Abstract
Products encoded in the trans-acting factor (TAF) region are necessary for the biosynthesis of anguibactin and for maximal expression of iron transport and biosynthesis genes in the plasmid-encoded iron-scavenging system of Vibrio anguillarum. Here we identify angB, a locus located in the TAF region, which encodes products essential for anguibactin biosynthesis. We demonstrate that a 287-amino-acid polypeptide, encoded by angB and designated AngB, has an isochorismate lyase activity necessary for the synthesis of 2,3-dihydroxybenzoic acid, an anguibactin biosynthesis intermediate. Complementation of various angB mutations provided evidence that an additional, overlapping gene exists at this locus. This second gene, designated angG, also has an essential biosynthetic function. The angG gene directs the expression of three polypeptides when overexpressed in Escherichia coli, all of which are translated in the same frame as AngB. The results of site-directed mutagenesis and in vivo phosphorylation experiments suggest that the carboxy-terminal end of AngB and the AngG polypeptide(s) function as aryl carrier proteins involved in the assembly of the anguibactin molecule. Our results also show that the regulatory functions of the TAF are encoded in a region, TAFr, which is distinct from and independent of the angB and angG genes.
Bacteria of the genus Vibrio are commonly found as etiologic agents of disease in both humans and animals (1). An important member of this group is Vibrio anguillarum, a marine bacterium which is responsible for both marine and freshwater fish epizootics throughout the world (10, 26). The disease caused by V. anguillarum has remarkable similarities to invasive-septicemic disease in humans, and for that reason the V. anguillarum-host fish system is an ideal paradigm for studying a native eukaryotic host-pathogen interaction leading to disease (9, 15–17, 19).
Iron is an essential element for nearly all microorganisms, yet in biological fluids it exists only as a complex with iron-binding proteins, making it essentially unavailable (9, 18). Therefore, invasive microorganisms must have the ability to use complexed iron in order to grow within their hosts. Highly virulent strains of the marine pathogen V. anguillarum possesses a 65-kb virulence plasmid encoding an iron-scavenging system which is essential for virulence in this bacterium (19–21, 54). This system consists of a low-molecular-weight iron-binding compound, anguibactin; once secreted, this compound competes for bound iron within the host fish. The iron-anguibactin complex is then internalized by an energy-dependent transport system which includes the FatA, -B, -C, and -D proteins (2–5, 27). The genes encoding proteins involved in the biosynthesis and transport of anguibactin lie on a 25-kb contiguous region of the virulence plasmid. However, to achieve maximal expression of the system, products from a noncontiguously located region of the plasmid are necessary (48). This poorly defined region, referred to as the trans-acting factor (TAF) region, encodes a product(s), TAF, which acts to increase the transcription of the anguibactin transport-biosynthesis operon, containing fatDCBAangRT, as well as to increase dramatically the rate of synthesis of the anguibactin molecule itself (37, 48). It has been suggested that rather than encoding a biosynthesis activity directly, TAF acts as an indirect regulator of anguibactin biosynthesis gene expression (37, 48). Further positive regulation of anguibactin biosynthesis and transport gene expression is mediated by the plasmid-encoded AngR (for anguibactin system regulator) protein (22). We were recently able to assign the regulatory functions of AngR to the amino-terminal end of the AngR protein (52). Recently, we have shown that AngR, in addition to being a regulator, possesses sequences indicative of nonribosomal peptide synthetases, suggesting that it may play a role as an anguibactin biosynthesis enzyme (25, 28, 39, 42, 52). Negative iron regulation of this system is mediated by the chromosomally encoded Fur protein, as well as the Fur-regulated cis-acting antisense RNA α molecule (11, 12, 29, 36, 44, 51, 53).
We report here the dissection of the TAF into two separable entities: an anguibactin biosynthesis function (TAFb) and a trans-acting regulatory function (TAFr). The TAFb region harbors two overlapping genes, angB and angG. The latter is located within the 3′ end of the angB coding region. The AngB protein appears to be bifunctional, with its amino end conferring an isochorismate lyase activity required for the production of 2,3-dihydroxybenzoic acid (DHBA) and its carboxyl end likely serving as an aryl carrier protein functioning in anguibactin assembly. We also demonstrate that the carboxy-terminal end of AngB can be synthesized from the angG gene as an independent polypeptide which, like AngB, also functions as an aryl carrier protein involved in anguibactin assembly.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
V. anguillarum 531A(pJHC1) harbors one plasmid, pJHC1, which is a pJM1-like plasmid containing the iron transport, siderophore biosynthesis, and TAF genes. This strain was used because it is recombination proficient, therefore allowing the placement of mutations directly on the pJHC1 plasmid (47). Table 1 shows the strains and plasmids used in this endeavor.
TABLE 1.
Bacterial strains and plasmids
| Bacterial strain or plasmid | Description | Source or reference |
|---|---|---|
| Bacterial strains | ||
| E. coli | ||
| DH5α | supE44 hsdR17 recA1 endA1 gyrA96 thi-1 ΔlacU169 relA1 (φ80lacZΔM15) | BRL |
| JM109λpir | recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 Δ(lac-proAB+) [F′ traD36, proAB lacIqlacZΔM15] λpir | Lab collection |
| V. anguillarum | ||
| 775(pJM1) | Wild type; Pacific Ocean prototype | 19 |
| 531A(pJHC1) | Wild type; Atlantic Ocean prototype | 49 |
| S531A-1 | Plasmidless derivative of 531A | This work |
| 531A(pJHC1δ) | pJHC1 deletion derivative; TAF− | This work |
| 531A(pJHC1::Ω) | pJHC1 derivative containing the Ω transcription-translation terminator at the BglII site in angB; B− G− | This work |
| 531A(pJHC1::K) | pJHC1 derivative containing a Klenow modification at the BglII site in angB; B− G+ | This work |
| 531A(pJHC1::ORFFδ) | pJHC1 derivative containing an in-frame deletion in ORF F | This work |
| Plasmids | ||
| pJM1 | Indigenous plasmid in strain 775 | 17 |
| pJHC1 | Indigenous plasmid in strain 531A | 49 |
| pJHC-S100 | Kmr-encoding cloning vector derived from pBR325 | 37 |
| pBluescript SK | Cloning vector | Stratagene |
| pBR322 | Cloning vector | 8 |
| pTW99 | pBR322 with kanamycin resistance gene from pUC4K cloned into SalI site | This work |
| pTW100 | TAFb region cloned as a BamHI-SalI fragment into pJHC-S100 | This work |
| pTW104 | Derivative of pTW100 containing ORF B as an EcoRV-SpeI fragment | This work |
| pTW105 | pTW104 with the Ω fragment cloned into the BglII site | This work |
| pTW200 | angB and angG cloned by PCR into pTW99 | This work |
| pTW201 | angB cloned by PCR into pTW99 | This work |
| pTW202 | EcoRV-BglII deletion derivative of pTW200 | This work |
| pTW203 | pTW200 containing a Klenow modification at the BglII site in angB | This work |
| pTW200S248L | pTW200 containing an S248L mutation introduced by site-directed mutagenesis | This work |
| pTW203S248L | pTW203 containing an S248L mutation introduced by site-directed mutagenesis | This work |
| pQE60 | T5 phage promoter expression vector | Qiagen |
| pQE60-B+ G+ | angB and angG cloned into pQE60 | This work |
| pQE60-B− G+::K | pQE60-B+ G+ containing a Klenow modification at the BglII site in angB | This work |
V. anguillarum was cultured at 28°C in trypticase soy broth supplemented with 1% NaCl or trypticase soy agar supplemented with 1% NaCl (TSAS). For experiments determining iron uptake characteristics, strains were grown in M9 minimal medium (19) supplemented with 0.2% Casamino Acids (CM9) (Difco Laboratories). To achieve iron-restrictive conditions, ethylenediamine-di-(o-hydroxyphenylacetic acid) (EDDA) was added to the medium. Escherichia coli strains were grown at 37°C in Luria broth or on Luria agar. When required, antibiotics (Sigma) were added at the following concentrations: ampicillin, 500 μg/ml for V. anguillarum and 100 μg/ml for E. coli; kanamycin, 200 μg/ml for V. anguillarum and 50 μg/ml for E. coli; tetracycline, 10 μg/ml; streptomycin, 100 μg/ml; and rifampin, 100 μg/ml.
Marker exchange-eviction mutagenesis.
To introduce mutations into the wild-type pJHC1 plasmid, we used a procedure that was based on a method described by Reid and Collmer (34), employing a suicide vector (pTW-MEV) which contained the R6K origin of replication, the sacB gene, and an ampicillin resistance gene. Mutation-containing suicide plasmids were transferred to V. anguillarum 531A(pJHC1) by conjugation. Plasmid integration was confirmed by restriction endonuclease analysis of purified pJHC1 DNA. Plasmid cointegrates were then plated on TSAS containing 10% sucrose in order to identify individuals that had undergone a second recombination event which led to sacB excision. Some of these sucrose-resistant clones were generated by a resolution event leading to the replacement of the wild-type allele on pJHC1 by the mutant allele. Successful replacement mutants were identified by restriction endonuclease analysis of purified pJHC1 DNA.
To generate the ∼20-kb deletion that resulted in plasmid pJHC1∂, we created a plasmid cointegrate which, after the first crossover event, carried both a kanamycin resistance-encoding fragment, inserted within open reading frame C (ORF C), and the sacB gene. pJHC1 harbors various repeated insertion sequences, and it was our goal to generate, via recombination events occurring between these sequences, large deletions, especially in the TAF region. Since the ORF C kanamycin resistance-encoding insertion did not affect anguibactin production, final selection with sucrose was done in the absence of kanamycin to allow the identification of a broader array of recombinants. Using this method, we were able to isolate several pJHC1∂-like deletions in multiple independent experiments. By restriction endonuclease analysis, PCR, and Southern blotting, we determined that pJHC1∂ was generated by recombination between two ∼1.2-kb repeated sequences designated RS-1 (data not shown).
Recombinant DNA procedures.
DNA purification, restriction endonuclease analysis, DNA ligations and transformations, PCRs, and agarose gel electrophoresis were performed according to standard protocols (7, 38). Transfer of DNA to V. anguillarum from E. coli strains was accomplished by conjugation as described previously (48). Automated DNA sequencing, primer synthesis, and protein microsequencing were carried out at the Department of Molecular Microbiology and Immunology Core Facility at Oregon Health Sciences University. DNA and protein sequence analysis were carried out at the National Center for Biotechnology Information, using the BLAST network service, and by using the sequence analysis software package of the University of Wisconsin Genetics Computer Group.
RNase protection assays.
RNAs were prepared as follows. An inoculum from an overnight culture was grown in minimal medium (1:100) with the appropriate antibiotics. The inoculated cultures were grown with EDDA supplemented to just below the MIC to achieve similar levels of iron-limiting stress for each strain tested. Total RNA was prepared by the hot-phenol method (50). RNase protections were carried out essentially as described previously (12).
Site-directed mutagenesis.
The S248L mutation was generated by site-directed mutagenesis, using a Quickchange site-directed mutagenesis kit (Stratagene) and synthetic mutagenic primers S248LU (5′-CTTGATTTTCCTTGGACTTGATTTGATACGCATAATGACACTACATAGC-3′) and S248LD (the reverse complement of S48LU). These mutant primers replace the C at the second position of codon 248 with a T, thereby changing the encoded amino acid from a serine to a leucine. This mutation was introduced into plasmids pTW200 and pTW203, resulting in plasmids pTW200S248L and pTW203S248L, respectively. These mutations were verified by DNA sequence analysis.
Analysis of polypeptides encoded within the angB region.
The complete angB ORF was cloned into the inducible overexpression vector pQE60 (Qiagen). In this construct, angB gene expression is placed under the control of the strong T5 phage promoter and a His6 tag is translationally fused to the C-terminal end of the ORF. By inducing the phage promoter and selectively purifying His-tagged polypeptides, it is possible to identify polypeptides whose synthesis is directed from the translational start signal of pQE60 as well as from translational start signals within this ORF, as long as they are translated in the same frame as those originating at the plasmid's translational start signal. His-tagged polypeptides were purified under denaturing conditions in a nickel-nitrilotriacetic acid-agarose column, using the protocol recommended by the supplier (Qiagen). Protein separation was carried out by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described previously (30). The resolving gel was 15.5% acrylamide (30:0.8 acrylamide:bisacrylamide ratio).
Detection of angB-encoded polypeptides in V. anguillarum.
To analyze angB-encoded polypeptides in V. anguillarum, a polyclonal antiserum was generated by injecting a New Zealand White rabbit with the His-tagged purified polypeptides produced as described above. The presence of angB-encoded polypeptides in V. anguillarum was assessed by radioimmunoprecipitation. Cultures of V. anguillarum cells were grown from early log phase to stationary phase in CM9 minimal medium under iron-limiting conditions in the presence of TRAN-35S-label (1,089 Ci/mmol, 50 μCi/ml; ICN Biomedicals Inc., Costa Mesa, Calif.). The cells were collected by centrifugation and boiled for 10 min in 25 μl of phosphate-buffered saline containing 2% SDS. The samples were then diluted 40-fold with NET gel buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 0.1% Nonidet P-40, 0.1% SDS, 1 mM EDTA, 0.25% gelatin) and precleared by centrifugation at 16,000 × g for 10 min. Polyclonal AngB antiserum was then added to the cleared lysates at a 1:200 dilution, and the mixtures were incubated for 2 h at 4°C. The immunocomplex was then captured by adding protein A-agarose (50 μl of packed beads) and gently mixing for 1 h at 4°C. The protein A-antigen-antibody complex was then collected by centrifugation for 1 min at 12,000 × g and washed three times with NET gel buffer. Immunoprecipitated proteins were analyzed by SDS-PAGE and fluorography.
To determine if AngB is phosphopantetheinylated, we grew V. anguillarum cells from early log phase to stationary phase in a low-phosphate minimal medium under iron-limiting conditions in the presence of 32Pi (285 Ci/mg; 500 μCi/ml; ICN Biomedicals Inc.). For this experiment, CM9 medium was modified by replacing phosphate with 100 mM MOPS (morpholinepropanesulfonic acid) as a buffer and KH2PO4 was added as a source of inorganic phosphate at a concentration of 0.3 mM. The labeled cells were processed for immunoprecipitation exactly as described above. Immunoprecipitated 32P-labeled proteins were analyzed by SDS-PAGE and autoradiography.
Determination of anguibactin synthesis and utilization and DHBA production.
Three methods were used to determine anguibactin synthesis. First, strains were tested for their ability to produce a halo on chrome azurol S (CAS) agar (40). Second, supernatants from iron-limited cultures were tested by anguibactin bioassay as previously described (48). Finally, strains were tested for sensitivity to the iron-chelating compound EDDA via growth assays in liquid medium. In all cases, a decrease in halo production on CAS agar correlated with a decrease in anguibactin level in the bioassay and an increase in sensitivity to EDDA. DHBA was assayed by employing the Arnow reaction (6) calibrated with a DHBA standard. In addition, a bioassay employing specific Salmonella strains that require DHBA to synthesize enterobactin (33) was used to confirm the presence of this catechol. Strains were tested for their ability to utilize anguibactin as previously described (48).
Nucleotide sequence accession number.
The TAFb locus nucleotide sequence data reported here have been deposited in the GenBank database under accession no. AF311973.
RESULTS
Identification of ORFs within the TAF region.
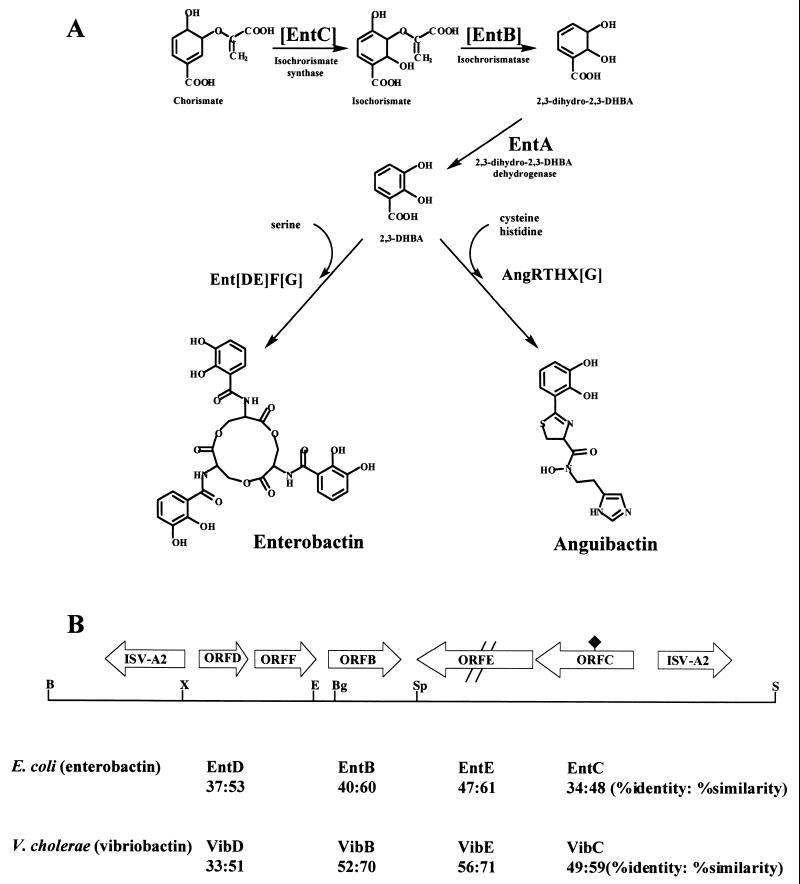
To identify the gene(s) responsible for the TAF activity, we performed exploratory DNA sequencing within a 20-kb region of the pJM1 plasmid previously shown to encode TAF activity (48) and searched for DNA sequences with putative functions consistent with the TAF activities. Using this approach, a cluster of five ORFs was identified. Four of the ORFs showed a high degree of homology to proteins necessary for early steps in the biosynthesis of the DHBA-containing siderophores enterobactin in E. coli and vibriobactin in Vibrio cholerae (31, 41, 55): ORF D (EntD, VibD), ORF B (EntB, VibB), ORF E (EntE, VibE), and ORF C (EntC, VibC) (Fig. 1). ORF D is homologous to a 5′-end-deleted version of EntD and VibD (the deletion corresponds to the first 37 amino acids, including the starting methionine) and therefore is probably nonfunctional. ORF E is homologous to EntE, but a frameshift mutation approximately in the middle of the gene causes premature termination of the predicted protein; therefore, this ORF is also probably nonfunctional. ORF C is also deranged compared to EntC due to two translational stop signals in the ORF. Figure 1 also shows that an additional ORF, ORF F, found within the cluster shows significant homology to members of a family of ATP-binding proteins which act as a subunit in ATP-binding cassette (ABC) transport systems (32). The most similar proteins within this family are those known to function in the import of ferrisiderophores. Therefore ORF B, with similarity to EntB, and ORF F, with similarity to ATP-binding proteins found in ABC transporters, were the only two ORFs within this region that were likely to encode functional proteins. These five ORFs lie within a 9-kb SalI-BamHI fragment and are flanked by identical inverted insertion sequence elements that are also identical to ISV-A2, previously found in another region of the pJM1 plasmid (43). These insertion sequences and the ORFs form a composite transposon-like structure (Fig. 1B). Figure 1A shows the pathway for enterobactin biosynthesis in E. coli and a proposed pathway for anguibactin biosynthesis. Only the AngR, AngH, and AngT components have been characterized to date (45, 52). Since anguibactin contains a DHBA moiety (4, 14), the predicted function of at least ORF B is consistent with a role in anguibactin biosynthesis. For the sake of simplicity, we have used the designation TAFb (b = biosynthesis) for the region containing the ORFs.
FIG. 1.
Siderophore biosynthesis pathways, physical map of the TAFb region, and homologs of ORFs with siderophore biosynthesis genes. (A) Putative pathways for the biosynthesis of enterobactin and anguibactin. Brackets designate enterobactin biosynthesis enzymes that are homologous to ORFs in the TAFb region. AngR, AngT, and AngH have been characterized previously (45, 52); AngG is characterized in this paper; and X represents other activities required for anguibactin biosynthesis that are still being characterized. (B) Physical map of the locations of ORFs found in the TAFb region and homologies to selected siderophore biosynthesis genes from E. coli and V. cholerae. The double slash symbolizes the frameshift mutation in ORF E; the black diamond corresponds to two translational stop signals in ORF C. B, BamHI; Bg, BglII; E, EcoRV; S, SalI; Sp, SpeI; X, XhoI.
Generation of a deletion derivative, pJHC1∂, lacking the TAFb region.
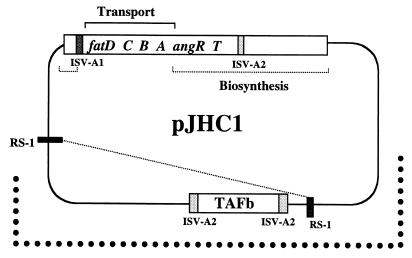
To test whether the TAFb region plays any role in anguibactin biosynthesis, we generated a deletion derivative of pJHC1 that lacked this region. We used pJHC1 instead of pJM1 because of the higher-level anguibactin production phenotype conferred by the former, which increases the sensitivity of detection of this siderophore in modified derivatives, and also because of the higher recombination proficiency of the strain harboring pJHC1 (46, 47, 49). This deletion of an approximately 20-kb segment was generated by marker exchange-eviction mutagenesis (34) employing 1.4-kb native repeated sequences other than the ISV-A2 elements (Fig. 2). These repeated sequences (RS-1) are missing from the pJM1 plasmid (reference 49 and unpublished data). To generate the ∂ mutation, we used a plasmid carrying the R6K origin of replication (which is nonfunctional in V. anguillarum), the sacB gene, and the TAFb region cloned as a 9-kb SalI-BamHI fragment with a kanamycin resistance-encoding fragment inserted within ORF C. This plasmid was mobilized to strain 531A(pJHC1), and plasmid cointegrates were identified by their kanamycin resistance. These cointegrates were then grown in TSAS with 10% sucrose. Since the sacB gene product is lethal in gram-negative bacteria, the only clones that grew were those that had undergone a second recombination event which excised sacB. These sucrose-resistant derivatives could have arisen by any of several possible resolution events involving either recombination between the two TAFb regions flanking the sacB gene, which can result in replacement of the wild-type ORF C by the mutated ORF C, or recombination between the two RS-1 sequences, generating a derivative with a deletion encompassing the region shown in Fig. 2. Final selection with sucrose was done in the absence of kanamycin to allow the identification of a broader array of recombinants. We describe in this section a deletion derivative, designated pJHC1∂, generated by recombination between the two RS-1 sequences (Fig. 2). The strain harboring this deletion derivative, 531A(pJHC1∂), did not produce anguibactin; however, it retained the ability to utilize this compound when supplied exogenously (data not shown). This biosynthesis defect was partially complemented by the addition of a recombinant plasmid, pTW100, which contained the TAFb region, while the plasmidless strain S531A-1 harboring only pTW100 neither produced anguibactin nor could it utilize it. Analysis of pTW100 deletion derivatives demonstrated that a fragment containing just ORF B (pTW104) was sufficient for this complementation. Furthermore, the insertion of the transcriptional-translational terminator Ω within ORF B (pTW105) completely abolished complementation (data not shown). Taken together with the mutagenesis and complementation data (shown in subsequent sections), these results demonstrate that this ORF is the only one encoded within this cluster which is absolutely necessary for anguibactin biosynthesis, and therefore we assigned it the anguibactin biosynthesis gene designation, angB.
FIG. 2.
Generation of a deletion derivative of the virulence plasmid pJHC1 by recombination between indigenous insertion sequences. Heavily dotted lines denote the TAF region as originally described (48). The fatDCBAangRT iron transport biosynthesis operon and the location of biosynthesis genes are also shown. ISV-A1 and ISV-A2 are insertion sequences in the iron uptake region that were previously characterized (43). The ISV-A2 sequences flanking the TAFb region and the two RS-1 repeated sequences are characterized in this work.
Generation of angB and ORF F mutants by allelic exchange.
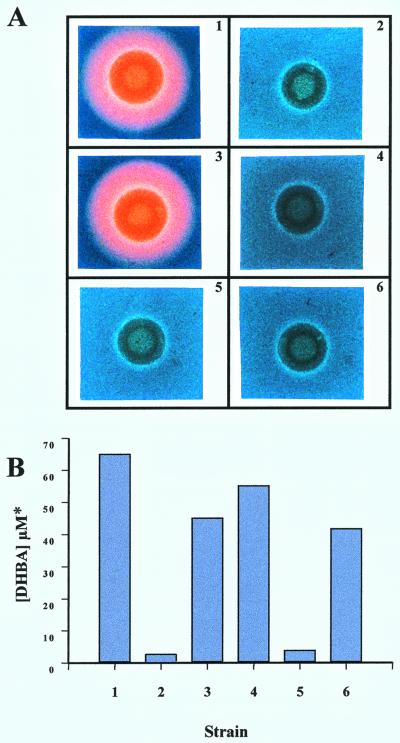
Clearly, angB must be the only gene within this cluster which is obligatory for the biosynthesis of anguibactin, although its activity restores only part of the maximum anguibactin biosynthesis potential. However, in the complementation analysis described above, we could not ensure that ORF F was properly expressed from plasmid pTW100. Therefore, ORF F, while not absolutely necessary for the synthesis of anguibactin, could contribute to it. To test this hypothesis, we generated by marker exchange-eviction mutagenesis an in-frame deletion that removed approximately 50% of this ORF. An in-frame deletion mutation was created to ensure that the mutation would not influence the expression of angB, because ORF F is located upstream of and in the same orientation as angB, which suggested that a potential transcriptional linkage between these determinants might exist. The angB gene was also mutated by marker exchange-eviction mutagenesis, resulting in the insertion of the Ω transcriptional-translational terminator (23) at the BglII site within the 5′ end of the gene. Of these mutations, the only modification found to affect anguibactin biosynthesis was the mutation introduced into angB in the strain designated 531A(pJHC1::Ω). In this strain, anguibactin production was completely abolished (see Fig. 4A). A strain carrying the mutation in ORF F was indistinguishable from the wild type in its behavior on CAS agar as well as its sensitivity to EDDA and production of anguibactin as measured by bioassay (data not shown). These findings confirm that in the TAFb region the angB gene is essential for anguibactin biosynthesis, and they demonstrate that ORF F has no function in anguibactin biosynthesis under the conditions tested here.
FIG. 4.
Genetic complementation of the 531A(pJHC1::Ω) mutant with the angB gene and its 3′-deleted derivative. (A) Detection of anguibactin by using CAS agar plates (40). The identity of anguibactin as the CAS-reactive product was verified by bioassays as previously described (49). Strains tested were as follows: panel 1, WT, 531A(pJHC1/pTW99); panel 2, 531A(pJHC1::Ω/pTW99); panel 3, 531A(pJHC1::Ω/pTW200); panel 4, 531A(pJHC1::Ω/pTW201); panel 5, S531A-1(pTW99); and panel 6, S531A-1(pTW201). Strains in panels 1, 2, and 5 carry pTW99 as a vector control. S531A-1 is a plasmidless derivative of 531A. (B) Production of DHBA by the same strains grown to stationary phase in complete minimal medium. DHBA production was analyzed by using the Arnow reaction (6) and calibrating the assay with a DHBA standard. The presence of DHBA was verified by bioassays with Salmonella strains containing enb1 and enb7 mutations according to the method of Pollack and Neilands (33).
Genetic and biochemical characterization of the angB functions.
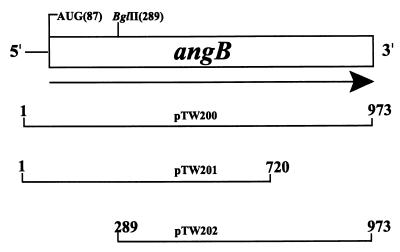
The angB gene possesses an ORF of 864 bp encoding a predicted 32.4-kDa protein of 287 amino acids. AngB shows sequence homology to the EntB protein of E. coli (Fig. 1B), which is a bifunctional protein necessary for a step in the biosynthesis of DHBA (B activity), a precursor for enterobactin biosynthesis and for enterobactin assembly (G activity). In EntB, the DHBA synthesis activity resides in the N-terminal 187 amino acids of the protein while the enterobactin assembly activity resides in the C-terminal portion (residues 188 to 285) of the protein (24, 35). To test for bifunctionality of AngB, we constructed a clone containing the complete angB gene (pTW200), which would be expected to impart both B and G activities, and an otherwise identical clone lacking the final 345 nucleotides (114 amino acids) of angB (pTW201), which would be expected to possess B activity and lack G activity (Fig. 3). These constructs were then tested for their ability to complement the angB mutant strain 531A(pJHC1::Ω). This strain, when carrying the full-length angB gene (pTW200), showed complete complementation for anguibactin synthesis (Fig. 4A). However, when this strain carried the deletion derivative pTW201, anguibactin was not produced. To distinguish potential B activity in these strains, they were also tested for their ability to produce DHBA under iron-limiting conditions. Figure 4B shows that the strain carrying pTW201 produced DHBA despite its inability to produce anguibactin. Therefore, the upstream region of the angB gene, approximately 75% of the ORF from the 5′ end, is sufficient for the production of DHBA; i.e., it has B activity. However, it does not confer the ability to produce anguibactin. Therefore, the angB gene, like entB, must also encode another essential anguibactin biosynthesis activity, in addition to the B activity, which must act somewhat after B activity in the anguibactin biosynthesis pathway. Furthermore, when introduced into a plasmidless V. anguillarum strain, S531A-1, pTW201 confers the ability to produce DHBA (Fig. 4B), as it did in the angB mutant strain, indicating that the EntC-like and EntA-like activities necessary for DHBA production (Fig. 1) must be encoded on the V. anguillarum chromosome.
FIG. 3.
Scheme of a DNA region containing the angB gene and 5′- and 3′-deletion derivatives cloned in pTW99. The angB gene initiation codon starts at bp 87, and the translational stop signal is located at bp 973. pTW200 harbors the complete angB gene, pTW201 contains a 3′-end-deleted derivative of angB, and pTW202 harbors a 5′-end-deleted derivative of angB. The angB gene and modified derivatives are placed under the control of the tetracycline resistance gene promoter from a pBR322-derived vector.
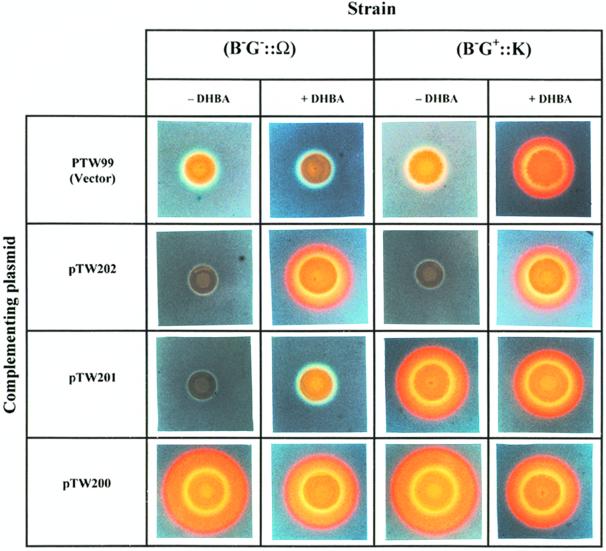
To further characterize angB-encoded functions, we created a frameshift mutation at the 5′ end of the gene by marker exchange mutagenesis. This mutant, 531A(pJHC1::K), was generated by cleaving the BglII site (located at codon 67) in angB (Fig. 3) and filling in the ends with the Klenow fragment of DNA polymerase I, resulting in a 4-bp insertion, which caused a frameshift leading to a predicted translational termination 13 amino acids after the BglII site. This modification was verified by DNA sequencing. Transcription through this modified site should not be affected, although premature termination of translation could influence transcription. In contrast, the angB mutant strain 531A(pJHC1::Ω) is clearly blocked for transcription (and therefore translation) at the same BglII site. The ability of each of these mutant strains to produce anguibactin was tested in the presence of 50 μM DHBA in order to bypass the need for B activity. Interestingly, these two mutants behaved very differently when grown in the presence of DHBA (Fig. 5, row 1). Neither of these two mutants produced anguibactin. However, only in strain 531A(pJHC1::K) could this defect be repaired by the addition of DHBA. This result further confirms that the angB locus encodes two functions. Furthermore, it also shows that while transcription of the gene is necessary for both functions, premature termination of the AngB polypeptide at the BglII site affects only the DHBA synthesis activity (B activity), leaving the G activity intact. Therefore, G activity can exist independently of the AngB polypeptide. This result is corroborated by the finding that a clone which contains only the 3′ end of angB, and thus has only G activity (Fig. 3), complements the mutation in strain 531A(pJHC1::Ω) when DHBA is provided (Fig. 5, row 2), further indicating that G activity is independent of B activity and is encoded within the 3′ end of the angB locus. Conversely, if a clone containing the 5′ end of the angB gene (pTW201), and thus providing B activity, is harbored by mutant strain 531A(pJHC1::K), which provides G activity, anguibactin is produced. However, this does not occur when pTW201 is harbored by mutant strain 531A(pJHC1::Ω), which does not possess either B or G activity (Fig. 5, row 3). In summary, the 5′ end of the angB gene is sufficient for B activity while the 3′ end of this gene is sufficient for G activity. Since these two activities are separable, and G activity can exist as an independent entity, we have designated the region encoding G activity as the angG gene.
FIG. 5.
Genetic and chemical complementation of strains with polar and nonpolar angB mutations. Detection of anguibactin by using CAS agar plates was carried out as described in the legend to Fig. 4. Strains are indicated by phenotype and mutation. Therefore, strain 531A(pJHC1::Ω) is represented by (B−G−::Ω) and strain 531A(pJHC1::K) is represented by (B−G+::K). The maps for the complementing plasmids are shown in Fig. 3. The siderophore assays were carried out in the presence (+) and in the absence (−) of 50 μM DHBA.
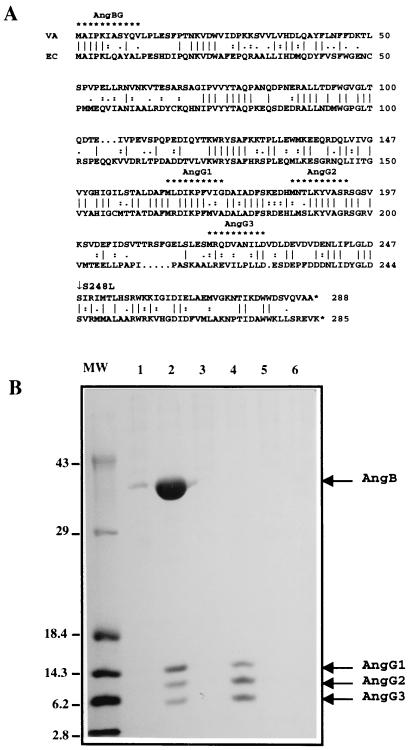
Figure 6A shows an alignment of the AngB and EntB amino acid sequences. These polypeptides align significantly throughout their sequences. Combining these results with our genetic analysis, we surmise that G activity in angB mutant backgrounds may be associated with a smaller angB-encoded polypeptide that is read in the same frame as the AngB polypeptide but is synthesized independently. This putative polypeptide, AngG, would therefore be synthesized from an intragenic translational start site within angB. To test this hypothesis, the complete angB gene was cloned into the inducible overexpression vector pQE60. In this construct, angB gene expression is placed under the control of the strong T5 phage promoter and a His6 tag is translationally fused to the C-terminal end of the ORF. By inducing the phage promoter and selectively purifying His-tagged polypeptides, it should be possible to identify polypeptides whose synthesis is directed from the translational start signal of pQE60 as well as from translational start signals within this ORF, as long as they are translated in the same frame as those originating in the plasmid translational start signal. Using this approach, we identified four His-tag-purified polypeptides under inducing conditions (Fig. 6B, lane 2). The N-terminal sequences of the four polypeptides was determined, and the sequences and positions within angB are presented in Fig. 6A. All four polypeptides are encoded by the same ORF; the molecular mass and N-terminal sequence of the largest agree with those of the complete AngB protein, while the other three (AngG1, AngG2, and AngG3) range in size from 15 to 5.5 kDa. All four polypeptides start with a methionine. Therefore, it is likely that they are the result of translational initiations. To demonstrate that AngG1, AngG2, and AngG3 were not breakdown products of the AngB polypeptide, a derivative of the angB gene carrying the BglII-Klenow frameshift mutation was also cloned in pQE60 for T5 phage promoter-directed expression. In this construct, translation of the full-length AngB protein is terminated at the BglII site (and thus the protein is not His tagged), yet AngG1, AngG2, and AngG3 should still be produced as His-tagged products if they are the products of intragenic translational initiations. Figure 6B, lane 4, shows that this is the case. The proteins AngG1, AngG2, and AngG3 are synthesized using the BglII-Klenow derivative cloned in pQE60, pQE60-B− G+::K, clearly showing that these proteins are not degradation products of AngB, and therefore they must be initiated at the sites deduced from the N-terminal sequence analysis (Fig. 6A).
FIG. 6.
Alignment of amino acid sequences of AngB and EntB and electrophoretic analysis of angB- and angG-encoded polypeptides expressed in an E. coli overexpression system. (A) Alignment of the AngB and EntB proteins was carried out by using the GAP program (Genetics Computer Group). The vertical lines show identical amino acids, dots are conserved amino acid substitutions, and two dots indicate a higher degree of conservation. Asterisks over the AngB protein sequence indicate the amino termini of the AngB, AngG1, AngG2, and AngG3 polypeptides identified in panel B. These amino terminal sequences were determined by protein microsequencing as described in Materials and Methods. A vertical arrow shows the position of the S248L mutation within the 243-FLGLDSI-249 sequence, which exhibits homology to the consensus sequence of the putative serine phosphopantetheinylation domain (24). (B) SDS-PAGE of the His-tagged, purified polypeptides expressed from the complete angB ORF (pQE60-B+G+) and a derivative containing the BglII-Klenow nonpolar mutation (pQE60-B−G+::K) cloned into the inducible overexpression vector pQE60. A His6 tag was translationally fused to the carboxy-terminal end of the ORF in these constructs. Therefore, the AngG polypeptides are also His-tag labeled because they have the same carboxy terminus as AngB. E. coli M15 cells harboring the angB derivatives cloned in pQE60 were grown to the mid-log phase at 37°C when isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the cultures to be induced (2 mM final concentration). Cells were allowed to grow for an additional 4 h, at which time they were harvested for analysis of the His-tag-labeled polypeptides, as described in Materials and Methods. Lanes: 1 and 2, pQE60-B+G+; 3 and 4, pQE60-B−G+::K; 5 and 6, pQE60. Cells in lanes 1, 3, and 5 were not induced, while those in lanes 2, 4, and 6 were IPTG induced. MW; molecular mass standards (in kilodaltons). VA, V. anguillarum; EC, E. coli.
A serine residue located within a domain found on aryl carrier proteins is essential for anguibactin biosynthesis.
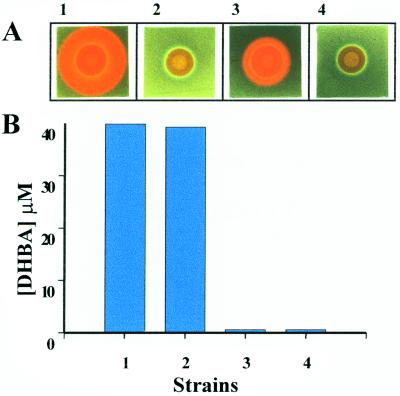
Inspection of the sequence at the carboxy terminus of the G region in AngB and in the AngG polypeptides revealed the presence of a domain found in aryl carrier proteins. In such proteins, this domain undergoes covalent phosphopantetheinylation prior to attachment of the assembling siderophore or antibiotic at this site (24). Since in most of these cases phosphopantetheinylation occurs at a specific serine residue within this domain, we decided to modify this amino acid to assess its influence on anguibactin production. By using site-directed mutagenesis, we generated an S248L mutation (Fig. 6A) in two plasmids, pTW200 and pTW203, thereby creating plasmids pTW200S248L and pTW203S248L, respectively. The latter plasmid also carries a Klenow modification of the BglII site within angB. Each of these plasmids was conjugated to the 531A(pJHC1::Ω) strain. Figure 7A shows that the S248L mutation leads to a complete abolishment of anguibactin production compared to the isogenic control. Yet DHBA production in this mutant is unaffected, further demonstrating the separability of the B and G activities (Fig. 7B). To determine whether G activity alone could be modified by the S248L mutation, we tested pTW203, which has only a functional angG gene, and an isogenic plasmid with the S248L mutation, pTW203S248L. The results, shown in Fig. 7A, demonstrated that the S248L mutation abolishes anguibactin biosynthesis. Because of the lack of B activity in these derivatives, DHBA was added in order to measure G activity in strains 3 and 4 (Fig. 7A). These results indicate that the serine residue at position 248 is essential for G activity regardless of whether it comes from the AngB or the AngG protein. It is therefore likely that AngG and the carboxy terminus of AngB are aryl carrier proteins that undergo phosphopantetheinylation at S248 in one of the steps in the anguibactin biosynthesis pathway.
FIG. 7.
Analysis of AngB and AngG functions in the S248L mutant. (A) Use of CAS agar plates, as described in the legend to Fig. 4, to detect anguibactin production by the following strains: panel 1, 531A(pJHC1::Ω/pTW200), phenotype B− G−/B+ G+; panel 2, 531A(pJHC1::Ω/pTW200S248L), phenotype B− G−/B+ G−; panel 3, 531A(pJHC1::Ω/pTW203), phenotype B− G−/B− G+; and panel 4, 531A(pJHC1::Ω/pTW203S248L), phenotype B− G−/B−G−. For the strains in panels 3 and 4, DHBA was added to a 50 μM final concentration to overcome the need for B activity. (B) Production of DHBA by the same strains, grown to stationary phase under conditions of iron limitation in complete minimal medium. DHBA was analyzed by using the Arnow reaction and calibrating the assay with a DHBA standard. The presence of DHBA was verified by bioassays with Salmonella strains containing enb1 and enb7 mutations according to the method of Pollack and Neilands (33).
Analysis of angB- and angG-encoded polypeptides in V. anguillarum.
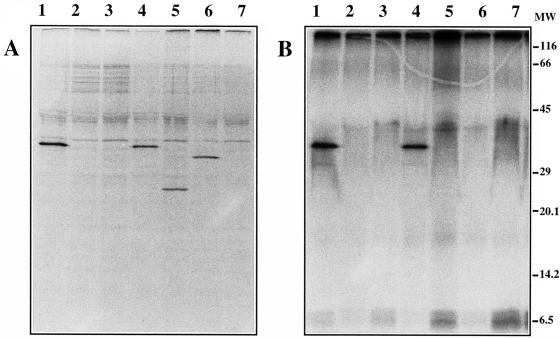
To confirm that the proteins that we identified genetically in V. anguillarum and biochemically in E. coli are synthesized in the V. anguillarum cytoplasm, we used a polyclonal antiserum, raised against purified AngB, AngG1, AngG2, and AngG3 polypeptides, in a radioimmunoprecipitation assay. Total proteins from various angB and angG mutants with and without complementing plasmids were steady-state labeled with [35S]methionine and then subjected to immunoprecipitation employing the AngB-AngG antiserum. Figure 8A shows that the AngB-AngG antiserum recognized a 37-kDa protein in wild-type V. anguillarum, but not in the 531A(pJHC1::Ω) and 531A(pJHC1::K) mutants (lanes 1 to 3). This protein was also present in the 531A(pJHC1::Ω) strain harboring complementing plasmid pTW200, which encodes both B and G activities (Fig. 8A, lane 4). We also examined the pJHC1::Ω-containing strain harboring plasmid pTW201, which encodes a 3′-end-deleted version of angB with B but not G activity. A protein of 22.5 kDa (Fig. 8A, lane 5), the expected molecular mass for a construct with a C-terminal truncation, was detected in this strain. When the S248L mutant plasmid pTW200S248L was tested in strain 531A(pJHC1::Ω), a protein which runs slightly faster than AngB on SDS-PAGE was detected (33 kDa). Since the S248L mutation is expected to abolish the putative phosphopantetheinylation site in AngB and in the AngG polypeptides, it is possible that the different mobilities of these two proteins on SDS-PAGE gels reflect the presence or absence of the phosphopantetheinylate moiety. To test this hypothesis, we performed a 32Pi labeling experiment in parallel with the [35S]methionine labeling experiment described above, with the intent of labeling the phosphate group in the phosphopantetheinylate moiety of AngB. 32P-labeled proteins were observed only in the wild type and the complemented mutant strains, and they were completely absent from mutant derivatives (Fig. 8B), demonstrating that AngB is modified by a phosphate-containing moiety. The absence of 32P label in the truncated AngB protein suggests that modification occurs at a site within the carboxy-terminal end of AngB. Furthermore, the absence of 32P label in the AngBS248L mutant protein demonstrates that this site is essential for modification and suggests that modification is necessary for G activity.
FIG. 8.
Radioimmunoprecipitation analysis of angB-encoded polypeptides in the V. anguillarum cytoplasm. (A) Cultures of V. anguillarum were grown from early log phase to stationary phase in CM9 minimal medium under iron-limiting conditions in the presence of [35S]methionine (50 μCi/ml; ICN Biomedicals Inc.). The cells were collected by centrifugation and processed for immunoprecipitation as described in Materials and Methods. Immunoprecipitated proteins were analyzed by SDS-PAGE and fluorography. Proteins from the following strains were analyzed: lane 1, 531A(pJHC1/pTW99), wild-type strain; lane 2, 531A(pJHC1::Ω/pTW99), phenotype B− G−; lane 3, 531A(pJHC1::K/pTW99), phenotype B− G+; lane 4, 531A(pJHC1::Ω/pTW200), phenotype B− G−/B+ G+; lane 5, 531A(pJHC1::Ω/pTW201), phenotype B− G−/B+ G− (truncated B protein); lane 6, 531A(pJHC1::Ω/pTW200S248L), phenotype B− G−/B+ G−; and lane 7, S531A-1(pTW99), phenotype B− G− (S531A-1 is the plasmidless 531A derivative). (B) Cultures of V. anguillarum were grown from early log phase to stationary phase in a low-phosphate minimal medium under conditions of iron limitation in the presence of 32Pi (500 μCi/ml; ICN Biomedicals Inc.). For this experiment, CM9 medium was modified by replacing phosphate with 100 mM MOPS as a buffer and KH2PO4 was added as a source of inorganic phosphate at a final concentration of 0.3 mM. The labeled cells were processed for immunoprecipitation exactly as described above. Immunoprecipitated 32P-labeled proteins were analyzed by SDS-PAGE and autoradiography. Proteins analyzed were from the same strains as those shown in panel A. MW, molecular mass standards (in kilodaltons).
Do angB and angG encode the regulatory activity attributed to the TAF factor?
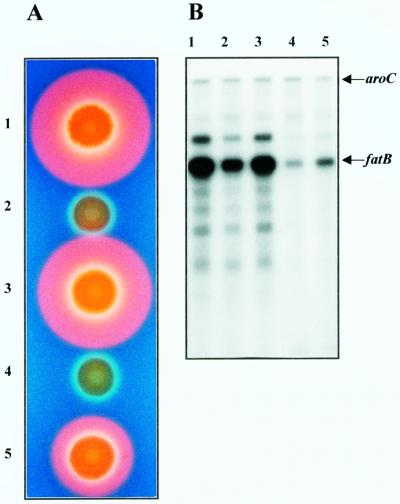
The TAF products were originally described as being essential for maximal production of anguibactin (48) and for maximal expression of anguibactin biosynthesis and iron transport genes (13, 37, 48). In previous sections, we defined a subregion of TAF, designated TAFb, in which we identified the angB and angG genes, found to be essential for anguibactin biosynthesis, therefore initiating the molecular dissection of the components of the TAF factor(s). Those results indicated that complementation of strain 531A(pJHC1∂) with clones encoding both AngB and AngG polypeptides restored anguibactin biosynthesis to only about half of the wild-type level. To understand the basis for this partial complementation, we examined a series of clones in various mutant strains. Figure 9A shows the anguibactin production phenotypes for these strains. Strain 531A(pJHC1::Ω) does not produce anguibactin. When this mutation is complemented with pTW200, resulting in strain 531A(pJHC1::Ω/pTW200), anguibactin production is restored to wild-type levels. However, introduction of the same clone into the 531A(pJHC1δ) strain results in the synthesis of about half of the wild-type level of anguibactin. These results indicate that the product(s) of the angB and/or angG gene could only restore partial anguibactin production when complementing strain 531A(pJHC1δ) but resulted in wild-type levels of anguibactin production if complementing the modification in strain 531A(pJHC1::Ω). Since the plasmid encoding AngB and AngG (pTW200) is the same in both cases, it is surmised that this difference in genotype must be associated with the large deletion in strain 531A(pJHC1δ). The deletion that generated this plasmid extended beyond the TAFb region (Fig. 2). It is reasonable to assume that there is another component in the missing region that is important for restoration of maximal anguibactin production. We have originally attributed two functions to the TAF factor, one associated with anguibactin production and the other associated with regulation of the expression of anguibactin biosynthesis and iron transport genes (37, 48). It is therefore possible that TAFb, which is essentially AngB and AngG, is only part of the TAF activity and that another TAF component, which is absent from pJHC1δ, is responsible for the regulatory phenomenon. This missing TAF regulatory component could influence anguibactin production via a regulatory effect on the synthesis of the biosynthetic genes angRT, which are encoded by the iron transport-biosynthesis operon fatDCBAangRT. To test this hypothesis, we analyzed the levels of expression of the polycistronic mRNA encoded by the fatDCBAangRT operon (Fig. 2) utilizing a fatB riboprobe in an RNase protection assay. The strains were grown under iron-limiting conditions at their MICs for the iron chelator EDDA. Figure 9B, lane 1, shows the fatB-specific mRNA levels in the wild-type strain 531A(pJHC1). Lane 2 shows that the level of fatB-specific mRNA in the 531A(pJHC1::Ω) strain is moderately reduced compared to the wild-type level. This is likely due to the loss of anguibactin's contribution to the regulation of the polycistronic mRNA encoded by fatDCBAangRT, a phenomenon described previously (13). Introduction of a clone harboring the angB- and angG-encoded activities (pTW200) into the 531A(pJHC1::Ω) strain restored production of anguibactin, and therefore the level of fatB-specific mRNA was also restored to that of the wild type. However, the level of fatB-specific mRNA in strain 531A(pJHC1δ) was dramatically lower than that in the 531A(pJHC1::Ω) strain (compare lanes 2 and 4). Since neither of these strains produces anguibactin, the mRNA levels are not influenced by anguibactin's contribution to fatB expression, indicating that a regulatory factor must be absent from the 531A(pJHC1δ) strain. Furthermore, when the plasmid harboring angB and angG was introduced into 531A(pJHC1δ), the fatB-specific mRNA levels were only slightly increased; this minor increase is again presumably due to the regulatory effect caused by the resumption of anguibactin production.
FIG. 9.
Contribution of TAFb and TAFr activities to anguibactin production and regulation of expression of the fatDCBAangRT operon. (A) Detection, using CAS agar plates as described in the legend to Fig. 4, of anguibactin produced by the following strains: panel 1, 531A(pJHC1/pTW99), wild-type strain; panel 2, 531A(pJHC1::Ω/pTW99), phenotype B− G−; panel 3, 531A(pJHC1::Ω/pTW200), phenotype B−G−/B+G+; panel 4, 531A(pJHC1δ/pTW99), phenotype TAF−; and panel 5, 531A(pJHC1δ/pTW200), phenotype TAFr−/B+ G+. The strains in lanes 1, 2, and 4 also carry cloning vector pTW99 as a control. (B) RNase protection analysis of mRNAs synthesized by the strains (lanes correspond to the panel numbers listed above). Total RNA was harvested from the strains grown under iron-limiting conditions at their respective MICs for EDDA to achieve identical levels of iron stress. Specific transcripts for fatB and aroC were detected by RNase protection, using the riboprobes for recognition of these transcripts as described in Materials and Methods. The aroC mRNA is constitutively expressed in V. anguillarum and was used as an internal control. Specific RNase-protected RNAs (aroC and fatB) are indicated by arrows. Bands seen above the fatB transcript in lanes 1 to 3 were the result of incomplete RNase treatment and were seen only when fatB transcript levels were exceptionally high.
Therefore, we have dissected the TAF factor into two components: one, TAFb, is associated with anguibactin production and is dependent on the presence of both the AngB and AngG polypeptides, while the other, designated TAFr, is a regulatory function, that is independent of anguibactin's effect and is encoded by the remaining region of TAF.
DISCUSSION
Genes encoding anguibactin biosynthesis and utilization functions lie within several operons which are located on a 25-kb region of the V. anguillarum virulence plasmid (48). However, to achieve maximal expression of this iron-scavenging system, products from a noncontiguous region of the plasmid are required. This region, designated TAF, encodes trans-acting factors which act to increase production of the siderophore anguibactin (48) as well as to enhance the expression of a polycistronic mRNA encoded by ferric anguibactin transport and biosynthesis genes (37).
In this work, we have begun the dissection of the TAF region and have determined that there are at least two components. One, TAFb, is directly associated with anguibactin biosynthesis, while the other, TAFr, plays a regulatory role in the expression of the polycistronic mRNA encoded by fatDCBAangRT. The fact that AngR is not only a regulatory protein itself but also a biosynthetic enzyme, as is AngT (52), suggests that TAFr plays an indirect role in anguibactin biosynthesis via its regulatory effect on the two biosynthesis genes present in the same polycistronic mRNA. Our results support this hypothesis: strains lacking the TAFr region were not fully complemented by TAFb for the expression of the fatDCBAangRT operon or for anguibactin production. However, TAFb clones do fully complement both for expression of the fatDCBAangRT operon and for anguibactin production when the strain harbors TAFr.
In this report, we have focused on the analysis of the TAFb component. Our analysis demonstrated that the TAFb region encodes several ORFs exhibiting high levels of homology to proteins involved in the biosynthesis and utilization of DHBA-containing siderophores, such as vibriobactin (55) and enterobactin (31, 41). Only two ORFs, B and F, were found to have the potential to encode functional proteins. However, we showed by generation of an internal deletion that ORF F is not necessary for anguibactin biosynthesis. Our mutagenesis and complementation experiments also demonstrated that two genes within ORF B, now designated angB and angG, are the only genes within this cluster which are necessary for anguibactin biosynthesis. The TAFb region containing this cluster is flanked by identical and inverted insertion sequence elements, forming a composite transposon-like structure. Similar insertion sequences flank the fatDCBAangRT operon (17, 43). These observations suggest that the V. anguillarum virulence plasmid may have acquired siderophore biosynthesis and utilization genes horizontally via transposition events. Chance and necessity may have prompted the acquisition of a mobile unit carrying angB and angG along with the other genes, even though angB and angG were the only two genes in this cluster needed for anguibactin production. These nonessential genes in the TAFb cluster may have accumulated mutations and become pseudogenes later in the evolution of this system. It is also of interest that the gene organization of the pseudogenes ORF C and ORF E, as well as the angB gene, in the TAFb cluster is identical to that of the chromosomally encoded vibC, vibE, and vibB genes of V. cholerae (55), and this organization is distinct from that of the same genes in E. coli (31, 41). It is tempting to speculate that the TAFb cluster originated in V. cholerae or in an ancestral organism and that it was acquired by the V. anguillarum virulence plasmid through transposition events and horizontal transfer.
Members of Earhardt's laboratory isolated transposon mutations in the 3′ end of the entB gene that resulted in truncated EntB proteins with the phenotype EntB+ Ent−, suggesting that an EntG activity was encoded in the 3′ end of the entB gene. Immunological and in vitro transcription-translation analysis identified only a protein with a molecular mass corresponding to that of EntB (41). Therefore, the experimental evidence gathered using the approaches described above strongly suggested that EntB is a bifunctional protein. However, these investigators pointed out that their studies did not completely eliminate the possibility that a separate EntG polypeptide exists (41).
Our genetic analysis of the TAFb component angB demonstrated that it is a single ORF encoding two distinct functions that are essential for anguibactin biosynthesis. Either of these two activities, B or G, can function independently of the other: a stop mutation at the BglII site located at 194 bp from the translational start of angB, or a deletion of these first 194 bp, destroys B activity without affecting the G activity. Furthermore, deletions of 345 bp from the 3′ end abolish G activity, while B activity is unaffected. The B activity encoded by the angB gene corresponds to a 37-kDa protein. The truncated version generated by the 3′-end deletion, which still possesses B activity, was also identified in V. anguillarum as a protein whose molecular mass corresponds to 172 amino acids. We believe that the B activity is an isochorismate lyase (24, 35) required for the conversion of isochorismate to 2,3-dihydro-2,3-DHBA, not only due to the fact that AngB and EntB exhibit significant homology, but also because deletion mutations in the 5′ end of the gene that results in the loss of B activity are rescued by the addition of DHBA. Conversely, strains carrying just the 5′ end of angB, possessing B activity but deficient in G activity, secrete DHBA.
Our present genetic evidence indicates that, at least in angB mutant backgrounds, an independent entity with G activity can exist in V. anguillarum, since a strain with a nonpolar mutation in the 5′ end of the angB gene exhibits G activity even when this mutation is introduced onto the virulence plasmid by allelic exchange. This activity correlates with the presence of three polypeptides that are encoded in the 3′ end of the angB gene and are all translated in the same frame as AngB, as demonstrated by using an E. coli overexpression system and protein microsequencing. Although the genetic evidence for the existence of a separate AngG polypeptide is very strong, we were unable to detect such a protein in any V. anguillarum strain, potentially due to low levels of this protein. Because of this we were unable to determine the biological significance of this protein. Our present efforts are directed at elucidating whether AngB and/or one or all of the smaller polypeptides encode the G activity in wild-type cells by creating silent mutations in the ribosome binding sites for the angG-encoded polypeptides.
Recently, Gehring et al. (24) demonstrated that in the bifunctional EntB protein the enterobactin assembly activity resides in the C-terminal portion (residues 188 to 285) and showed that EntB must have a C-terminal apo-aryl carrier protein domain that undergoes covalent phosphopantetheinylation, most probably at serine 245, located in a consensus sequence. Comparison of the carboxy-terminal end of AngB with the AngG polypeptides and the carboxy-terminal end of EntB identified a similar domain (FLGLDSI) with a serine residue located at position 248. In AngB this domain extends from amino acids 243 to 249, and it is also present in the corresponding portions of the AngG proteins. Our results demonstrated that an S248L mutation results in the loss of the G activity while conserving the B activity, indicating that this serine residue is essential for G activity. However, we could not determine whether the loss of G activity caused by the mutation in AngB is due to an effect on the AngG polypeptides only or also in the corresponding sequence in the carboxy-terminal end of AngB. What is clear is that the same mutation in the nonpolar mutant B− G+::K, which should only synthesize the AngG polypeptides, results in the loss of G activity. Since this mutation was a single nucleotide change resulting in the S248L mutation, it is likely that the change is occurring only at the protein level, thus providing further proof of the existence of the AngG polypeptides in these strains. The results of in vivo phosphorylation experiments suggest that the carboxy-terminal end of AngB functions as an aryl carrier protein involved in the assembly of the anguibactin molecule. Since the S248L mutation abolishes G activity in a strain that should synthesize only the AngG polypeptides, it is reasonable to think that the S248 site in AngG is also modified and that AngG is an aryl carrier protein, although these modified polypeptides were not detected in V. anguillarum.
Our present efforts are directed to elucidate whether one or all of the smaller polypeptides encode the G activity. We have also identified open reading frames within the TAFr region which might encode the functions responsible for the TAFr activity. Analyses of these will contribute to the understanding of the molecular nature of the TAFr regulatory activity encoded by the V. anguillarum virulence plasmid.
ACKNOWLEDGMENTS
This work was supported by Public Health grant AI19018 from the NIH to J.H.C. T.J.W. was the recipient of an NIH postdoctoral training fellowship.
We gratefully acknowledge Lisa Welch's assistance with antibody production and Paul Shapiro's advice regarding radioimmunoprecipitation experiments.
REFERENCES
- 1.Actis L A, Tolmasky M E, Crosa J H. Vibrosis. In: Woo P T K, Bruno D W, editors. Fish diseases and disorders. 3. Viral, bacterial and fungal infections. Wallingford, United Kingdom: CAB International Publishing; 1999. pp. 523–557. [Google Scholar]
- 2.Actis L A, Tolmasky M E, Crosa L M, Crosa J H. Characterization and regulation of the expression of FatB, an iron transport protein encoded by the pJM1 virulence plasmid. Mol Microbiol. 1995;17:197–204. doi: 10.1111/j.1365-2958.1995.mmi_17010197.x. [DOI] [PubMed] [Google Scholar]
- 3.Actis L A, Tolmasky M E, Farrell D H, Crosa J H. Genetic and molecular characterization of essential components of the Vibrio anguillarum plasmid-mediated iron-transport system. J Biol Chem. 1988;263:2853–2860. [PubMed] [Google Scholar]
- 4.Actis L A, Fish W, Crosa J H, Kellerman K, Ellenberger S R, Hauser F M, Sanders-Loehr J. Characterization of anguibactin, a novel siderophore from Vibrio anguillarum 775(pJM1) J Bacteriol. 1986;167:57–65. doi: 10.1128/jb.167.1.57-65.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Actis L A, Potter S, Crosa J H. Iron-regulated outer membrane protein OM2 of Vibrio anguillarum is encoded by virulence plasmid pJM1. J Bacteriol. 1985;161:736–742. doi: 10.1128/jb.161.2.736-742.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arnow E L. Colorimetric determination of the components of 2,3-dihydroxy-phenylalanine tyrosine mixtures. J Biol Chem. 1937;118:531–537. [Google Scholar]
- 7.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolivar F, Rodriguez R L, Greene P J, Betlach M C, Heyneker H L, Boyer H W. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 9.Bullen J J, Griffiths E, editors. Iron and infection. 2nd ed. West Sussex, United Kingdom: John Wiley and Sons Ltd.; 1999. [Google Scholar]
- 10.Canestrini G. La malattia dominante delle anguille. Atti Ist Veneto Sci Lett Arti Cl Sci Mat Nat. 1893;7:809–814. [Google Scholar]
- 11.Chai S, Welch T, Crosa J H. Characterization of the interaction between Fur and the iron transport promoter of the virulence plasmid in Vibrio anguillarum. J Biol Chem. 1998;273:33841–33847. doi: 10.1074/jbc.273.50.33841. [DOI] [PubMed] [Google Scholar]
- 12.Chen Q, Crosa J H. Antisense RNA, Fur, iron, and the regulation of iron transport genes in Vibrio anguillarum. J Biol Chem. 1996;271:1885–1891. doi: 10.1074/jbc.271.31.18885. [DOI] [PubMed] [Google Scholar]
- 13.Chen Q, Wertheimer A M, Tolmasky M E, Crosa J H. The AngR protein and the siderophore anguibactin positively regulate the expression of iron-transport genes in Vibrio anguillarum. Mol Microbiol. 1996;22:127–134. doi: 10.1111/j.1365-2958.1996.tb02662.x. [DOI] [PubMed] [Google Scholar]
- 14.Chen Q, Actis L A, Tolmasky M E, Crosa J H. Chromosome-mediated 2,3-dihydroxybenzoic acid is a precursor in the biosynthesis of the plasmid-mediated siderophore anguibactin in Vibrio anguillarum. J Bacteriol. 1994;176:4226–4234. doi: 10.1128/jb.176.14.4226-4234.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crosa J H. Molecular genetics of iron transport as a component of bacterial virulence. In: Bullen J J, Griffiths E, editors. Iron and infection. 2nd ed. West Sussex, United Kingdom: John Wiley and Sons Ltd.; 1999. pp. 255–288. [Google Scholar]
- 16.Crosa J H. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol Mol Biol Rev. 1997;61:319–336. doi: 10.1128/mmbr.61.3.319-336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crosa J H. Genetics and molecular biology of siderophore-mediated iron transport in bacteria. Microbiol Rev. 1989;53:517–530. doi: 10.1128/mr.53.4.517-530.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crosa J H. The relationship of plasmid-mediated iron transport and bacterial virulence. Annu Rev Microbiol. 1984;38:69–89. doi: 10.1146/annurev.mi.38.100184.000441. [DOI] [PubMed] [Google Scholar]
- 19.Crosa J H. A plasmid associated with virulence in the marine fish pathogen Vibrio anguillarum specifies an iron-sequestering system. Nature. 1980;284:566–568. doi: 10.1038/284566a0. [DOI] [PubMed] [Google Scholar]
- 20.Crosa J H, Hodges L L, Schiewe M H. Curing of a plasmid is correlated with an attenuation of virulence in the marine fish pathogen Vibrio anguillarum. Infect Immun. 1980;27:897–902. doi: 10.1128/iai.27.3.897-902.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crosa J H, Schiewe M H, Falkow S. Evidence for plasmid contribution to the virulence of the fish pathogen Vibrio anguillarum. Infect Immun. 1997;18:509–513. doi: 10.1128/iai.18.2.509-513.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farrell D H, Mikesell P, Actis L A, Crosa J H. A regulatory gene, angR, of the iron uptake system of Vibrio anguillarum: similarity with phage P22 cro and regulation by iron. Gene. 1990;86:45–51. doi: 10.1016/0378-1119(90)90112-5. [DOI] [PubMed] [Google Scholar]
- 23.Frey J, Krisch H M. Ω mutagenesis in gram-negative bacteria: a selectable interposon which is strongly polar in a wide range of bacterial species. Gene. 1985;36:143–150. doi: 10.1016/0378-1119(85)90078-2. [DOI] [PubMed] [Google Scholar]
- 24.Gehring A M, Bradley K A, Walsh C T. Enterobactin biosynthesis in Escherichia coli: isochorismate lyase (EntB) is a bifunctional enzyme that is phosphopantetheinylated by EntD and then acylated by EntE using ATP and 2,3-dihydroxybenzoate. Biochemistry. 1997;36:8495–8503. doi: 10.1021/bi970453p. [DOI] [PubMed] [Google Scholar]
- 25.Guilvout I, Mercereau-Puijalon O, Bonnefoy S, Pugsley A P, Carniel E. High-molecular-weight protein 2 of Yersinia enterocolitica is homologous to AngR of Vibrio anguillarum and belongs to a family of proteins involved in nonribosomal peptide synthesis. J Bacteriol. 1993;175:5488–5504. doi: 10.1128/jb.175.17.5488-5504.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harbell S O, Hodgins H O, Schiewe M H. Studies on the pathology of vibriosis in coho salmon. J Fish Dis. 1979;2:527–535. [Google Scholar]
- 27.Jalal M, Hossain V, van der Helm D, Sanders-Loehr J, Actis L A, Crosa J H. Structure of anguibactin, a unique plasmid-related bacterial siderophore from the fish pathogen Vibrio anguillarum. J Am Chem Soc. 1989;111:292–296. [Google Scholar]
- 28.Konz D, Klens A, Schorgendorfer K, Marahiel M A. The bacitracin biosynthesis operon of Bacillus licheniformis ATCC 10176: molecular characterization of three multi-modular synthetases. Chem Biol. 1997;4:927–937. doi: 10.1016/s1074-5521(97)90301-x. [DOI] [PubMed] [Google Scholar]
- 29.Köster W L, Actis L A, Waldbeser L S, Tolmasky M E, Crosa J H. Molecular characterization of the iron transport system mediated by the pJM1 plasmid in Vibrio anguillarum. J Biol Chem. 1991;266:23829–23833. [PubMed] [Google Scholar]
- 30.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Nahlik M S, Brickman T J, Ozenberger B A, McIntosh M A. Nucleotide sequence and transcriptional organization of the Escherichia coli enterobactin biosynthesis cistrons entB and entA. J Bacteriol. 1989;171:784–790. doi: 10.1128/jb.171.2.784-790.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nikaido H, Hall J A. Overview of bacterial ABC transporters. Methods Enzymol. 1998;292:3–20. doi: 10.1016/s0076-6879(98)92003-1. [DOI] [PubMed] [Google Scholar]
- 33.Pollack J R, Neilands J B. Enterobactin, an iron transport compound from Salmonella typhimurium. Biochem Biophys Res Commun. 1970;39:989–992. doi: 10.1016/0006-291x(70)90819-3. [DOI] [PubMed] [Google Scholar]
- 34.Reid J L, Collmer A. An nptI-sacB-sacR cartridge for constructing directed, unmarked mutations in gram-negative bacteria by marker exchange-eviction mutagenesis. Gene. 1987;57:239–246. doi: 10.1016/0378-1119(87)90127-2. [DOI] [PubMed] [Google Scholar]
- 35.Rusnak F, Liu J, Quinn N, Berchtold G A, Walsh C T. Subcloning of the enterobactin biosynthetic gene entB: expression, purification, characterization, and substrate specificity of isochorismatase. Biochemistry. 1990;29:1425–1435. doi: 10.1021/bi00458a013. [DOI] [PubMed] [Google Scholar]
- 36.Salinas P, Waldbeser L S, Crosa J H. Regulation of the expression of bacterial iron transport genes: possible role of an antisense RNA as a repressor. Gene. 1993;123:33–38. doi: 10.1016/0378-1119(93)90535-b. [DOI] [PubMed] [Google Scholar]
- 37.Salinas P C, Tolmasky M E, Crosa J H. Regulation of the iron uptake system in Vibrio anguillarum: evidence for a cooperative effect between two transcriptional activators. Proc Natl Acad Sci USA. 1989;86:3529–3533. doi: 10.1073/pnas.86.10.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 39.Scholten J D, Chang K, Babbit P, Charest H, Sylvestre M, Dunaway-Mariano D. Novel enzymatic hydrolytic dehalogenation of a chlorinated aromatic. Science. 1991;253:182–185. doi: 10.1126/science.1853203. [DOI] [PubMed] [Google Scholar]
- 40.Schwyn B, Neilands J B. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 41.Staab J F, Earhart C F. EntG activity of Escherichia coli enterobactin synthetase. J Bacteriol. 1990;172:6403–6410. doi: 10.1128/jb.172.11.6403-6410.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stachelhaus T, Marahiel M A. Modular structure of peptide synthetases revealed by dissection of the multifunctional enzyme GrsA. J Biol Chem. 1995;270:6163–6169. doi: 10.1074/jbc.270.11.6163. [DOI] [PubMed] [Google Scholar]
- 43.Tolmasky M E, Crosa J H. Iron transport genes of the pJM1-mediated iron uptake system of Vibrio anguillarum are included in a transposon-like structure. Plasmid. 1995;33:180–190. doi: 10.1006/plas.1995.1019. [DOI] [PubMed] [Google Scholar]
- 44.Tolmasky M E, Wertheimer A M, Actis L A, Crosa J H. Characterization of the Vibrio anguillarum fur gene: role in regulation of expression of the FatA outer membrane protein and catechols. J Bacteriol. 1994;176:213–220. doi: 10.1128/jb.176.1.213-220.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tolmasky M E, Actis L A, Crosa J H. A histidine decarboxylase gene encoded by the Vibrio anguillarum plasmid pJM1: histamine is a precursor in the biosynthesis of anguibactin. Mol Microbiol. 1994;15:87–95. doi: 10.1111/j.1365-2958.1995.tb02223.x. [DOI] [PubMed] [Google Scholar]
- 46.Tolmasky M E, Actis L A, Crosa J H. A single amino acid change in AngR, a protein encoded by pJM1-like virulence plasmids, results in hyperproduction of anguibactin. Infect Immun. 1993;61:3228–3233. doi: 10.1128/iai.61.8.3228-3233.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tolmasky M E, Gammie A E, Crosa J H. Characterization of the recA gene of Vibrio anguillarum. Gene. 1992;110:41–48. doi: 10.1016/0378-1119(92)90442-r. [DOI] [PubMed] [Google Scholar]
- 48.Tolmasky M E, Actis L A, Crosa J H. Genetic analysis of the iron uptake region of the Vibrio anguillarum plasmid pJM1: molecular cloning of genetic determinants encoding a novel trans activator of siderophore biosynthesis. J Bacteriol. 1988;170:1913–1919. doi: 10.1128/jb.170.4.1913-1919.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tolmasky M E, Salinas P C, Actis L A, Crosa J H. Increased production of the siderophore anguibactin mediated by pJM1-like plasmids in Vibrio anguillarum. Infect Immun. 1988;56:1608–1614. doi: 10.1128/iai.56.6.1608-1614.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.von Gabain A, Belasco J, Schottel J, Chang A, Cohen S. Decay of mRNA in Escherichia coli: investigation of the fate of specific segments of transcripts. Proc Natl Acad Sci USA. 1983;80:653–657. doi: 10.1073/pnas.80.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Waldbeser L S, Tolmasky M E, Actis L A, Crosa J H. Mechanisms for negative regulation by iron of the FatA outer membrane protein gene expression in Vibrio anguillarum 775. J Biol Chem. 1993;268:10433–10439. [PubMed] [Google Scholar]
- 52.Wertheimer A M, Verweij W, Chen Q, Crosa L M, Nagasawa M, Tolmasky M E, Actis L A, Crosa J H. Characterization of the angR gene of Vibrio anguillarum: essential role in virulence. Infect Immun. 1999;67:6496–6509. doi: 10.1128/iai.67.12.6496-6509.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wertheimer A M, Tolmasky M E, Actis L A, Crosa J H. Structural and functional analyses of mutant Fur proteins with impaired regulatory function. J Bacteriol. 1994;176:5116–5122. doi: 10.1128/jb.176.16.5116-5122.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolf M, Crosa J H. Evidence for the role of a siderophore in promoting Vibrio anguillarum infections. J Gen Microbiol. 1986;132:2949–2952. doi: 10.1099/00221287-132-10-2949. [DOI] [PubMed] [Google Scholar]
- 55.Wyckoff E E, Stoebner J A, Reed K E, Payne S M. Cloning of a Vibrio cholerae vibriobactin gene cluster: identification of genes required for early steps in siderophore biosynthesis. J Bacteriol. 1997;179:7055–7062. doi: 10.1128/jb.179.22.7055-7062.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]