Abstract
The pilus subunit, the pilin, of conjugative IncP pili is encoded by the trbC gene. IncP pilin is composed of 78 amino acids forming a ring structure (R. Eisenbrandt, M. Kalkum, E.-M. Lai, C. I. Kado, and E. Lanka, J. Biol. Chem. 274:22548–22555, 1999). Three enzymes are involved in maturation of the pilin: LepB of Escherichia coli for signal peptide removal and a yet-unidentified protease for removal of 27 C-terminal residues. Both enzymes are chromosome encoded. Finally, the inner membrane-associated IncP TraF replaces a four-amino-acid C-terminal peptide with the truncated N terminus, yielding the cyclic polypeptide. We refer to the latter process as “prepilin cyclization.” We have used site-directed mutagenesis of trbC and traF to unravel the pilin maturation process. Each of the mutants was analyzed for its phenotypes of prepilin cyclization, pilus formation, donor-specific phage adsorption, and conjugative DNA transfer abilities. Effective prepilin cyclization was determined by matrix-assisted laser desorption-ionization–mass spectrometry using an optimized sample preparation technique of whole cells and trans-3-indolyl acrylic acid as a matrix. We found that several amino acid exchanges in the TrbC core sequence allow prepilin cyclization but disable the succeeding pilus assembly. We propose a mechanism explaining how the signal peptidase homologue TraF attacks a C-terminal section of the TrbC core sequence via an activated serine residue. Rather than cleaving and releasing hydrolyzed peptides, TraF presumably reacts as a peptidyl transferase, involving the N terminus of TrbC in the aminolysis of a postulated TraF-acetyl-TrbC intermediate. Under formal loss of a C-terminal tetrapeptide, a new peptide bond is formed in a concerted action, connecting serine 37 with glycine 114 of TrbC.
Horizontal gene transfer by bacterial conjugation between a donor bacterium and recipient cell(s) of distinct species requires the formation of physically stable contacts, conjugative junctions (44), before DNA transfer can occur. The formation of such contacts among gram-negative bacteria, known as mating-pair formation (Mpf), is initiated by conjugative pili (for a recent review, see reference 56). Pili are extracellular filaments extending from the surface of donor cells. These filaments are tube-like structures about 10 nm in outer diameter with a 2-nm central, hydrophilic lumen (23) composed of at least one major subunit protein, the pilin. Although minor structural components have been proposed (14), none of these have been identified to date (1). In the case of self-transmissible broad-host-range IncP plasmids, the process of pilus production requires each of 11 plasmid-encoded components of the Mpf system (20). Two processes are known to be maintained by these pili: DNA transfer and donor-specific phage reproduction. The pilus is an essential prerequisite for conjugation, since functional dissection of DNA transfer systems has shown that nonpolar inactivation of the pilin precursor gene or any gene of the pilus assembly machinery does not allow DNA transfer (20). Pili may also function as phage receptors. Examples are the bacterial viruses M13 and R17 attaching to the F pilus (24, 41), whereas Pf3 and PRR1 dock to the IncP pilus (8). Adsorption of the phages to the pilus provides the initial step for the process of phage infection.
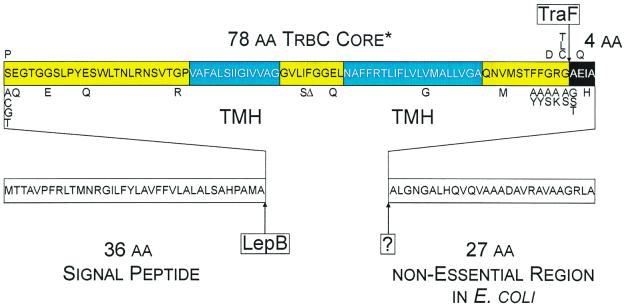
IncP pilin maturation is a multistep process, involving at least three components (Fig. 1). Two of them are encoded by the host chromosome. A yet-unidentified protease is responsible for removal of a 27-amino-acid (aa) C-terminal peptide from the original 145-aa gene product PreProTrbC (13). Second, LepB, the signal peptidase I of Escherichia coli, cleaves a leader peptide at the N terminus of ProTrbC (19), resulting in TrbC*, the prepilin. The final step in this maturation cascade is catalyzed by a plasmid-encoded function (19): TraF (13). This protease not only removes four additional C-terminal aa from TrbC* but also forms a cyclic product, the pilin, by introducing a new peptide bond between S37 and G114 of TrbC*.
FIG. 1.
Processing scheme of RP4 TrbC. The protein is shown as a box with its sequence in one-letter code inside. The 36-aa signal peptide and the 27-aa C-terminal cleaved peptide are shown as white boxes. The core sequence is shaded yellow with two predicted transmembranal helices (TMH) shown in blue. The 4-aa residues removed by TraF are inverted. Point mutations are annotated below or above the original sequence. Letters below indicate point mutants that are still cyclized by TraF; letters above indicate point mutants that are not processed by TraF. LepB and TraF are the enzymes that cleave TrbC, where “?” is the as-yet-unidentified host-encoded protease.
The mechanism of the maturation reaction and the positioning of the peptidase domain in the periplasm resemble those of host cell-encoded signal peptidases such as LepB of E. coli or SipS of Bacillus subtilis (19). This proposal derives from the pattern of conserved amino acid residues in TraF and TraF-like proteins from other conjugative systems compared to that of various signal peptidases. These signal or leader peptidases differ from the classical serine proteases by utilizing a catalytic-dyad-like mechanism instead of a catalytic triad (10, 51). The purpose of this report is to elucidate the mechanism of prepilin cyclization, catalyzed by TraF of the broad-host-range IncPα plasmid RP4. In analogy to the catalytic dyad-like mechanism of leader peptidases a peptide bond is broken. In contrast, however, the energy of the scissile bond is conserved and used for the formation of a new peptide bond, yielding the circular pilin.
MATERIALS AND METHODS
Strains, phages, and plasmids.
E. coli K-12 strains used in this study were SCS1 (a DH1 derivative [21]) and HB101 (7) as hosts for plasmids. The nalidixic acid-resistant derivative HB101 Nxr was used as the recipient in conjugation experiments, and JE2571 (leu thr fla pil str [9]) was used for phage sensitivity assays and electron microscopy. Cells were grown in YT medium (32) buffered with 25 mM 3-(N-morpholino)propanesulfonic acid (sodium salt, pH 8.0) and supplemented with 0.1% glucose and 25 μg of thiamine hydrochloride per ml. When appropriate, antibiotics were added as follows: ampicillin (sodium salt), 100 μg/ml; chloramphenicol, 10 μg/ml; tetracycline hydrochloride, 10 μg/ml; nalidixic acid (sodium salt), 30 μg/ml. Phages PRD1, PRR1, and Pf3 (4, 35) were propagated as described previously (47). The plasmids used in this study are listed in Table 1.
TABLE 1.
Plasmids used in this study
| Plasmid | Descriptiona | Relevant genotype | Selective markerb | Replicon | Reference or source |
|---|---|---|---|---|---|
| pDB126 | pML123Ω[BamHI; RP4 BfaI 45,893–53,462 bp] | (trbB-trbM)+ (traF-traM)+oriT+ | Cm | ColD | 2 |
| pDB126ΔtrbC | pDB126Δ[RP4 19,803–20,226 bp] | (trbB trbD-trbM)+ (traF-traM)+oriT+trbC0 | Cm | ColD | This work |
| pDB129 | pDB126Δ[RP4 45,893–46,583 bp] | (trbB-trbM)+ (traG-traM)+oriT+ traF0 | Cm | ColD | 19 |
| pDJ100c | pRE100Δ[BsrGI-AatII]Ω[RP4 BsrGI-AatII, 18,190–19,441 bp] | Ap | pMB1 | This work | |
| pDJ100trbCX00Zd | pRE100Δ[BsrGI-AatII]Ω[RP4 BsrGI-AatII, 18,190–19,441 bp] | Mutation indicated | Ap | pMB1 | This work |
| pGZ119EH, HE | Cloning vector; Ptac/lacIq | Cm | ColD | 27 | |
| pJH472 | pGZ119EHΔ[KpnI-EcoRI]Ω[RP4 46,060–46,591 bp] | traF+ | Cm | ColD | 13 |
| pJH472X00Zd | pGZ119EHΔ[KpnI-EcoRI]Ω[RP4 46,060–46,591 bp] | traF+, mutation indicated | Cm | ColD | This work |
| pJJ178 | pMS119EHΔ[NdeI-HindIII]Ω[R751 18,511–19,288 bp] | trbC+ R751 | Ap | pMB1 | This work |
| pML123 | pGZ119EHΔ[EcoRI-BamHI]Ω[EcoRI-XmnI adapter, RP4 XmnI-NotI, 18,841–30,042 bp] | (trbB-trbM)+ | Cm | ColD | 28 |
| pML123mtrbC45 | pML123Ω[dCTAGTCTAGACTAG at position RP4 19,938] | (trbB trbD-trbM)+trbC0 | Cm | ColD | 20 |
| pML123ΔtrbC | pML123Δ[RP4 19,803–20,226 bp] | (trbB, trbD-trbM)+trbC0 | Cm | ColD | This work |
| pML123trbCX00Zd | pGZ119EHΔ[EcoRI-BamHI]Ω[EcoRI-XmnI adapter, RP4 XmnI-NotI, 18,841–30,042 bp] | (trbB trbC+, mutation indicated, trbD-trbM)+ | Cm | ColD | This work |
| pMS119EH | Cloning vector; Ptac/lacIq | Ap | pMB1 | 49 | |
| pMS470Δ8 | pMS119EHΔ[XbaI-PstI]Ω[pT7-7 XbaI-NdeI 40-bp fragment, R751 traC AvaI-SphI 1.4 kb] cloning vector; Ptac/lacIq | Ap | pMB1 | 3 | |
| pPL178 | pMS119EHΔ[NdeI-EcoRI]Ω[tra 1,755–2,156 bp] | trbC+ pTiC58 | Ap | pMB1 | This work |
| pPLtraF | pKK38ASHΔ[NcoI-PstI]Ω[tra 2,255–2,523 bp] | traF+ pTiC58 | Tc | This work | |
| pRE100 | pMS119HEΔ[HindIII-EcoRI]Ω[HindIII-BsrGI-XbaI-AatII-EcoRI adapter] cloning vector; Ptac/lacIq | Ap | pMB1 | This work | |
| pRE178 | pMS119EHΔ[EcoRI-HindIII]Ω[RP4 19,797–20,244 bp] | trbC+ | Ap | pMB1 | 13 |
| pRE178X00Zd | pMS119EHΔ[EcoRI-HindIII]Ω[RP4 19,797–20,244 bp] | trbC+, mutation indicated | Ap | pMB1 | This work |
| pRE178Δ3 | pMS119EHΔ[EcoRI-HindIII]Ω[RP4 19,797–20,163 bp] | trbCΔ3+ | Ap | pMB1 | This work |
| pRE178Δ3.05 | pMS119EHΔ[EcoRI-HindIII]Ω[RP4 19,797–20,160 bp] | trbCΔ3.05+ | Ap | pMB1 | This work |
| pRE178Δ3.1 | pMS119EHΔ[EcoRI-HindIII]Ω[RP4 19,797–20,157 bp] | trbCΔ3.1+ | Ap | pMB1 | This work |
| pRE178Δ4 | pMS119EHΔ[EcoRI-HindIII]Ω[RP4 19,797–20,151 bp] | trbCΔ4+ | Ap | pMB1 | This work |
| pWP471 | pJF119EHΩ[RP4 NspI-HaeII 45,909–46,577 bp] | traF+ | Ap | pMB1 | 54 |
| pWP471X00Zd | pJF119EHΩ[RP4 NspI-HaeII 45,909–46,577 bp] | traF+, mutation indicated | Ap | pMB1 | This work |
RP4 (M93696), R751 (U67194), and pTiC58 (AF057718, trbC; U40389 traF; X53264, virB2) sequence coordinates of inserted fragments are given according to the published sequence data; GenBank accession numbers are in parentheses.
Ap, ampicillin resistance; Cm, chloramphenicol resistance; Tc, tetracycline resistance.
pDJ100 contains the indicated Tra2 fragment in an antitranscriptive orientation. The fragment starts inside the trbB gene and ends within the first third of trbE.
For mutagenesis strategy, see details in Materials and Methods.
DNA techniques.
Standard molecular cloning techniques were performed as described by Sambrook et al. (43).
Generation of trbC and traF mutants.
The structural genes of trbC and traF were directly mutagenized using the site-directed-mutagenesis kit from Stratagene. Mutagenesis of trbC was done on plasmid pRE178 or pDH100. From the latter plasmid an AatII-BsrGI (1.3-kb) fragment, one containing the site of mutation in trbC, was isolated and inserted into the corresponding site of either pML123 or pDB126. Mutants of traF were generated on pJH472, digested with EcoRI-HindIII, and inserted into EcoRI-HindIII-digested vectors pMS119HE/pGZ119HE. The 22-mer primers used to introduce point mutations were designed by changing as few base pairs as possible, with a maximum of two bases changed per mutation (see Tables 3 and 4). The alleles of trbC and traF described in this study are indicated as X00Z, where “X” represents the wild-type residue, “00” indicates the residue number(s) corresponding to the full-length protein, and “Z” indicates the newly introduced residue. Following mutagenesis, the nucleotide sequence of each trbC and traF mutant was verified by DNA sequencing using the dideoxy-chain termination method according to the method of Sanger et al. (45).
TABLE 3.
TrbC mutant phenotypes
| trbC allele | Phenotype
|
Primer usede (bp coordinates) | |||||
|---|---|---|---|---|---|---|---|
| TrbC maturationa | Transfer frequencyb | Dpsc
|
Pilus formationd | ||||
| PRD1 | PRR1 | Pf3 | |||||
| Wild type | C | 0.9 × 10−1 | + | + | + | + (b) | GAGCTCGGTACCCGGGGATCC, ATGGAAGCTTGATTAGGCGAG CCGTCCAGCCGC (20,214–20,238) |
| mtrbC45 | NA | <1 × 10−7 | − | − | − | − | Reference 20 |
| ΔtrbC | NA | <1 × 10−7 | − | − | − | − | GTATTTCCAATGACAACGGCGGTACCGTTC (19,788–19,801), GAGC CGTCCAGCCGGTACCGCACGCACGGC (20,199–20,228) |
| trbCS37A | C | 0.9 × 10−1 | + | + | + | + (b) | GGCGATGGCCGCGGAAGGCACC (19,895–19,916) |
| trbCS37C | C/L | 0.9 × 10−1 | − | − | − | − | GGCGATGGCCTGCGAAGGCACC (19,895–19,916) |
| trbCS37G | C | 0.9 × 10−1 | + | + | + | + (b) | GGCGATGGCCGGGGAAGGCACC (19,895–19,916) |
| trbCS37P | NAf | <1 × 10−7 | − | − | − | − | GGCGATGGCCCCGGAAGGCACC (19,895–19,916) |
| trbCS37T | C | 0.8 × 10−1 | + | + | + | + (b) | GGCGATGGCCACGGAAGGCACC (19,895–19,916) |
| trbCE38Q | C | 0.9 × 10−1 | + | + | + | + (b) | GATGGCCTCGCAAGGCACCGGC (19,898–19,919) |
| trbCG42E | C | <1 × 10−7 | − | − | − | − | AGGCACCGGCGAAAGCTTGCC (19,911–19,934) |
| trbCE47Q | C | <1 × 10−7 | − | − | − | − | CTTGCCATATCAGAGCTGGCTG (19,926–19,948) |
| trbCG59R | C | <1 × 10−7 | − | − | − | − | CTCCGTAACCCGCCCGGTGGCC (19,961–19,982) |
| trbCI78S | C | <1 × 10−7 | − | − | − | − | Reference 18 |
| trbCF79Δ | C | <1 × 10−7 | − | − | − | − | CGGCGTGCTGATCΔGGCGGCGAACTCA (20,018–20,046) |
| trbCE82Q | C | 0.7 × 10−1 | + | + | + | + (b) | CTTCGGCGGCCAACTCAACGCC (20,030–20,051) |
| trbCV96G | C | <1 × 10−7 | − | − | − | − | CCTGGTTCTGGGCATGGCGCTG (20,072–20,096) |
| trbCV106M | C | 0.9 × 10−1 | + | + | + | + (b) | CGCGCAGAACATGATGAGCACC (20,102–20,123) |
| trbCF110A | C | 0.9 × 10−3 | − | − | − | − | GATGAGCACCGCCTTCGGTCGTG (20,117–20,138) |
| trbCF110Y | C | <1 × 10−7 | − | − | − | − | GATGAGCACCTACTTCGGTCGTG (20,117–20,138) |
| trbCF111A | C | <1 × 10−7 | + | + | + | − | GATGAGCACCTTCGCCGGTCGTG (20,117–20,138) |
| trbCF111Y | C | 0.3 × 10−7 | + | + | + | − | GATGAGCACCTTCTACGGTCGTG (20,117–20,138) |
| trbCG112A | C | 0.3 × 10−7 | + | + | + | + (b) | CACCTTCTTCGCTCGTGGTGCC (20,120–20,141) |
| trbCG112D | L | <1 × 10−7 | − | − | − | − | Reference 18 |
| trbCG112S | C | 0.8 × 10−1 | + | + | + | + (b) | CACCTTCTTCAGTCGTGGTGCC (20,120–20,141) |
| trbCR113A | C | 0.9 × 10−1 | + | − | − | − | CTTCTTCGGTGCTGGTGCCGAA (20,123–20,144) |
| trbCR113K | C | 0.9 × 10−1 | + | − | − | + (b) | CTTCTTCGGTAAGGGTGCCGAA (20,123–20,144) |
| trbCG114A | C | <1 × 10−7 | + | + | + | + (b) | CTTCGGTCGTGCGGCCGAAATC (20,126–20,147) |
| trbCG114C | L | <1 × 10−7 | − | − | − | − | CTTCGGTCGTTGTGCCGAAATC (20,126–20,147) |
| trbCG114L | L | <1 × 10−7 | − | − | − | − | CTTCGGTCGTCTTGCCGAAATC (20,126–20,147) |
| trbCG114S | C/L | 0.9 × 10−1 | + | − | − | + (b) | CTTCGGTCGTAGTGCCGAAATC (20,126–20,147) |
| trbCG114T | L | <1 × 10−7 | − | − | − | − | CTTCGGTCGTACTGCCGAAATC (20,126–20,147) |
| trbCA115G | C/L | 0.7 × 10−1 | − | − | − | + (i) | GGTCGTGGTGGCGAAATCGCGG (20,129–20,150) |
| trbCA115T | C | 0.9 × 10−1 | + | + | + | + (b) | GGTCGTGGTACCGAAATCGCGG (20,129–20,150) |
| trbCE116Q | L | <1 × 10−7 | − | − | − | − | CGTGGTGCCCAAATCGCGGCC (20,132–20,153) |
| trbCI117H | C | 0.8 × 10−1 | NA | NA | NA | NA | GAGCTCGGTACCCGGGGATCC, ATGGAAGCTTGATTAGTGTTC GGCACCACGACC (20,130–20,144) |
| trbCΔ3 | C | NA | NA | NA | NA | NA | GAGCTCGGTACCCGGGGATCC, ATGGAAGCTTGATTACGCGAT TTCGGCACC (20,136–20,150) |
| trbCΔ3.05 | C | NA | NA | NA | NA | NA | GAGCTCGGTACCCGGGGATCC, ATGGAAGCTTGATTAGATTTC GGCACCACG (20,133–20,147) |
| trbCΔ3.1 | L | NA | NA | NA | NA | NA | GAGCTCGGTACCCGGGGATCC, ATGGAAGCTTGATTAGATTTC GGCACCACG (20,130–20,144) |
| trbCΔ4 | L | NA | NA | NA | NA | NA | GAGCTCGGTACCCGGGGATCC, ATGGAAGCTTGATTAACCACG ACCGAAGAA (20,124–20,138) |
Detection by Western blotting and MS as described in Materials and Methods. Pilin maturation is characterized as follows: C, circular pilin; L, linear TrbC*; C/L, circular pilin and linear TrbC* were detectable simultaneously; NA, not applicable.
Transconjugants per donor cell after 1 h of incubation with recipient HB101 Nxr at 37°C as described in Materials and Methods. The given frequencies represent the average values of three independent experiments.
Donor phage specificity (Dps) is characterized as follows: +, Dps positive; −, Dps negative; NA, not applicable. Data were derived from standard phage plaque assays and electron microscopy; for details, see Materials and Methods.
Pilus formation is characterized as follows: + (b), great amounts of pili were only detectable as bundles; + (i), few pili were only detectable as individuals; −, no pili were detectable; NA, not applicable.
For point mutations, only the transcriptive strand primer is given, and for deletion mutants and the wild type, both the primers used are given. Nucleotides derived from the original sequence are written in italics; mutagenized nucleotides are in boldface letters. RP4 coordinates of the nucleotides are given in parentheses according to published sequence data (GenBank accession no. M93696).
For details, see Results.
TABLE 4.
TraF mutant phenotypes
| traF allele | Phenotype
|
Primer usede (bp coordinates) | |||||
|---|---|---|---|---|---|---|---|
| TrbC maturationa | Transfer frequencyb | Dpsc
|
Pilus formationd | ||||
| PRD1 | PRR1 | Pf3 | |||||
| Wild type | C | 0.9 × 10−1 | + | + | + | + (b) | Reference 14 |
| ΔtraF | L | <1 × 10−7 | − | − | − | − | Reference 19 |
| traFS37A | C/L | 0.8 × 10−4 | − | − | − | − | Reference 19 |
| traFC59A | C/L | 0.8 × 10−1 | + | + | + | + (i) | GTCATGTTCGCCCCGCCGCAAG (46,405–46,426) |
| traFC80A | C/L | 0.7 × 10−1 | + | + | + | + (i) | GGCGGTTTCGCCCCCGGCGA (46,344–46,363) |
| traFK89Q | C/L | 0.5 × 10−3 | − | − | − | − | Reference 19 |
| traFK89L | C/L | 1.6 × 10−4 | − | − | − | − | CTACATGATGCTGCGAGTTTTAG (46,315–46,337) |
| traFK89R | C/L | 0.8 × 10−2 | − | − | − | − | CTACATGATGAGGCGAGTTTTAG (46,315–46,337) |
| traFR90L | C/L | 0.8 × 10−1 | + | + | + | + (b) | CATGATGAAGCTAGTTTTAGCCG (46,312–46,334) |
| traFP129I | C | 0.9 × 10−1 | + | + | + | + (b) | CGGCCGCTGATTCGTTATCAG (46,196–46,216) |
| traFD155I | L | <1 × 10−7 | − | − | − | − | CACGTCTTTCCTCGGCCGCTAC (46,118–46,139) |
| traFD155N | C/L | 3.1 × 10−3 | − | − | − | − | CACGTCTTTCAACGGCCGCTAC (46,118–46,139) |
| traFR157A | C/L | 3.6 × 10−2 | + | + | + | + (i) | CTTTCGACGGCGCCTACTTCGG (46,112–46,133) |
| traFY158F | C | 0.9 × 10−1 | + | + | + | + (b) | CGACGGCCGCTTCTTCGGGCC (46,109–46,130) |
Detection by Western blotting and MS as described in Materials and Methods. Pilin maturation is characterized as follows: C, circular pilin; L, linear TrbC*; C/L, circular pilin and linear TrbC* were detectable simultaneously.
Transconjugants per donor cell after 1 h of incubation with recipient HB101 Nxr at 37°C as described in Materials and Methods. The given frequencies represent the average values of three independent experiments.
Donor phage specificity (Dps) is characterized as follows: +, Dps positive; −, Dps negative. Data were derived from standard phage plaque assays and electron microscopy; for details, see Materials and Methods.
Pilus formation is characterized as follows: + (b), great amounts of pili were only detectable as bundles; + (i), few pili were only detectable as individuals; −, no pili were detectable.
Only the transcriptive strand primers used are given. Nucleotides derived from the original sequence are written in italics; mutagenized nucleotides are in boldface letters. RP4 coordinates of the nucleotides are given in parentheses according to published sequence data (GenBank accession no. M93696).
Conjugation assays.
For quantitative filter matings appropriate amounts of donor (0.5 ml, A600 = 0.3) and recipient cells (5.0 ml, A600 = 0.3) were mixed and collected onto a Millipore filter (0.45-μm pore size, 25 mm in diameter). Each filter was incubated for 1 h at 37°C on a nutrient agar plate without selection. Cells were resuspended, and transconjugants were grown on YT agar plates containing nalidixic acid (sodium salt) and chloramphenicol for selection of pDB126, pDB129, or its derivatives (Tables 2 and 3).
TABLE 2.
Classification of phenotypes
| Strain | Relevant genotype | Relevant phenotypea |
|---|---|---|
| HB101(pDB126) | (trbB-trbM)+ (traF-traM)+oriT+ | Transfer + |
| HB101(pDB126ΔtrbC) | (trbB trbD-trbM)+ (traF-traM)+oriT+, trbC0 | Transfer − |
| HB101(pDB126trbCX00Z) | (trbB trbC+, mutation indicated, trbD-trbM)+ (traF-traM)+oriT+ | Transfer +/− |
| HB101(pWP471/pDB129) | traF+/(trbB-trbM)+ (traG-traM)+oriT+ traF0 | Transfer + |
| HB101(pDB129) | (trbB-trbM)+ (traG-traM)+oriT+ traF0 | Transfer − |
| HB101(pWP471X00Z/pDB129) | traF+, mutation indicated/(trbB-trbM)+ (traG-traM)+oriT+ traF0 | Transfer +/− |
| JE2571(pWP471/pML123) | traF+/(trbB-trbM)+ | Dps/pilus formation + |
| JE2571(pWP471/pML123ΔtrbC) | traF+/(trbB trbD-trbM)+trbC0 | Dps/pilus formation − |
| JE2571(pWP471/pML123trbCX00Z) | traF+/(trbB trbC+, mutation indicated, trbD-trbM)+ | Dps/pilus formation +/− |
| JE2571(pWP471/pML123) | traF+/(trbB-trbM)+ | Dps/pilus formation + |
| JE2571(pML123) | (trbB-trbM)+ | Dps/pilus formation − |
| JE2571(pWP471X00Z/pML123) | traF+, mutation indicated/(trbB-trbM)+ | Dps/pilus formation +/− |
| SCS1(pJH472/pRE178) | traF+/trbC+ | Pilin detection + |
| SCS1(pJH472) | traF+ | Pilin detection − |
| SCS1(pJH472/pRE178X00Z) | traF+/trbC+, mutation indicated | Pilin detection +/− |
| SCS1(pJH472X00Z/pRE178) | traF+, mutation indicated/trbC+ | Pilin detection +/− |
Transfer, DNA transfer abilities determined by conjugation experiments as described in Materials and Methods. Transfer +, transfer positive; transfer −, transfer negative. Donor-specific phage propagation (Dps) and pilus formation are characterized as positive (+) for Dps and pilus formation or negative (−) for Dps and pilus formation. Detection of TrbC precursors or pilin by Western blotting and MALDI-TOF-MS is indicated as positive (+, detection) or negative (−, no detection).
Protein expression and Western blotting.
Extracts of E. coli SCS1 cells were electrophoresed on Tricine-sodium dodecyl sulfate (17%)-polyacrylamide gels, electroblotted onto nitrocellulose (BA85; Schleicher & Schuell) membranes, and incubated with the immunoglobulin G (IgG) fraction (dilution of 1:2,000 with respect to the IgG concentration of the original serum) of purified anti-RP4-pilus serum as described previously (19). The IgG fraction of rabbit anti-pilus serum was preabsorbed by incubation with nondenatured cell extract of SCS1(pMS119EH).
Assay for phage sensitivity.
Standard phage plaque assays were performed as described previously (20) with E. coli JE2571 as the host strain (Tables 2 and 3).
Electron microscopy.
Phage adsorption to pili was investigated by electron microscopy. Phages and pili were visualized as described previously (9, 19). In brief, cells of the nonpiliated strain JE2571 carrying suitable plasmids were grown overnight on YT agar plates. Using a sterile loop a small portion of cells was scraped off the plate and gently suspended in a 50-μl drop of 50 mM ammonium acetate (pH 7.0). Phage particles were added in the appropriate dilution, and the mixture was incubated for 30 min at room temperature. Copper grids coated with Butvar B98 support film (48) and stabilized with a thin layer of carbon were floated for about 1 min on the cell suspension and then washed three times by floating them on 50-μl drops of 50 mM ammonium acetate. Pili and phages were then negatively stained with 1% sodium phosphotungstate.
MS.
TrbC was detected, using matrix-assisted laser desorption-ionization time-of-flight mass spectrometry (MALDI-TOF-MS) as described previously (13). Sample preparations containing whole E. coli cells were cocrystallized with trans-3-indolyl acrylic acid and measured on a Bruker Reflex II MALDI-TOF mass spectrometer.
RESULTS
Phenotypes of trbC and traF mutants.
A collection of defined TrbC point mutations was generated for the purpose of studying (i) the removal of the signal peptide, (ii) the cyclization reaction catalyzed by TraF, and (iii) pilus assembly from subunits. Therefore, mutations were clustered at the signal peptide cleavage site, in the area of the proposed TraF recognition site, and in the core sequence of the TrbC precursor. The basis for traF mutagenesis was multiple sequence alignments showing extensive similarities between several TraF-like proteins and signal peptidases of various bacterial species (Fig. 2). Mutant phenotypes were determined by complementation, including quantification of transfer frequency, pilus production analyzed by electron microscopy, and donor-specific phage propagation. Cyclization, the key maturation step, was monitored in the presence of traF directly by MS using a sample preparation technique with whole bacteria (13). In conjugation experiments a nonpolar trbC deletion mutant could be complemented for plasmid transfer when trbC was provided in trans, but neither pilus production nor phage propagation was observed on donor cells (Table 3). The different systems set up to study mutational effects of trbC (in cis) and traF (in trans) are described in Table 2. Sequence alterations were introduced as described in Materials and Methods and then analyzed with the HB101 system for efficient DNA transfer, the JE2571 system for pilus overproduction and donor phage specificity (Dps), and finally the SCS1 system for protein overproduction (Table 2). trbC and traF mutagenesis data are summarized in Tables 3 and 4.
FIG. 2.
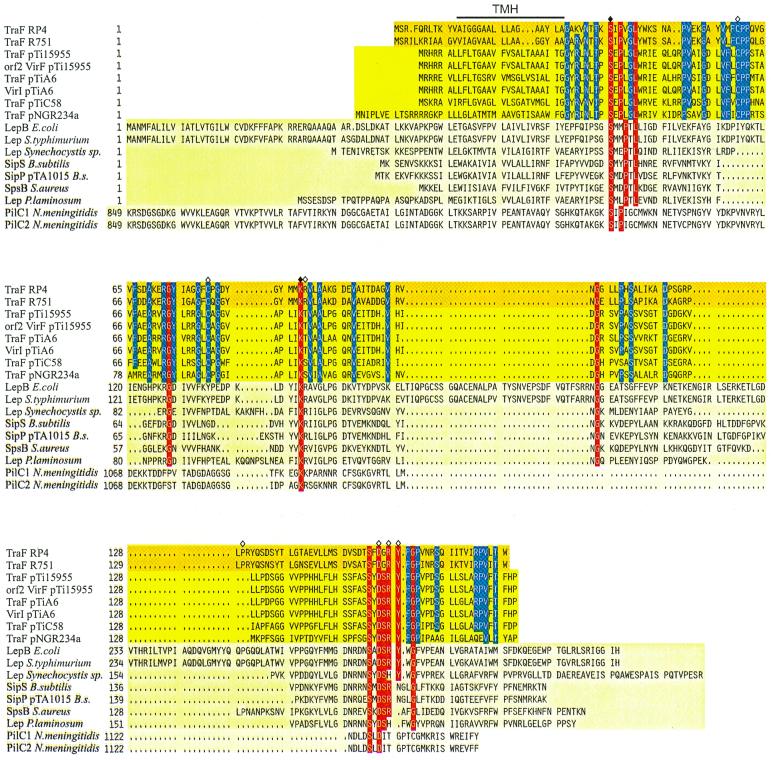
Sequence alignment of TraF-like proteins. Amino acid compositions of the different gene products are arranged in families; GenBank accession numbers are given in parentheses. Dark yellow background, bacterial conjugative plasmids' RP4 (L27758) and R751 (M94367) TraF. Yellow background, A. tumefaciens TraF homologues from pTi15955 (P15595), pTiA6 (U43674), pTiC58 (U40389), and putative pTi15955 protein Orf2 (S15913), pTiA6NC VirI (2773263), as well as Rhizobium Ti plasmid homologue pNGR234a TraF (P55417). Light yellow background, leader peptidases LepB of E. coli (K00426), Lep of Salmonella enterica serovar Typhimurium (X54933), Lep of Synechocystis sp. (PCC6803), SipS of B. subtilis (Z11847), SipP of B. subtilis plasmid pTA1015 (AAC44415), SpsB of Staphylococcus aureus (U65000), and Lep of Phormidium laminosum (S51921). Pale yellow background, N. meningitidis PilC1 and PilC2 (Y13020 and Y13021, respectively). Identical amino acid residues in at least 11 sequences are shown with a red background. The catalytically active residues shown for E. coli leader peptidase I are marked with a filled rhombus, the respective amino acids in RP4 TraF have been mutated, and further mutation sites in RP4 TraF are marked with an open rhombus, whereas the predicted transmembranal helix (TMH) for RP4/R751 TraF is indicated with a line above the alignment. Gaps introduced to maximize alignment are indicated by dots. To indicate the high conservation of TraF-like proteins in the uppermost two families, identical amino acid residues of these proteins are indicated by a blue background.
Signal peptide cleavage is essential for prepilin cyclization.
The translation product of trbC, consisting of 145 aa, is processed at the N terminus by removal of a 36-aa signal peptide and cleavage of 27 residues at the C terminus (13, 19). Inhibition of the N-terminal signal peptide removal in TrbCS37P should hamper targeting of the mutant protein to the cell surface but should not influence the initial C-terminal processing step of TrbC (Fig. 3A, lane c). TrbCS37P is the only trbC mutant protein described in this study from which no signal was obtained by MS using whole-cell preparations. Thus, we conclude that the cellular localization of TrbCS37P must differ from the wild-type situation considerably. Western blot analysis showed that TrbCS37P maturation was arrested after the initial truncation at the C terminus. This indicates that the cleavage reaction at the C terminus might take place in the cytoplasm, since targeting to the inner membrane is obviously not required. However, cyclization of TrbCS37P by TraF does not occur under these conditions (Fig. 3A, lane c).
FIG. 3.
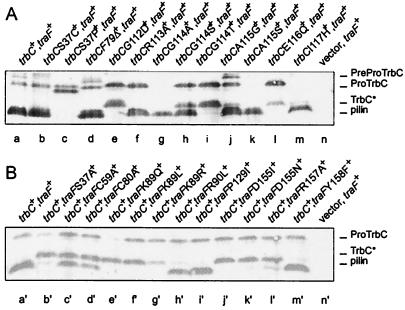
Western blot analysis (for conditions, see Materials and Methods) of E. coli SCS1 cell extracts (2 μl/lane) containing the plasmids indicated. (A) Mutations in trbC. Lane a, pRE178 (trbC+) and pJH472 (traF+); lane b, pRE178S37C and pJH472; lane c, pRE178S37P and pJH472; lane d, pRE178F79Δ and pJH472; lane e, pRE178G112D and pJH472; lane f, pRE178R113A and pJH472; lane g, pRE178G114A and pJH472; lane h, pRE178G114S and pJH472; lane i, pRE178G114T and pJH472; lane j, pRE178A115G and pJH472; lane k, pRE178A115S and pJH472; lane l, pRE178E116Q and pJH472; lane m, pRE178I117H and pJH472; lane n, pMS119 (vector) and pJH472. (B) Mutations in traF. Lane a′, pRE178 (trbC+) and pJH472 (traF+); lane b′, pRE178 and pJH472S37A; lane c′, pRE178 and pJH472C59A; lane d′, pRE178 and pJH472C80A; lane e′, pRE178 and pJH472K89Q; lane f′, pRE178 and pJH472K89L; lane g′, pRE178 and pJH472K89R; lane h′, pRE178 and pJH472R90L; lane i′, pRE178 and pJH472P129I; lane j′, pRE178 and pJH472D155I; lane k′, pRE178 and pJH472D155N; lane l′, pRE178 and pJH472R157A; lane m′, pRE178 and pJH472Y158F; lane n′, pMS119EH (vector) and pJH472. Positions of the 145-aa PreProTrbC, N-terminally cleaved ProTrbC, N- and C-terminally processed TrbC∗, and (circular) pilin are indicated on the right side of the figure.
A total of 28 C-terminal residues of the TrbC-precursor are dispensable for pilin maturation.
A trbC deletion mutant analysis was carried out to evaluate the C-terminal processing step. Characterization of truncated trbC derivatives revealed that polypeptides ending with A118 (trbCΔ3) or I117 (trbCΔ3.05) were still converted to pilin and served as substrates for pilus assembly (Fig. 4, lanes c′ and d′). However, further truncation of TrbC showed that a peptide ending with E116 (trbCΔ3.1) no longer functioned as a substrate for TraF (Fig. 4, lane e′). The determined mass of this peptide (m/z 8338) and its Western blot data proved that it was only processed at the N terminus. Accordingly, the product remained linear. TrbC ending with G114 (trbCΔ4), the residue which forms the intramolecular peptide bond with residue S37, also does not serve as a substrate for the cyclization reaction (Fig. 4, lane f′). These data demonstrated that 28 C-terminal residues of TrbC are dispensable for cyclization in E. coli. As we show here, the shortest functional substrate for pilin formation consists of 81 aa, beginning with S37 and ending with I117. We should emphasize that a short tail of three residues, positions 115 to 117, is sufficient and absolutely essential for the cyclization reaction. These residues are removed during peptide bond formation between S37 and G114.
FIG. 4.
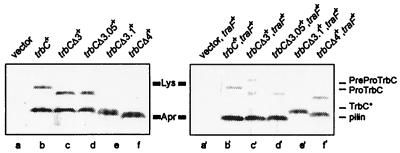
Western blot analysis (for conditions see Materials and Methods) of E. coli SCS1 cell extracts (2 μl/lane) in the absence (left) or the presence (right) of traF. Plasmids used are as follows. Lane a, pMS119EH (vector); lane b, pRE178 (trbC+); lane c, pRE178Δ3 (trbCΔ3+); lane d, pRE178Δ3.05 (trbCΔ3.05+); lane e, pRE178Δ3.1 (trbCΔ3.1+); lane f, pRE178Δ4 (trbCΔ4+); lane a′, pJH472 (traF+) and pMS119EH (vector); lane b′, pJH472 (traF+) and pRE178 (trbC+); lane c′, pJH472 (traF+) and pRE178Δ3 (trbCΔ3+); lane d′, pJH472 (traF+) and pRE178Δ3.05 (trbCΔ3.05+); lane e′, pJH472 (traF+) and pRE178Δ3.1 (trbCΔ3.1+); lane f′, pJH472 (traF+) and pRE178Δ4 (trbCΔ4+). Standard molecular mass markers (Rainbow labeled markers, low range; Amersham Pharmacia Biotech): Lys, lysozyme (14.3 kDa), and Apr, aprotinin (6.5 kDa). Positions of the 145-aa PreProTrbC, N-terminally cleaved ProTrbC, N- and C-terminally processed TrbC∗, and (circular) pilin (TrbC) are indicated on the right side of the figure.
Several mutations at the signal peptide cleavage site in the TrbC core are tolerated by LepB and do not affect cyclization by TraF.
Serine at position 37 can be replaced by threonine, alanine, glycine, or cysteine (Fig. 3A) without a recognizable effect on the cyclization reaction of TrbC. In addition, these mutations did not influence the transfer frequency. A proline in the +1 position after the cleavage site terminates processing of the leader peptide (40). Hence, the substitution of S37 by proline was deleterious, probably because of a strong structural disturbance of the signal peptidase moiety (vide supra). Since TrbC cyclization was unaffected by the replacement of residue S37 in most cases, the substrate's specificity was thought to reside someplace else in the TrbC molecule, most likely in its C-terminal processing region.
Specificity for TrbC cyclization resides in residues G112 to I117.
To investigate the C-terminal residues which are important for recognition and/or catalysis of cyclization, each amino acid from position 110 to 117 was replaced individually. No influence on cyclization was detected for residues 110 (F110A/Y), 111 (F111A/Y), 113 (R113A/K), 115 (A115G/S/T), and 117 (I117H) (Fig. 3 and Table 3). Furthermore, mutation of glycines 112 and 114 to alanine or serine had no influence on TrbC processing (Table 3). In contrast, when mutations G112D and G114C/L/T or the alteration of the functional group at position 116 (E116Q) were introduced, these TrbC polypeptides remained at the stage of TrbC* in the presence of TraF (Fig. 3 and Table 3). From the deletion study of the TrbC C-terminal end we knew that residues up to position I117 are required for maturation. Hence, it was unexpected that the replacement of position I117 by histidine would be silent, i.e., that a wild-type cyclic product would be obtained. These data suggest that specificity of the cyclization reaction resides mainly in residues G112 to I117 of TrbC.
Further support for the importance of the specificity residues was found by analysis of highly related TrbC-like proteins as substrates for RP4 TraF. TrbC of the IncPβ plasmid R751, which is conserved to the RP4 TrbC sequence in residues F110 to A118, was processed by RP4 TraF properly, leading to a Tra+ phenotype, whereas TrbC of Agrobacterium tumefaciens Ti plasmid pTiC58, differing from RP4 TrbC in positions F111 to R113 and E113, remained linear (data not shown).
Mutations in the TrbC core affect pilus formation but not cyclization.
Most mutations generated in the TrbC core abolish conjugative transfer, phage propagation, and pilus formation (G42E, G59R, I78S, F79Δ, and V96G; Table 3). However, cyclization of the mutant proteins still takes place, indicating that the cyclized proteins no longer function as substrates for the pilus assembly machinery but do not interfere with the formation of a circular product (Table 3). The TrbC derivatives with glutamine residues instead of glutamates (E38Q, E47Q, and E82Q) displayed wild-type TrbC processing and activity (Table 3). For pilus formation and DNA transfer only the carboxyl group of the glutamate E47 was essential. Exchanging the V at position 106 with M did not affect phenotypic behavior.
Since cyclization of TrbC occurs only when TraF is present (13), it is likely that TraF indeed acts as the enzyme responsible for the formation of the intramolecular peptide bond between S37 and G114 of TrbC. This notion was strongly supported by the differential activity of a series of TraF mutations.
TraF catalyzes intramolecular TrbC cyclization.
Database search and sequence alignment suggested structural and functional relationship of TraF to signal peptidases (19). The structural knowledge of E. coli signal peptidase I (LepB) was exploited to substantiate the proposed functional similarity of TraF to signal peptidases (37). Accordingly, two cysteine residues (C59 and C80) that potentially form a disulfide bridge and a conserved proline at position 129 (P129) were changed. Further, residues in three conserved regions of TraF, those that share significant similarities with the catalytic domain of LepB and its strongly conserved carboxy terminus, were chosen for mutagenesis (Fig. 2, Table 4). The phenotypes of the traF mutants were evaluated in analogy to those of trbC mutants, i.e., transfer frequency, phage propagation, pilus production, and TrbC processing, determined by Western blot analysis and MALDI-TOF-MS (Table 4). The three regions in TraF apparently have functional relevance because defined mutations show reduced or diminished transfer frequencies (S37A, K89Q, K89L, K89R, D155I, D155N, and R157A) (Fig. 3B, Table 4). Two other highly conserved residues (R90L and P129I) among TraF analogs do not seem to be essential for TraF activity, since their mutant phenotypes do not differ from the wild type. Two cysteines (C59A and C80A), which may have structural importance, affect the activity for cyclization of TrbC strongly. For these mutants, simultaneous detection of TrbC* and pilin by MS was possible. The ratio of both species (TrbC*/pilin, as determined by Western blot analysis) was about 1. The highest ratio (3/1) for which both signals could be detected by MS was found in TraFR157A. Further lowering of pilin production, as seen with several TraF mutants (TraFS37A, K89Q/L/R, and D155N), resulted in detection of cyclic, fully processed TrbC by Western blot analysis only. The significantly larger amount of linear TrbC* rather than that of processed pilin suggested a strong effect of these particular mutations in TraF on the final maturation step. However, while this amount of pilin was still sufficient to support conjugation, the formation of pili and propagation of donor specific phages was not detectable.
Donor-specific phage propagation and pilus formation depend on threshold levels of pilin.
Propagation of three IncP-specific bacterial viruses (PRD1, Pf3, and PRR1) was determined for TrbC and TraF mutants. The ratio of TrbC* and pilin was estimated by Western blot analysis. TraF and TrbC mutants which produce fully processed TrbC in comparable amounts to the wild type show normal donor-specific phage propagation and pilus formation. About one-third of TrbC* must be converted to pilin at least, as in derivatives carrying TraFC59A or C80A, before donor phage sensitivity and pilus formation are detectable. If, however, maturation of TrbC is lowered but to a level still sufficient for DNA transfer (TraFS37A, K89Q, K89L, K89R, and D155N) or is lost completely (TraFD155I, TrbCG112D, G114C/L/T, and E116Q), propagation of phages and formation of pili detectable by electron microscopy is completely abolished (Fig. 3B, Table 4). Thus, phage propagation and pilus formation only take place when a certain threshold amount of pilin is produced, i.e., if the TrbC processing cascade proceeds efficiently.
Uncoupling of conjugative DNA transfer from phage propagation.
DNA transfer, pilus assembly, and propagation of phages require the complete TrbC processing cascade to take place. In contrast to a diminished pilin production in several TraF mutants, single-amino-acid exchanges in TrbC specifically inhibit one of the pilin-dependent processes. Detectable pilus assembly and phage plaque formation require more pilin than conjugative DNA transfer, indicating that uncoupling of the phenotypes is possible on a quantitative basis. Mutations trbCF111A and trbCG114A led to a DNA transfer-deficient phenotype but still allowed production of pilin and propagation of PRD1, PRR1, and Pf3 (Table 3). For TrbCG114A, bundles of pili could be detected by electron microscopy. Thus, DNA transfer is independent from pilus formation. In contrast, TrbCF110A and TrbCA115G abolished phage plaque formation completely, although pilin production and DNA transfer were comparable to those of the wild type (Table 3). Hence, formation of a pilus consisting of mutagenized pilin (TrbCA115G, F110A) might not be sufficient for propagation of phages in these cases. The two phenotypes, Tra+ Dps− and Tra− Dps+, demonstrated that conjugative DNA transfer and phage propagation can be uncoupled from each other by mutagenesis of TrbC.
Propagation of PRD1 and PRR1-Pf3 utilize two different Tra2-encoded receptor structures.
PRD1 needs 11 components of the Mpf system for propagation (18, 20). Nonetheless, the receptor protein on the host cell has not been identified (16, 20). Earlier studies of PRR1 and Pf3 showed the adsorption of these phages to IncP pili (8).
In phage adsorption experiments traced by electron microscopy, we were able to distinguish between direct cell surface attachment of PRD1 and binding of PRR1-Pf3 to the extended pilus (data not shown). Although the pilus is not the receptor for PRD1, some mutations in trbC abolish PRD1 propagation. Such mutations always result in an additional PRR1-Pf3-negative phenotype (Table 3 and reference 18). On the other hand, three mutations in trbC (R113A/K, G114S) have been found that abolish PRR1 or Pf3 attachment but still allow the adsorption of PRD1 to the host cell. The TrbCR113A mutant no longer assembles pili, whereas the TrbCR113K and TrbCG114S mutants still show bundles of pili. Thus, for PRR1 or Pf3 the receptor is the pilin assembled in a conjugative pilus, whereas for PRD1 it remains an open question if a special TrbC structure functions as the target on the cell surface.
DISCUSSION
In this study we show that RP4 traF encodes an enzyme responsible for a highly specialized cutting-joining reaction, producing a cyclic polypeptide. TraF's enzymatic activity is essential for RP4-mediated conjugative transfer, for the assembly of functional receptors of donor-specific phages PRD1, PRR1, and Pf3, and for synthesis of conjugative pili. TraF processes the pilus subunit TrbC in nonpolar Tra2 mutants, as well as in several TrbC point mutants. Less pilin is needed to lead to a transfer-positive phenotype than for formation of visible pili and adsorption of bacterial viruses (Fig. 3B). The assembly of pilin into the pilus structure is strongly dependent on the configuration of TrbC. Each of the point mutations in the TrbC core (G42E, E47Q, G59R, I78S, F79Δ, and V96G) yielded a circular product, but none of these mutants produced detectable amounts of pili. Moreover, each of these mutants was transfer deficient. Assembly of a pilus seems to be another highly specific process coordinated by the remaining essential functions (TrbB, -D, -E, -F, -G, -H, -I, -J, and -L) of the Mpf system (17). This specificity might depend on translational coupling of trbC with the preceding gene trbB, the product of which is the hexameric NTPase TrbB, a protein belonging into the VirB11 group (25, 26). The weak NTPase activity of the ring-shaped molecule which is associated with the inner membrane could serve as a chaperone-like function in the pilus assembly process.
Mutations in the C-terminal processing region of TrbC led to different phenotypes. Three mutants (TrbCR113A/K and G114S) lost Pf3 and PRR1 adsorption activity but supported DNA transfer and PRD1 propagation. Moreover, in the TrbCG114A mutant, bundles of pili are detectable and phages PRR1 and Pf3 still infect the cells, but no conjugative transfer of DNA occurred (Table 3). When TrbCG114S is expressed in the absence of TraF, a detectable amount of protein showed the same mobility as fully processed pilin in Western blot analysis (data not shown); only the unprocessed form was detected by MS. This indicated a possible TrbC truncation completely independent of TraF. Due to missing phage adsorption, missing pilus formation, and no DNA transfer abilities of this mutant in the absence of TraF, a functional (circular) pilin was excluded but cannot be ruled out completely, in particular because Edman sequencing was unsuccessful. Since no functional influence on phenotypes could be assigned, our studies did not focus further on this mutant. Nonetheless, position G114 inhabits a key role in the processing reaction of TrbC; thus, alterations are only tolerated in a limited manner.
From the pattern of conserved amino acid residues in TraF and TraF analogs compared to that of the various signal peptidases and from the results obtained in this study, we concluded that the mechanism of the cleavage reaction and the targeting of the peptidase domain in the periplasm resemble those of signal peptidases (Fig. 2). The N termini of prokaryotic and eukaryotic signal peptidases are anchored in the cytoplasmic membrane by one or more hydrophobic transmembrane helices (10, 12, 19). These segments are not directly involved in catalysis but are important for the correct localization of the catalytic domain at the periplasmic side of the cytoplasmic membrane (for a review, see reference 12). No cyclization of TrbC* could be observed when the proposed transmembranal helix of RP4 TraF (residues 10 to 28 [19]) was deleted. The truncated mutant protein probably remains in the cytoplasm and thus cannot fulfill its function in the processing cascade of TrbC.
Signal peptidases belong to a unique class of serine proteases (6, 36, 50). Similar to LexA-like proteases, their catalytic activity depends on a serine-lysine dyad-like mechanism (5, 53). In analogy to chromosome-encoded signal peptidases of E. coli and B. subtilis, site-directed mutagenesis of the proposed active site residues S37 and K89 of TraF leads to gene products that display reduced activity and that do not support the synthesis of conjugative pili. However, all mutations introduced into the proposed catalytically active center of TraF still had a residual transfer activity of at least 1 in 1,000 donor cells. This could be explained by a very low proteolytic cleavage activity of mutant TraF, demonstrated by Western blot analysis. Evidence for the existence of very short, rod-like pilus stumps on the cell surface is not given at present. Rod-like stumps might be sufficient to establish the cell-to-cell contact between donor and recipient, especially in the high cell density of the filter assay.
Since S37 and K89 of RP4 TraF are conserved in all known TraF-like proteins (Fig. 2) and almost all known prokaryotic type I signal peptidases, we hypothesize that all of these enzymes function on the base of a catalytic serine-lysine dyad-like mechanism. The putative active site lysine residue could not be replaced by histidine in SipS of B. subtilis (53) or in LepB (51) and LexA of E. coli (29) without substantial loss of activity. Likewise, the structurally related eukaryotic type I signal peptidases Sec11, Spc18, and Spc21, which contain a conserved histidine residue at the position of the catalytic lysine residue of the prokaryotic type I signal peptidases, catalyze signal peptide cleavage by a different mechanism (52).
Aspartic acid 153 of B. subtilis SipS (D155 in RP4 TraF) is important for catalysis (53). This residue is conserved in other signal peptidases and in each of the TraF analogs. Changing the respective aspartate in RP4 TraF to isoleucine (D155I) led to a phenotype totally defective in TrbC maturation. No pilus production, phage adsorption, or transfer of DNA could be observed. When the aspartate was replaced by an asparagine (D155N), no pili were visible, phage adsorption was lost, and the transfer rate was reduced dramatically (10−3, Table 4). Thus, D155 fulfills an important role in the process of cyclization and represents, in addition to S37 and K89, a third essential residue of TraF. However, it is conceivable that this residue is specific for TraF-like proteases since it is highly conserved in this group of proteins (Fig. 2), but its function remains speculative. Possibly, D155 is part of an important structural element which might also include other residues of the conserved region at the C-terminal end of the TraF-like proteins. The latter hypothesis is supported by several observations. First, replacement of R157 by alanine caused a drastic reduction of TraF activity. Second, S153, Y158, F159, and G160 are highly conserved residues in all TraF-like proteases (Fig. 2).
The proposed mechanism for proteolytic processing by signal peptidases is based on the data of the autoproteolysis of E. coli LexA, an intramolecular process, catalyzed by S119 and K156 (30). Upon self-cleavage, LexA loses repressor activity and some 20 SOS functions of the cell are derepressed. The nucleophilic hydroxyl group of residue S119 attacks the carbonyl carbon of the scissile peptide bond, while the amino group of lysine 156 acts as a general base. Thus, the self-cleavage reaction of LexA would proceed through a covalent tetrahedral intermediate and an acyl-enzyme intermediate, as shown for the hydrolysis of peptide bonds by serine proteases (36). However, the proposed mechanism for LexA differs from that of the classical serine proteases because general base catalysis is carried out by a lysine side chain instead of the imidazole ring of the histidine. Based on the present data, we suggest that TraF also makes use of a serine-lysine catalytic dyad-like mechanism: the hydroxyl group of residue S37 of TraF acts as the nucleophile attacking the carbonyl carbon of the scissile peptide bond at the C-terminal end of the pilin precursor (Fig. 5). The deprotonated form of the ɛ-amino group of K89 would serve to activate the hydroxyl group of S37. The lysine's ɛ-amino group must be deprotonated to act as a general base. The microenvironment surrounding the lysine could include either a local positive charge or a hydrophobic moiety, thus lowering its pKa. There are many examples of lysine residues having pKa values significantly lower than 10.5 (39). The low pKa would allow the lysine to act as the general base and promote catalysis. We propose the formation of an oxyanion hole which might be stabilized by hydrogen bridge formation to a proximal arginine or any other proton bridge donor. However, crystallographic and mutational data indicated that the proximal arginine in LepB does not support such a formation (36). To exclude the possible importance of the proximal arginine R90 in RP4 TraF, this residue was changed to leucine. Comparable to LepB no decrease in activity resulted from this mutation, indicating that R90 does not influence the putative oxyanion hole.
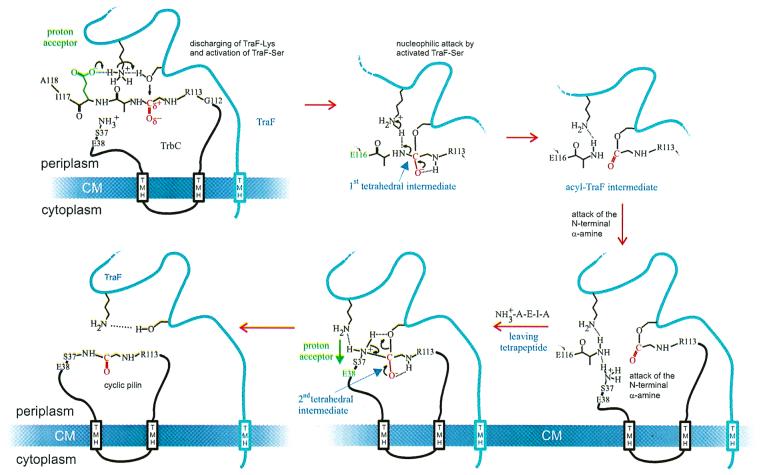
FIG. 5.
Proposed mechanism for the TraF-catalyzed formation of an internal peptide bond in TrbC. For details, see Discussion.
The TraF-like proteins and the type I signal peptidases differ fundamentally in the formation of a new peptide bond in the target protein, namely, the pilin. This unique type of reaction is likely to be coupled with the removal of the tetra peptide (A115 to A118), since no intermediate of TrbC was found in our analyses of all TrbC and TraF mutants. Either TrbC is processed and cyclized or the protein remains unprocessed and linear. Our proposed mechanism for TraF catalysis of pilin maturation diverges from the model proposed for signal peptidases after the first tetrahedral intermediate is formed (Fig. 5). The resulting TraF acyl intermediate might conserve the energy, which would be freed by breaking the peptide bond between G114 and A115, explaining why the C-terminal deletion mutant of TrbC ending at position G114 could not be cyclized by TraF. Deletions ending with I117 or H117 or else A118 were indeed cyclized (Fig. 4), showing that at the least, a triple peptide must be present. In the reaction mechanism described for LepB, the energy is set free by loss of a water molecule to the environment (11), resulting in the hydrolysis of the acyl intermediate. It is known that these acyl intermediates can react with other nucleophiles in hydrophilic environments (15, 42). Most probably a comparable reaction is driven by TrbC-acyl-TraF.
Binding of S37 to G114 of the immobilized and activated TrbC would be the next step of the reaction. The pKa of the serine α-amino group should be 9.21 and thus protonated under physiological conditions. Therefore, similar to the mechanism for protein splicing (38), the binding to the C terminus could occur by formation of an ester bond with the serine side chain which afterward is transformed to a peptide bond by acyl replacement. We showed that the α-amino group of S37 and not the hydroxyl function is involved in ring formation. Since each of the mutants TrbCS37A/C/G lacking a side chain hydroxyl yielded cyclic peptides, we conclude that the ɛ-amino group of S37 and not the hydroxyl group is involved in ring formation. A protonated N terminus would transfer its proton to the removed tetrapeptide and at the same time attack the acylic function, while forming a second tetrahedral intermediate as shown in Fig. 5. This would result in cyclic TrbC and the restored TraF. When S37 of TrbC was replaced by bulkier residues such as threonine and cysteine, the cyclization of TrbC still took place, but much less effectively, indicating possible steric hindrance.
Whatever mechanism takes place, the biogenesis of the mature pilin appears to be a concerted reaction involving cleavage and rejoining of a linear, doubly truncated peptide. The absence of a circular peptide in the case of the TrbC G114 deletion mutant supports this notion, suggesting that the removal of four amino acid residues (A115 to A118) by TraF peptidase is an intrinsic step in cyclization of the pilin. However, at present, our data may not exclude the possibility that an additional chromosome-encoded enzyme is involved in forming the peptide linkage, especially since the identity of the chromosome-encoded enzyme which performs the first C-terminal cleavage on TrbC remains unknown. Potential candidates to carry out this task are cytoplasmic peptidases such as Lon, which are specific for maturation between hydrophilic residues (31), or the tail-specific protease Prc, which is located in the cytoplasmic membrane (22, 31).
Circular peptides used as subunits of extrafilamentous structures might be found not only in conjugative systems but also in some of the proposed macromolecular secretion systems of human and animal pathogens (Fig. 6). Three sequences, the pertussis toxin operon of Bordetella pertussis (55) and two recently identified similar virulence operons of Brucella suis (34) and Brucella abortus (46), contain potential pilin subunits, PtlA and VirB2, respectively (Fig. 6). The percentage of identity with TrbC of RP4 is, for the overall protein sequence, rather low. A more detailed sequence comparison revealed that two alanine residues directly preceding the signal peptide cleavage site in TrbC are found as well at comparable positions in PtlA and VirB2 (Fig. 6). Signal peptide predictions for the latter two proteins using the SignalP V1.1 program (http://www.cbs.dtu.dk/services/SignalP/) revealed a potential signal peptide cleavage site following the second of these alanines for both sequences (33). Moreover, a sequence identical to the sequence required for the leaving tetrapeptide in TrbC (AEIA) is found at similar positions relative to TrbC in PtlA and in VirB2 (Fig. 6). Future studies are required to evaluate whether circular pilin-like proteins are present in these secretion systems.
FIG. 6.
Potential common processing mechanism of RP4 TrbC (M93696), Bordetella pertussis PtlA (L10720), Brucella suis VirB2 (AF141604), and Brucella abortus VirB2 (AF226278). The latter two sequences are identical and shown once only. Databank GenBank accession numbers are given in parentheses. Only two short portions of the respective sequences are shown. The first and last amino acid positions in each line are numbered according to the original sequences. Lines correspond to conserved residues in all sequences; colons in only two of the sequences. Residues of a proposed common maturation process are printed in white. Arrows mark sites of RP4 TrbC processing.
ACKNOWLEDGMENTS
We thank Hans Lehrach for generous support. We also thank Stephen K. Farrand for providing plasmids pPLtraF and pPLtrbC and for critical reading of the manuscript. The project was stimulated by discussions within the EU-BIOTECH concerted action BIO4-CT-0099, Mobile genetic Elements' Contribution to Bacterial Adaptability and Diversity (MECBAD). The expert technical assistance of Marianne Schlicht is greatly appreciated.
Work in E. Lanka's laboratory was supported by Sonderforschungsbereich grant 344/A8 of the Deutsche Forschungsgemeinschaft. M. Kalkum was supported by BMBF grant 0311018.
REFERENCES
- 1.Anthony K G, Sherburne C, Sherburne R, Frost L S. The role of the pilus in recipient cell recognition during bacterial conjugation mediated by F-like plasmids. Mol Microbiol. 1994;13:939–953. doi: 10.1111/j.1365-2958.1994.tb00486.x. [DOI] [PubMed] [Google Scholar]
- 2.Balzer D, Pansegrau W, Lanka E. Essential motifs of relaxase (TraI) and TraG proteins involved in conjugative transfer of plasmid RP4. J Bacteriol. 1994;176:4285–4295. doi: 10.1128/jb.176.14.4285-4295.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balzer D, Ziegelin G, Pansegrau W, Kruft V, Lanka E. KorB protein of promiscuous plasmid RP4 recognizes inverted sequence repetitions in regions essential for conjugative plasmid transfer. Nucleic Acids Res. 1992;20:1851–1858. doi: 10.1093/nar/20.8.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bamford D H, Rouhiainen L, Takkinen K, Söderlund H. Comparison of the lipid-containing bacteriophages PRD1, PR3, PR4, PR5 and L17. J Gen Virol. 1981;57:365–373. doi: 10.1099/0022-1317-57-2-365. [DOI] [PubMed] [Google Scholar]
- 5.Black M T. Evidence that the catalytic activity of prokaryote leader peptidase depends upon the operation of a serine-lysine catalytic dyad. J Bacteriol. 1993;175:4957–4961. doi: 10.1128/jb.175.16.4957-4961.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black M T, Munn J G R, Allsop A E. On the catalytic mechanism of prokaryotic leader peptidase I. Biochem J. 1992;282:539–543. doi: 10.1042/bj2820539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 8.Bradley D E. Adsorption of bacteriophages specific for Pseudomonas aeruginosaR factor RP1 and R1822. Biochem Biophys Res Commun. 1974;57:893–900. doi: 10.1016/0006-291x(74)90630-5. [DOI] [PubMed] [Google Scholar]
- 9.Bradley D E. Determination of pili by conjugative bacterial drug resistance plasmids of incompatibility groups B, C, H, J, K, M, V, and X. J Bacteriol. 1980;141:828–837. doi: 10.1128/jb.141.2.828-837.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dalbey R E. Leader peptidase. Mol Microbiol. 1991;5:2855–2860. doi: 10.1111/j.1365-2958.1991.tb01844.x. [DOI] [PubMed] [Google Scholar]
- 11.Dalbey R E, Lively M O, Bron S, van Dijl J M. The chemistry and enzymology of the type I signal peptidases. Protein Sci. 1997;6:1129–1138. doi: 10.1002/pro.5560060601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalbey R E, von Heijne G. Signal peptidases in prokaryotes and eukaryotes—a new protease family. Trends Biochem Sci. 1992;17:474–478. doi: 10.1016/0968-0004(92)90492-r. [DOI] [PubMed] [Google Scholar]
- 13.Eisenbrandt R, Kalkum M, Lai E-M, Lurz R, Kado C I, Lanka E. Conjugative pili of IncP plasmids, and the Ti plasmid T pilus are composed of cyclic subunits. J Biol Chem. 1999;274:22548–22555. doi: 10.1074/jbc.274.32.22548. [DOI] [PubMed] [Google Scholar]
- 14.Frost L S, Lee J S, Scraba D G, Paranchych W. Two monoclonal antibodies specific for different epitopes within the amino-terminal region of F pilin. J Med Microbiol. 1986;168:192–198. doi: 10.1128/jb.168.1.192-198.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaertner H, Watanabe T, Sinisterra J V, Puigserver A. Peptide synthesis catalyzed by modified α-chymotrypsin in low-water organic media. J Org Chem. 1991;56:3149–3153. [Google Scholar]
- 16.Grahn A M, Caldentey J, Bamford J K H, Bamford D H. Stable packaging of phage PRD1 DNA requires adsorption protein P2, which binds to the IncP plasmid-encoded conjugative transfer complex. J Bacteriol. 1999;181:6689–6696. doi: 10.1128/jb.181.21.6689-6696.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grahn A M, Haase J, Bamford D H, Lanka E. Components of the RP4 conjugative transfer apparatus form an envelope structure bridging inner and outer membranes of donor cells: implications for related macromolecule transport systems. J Bacteriol. 2000;182:1564–1574. doi: 10.1128/jb.182.6.1564-1574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grahn A M, Haase J, Lanka E, Bamford D H. Assembly of a functional phage PRD1 receptor depends on 11 genes of the IncP plasmid mating pair formation complex. J Bacteriol. 1997;179:4733–4740. doi: 10.1128/jb.179.15.4733-4740.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haase J, Lanka E. A specific protease encoded by the conjugative DNA transfer system of IncP and Ti plasmids is essential for pilus synthesis. J Bacteriol. 1997;179:5728–5735. doi: 10.1128/jb.179.18.5728-5735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haase J, Lurz R, Grahn A M, Bamford D H, Lanka E. Bacterial conjugation mediated by plasmid RP4: RSF1010 mobilization, donor-specific phage propagation, and pilus production require the same Tra2 core components of a proposed DNA transport complex. J Bacteriol. 1995;177:4779–4791. doi: 10.1128/jb.177.16.4779-4791.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanahan D. Studies on transformation of Escherichia coliwith plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 22.Hara H, Nishimura Y, Kato J, Suzuki H, Nagasawa H S A, Hirota Y. Genetic analyses of processing involving C-terminal cleavage in penicillin-binding protein 3 of Escherichia coli. J Bacteriol. 1989;171:5882–5889. doi: 10.1128/jb.171.11.5882-5889.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ippen-Ihler K A, Minkley E G J. The conjugation system of F, the fertility factor of Escherichia coli. Annu Rev Genet. 1986;20:593–624. doi: 10.1146/annurev.ge.20.120186.003113. [DOI] [PubMed] [Google Scholar]
- 24.Jacobson A. Role of F pili in the penetration of bacteriophage F1. J Virol. 1972;10:835–843. doi: 10.1128/jvi.10.4.835-843.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krause S, Bárcena M, Pansegrau W, Lurz R, Carazo J M, Lanka E. Sequence related protein export NTPases encoded by the conjugative transfer region of RP4 and by the cag pathogenicity island of Helicobacter pylorishare similar hexameric ring structures. Proc Natl Acad Sci USA. 2000;97:3067–3072. doi: 10.1073/pnas.050578697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krause S, Pansegrau W, Lurz R, de la Cruz F, Lanka E. Enzymology of type IV macromolecule secretion systems; the conjugative transfer regions of plasmids RP4 and R388 and the cag pathogenicity island of Helicobacter pyloriencode structurally and functionally related nucleoside triphosphate hydrolases. J Bacteriol. 2000;182:2761–2770. doi: 10.1128/jb.182.10.2761-2770.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lessl M, Balzer D, Lurz R, Waters V L, Guiney D G, Lanka E. Dissection of IncP conjugative plasmid transfer: definition of the transfer region Tra2 by mobilization of the Tra1 region in trans. J Bacteriol. 1992;174:2493–2500. doi: 10.1128/jb.174.8.2493-2500.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lessl M, Balzer D, Weyrauch K, Lanka E. The mating pair formation system of plasmid RP4 defined by RSF1010 mobilization and donor-specific phage propagation. J Bacteriol. 1993;175:6415–6425. doi: 10.1128/jb.175.20.6415-6425.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin L L, Little J W. Autodigestion and RecA-dependent cleavage of Ind-mutant LexA proteins. J Mol Biol. 1989;210:439–452. doi: 10.1016/0022-2836(89)90121-6. [DOI] [PubMed] [Google Scholar]
- 30.Little J W. LexA cleavage and other self-processing reactions. J Bacteriol. 1993;175:4943–4950. doi: 10.1128/jb.175.16.4943-4950.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller C G. Protein degradation and proteolytic modification. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 938–954. [Google Scholar]
- 32.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. pp. 431–433. [Google Scholar]
- 33.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Prot Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 34.O'Callaghan D, Cazevieille C, Allardet-Servent A, Boschiroli M L, Bourg G, Foulongne V, Frutos P, Kulakov Y, Ramuz M. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol Microbiol. 1999;33:1210–1220. doi: 10.1046/j.1365-2958.1999.01569.x. [DOI] [PubMed] [Google Scholar]
- 35.Olsen R H, Siak J-S, Gray R H. Characteristics of PRD1, a plasmid-dependent broad host range DNA bacteriophage. J Virol. 1974;14:689–699. doi: 10.1128/jvi.14.3.689-699.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paetzel M, Dalbey R E. Catalytic hydroxyl/amine dyads within serine proteases. Trends Biochem Sci. 1997;22:28–31. doi: 10.1016/s0968-0004(96)10065-7. [DOI] [PubMed] [Google Scholar]
- 37.Paetzel M, Dalbey R E, Strynadka N C J. Crystal structure of a bacterial signal peptidase in complex with a β-lactam inhibitor. Nature. 1998;396:186–190. doi: 10.1038/24196. [DOI] [PubMed] [Google Scholar]
- 38.Perler F B. Protein splicing of inteins and hedgehog autoproteolysis: structure, function, and evolution. Cell. 1998;92:1–4. doi: 10.1016/s0092-8674(00)80892-2. [DOI] [PubMed] [Google Scholar]
- 39.Planas A, Kirsch J F. Reengineering the catalytic lysine of aspartate aminotransferase by chemical elaboration of a genetically introduced cysteine. Biochemistry. 1991;30:8268–8276. doi: 10.1021/bi00247a023. [DOI] [PubMed] [Google Scholar]
- 40.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riechmann L, Holliger P. The C-terminal domain of TolA is the coreceptor for filamentous phage infection of E. coli. Cell. 1997;90:351–360. doi: 10.1016/s0092-8674(00)80342-6. [DOI] [PubMed] [Google Scholar]
- 42.Rose K, Stöcklin R, Savoy L-A, Regamey P-O, Offord R E, Vuagnat P, Markussen J. Reaction mechanism of trypsin-catalysed semisynthesis of human insulin studied by fast atom bombardment mass spectrometry. Prot Eng. 1991;4:409–412. doi: 10.1093/protein/4.4.409. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 44.Samuels A L, Lanka E, Davies J E. Conjugative junctions in RP4-mediated mating of Escherichia coli. J Bacteriol. 2000;182:2709–2715. doi: 10.1128/jb.182.10.2709-2715.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sieira R, Comerci D J, Sanchez D O, Ugalde R A. A homologue of an operon required for DNA transfer in Agrobacterium is required in Brucella abortusfor virulence and intracellular multiplication. J Bacteriol. 2000;182:4849–4855. doi: 10.1128/jb.182.17.4849-4855.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stanisich V A. The properties and host range of male-specific bacteriophages of Pseudomonas aeruginosa. J Gen Microbiol. 1974;84:332–342. doi: 10.1099/00221287-84-2-332. [DOI] [PubMed] [Google Scholar]
- 48.Stoops S C, Momany C, Ernst S R, Oliver R M, Schroeter J P, Bretaudiere J-P, Hackert M L. Comparison of the low-resolution structures of ornithine decarboxylase by electron microscopy and X-ray crystallography: the utility of methylamine tungstate stain and Butvar support film in the study of macromolecules by transmission electron microscopy. J Electron Microsc Tech. 1991;18:157–166. doi: 10.1002/jemt.1060180210. [DOI] [PubMed] [Google Scholar]
- 49.Strack B, Lessl M, Calendar R, Lanka E. A common sequence motif, -E-G-Y-A-T-A-, identified within the primase domains of plasmid-encoded I- and P-type DNA primases and the α protein of the Escherichia colisatellite phage P4. J Biol Chem. 1992;267:13062–13072. [PubMed] [Google Scholar]
- 50.Sung M, Dalbey R E. Identification of potential active-site residues in the Escherichia colileader peptidase. J Biol Chem. 1992;267:13154–13159. [PubMed] [Google Scholar]
- 51.Tschantz W R, Sung M, Delgado-Partin V M, Dalbey R E. A serine and a lysine residue implicated in the catalytic mechanism of the Escherichia colileader peptidase. J Biol Chem. 1993;268:27349–27354. [PubMed] [Google Scholar]
- 52.van Dijl J M, de Jong A, Vehmaanperä J, Venema G, Bron S. Signal peptidase I of Bacillus subtilis: pattern of conserved amino acids in prokaryotic and eukaryotic type I signal peptidases. EMBO J. 1992;11:2819–2828. doi: 10.1002/j.1460-2075.1992.tb05349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Dijl J M, de Jong A, Venema G, Bron S. Identification of the potential active site of the signal peptidase SipS of Bacillus subtilis: structural and functional similarities with LexA-like proteases. J Biol Chem. 1995;270:3611–3618. doi: 10.1074/jbc.270.8.3611. [DOI] [PubMed] [Google Scholar]
- 54.Waters V L, Strack B, Pansegrau W, Lanka E, Guiney D G. Mutational analysis of essential IncPα plasmid transfer genes traF and traG and involvement of traFin phage sensitivity. J Bacteriol. 1992;174:6666–6673. doi: 10.1128/jb.174.20.6666-6673.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weiss A A, Johnson F D, Burns D L. Molecular characterization of an operon required for pertussis toxin secretion. Proc Natl Acad Sci USA. 1993;90:2970–2974. doi: 10.1073/pnas.90.7.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zechner E L, de la Cruz F, Eisenbrandt R, Grahn A M, Koraimann G, Lanka E, Muth G, Pansegrau W, Thomas C M, Wilkins B M, Zatyka M. Conjugative DNA transfer processes. In: Thomas C M, editor. The horizontal gene pool, bacterial plasmids and gene spread. Amsterdam, The Netherlands: Harwood Academic Publishers GmbH; 2000. pp. 87–174. [Google Scholar]