatsu Nanozoomer whole slide scanner. Subsequently, the tissue images within the annotated malignant region, identified by a board-certified pathologist (L.J.), were continuously cropped into small image patches (1024×1024 pixels) for automatic H-score quantification.
Independent validation dataset 2
The second validation set consisted of 87 breast cancer TMA samples. The TMA slides were prepared from formalin-fixed, paraffin-embedded (FFPE) de-identified breast cancer tissue sections mounted on positively charged slides. These samples were processed for IHC staining at a CLIA-licensed clinical laboratory with the human epidermal growth factor receptor 2 (HER2) antibody by FDA-approved companion diagnostic assay and instrumentation. IHC images of these samples were captured at 10× magnification using an Aperio AT2 whole slide scanner, with each image encompassing a complete TMA core for H-score quantification.
Manual H-score assessment of varied sets
Development set
For the development set, two board-certified pathologists (B.M.E. and E.V.D.) were invited to independently assess the H-score of eIF5B in NSCLC tumor cells on IHC images. They categorized these tumor cells into four groups (negative, weak, moderate, and strong) based on the expression level of eIF5B within cell cytoplasm. Then, they calculated the H-score as a weighted average of the percentages of weak staining (weight = 1), moderate staining (weight = 2), and strong staining (weight = 3).
Independent validation set 1
For the first validation set, a board-certified pathologist (L.J.) evaluated the H-score of AKT3 on IHC images of PDAC using the similar method. She categorized tumor cells into four groups (negative, weak, moderate, and strong), according to the expression level of AKT3 throughout the entire cell region (including nucleus, cytoplasm, and membrane). The H-score was then calculated using the formula: H-score = (1×percentage of weak staining) + (2×percentage of moderate staining) + (3×percentage of strong staining).
Independent validation set 2
For the second validation set, a board-certified subspecialty breast pathologist (Y.F.) classified tumor cells on IHC images of breast cancer based on the membrane expression level and staining pattern of HER2 as follows: 1) score 0: no staining observed or incomplete faint/barely perceptible membrane staining of ≤10% tumor cells; 2) score 1+: incomplete faint/barely perceptible membrane staining of >10% tumor cells; 3) score 2+: weak to moderate complete membrane staining of >10% tumor cells or intense complete membrane staining of ≤10% tumor cells; 4) score 3+: intense complete membrane staining of >10% tumor cells, according to ASCO-CAP clinical practice guidelines. The H-score was then determined by: (1×percentage of 1+ tumor cells) + (2×percentage of 2+ tumor cells) + (3×percentage of 3+ tumor cells).
Separation of hematoxylin staining and DAB staining through color deconvolution
Original IHC images are composed of red, green, and blue (RGB) channels, which are contributed by both hematoxylin staining and DAB staining. To separate the hematoxylin signal and the DAB signal, color deconvolution was applied to the original IHC images32. The intensity of individual RGB channels was initially transformed to the optical density (OD) by the formula: ODC=−log10(IC/I0,C), where subscript C indicates one of three channels, IC indicates the intensity detected in this channel, and I0, C indicates the intensity of light entering the specimen, which was 255 in our case. Next, with the OD value for each channel, hematoxylin staining and DAB staining were quantified by reversing matrix multiplication: [ODR ODG ODB] = [SHematoxylin SDAB SBackground][Stain Matrix], where [Stain Matrix] is a 3×3 matrix with each row depicting a specific stain and each column depicting the OD value detected in individual RGB channels for each stain alone, . In the following analysis, the isolated signal of hematoxylin staining would be utilized for region recognition and nuclei segmentation, and the isolated signal of DAB staining would be used to quantify the protein expression level.
Region recognition of IHC images using a UNet-MobileNet model
Preparation of training, validation, and testing sets
Sixty-six IHC images (1024×1024 pixels) were randomly extracted from the development set and processed by color deconvolution for hematoxylin staining. Using the original IHC image and hematoxylin staining as references, the ground truth mask (based on pathology knowledge) for each image was manually labeled according to histological characteristics, where every pixel was specified to one of five classes: tumor region, stroma region, necrosis region, lymphocyte region, and background. The accuracy of these annotations was confirmed by two experienced board-certified pathologists (B.M.E. and E.V.D.). These prepared images were then randomly divided into training, validation, and testing sets in the proportion of 8:1:1.
Training process
To save computational cost, we trained a model combining U-Net backbone30 with depth-wise separable filters from MobileNet31 on the images of hematoxylin staining. Several additional steps were applied to the training set at the pre-processing stage, in order to reduce the batch effect of images from different sources and improve our model’s ability to generalize. First, pixel intensity on each image was globally normalized to a variable in the range of 0 to 1 and underwent random shifting by linear transformation. Second, random image augmentations such as flip and projective transformations were applied to all images in the training set and their masks in step. These processed images were then fed into the neural network to be trained for 100 epochs with a learning rate of 0.001. In other words, the learning algorithm worked through the entire training set 100 times and updated internal model parameters to improve the prediction accuracy. During the training process, the differences between the ground truth and the prediction were measured by a loss function based on the dice coefficient. As there was no significant change in the loss value at the end of the training process, the model trained at the 100th epoch was selected and used in subsequent analysis.
Evaluation of model performance
With the developed UNet-MobileNet model, the class of every pixel on the image could be automatically determined. To visualize the classification result, the input image was labeled with different colors according to the predicted classes. Pathologists (B.M.E. and E.V.D.) qualitatively assessed the model performance by examining the prediction results of the testing images. The pixel accuracy for each class was calculated to measure the model’s accuracy quantitatively. To assess the model’s stability, the model was applied to IHC images that had been processed with the white patch algorithm33 to various degrees. Furthermore, the model’s efficiency was evaluated by the analysis time for a single image.
Segmentation of nuclear region by hematoxylin intensity
Segmentation by Otsu’s thresholding
Cell nuclei were automatically segmented by comparing hematoxylin intensity to a threshold obtained using Otsu’s thresholding34 (selecting a cutoff value to minimize the intra-class intensity variance and maximize the inter-class intensity variance). We eliminated tiny objects smaller than 10 pixels. We then removed dark holes smaller than 10 pixels within the identified nuclear region, to avoid uneven staining and improve the robustness of the segmentation results.
Segmentation by an ISC-GAN model
The ISC-GAN model has demonstrated high accuracy in segmenting and classifying cell nuclei on IHC images35. In our study, the 10× IHC images were upscaled to 40× magnification and subsequently cropped into small image patches (512×512 pixels) for ISC-GAN prediction. All the nuclei masks predicted for the small image patches, derived from the same IHC image, were arranged according to spatial relationships, forming a large composite nuclei mask. This composite mask was then downscaled to 10× magnification for subsequent H-score analysis.
Comparison between Otsu’s thresholding results and ISC-GAN results
After generating nuclei masks for all the IHC images in the development set using Otsu’s thresholding or the ISC-GAN model, the Structural Similarity Index Measure (SSIM)36 was calculated to compare their corresponding nuclear density maps.
Determination of thresholds for DAB intensity classification
To classify DAB intensity into four ordinal categories: negative staining, weak staining, moderate staining, and strong staining, three threshold values are needed. Their initial values were set through a rough estimation based on DAB intensity from color deconvolution. By comparing corresponding DAB intensity with the primary thresholds, each pixel on IHC images was classified and labeled with different colors respectively (negative staining – white, weak staining – yellow, moderate staining – orange, strong staining – red). Two board-certified pathologists (B.M.E. and E.V.D.) confirmed these heat maps and adjusted the threshold values accordingly. The final determined thresholds would be utilized for future H-score analysis.
Automatic H-score quantification of IHC images
Color deconvolution was first applied to the original IHC image to separate hematoxylin staining and DAB staining. Based on hematoxylin staining alone, the trained UNet-MobileNet model would recognize and segment five different regions: tumor region, stroma region, necrosis region, lymphocyte region, and background. Meanwhile, Otsu’s thresholding or the ISC-GAN model was also used on hematoxylin staining to isolate all nuclei appearing on the image. With this information, nuclear region and cytoplasmic region of the target cells were identified respectively. Among these regions, the target area would be determined based on the distribution of a specific protein (cytoplasmic region for eIF5B, all regions for AKT3, cytoplasmic region for HER2). The DAB intensity of each pixel within the target area was classified as negative staining, weak staining, moderate staining, or strong staining according to the predefined classification standard. The final H-score was calculated by: H-score = (1×percentage of weak staining) + (2×percentage of moderate staining) + (3×percentage of strong staining).
Calibration of H-scores for membrane protein
For IHC images with negative or weak staining for membrane protein, it can be challenging for automatic algorithms to specifically target the membrane region for H-score quantification. Additionally, when IHC images stained for membrane protein, the staining intensity showing in cytoplasmic region changes in accordance with the protein expression level (Supplementary Figure 2). Therefore, in our algorithm, all pixels within non-nuclear region (i.e., cytoplasmic region) were included in H-score quantification for membrane protein. However, this approach may result in lower H-scores measured by our algorithm (machine H-scores) than those measured by pathologists (pathologist H-scores). To address this issue, we calibrated the membrane H-scores output from our algorithm. For this purpose, one point with the pathologist H-score between 200–250 and one point with the pathologist H-score between 250–300 were randomly selected from the 87 samples in the second validation set. For each selected point, the calibration parameter was calculated as follows: calibration parameter = pathologist H-score ÷ machine H-score. The average of these two calibration parameters was then used to calibrate the remaining 85 points in the second validation set: calibrated machine H-score = machine H-score × calibration parameter. The calibrated machine H-scores were capped at 300. This process was repeated 100 times, and the pathologist H-scores and calibrated machine H-scores were compared in each iteration. The mean correlation coefficient, slope, and intercept obtained from these 100 iterations were calculated and used to represent the calibration effect. Besides, for each point, its calibrated machine H-score would be recorded during the iteration when it was not involved in calculating the calibration parameter. The mean and standard deviation for each point were calculated based on these recorded H-scores and utilized to create a scatter plot with error bar.
RESULTS
The UNet-MobileNet model for recognizing five different regions based on hematoxylin staining
After applying the trained UNet-MobileNet model to hematoxylin staining of IHC images, five different regions were identified, including tumor region, stroma region, necrosis region, lymphocyte region, and background. Examples of region recognition results are shown in Figure 2 and Supplementary Figures 3–5. The model’s accuracy was confirmed at the pixel level. In the validation set, the pixel accuracy for tumor region, stroma region, necrosis region, lymphocyte region, and background was 0.93, 0.93, 0.97, 0.98, and 0.99, respectively. Moreover, in the testing set, the pixel accuracy for these five regions was 0.92, 0.93, 0.96, 0.97, and 0.99, respectively. For IHC images with varying degrees of white balancing, the UNet-MobileNet model demonstrated stable performance for region recognition, particularly in identifying tumor region, as shown in Supplementary Figure 6. This stability could be beneficial in making reliable H-score predictions for IHC images from diverse sources. As for the analysis speed of this developed model, region recognition of a single image could be accomplished in less than a second. In sum, our UNet-MobileNet model was able to accurately and efficiently segment five different regions based on hematoxylin staining.
Figure 2.
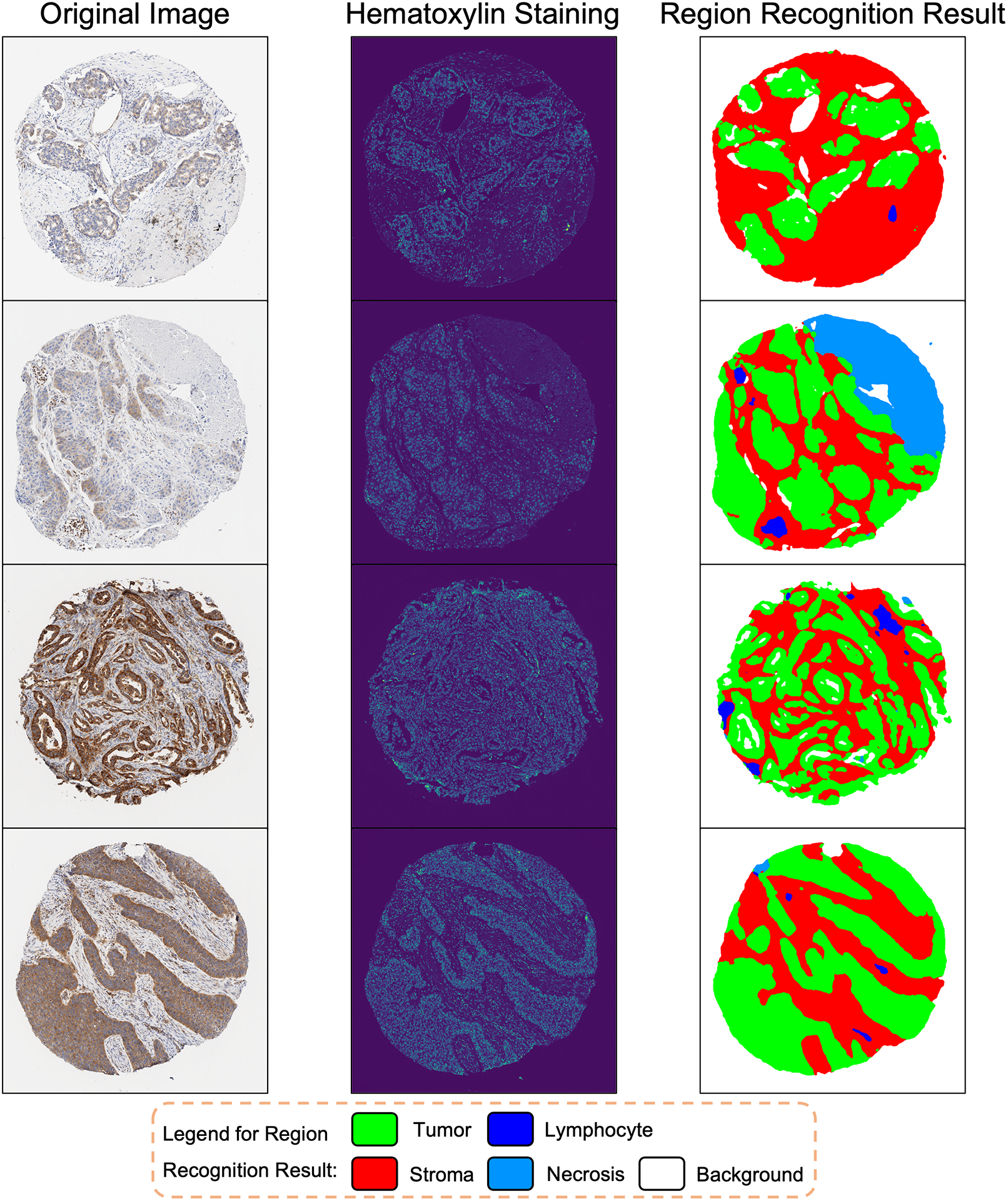
Examples of Region Recognition by the UNet-MobileNet Model Based on Hematoxylin Staining in IHC Images.
Separation of nuclear region and cytoplasmic region of the target cells
Based on hematoxylin intensity from color deconvolution, a threshold was automatically determined to isolate nuclei from background by Otsu’s thresholding. With this selected threshold value and morphological processing, we could identify nuclei on IHC images and separate nuclear region from cytoplasmic region of the target cells. For the development set, since we are interested in the expression level of eIF5B inside tumor cells and eIF5B is only expressed in cytoplasm, tumor cytoplasm was the target area for H-score quantification. Figure 3 and Supplementary Figure 7 show some examples of the identified nuclei and tumor cytoplasm. In addition to Otsu’s thresholding, the ISC-GAN model—a state-of-the-art deep learning approach—was also employed for nuclear segmentation of IHC images, as illustrated in Supplementary Figure 8. The segmentation outcomes from both methods demonstrated a high degree of similarity, with a median SSIM of 0.88 (Supplementary Figure 9). Given that Otsu’s thresholding demands substantially less time and computational resources, it was chosen as the preferred method for future H-score analysis.
Figure 3.
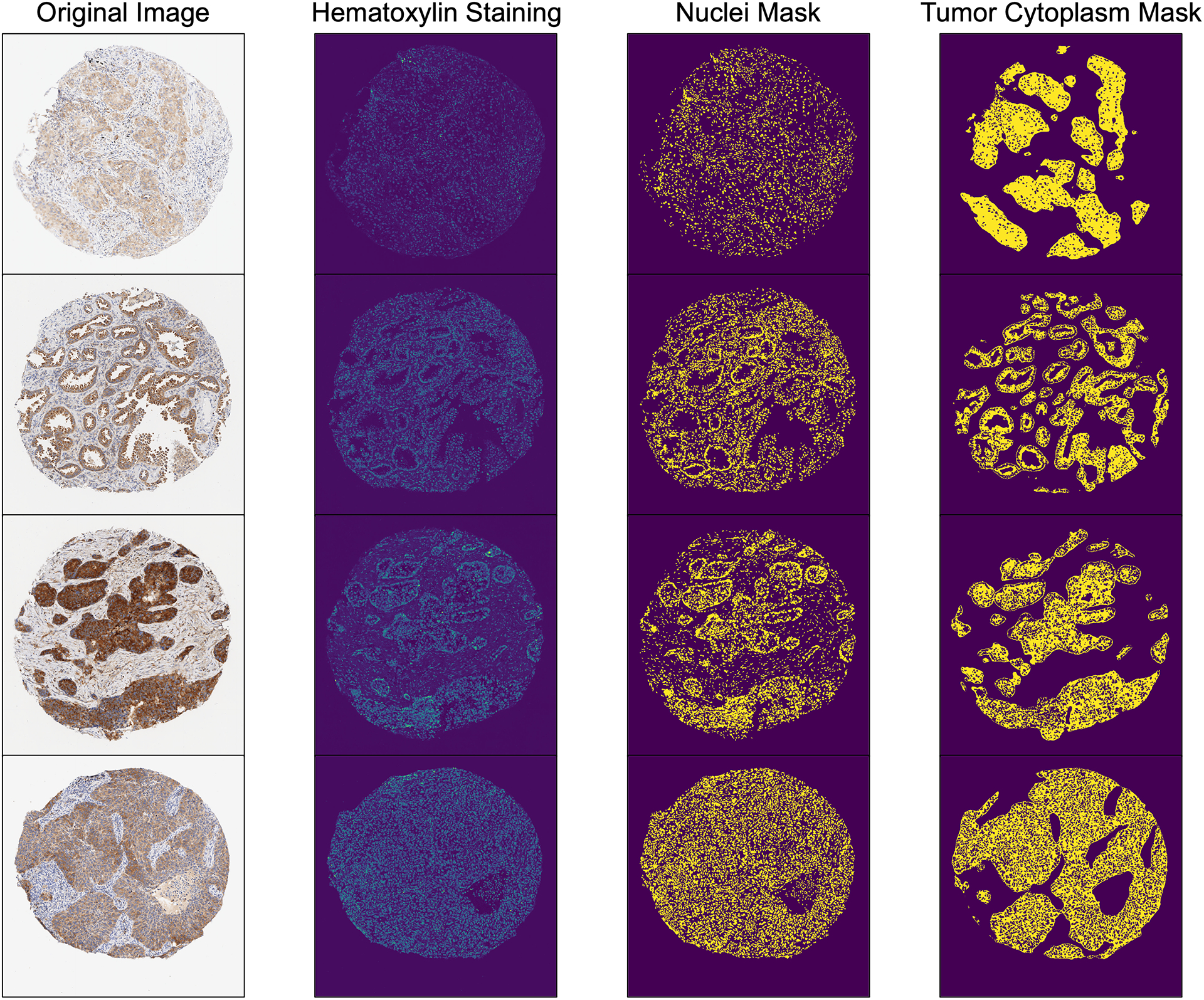
Examples of Nuclei Masks Generated by Otsu’s Thresholding for Hematoxylin Staining in IHC Images, Along with Corresponding Tumor Cytoplasm Masks.
Classification standard for DAB intensity
After confirming with two pathologists (B.M.E. and E.V.D.), the thresholds for classifying DAB staining were set at 0.2, 0.4, and 1, respectively, based on the intensity from color deconvolution. Therefore, pixels with DAB intensity lower than 0.2 would be classified as negatively stained, between 0.2 and 0.4 as weakly stained, between 0.4 and 1 as moderately stained, and higher than 1 as strongly stained. With this classification standard, the intensity category of any pixel on IHC images could be determined consistently. Heat maps were drawn to demonstrate the distribution of each DAB intensity category on the whole IHC images, as shown in Figure 4.
Figure 4.
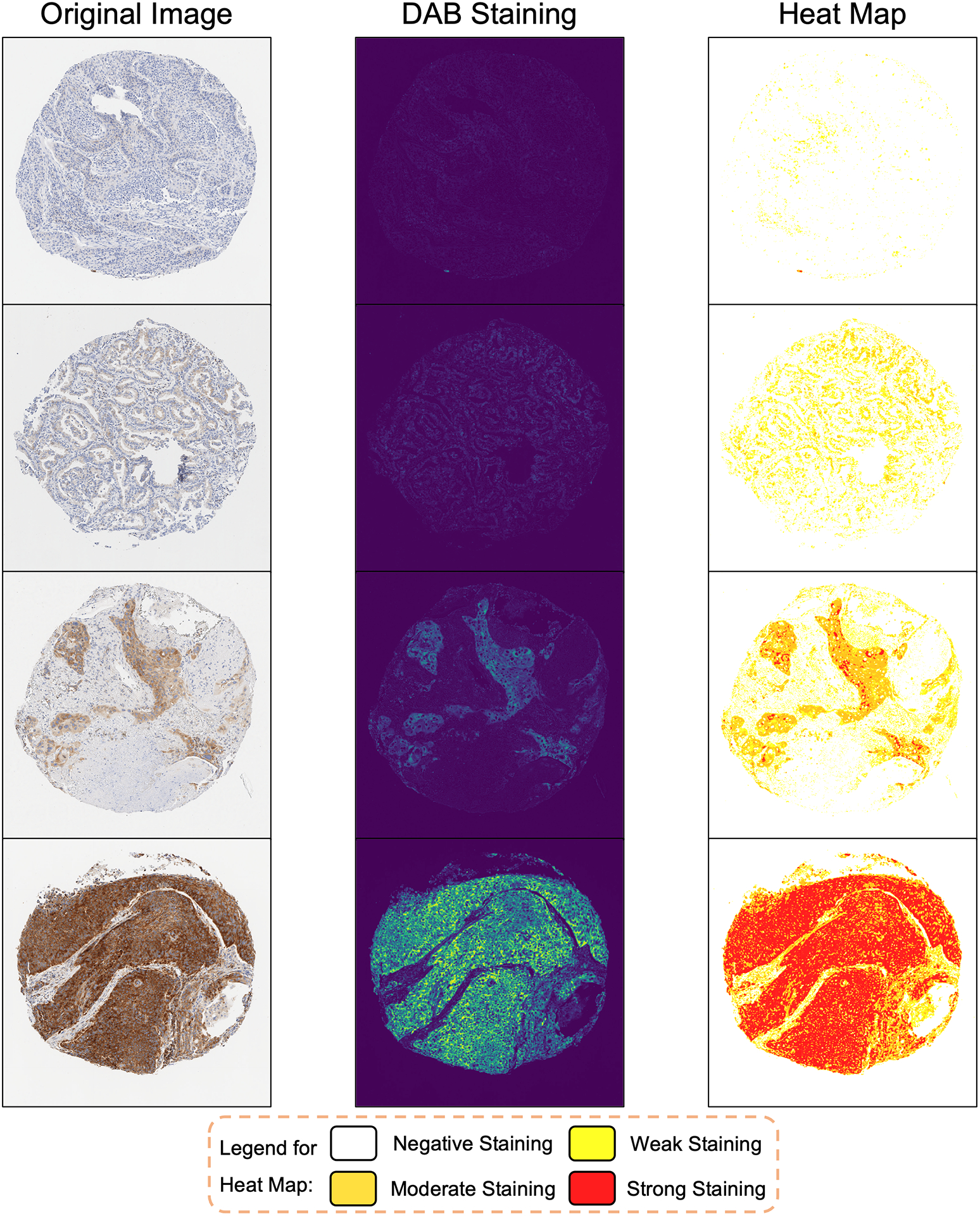
Examples of Heat Maps Depicting the Distribution of DAB Intensity Categories Across Entire IHC Images, Determined by Selected Threshold Values.
Comparison between H-scores by pathologists and by the automated algorithm for the development set
To measure the expression level of eIF5B in tumor regions, the H-score of each IHC image in the development set was quantified by two experienced board-certified pathologists (B.M.E. and E.V.D.) independently and by our developed algorithm. A scatter plot was drawn to show the relationship between the mean H-score produced by two pathologists and the H-score calculated by our algorithm of the same IHC image (Figure 5A). The Pearson correlation coefficient between the H-scores measured by these two methods was 0.83. For reference, the distribution of H-scores assessed by these two pathologists is displayed in Figure 5B (with a Pearson correlation coefficient of 0.84). Next, to compare the stability of manual H-score quantification and automatic H-score quantification, the coefficient of variation (CV) was calculated based on the H-scores for the IHC images from the same patient, by the formula: coefficient of variation (CV)=standard deviation / (median+1). The lower the CV of the H-scores from the same patient, the more stable the method is. As shown in Figure 5C, the cumulative distribution function (CDF) of CVs generated by our algorithm was very close to that produced by pathologists, indicating similar stability between these two methods. In conclusion, these results demonstrated that our developed algorithm could perform as well as proficient pathologists in H-score quantification.
Figure 5. Comparison Between Manually and Automatically Calculated H-scores for the Development Set.
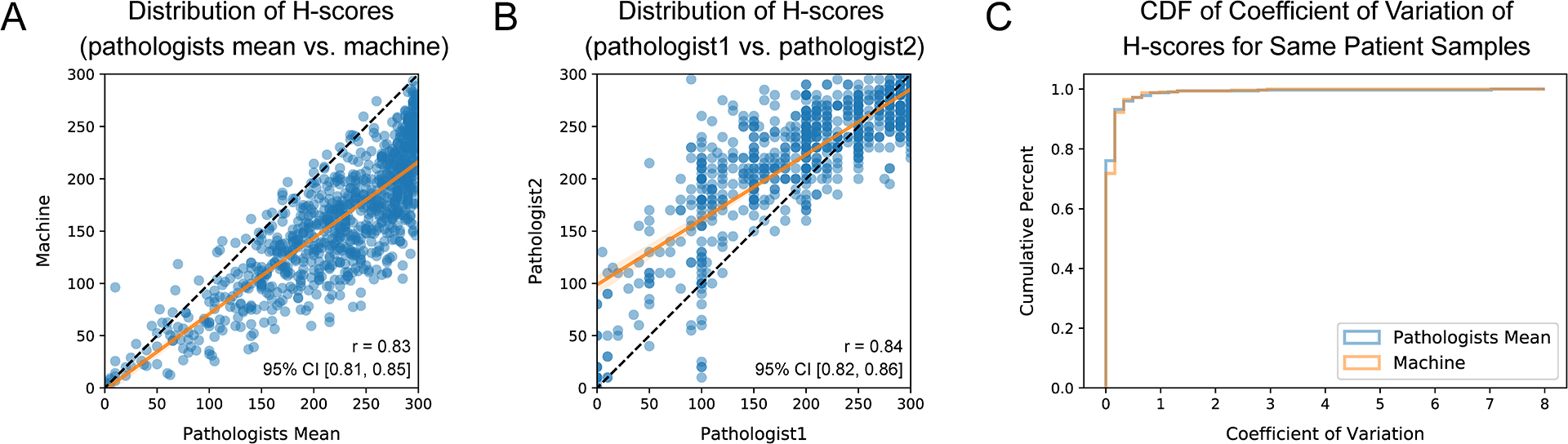
(A) Scatter plot demonstrating the correlation between the H-scores obtained through manual quantification by pathologists and those generated by our developed algorithm for the same IHC image. The black line represents the identity line, while the orange line is the regression line with a slope of 0.73 (95% CI: [0.70, 0.76]) and an intercept of −1.86 (95% CI: [−9.25, 5.52]). (We observed that the machine H-score tended to be lower than the pathologist H-score for the same IHC image, as explained in the discussion section.) (B) Scatter plot illustrating the correlation between the H-scores measured by two different pathologists for the same IHC image. The black line represents the identity line, while the orange line is the regression line with a slope of 0.62 (95% CI: [0.60, 0.65]) and an intercept of 98.75 (95% CI: [92.89, 104.60]). (C) Cumulative distribution function (CDF) of the coefficient of variation of H-scores for the IHC images assessed by pathologists or our algorithm from the same patient.
Comparison between H-scores by pathologists and by the automated algorithm for the independent validation sets
For the first validation set, the expression level of AKT3 in pancreatic tumor cells on each IHC image was quantified as a H-score by a pathologist (L.J.) and by our algorithm independently. The scatter plot in Figure 6A depicts the correlation between pathologist H-scores and machine H-scores for these 69 pancreatic cancer tissue samples (correlation coefficient: 0.78, slope: 0.44, intercept: 71.31). Regarding the second validation set, the expression level of HER2 in breast tumor cells was assessed by a pathologist (Y.F.) and by our algorithm independently. The H-scores generated by these two methods exhibited a correlation coefficient of 0.98, while the slope and intercept were 0.62 and −0.39, respectively (Figure 6B). After calibration, the mean correlation coefficient remained the same, but there was a significant improvement in the mean slope and intercept, which became 0.94 and −0.34, respectively. The scatter plot with error bar in Figure 6C demonstrates the stability of this calibration approach. The mean calibration parameter over the 100 iterations was 1.55 and would be utilized for future calibration.
Figure 6. Comparison Between Manually and Automatically Calculated H-scores for Two Independent Validation Sets.
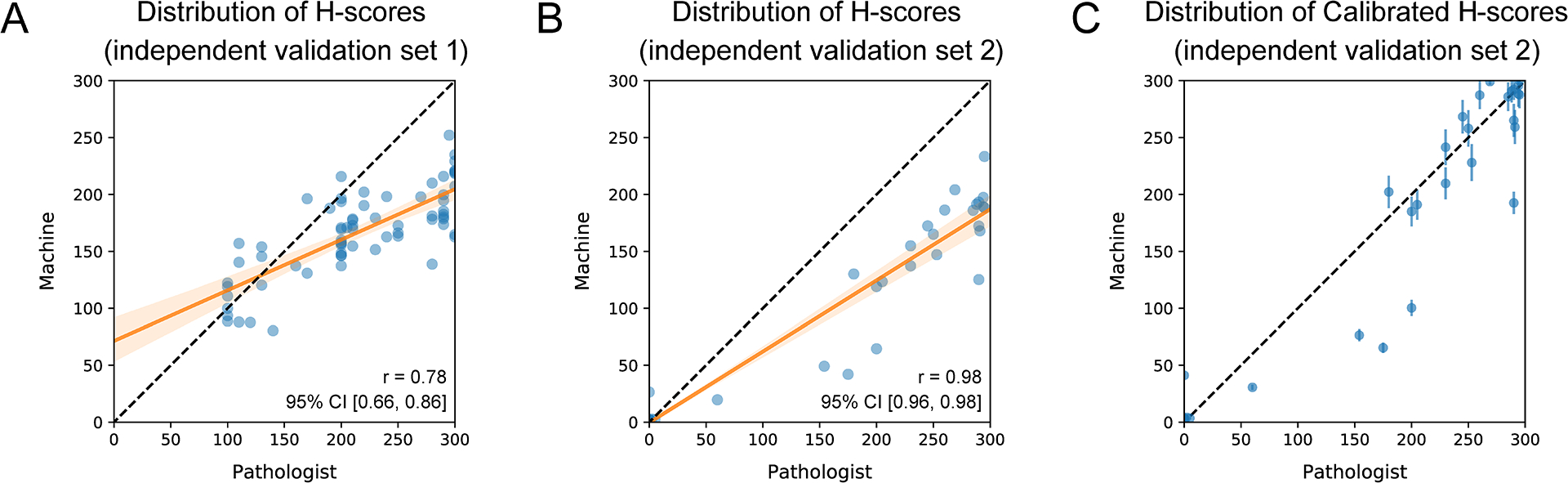
(A) Scatter plot depicting the correlation between pathologist H-scores and machine H-scores of AKT3 for the 69 pancreatic cancer tissue samples in the first validation set. The black line represents the identity line, while the orange line is the regression line with a slope of 0.44 (95% CI: [0.36, 0.53]) and an intercept of 71.31 (95% CI: [51.48, 91.15]). (B) Scatter plot illustrating the correlation between pathologist H-scores and machine H-scores of HER2 for the 87 breast cancer tissue samples in the second validation set. The black line represents the identity line, while the orange line is the regression line with a slope of 0.62 (95% CI: [0.60, 0.65]) and an intercept of −0.39 (95% CI: [−4.27, 3.49]). (C) Scatter plot with error bars displaying the mean and standard deviation of the calibrated machine H-score for each tissue sample in the second validation set over 100 iterations. The black line represents the identity line.
Website for H-score quantification of IHC images
To facilitate the research community’s use of this developed method, we built a website for automatic H-score quantification (AHSQ): https://lce.biohpc.swmed.edu/ahsq/ (Supplementary Figure 1). After the user selects the target cell region (tumor, stroma, necrosis, or lymphocyte) and the target cell structure (cytoplasm, nucleus, or all), this online tool takes a 10× IHC image (in .png or .tif file format) as input and directly outputs the H-score measured in the target area. For reference purposes, the results of the intermediate steps are also displayed on the page, including the predicted result of different regions, the identified target area, and the heat map for DAB intensity classification within the identified target area. The thresholds for classifying DAB intensity are initially set at 0.2, 0.4, and 1 by default. These threshold values were established in collaboration with pathologists (B.M.E. and E.V.D.). Considering that these values might not be suitable for all studies, our website allows users the flexibility to adjust them. The heat map for DAB intensity classification could visually assist users in determining the thresholds desirable for their specific application.
DISCUSSION
In this manuscript, we delineated the development and testing of a new method for accurately quantifying IHC staining through the calculation of the H-score. There are several new developments in this method. First, it is the first study to train a deep learning model for region recognition solely based on hematoxylin staining rather than on original IHC images, to avoid interference by DAB staining. Second, separating subcellular structures allows the H-score analysis to focus on the desired area and improves the compatibility of the developed algorithm with studies on nuclear proteins, cytoplasmic proteins, and membrane proteins. Third, the classification of DAB intensity is determined by definitive criteria and intuitively visualized by heat maps, thus providing the users with details of the computational process. Fourth, by working closely with experienced pathologists during the development process, we increased the credibility and reliability of our algorithm. Fifth, a website hosting our algorithm was created, enabling clinicians and researchers to use this automatic H-score quantification tool conveniently. With all of these improvements, we can accomplish H-score analysis for diverse proteins using a single software program, which takes an original IHC image as input and immediately outputs the calculated H-score, together with the predicted result of region recognition, the mask for the target area, and the heat map for DAB intensity classification inside the target area as references. This newly developed algorithm overcomes the shortcomings of prior algorithms and promisingly contributes to the high-throughput H-score quantification. It makes H-score analysis more reliable and accessible, benefiting both pathologists and biomedical researchers. The logic of this algorithm can also be integrated into the implementation of other software (e.g., QuPath29, HALO) for H-score quantification.
During the implementation, we noticed that the H-score measured by our automated algorithm tended to be lower than that assessed by pathologists (Figure 5A). This is due to machines having a superior capability to discern minuscule differences in staining intensity without interference by neighboring pixels. As shown in Supplementary Figure 10, the H-scores of these three IHC images given by pathologists were all nearly 300. However, according to the heat maps of DAB intensity classification within the tumor cytoplasm, there were some pixels in the target area that were weakly stained or moderately stained, which were better depicted by the H-scores output from our automated algorithm (161.54, 207.66, and 237.89). Thus, our developed algorithm could be a powerful tool to assist pathologists in performing accurate and efficient H-score quantification in a fraction of the time.
Finally, this automatic H-score quantification tool also has some limitations. First, the UNet-MobileNet model for region recognition was trained on non-small cell lung cancer (NSCLC) patient samples. However, for tumor cells deriving from different cell types (i.e., sarcomas, gliomas, lymphomas), the morphological characteristics may have less in common. If the performance of our model on other tumor samples is not satisfactory, a more comprehensive training set will be desired to integrate more images of different tumor types. Second, in this project, we have introduced an automated approach for calibrating H-scores, particularly for membrane proteins. While our approach has demonstrated favorable performance on our dataset, notably enhancing concordance between pathologist H-scores and machine-generated H-scores, it is important to acknowledge that its effectiveness can be influenced by the selection of calibration data points. Going forward, there is a need for the development of a more robust and standardized method to alleviate potential variability in the calibration process. Third, for calculating the H-score, the intensity of DAB staining was classified into four categories, including negative staining, weak staining, moderate staining, and strong staining, in order to mimic the current practice by pathologists. However, in theory, continuous values should carry more information than categorical values37. It would be more reasonable and objective to define a score determined directly from the continuous values of DAB intensity to characterize the protein expression. This way, the DAB intensity classification step can be eliminated from the analysis, since the classification step relies on the subjectively defined thresholds and may cause some loss of precision.
Supplementary Material
Funding
K.A.O. is supported by the NCI (R01CA207763, R01CA273585-01A1, and P50CA70907), the Cancer Prevention and Research Institute of Texas (CPRIT RP190610 and RP200327), and the Welch Foundation (I-1881). Y.X. is supported by the National Institutes of Health (P50CA70907, R01GM115473, and P30CA142543) and the Cancer Prevention and Research Institute of Texas (CPRIT RP180805). G.X. is supported by the National Institutes of Health (1R01GM140012, 1R01GM141519, 1R01DE030656, 1U01CA249245, and 2P30CA142543) and the Cancer Prevention and Research Institute of Texas (RP230330). The funding bodies had no role in this study’s design, collection, analysis, or interpretation of data.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Ethics Approval and Consent to Participate
All patient data used in this study have been de-identified. Additionally, as this research does not involve the collection of patient samples, according to the NIH Human Subjects Decision Tool, it is determined to be outside the scope of human subject research.
Data Availability Statement
The datasets used and analyzed during the current study are available upon reasonable request.
References
- 1.Graham RC Jr, Karnovsky MJ. THF EARLY STAGES OF ABSORPTION OF INJECTED HORSERADISH PEROXIDASE IN THE PROXIMAL TUBULES OF MOUSE KIDNEY: ULTRASTRUCTURAL CYTOCHEMISTRY BY A NEW TECHNIQUE. J Histochem Cytochem. 1966;14(4):291–302. [DOI] [PubMed] [Google Scholar]
- 2.Kim S-W, Roh J, Park C-S. Immunohistochemistry for pathologists: protocols, pitfalls, and tips. Journal of pathology and translational medicine. 2016;50(6):411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Matos LL, Trufelli DC, De Matos MGL, da Silva Pinhal MA. Immunohistochemistry as an important tool in biomarkers detection and clinical practice. Biomark Insights. 2010;5:BMI. S2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duraiyan J, Govindarajan R, Kaliyappan K, Palanisamy M. Applications of immunohistochemistry. J Pharm Bioallied Sci. 2012;4(Suppl 2):S307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramos-Vara J, Miller M. When tissue antigens and antibodies get along: revisiting the technical aspects of immunohistochemistry—the red, brown, and blue technique. Vet Pathol. 2014;51(1):42–87. [DOI] [PubMed] [Google Scholar]
- 6.McCarty JK, Miller L, Cox E, Konrath J, McCarty SK. Estrogen receptor analyses. Correlation of biochemical and immunohistochemical methods using monoclonal antireceptor antibodies. Arch Pathol Lab Med. 1985;109(8):716–721. [PubMed] [Google Scholar]
- 7.Aye Thike MJC, Fook-Chong Stephanie, Tan Puay Hoon, Aye. Immunohistochemical expression of hormone receptors in invasive breast carcinoma: correlation of results of H-score with pathological parameters. Pathology. 2001;33(1):21–25. [PubMed] [Google Scholar]
- 8.Parris TZ, Aziz L, Kovács A, et al. Clinical relevance of breast cancer-related genes as potential biomarkers for oral squamous cell carcinoma. BMC Cancer. 2014;14(1):324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azim HA, Peccatori FA, Brohée S, et al. RANK-ligand (RANKL) expression in young breast cancer patients and during pregnancy. Breast Cancer Res. 2015;17(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Souza AA, Altemani A, de Araujo NS, Texeira LN, de Araújo VC, Soares AB. Estrogen Receptor, Progesterone Receptor, and HER-2 Expression in Recurrent Pleomorphic Adenoma. Clinical Pathology. 2019;12:2632010X19873384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Starzyńska A, Sobocki BK, Sakowicz-Burkiewicz M, et al. VISTA H-Score Is Significantly Associated with a 5-Year DFS Rate in Oral Squamous Cell Carcinoma. Journal of Clinical Medicine. 2023;12(4):1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vougiouklakis T, Belovarac BJ, Lytle A, Chiriboga L, Ozerdem U. The diagnostic utility of EZH2 H-score and Ki-67 index in non-invasive breast apocrine lesions. Pathology-Research and Practice. 2020;216(9):153041. [DOI] [PubMed] [Google Scholar]
- 13.Derangère V, Lecuelle J, Lepage C, et al. Combination of CDX2 H-score quantitative analysis with CD3 AI-guided analysis identifies patients with a good prognosis only in stage III colon cancer. Eur J Cancer. 2022;172:221–230. [DOI] [PubMed] [Google Scholar]
- 14.Gazdar AF. Epidermal growth factor receptor inhibition in lung cancer: the evolving role of individualized therapy. Cancer Metastasis Rev. 2010;29(1):37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Avilés-Salas A, Muñiz-Hernández S, Maldonado-Martínez HA, et al. Reproducibility of the EGFR immunohistochemistry scores for tumor samples from patients with advanced non-small cell lung cancer. Oncol Lett. 2017;13(2):912–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allott EH, Cohen SM, Geradts J, et al. Performance of three-biomarker immunohistochemistry for intrinsic breast cancer subtyping in the AMBER consortium. Cancer Epidemiology and Prevention Biomarkers. 2016;25(3):470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rimm DL, Giltnane JM, Moeder C, et al. Bimodal population or pathologist artifact? J Clin Oncol. 2007;25(17):2487–2488. [DOI] [PubMed] [Google Scholar]
- 18.Jaraj SJ, Camparo P, Boyle H, et al. Intra-and interobserver reproducibility of interpretation of immunohistochemical stains of prostate cancer. Virchows Archiv. 2009;455(4):375–381. [DOI] [PubMed] [Google Scholar]
- 19.Hall BH, Ianosi-Irimie M, Javidian P, Chen W, Ganesan S, Foran DJ. Computer-assisted assessment of the human epidermal growth factor receptor 2 immunohistochemical assay in imaged histologic sections using a membrane isolation algorithm and quantitative analysis of positive controls. BMC Med Imaging. 2008;8(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masmoudi H, Hewitt SM, Petrick N, Myers KJ, Gavrielides MA. Automated quantitative assessment of HER-2/neu immunohistochemical expression in breast cancer. IEEE transactions on medical imaging. 2009;28(6):916–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8(11):1323–1328. [DOI] [PubMed] [Google Scholar]
- 22.Bolton KL, Garcia-Closas M, Pfeiffer RM, et al. Assessment of automated image analysis of breast cancer tissue microarrays for epidemiologic studies. Cancer Epidemiology and Prevention Biomarkers. 2010;19(4):992–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cass JD, Varma S, Day AG, et al. Automated quantitative analysis of p53, Cyclin D1, Ki67 and pERK expression in breast carcinoma does not differ from expert pathologist scoring and correlates with clinico-pathological characteristics. Cancers (Basel). 2012;4(3):725–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rizzardi AE, Johnson AT, Vogel RI, et al. Quantitative comparison of immunohistochemical staining measured by digital image analysis versus pathologist visual scoring. Diagn Pathol. 2012;7(1):42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rizzardi AE, Zhang X, Vogel RI, et al. Quantitative comparison and reproducibility of pathologist scoring and digital image analysis of estrogen receptor β2 immunohistochemistry in prostate cancer. Diagn Pathol. 2016;11(1):63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Xu B, Zheng C, et al. An end-to-end deep learning histochemical scoring system for breast cancer TMA. IEEE transactions on medical imaging. 2018;38(2):617–628. [DOI] [PubMed] [Google Scholar]
- 27.Choudhury KR, Yagle KJ, Swanson PE, Krohn KA, Rajendran JG. A robust automated measure of average antibody staining in immunohistochemistry images. J Histochem Cytochem. 2010;58(2):95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ram S, Vizcarra P, Whalen P, et al. Pixelwise H-score: A novel digital image analysis-based metric to quantify membrane biomarker expression from immunohistochemistry images. PLoS One. 2021;16(9):e0245638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bankhead P, Loughrey MB, Fernández JA, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep. 2017;7(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. Paper presented at: International Conference on Medical image computing and computer-assisted intervention2015. [Google Scholar]
- 31.Howard AG, Zhu M, Chen B, et al. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv preprint arXiv:170404861. 2017.
- 32.Ruifrok AC, Johnston DA. Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol. 2001;23(4):291–299. [PubMed] [Google Scholar]
- 33.Banić N, Lončarić S. Improving the white patch method by subsampling. Paper presented at: 2014 IEEE International Conference on Image Processing (ICIP)2014. [Google Scholar]
- 34.Otsu N A threshold selection method from gray-level histograms. IEEE transactions on systems, man, and cybernetics. 1979;9(1):62–66. [Google Scholar]
- 35.Wang S, Rong R, Gu Z, et al. Unsupervised domain adaptation for nuclei segmentation: Adapting from hematoxylin & eosin stained slides to immunohistochemistry stained slides using a curriculum approach. Computer Methods and Programs in Biomedicine. 2023;241:107768. [DOI] [PubMed] [Google Scholar]
- 36.Sara U, Akter M, Uddin MS. Image quality assessment through FSIM, SSIM, MSE and PSNR—a comparative study. Journal of Computer and Communications. 2019;7(3):8–18. [Google Scholar]
- 37.Altman DG, Royston P. The cost of dichotomising continuous variables. BMJ. 2006;332(7549):1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and analyzed during the current study are available upon reasonable request.


