Abstract
Glycated hemoglobin (HbA1c) indicates average glucose levels over three months and is associated with insulin resistance and type 2 diabetes (T2D). Longitudinal changes in HbA1c (ΔHbA1c) are also associated with aging processes, cognitive performance, and mortality. We analyzed ΔHbA1c in 1,886 non-diabetic Europeans from the Long Life Family Study to uncover gene variants influencing ΔHbA1c. Using growth curve modeling adjusted for multiple covariates, we derived ΔHbA1c and conducted linkage-guided sequence analysis. Our genome-wide linkage scan identified a significant locus on 17p12. In-depth analysis of this locus revealed a variant rs56340929 (explaining 27% of the linkage peak) in the ARHGAP44 gene that was significantly associated with ΔHbA1c. RNA transcription of ARHGAP44 was associated with ΔHbA1c. The Framingham Offspring Study data further supported these findings on the gene level. Together, we found a novel gene ARHGAP44 for ΔHbA1c in family members without T2D. Follow-up studies using longitudinal omics data in large independent cohorts are warranted.
Introduction
Glycated hemoglobin (HbA1c) is utilized both for the diagnosis and monitoring of type 2 diabetes (T2D), indicative of glycemic control and long-term complication risks in T2D management (Sherwani et al., 2016). HbA1c levels have a genetic basis. More than 120 loci associated with HbA1c have been identified in individuals without T2D through genome-wide association studies (GWAS) (Chen et al., 2021). Linkage scans have also revealed significant genomic regions influencing HbA1c (Meigs et al., 2002, 2007). Some of those regions and genes have been confirmed in multi-ancestry cohorts (Sarnowski et al., 2019).
Despite the acknowledged importance of HbA1c for T2D diagnosis and management, there remains a lack of longitudinal studies focusing on the long-term changes in HbA1c levels (ΔHbA1c). Most existing research tends to focus on the short-term fluctuations and control of HbA1c in T2D patients. However, understanding the long-term trends and changes in HbA1c levels, especially in populations without T2D, is crucial for developing more effective strategies for pre-diabetes diagnosis and improving healthy aging.
A previous GWAS from the Long Life Family Study (LLFS) confirmed two known common loci at GCK and HK1 and uncovered 25 suggestive loci for influencing baseline HbA1c among non-diabetic participants of the LLFS (An et al., 2014). In the present study, we conducted a GWAS of ΔHbA1c followed by a linkage-guided sequence analysis under a significant linkage peak on 17p12 using the latest available whole genome sequencing and omics data, and further pursued replication using the Framingham Offspring Study (FOS) data.
Design and Methods
Cohort Populations
The LLFS is a comprehensive, international, longitudinal study that spans two European ancestry generations, focusing on longevity and the factors underlying healthy aging (Wojczynski et al., 2022). The LLFS was conducted at four field centers, three in the United States (Boston, Pittsburgh, New York) and one in Denmark. It enrolled 4,953 participants from 539 families between 2006 and 2009. The visit 1 collected data on anthropometrics, blood pressure, physical performance, pulmonary function, and various blood tests. A second visit from 2014 to 2017 replicated the initial protocols and added carotid ultrasonography measures. HbA1c levels were measured at the University of Minnesota’s Advanced Research and Diagnostics Laboratory using high-performance liquid chromatography (HPLC). The measurements employed Tosoh analyzers calibrated to the National Glycohemoglobin Standardization Program’s standards. The laboratory’s precision for HbA1c values showed a coefficient of variation ranging from 1.4% to 1.9%. ΔHbA1c, derived by growth curve modeling using HbA1c collected from two exams seven years apart, was adjusted for age, sex, BMI, smoking, field centers and 20 principal components, and blom-transformed to approximate normality prior to genetic testing. Subjects with clinical diagnosis of T2D or T2D treatment and undiagnosed T2D cases whose fasting glucose ≥ 126 mg/dl or HbA1c ≥ 6.5% were excluded from this analysis.
Sequencing Data Preparation
Genotyping was performed on participants using the Illumina Human Omni 2.5 v1 chip by the Center for Inherited Disease Research (CIDR), leading to 1,421,289 SNPs after applying quality controls for call rate<98%, minor allele frequency (MAF) < 1%, p value Hardy–Weinberg equilibrium < 1e−6, and correct correspondence with the 1000 Genomes Project.
Whole genome sequencing (WGS) was executed using Illumina platforms at the McDonnell Genome Institute (MGI), Washington University, with reads aligned to GRCh38. Variant calling followed a four-step process using GATK tools, with additional QC to eliminate contaminated samples and those with unsuitable coverage or high Mendelian errors.
The visit 1 RNA sequencing was performed on extracted RNA from PAXgene™ Blood RNA tubes using the PAXgene microRNA extraction kit. Library preparation and quality control was managed by the Division of Computation & Data Sciences, Washington University. The nf-core/rnaseq 3.14.0 pipeline facilitated read alignment, duplication marking, and transcript quantification. Post-processing included filtering out samples with a high fraction of reads mapping to intergenic regions, filtering out genes with very low expression levels, normalizing gene counts with variance stabilizing transformation in DESeq2, leading to a final selection of 1,810 samples and 16,418 genes.
Lipid metabolomic profiling at visit 1 was executed at Washington University’s Biomedical Mass Spectrometry Lab. The laboratory implemented LC/MS for untargeted lipid detection, matched polar metabolites with internal and online databases, and annotated MS/MS lipid data. After rigorous QC and batch effect correction using a pooled QC sample, the analysis yielded data on 188 lipids across 13 compound classes.
Framingham Heart/Offspring Study for Replication
The Framingham Heart/Offspring Study (FHS/FOS) is a longitudinal cohort study tracking three generations for up to 65 years to assess cardiovascular disease risk factors. HbA1c measurements were taken from exam 5 and exam 7, selected for their similarity with LLFS intervals. Longitudinal HbA1c employed the same growth curve modeling adjusted for age and sex. Its genomic research, including whole genome sequencing (WGS) and related phenotypic analysis, is incorporated into the NHLBI’s Trans-Omics for Precision Medicine (TOPMed) Whole Genome Sequencing Program. WGS (freeze 9b) data from the Framingham Offspring Generation was used for replication. Bi-allelic single nucleotide variants (SNVs) that pass all QC filters were kept, resulting in ~52 million variants. Samples were excluded if they were either sequence controls, not sequenced in blood, or had FREEMIX percentage > 3%. Additionally, samples were removed if they had a mean depth of < 30x or < 95% of sites covered at 10x or < 80% at 20x. A total of 2,186 participants, derived from the offspring cohort’s sequencing data, were queried for replication.
Statistical Analysis
The Sequential Oligogenic Linkage Analysis Routines (SOLAR) program is designed to accommodate familial relatedness by employing maximum-likelihood based methods. These methods are utilized to estimate the residual genetic heritability of outcome measures, as well as to discern the variance that can be attributed to fixed covariate effects (Almasy & Blangero, 1998). We used SOLAR to select linked families with PEDLOD > 0.1 or PEDLOD/N > 0.01, where N was family members included in the pedigree trait count, are defined as the “potentially linked” of families. Then we ranked the families by LOD and the minimum number of families with the highest LODs that add up to at least 9 were defined as the “top linked” families. We performed GWAS analyses using a linear mixed model for additive dosage of the variants. Familial relationships were accounted for by including a kinship matrix, estimated via the “kinship” R package, as a random effect in the “lmekin” R package. LODs > 3 for GWLS and p < 5e-8 for GWAS association were used to declare significance. Associations between phenotype and RNASeq or phenotype and metabolites were analyzed using similar linear models adjusted for age, sex, and field centers.
RESULTS
Demographic Characteristics
This analysis included a total of 1,886 family members (826 men and 1,060 women) from LLFS with complete phenotypic data at both visits and genotypic information (Table 1). Similar inclusions and exclusions were applied in the replication cohorts and a total of 1,739 (752 men and 987 women) from the FOS with complete phenotypic data at both visits and genotypic information were selected for replication (Table 1). Detailed characteristics of the LLFS linkage-enriched group and others are also given in Table 2. We identified 176 subjects from 16 linkage-enriched families (“top linked” families with cumulative LODs over 9) of in the LLFS. Significant mean differences in characteristics were noted both between the study group and the others, as well as within each group across sexes. Additionally, the two cohorts differ significantly in terms of HbA1c changes; the LLFS shows minimal change in HbA1c levels, whereas the FOS exhibits a more substantial change (Table 1).
Table 1.
Sample characterisQcs of the LLFS and FOS cohorts
| Age | HbA1c | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| N | Mean (SD) | Range | N | Mean (SD) | |||
| Visit 1 | M | 856 | 64.3 (11.6) | 36–97 | 856 | 5.5 (0.3) | |
| F | 1090 | 63.8 (12.3) | 38–102 | 1090 | 5.5 (0.3) | ||
| LLFS | Visit2 | M | 856 | 72.1 (11.3) | 42–104 | 856 | 5.6 (0.3) |
| F | 1090 | 71.6 (12.0) | 45–110 | 1090 | 5.6 (0.3) | ||
| Change* | M | 856 | 7.8 (1.1) | 5–10 | 856 | −0.01 (1.0) | |
| F | 1090 | 7.8 (1.1) | 5–11 | 1090 | 0.0004 (1.0) | ||
| Visit 1 | M | 752 | 53.9 (9.9) | 30–82 | 752 | 5.2 (0.5) | |
| F | 987 | 53.6 (9.6) | 26–81 | 987 | 5.2 (0.5) | ||
| FOS | Visit2 | M | 752 | 60.6 (9.8) | 37–89 | 752 | 5.4 (0.4) |
| F | 987 | 60.3 (9.5) | 33–87 | 987 | 5.4 (0.4) | ||
| Change* | M | 752 | 6.7 (0.9) | 4–9 | 752 | 0.0034 (0.07) | |
| F | 987 | 6.7 (0.8) | 4–10 | 987 | 0.0036 (0.07) | ||
Absolute mean change
Table 2.
Sample characteristics of the linkage-enriched families in the LLFS
| Men | Women | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| N | Mean | SD | N | Mean | SD | ||
|
| |||||||
| Study Group (N=176) | Age (years) | 78 | 60.89† | 10.64 | 98 | 61.59† | 12.05 |
| Fasting Glucose (mg/dL) | 78 | 96.19*† | 9.68 | 98 | 92.22*† | 8.15 | |
| HbA1c (%) | 78 | 5.42 | 0.34 | 98 | 5.38† | 0.3 | |
| BMI (kg/m2) | 77 | 28.16*† | 3.35 | 98 | 26.44* | 4.81 | |
| ΔHbA1c | 78 | −0.04 | 1.13 | 98 | −0.05 | 1.13 | |
| HOMA-B | 70 | −0.04 | 0.92 | 93 | 0.01 | 0.89 | |
| HOMA-IR | 70 | −0.05 | 0.94 | 93 | 0.01 | 0.88 | |
| TG/HDL Ratio Growth Curve | 57 | 0.04 | 0.97 | 92 | −0.19 | 0.85 | |
| Other (N=1710) | Age (years) | 748 | 64.79† | 11.78 | 962 | 64.37† | 12.34 |
| Fasting Glucose (mg/dL) | 743 | 93.05*† | 11 | 959 | 89.26*† | 10.4 | |
| HbA1c (%) | 743 | 5.47 | 0.31 | 962 | 5.47† | 0.3 | |
| BMI (kg/m2) | 728 | 27.16*† | 3.46 | 944 | 26.22* | 4.56 | |
| ΔHbA1c | 748 | −0.01 | 0.99 | 962 | −0.0009 | 0.96 | |
| HOMA-B | 674 | −0.02 | 1.02 | 881 | −0.05 | 0.96 | |
| HOMA-IR | 674 | −0.06 | 1.01 | 881 | −0.09 | 0.98 | |
| TG/HDL Ratio Growth Curve | 420 | 0.03 | 0.99 | 632 | −0.004 | 1.01 | |
Significant (p < 0.05) mean differences between sexes
Significant (p < 0.05) mean differences (within sex) between subsets
HOMA-B, HOMA-IR: HOMA derived of β-cell function, insulin resistance
Discovery in LLFS
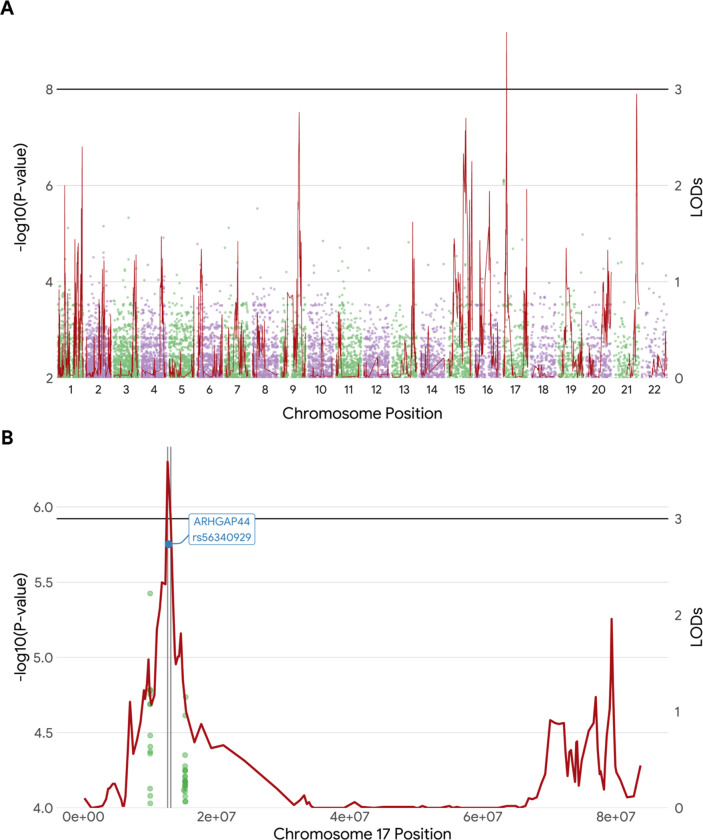
Genome-wide linkage analysis yielded a significant region on 17p12 (Fig. 1A. LODs = 3.59, 13 Mb). Heritability reached 36.8% with a p-value of 1.4e-9 (standard error = 6.8%). Whole genome sequencing association analysis did not find any significant variants with the p < 5e-8 (Fig. 1A), but it did reveal 19 suggestive variants with 5e-8 < p < 1e-5, including one in HK1 (rs16926246, p = 3.05e-7) that was previously reported for baseline HbA1c in the LLFS. The suggestive variant in the RAP1GAP2 (rs143842515, p = 2.51e-7), located 1Mb upstream of the linkage peak, was identified. A variant in the ARHGAP44 fell in 1-LOD-support interval (Fig. 1B rs56340929, p = 1.77E-06, MAF=6%, accounting for linkage = 26.5%).
Figure 1.
A. An overlay of Manhattan plot from the GWAS and GWLS results. The illustration synthesizes our findings in the overall data. The horizonal black reference line denotes the established criteria for genome-wide significance (p < 5e-8), and linkage scan significance (LODs > 3.0).
B. Highlights of the identified lead SNP rs56340929 under the right linkage peak (17p12) in the linkage-enriched families. The black line denotes a threshold at LODs > 3.0, while vertical gray lines represent the 1-LOD support interval. SNP rs56340929 is located within this interval.
Taking advantage of our available transcriptome sequencing (RNAseq) data, we assessed the association between quantification of the ARHGAP44 RNA transcript and ΔHbA1c among 176 subjects, and we found they were significantly associated (β = −0.0002, SE = 0.002, p = 0.02). We further explored the association of the ARHGAP44 RNA transcript and its corresponding SNP rs56340929, we found that the association was only marginally missed (β = −0.21, SE = 0.16, p = 0.07). We assessed currently available metabolomics data (188 metabolites in 13 compound classes) among the 16 linkage-enriched families. We found that triacylglycerol (p = 0.024) and sphingomyelin (p = 0.036) appeared to be marginally associated with ΔHbA1c. These however were non-significant after the p-values were corrected for multiple testing (p < 3e-4 for metabolites or p < 0.004 for compound classes). We also assessed rs56340929 associations with the currently available lipidomic data; and we found no significant associations after correction for multiplicity. This would not suggest at least from this analysis that the ARHGAP44 gene is directly engaged in regulating lipid metabolism.
Replication in the FOS
A total of 1,739 non-diabetic subjects from the FOS with complete phenotypic data at both visits and genotypic information were used for replication. Another SNP rs140270267 (830 bp downstream of the rs56340929, p = 0.0002, MAF = 2%) was identified, which indicated that the ARHGAP44-rs56340929 was only replicated at a nearby site (rs140270267, D’ = 1, R2 = 0.002, not in linkage disequilibrium, not at an exact SNP site) or at the ARHGAP44 gene level (SimpleM, Neff = 148, p = 0.00029 < 0.00034).
DISCUSSION
This analysis found an appreciable and significant genetic component (heritability of 37%) for ΔHbA1c among non-diabetic subjects from the LLFS. The heritability appears to be compatible with or slightly lower than our previously reported estimate of 42% in the LLFS (An et al., 2014) and estimates of 47–59% in other family studies (Meigs et al., 2002; Pilia et al., 2006; Soranzo, 2011) for HbA1c at baseline. No other familial aggregation reports of ΔHbA1c are noted. Interestingly, this analysis identified a significant linkage peak on 17p12 (LODs = 3.6) for ΔHbA1c. Several studies found suggestive linkage evidence on this region for relevant traits including fasting glucose (Loos et al., 2003), circulating leptin (Kissebah et al., 2000), T2D (Lindgren et al., 2002), coronary artery disease (Gao et al., 2014), and metabolism syndrome (Kissebah et al., 2000; Zhang et al., 2013). While no previous linkage scans were found for ΔHbA1c, Meigs and colleagues reported a suggestive linkage on chromosome 1 for baseline HbA1c in the FHS (Meigs et al., 2002). Interestingly, in this analysis, we found that our linkage peak on 17p12 was substantially attenuated (LODs from 3.6 to 1.0) when ΔHbA1c was corrected for baseline HbA1c, whereas it only modestly changed when ΔHbA1c was corrected for baseline fasting glucose levels (LODs from 3.6 to 3.4) or baseline hemoglobin levels (LODs from 3.6 to 3.8). This observation would suggest that the linkage peak for ΔHbA1c may be partially conditional on baseline HbA1c levels, but it does not clearly distinguish between glycemic and non-glycemic (erythrocytic) pathways.
Here, our best GWAS finding of HK1-rs16926246 (p = 3e-7) for ΔHbA1c did not reach genome-wide significance (p < 5e-8). To our knowledge, there are no previous GWAS reports for ΔHbA1c. For baseline HbA1c, the HK1 is a confirmed locus in the Women’s Genome Health Study (rs7072268, r2 = 0.13, MAF = 0.50, Pare et al 2008), the MAGIC consortium (rs16926246, MAF = 0.10, Soranzo et al 2010), and the LLFS (rs17476364, r2 = 0.62, MAF = 0.10, An 2014). These common SNPs seem to be not in perfect linkage disequilibrium, and likely represent independent HK1 variants. The HK1 gene encodes hexokinase 1, the enzyme catalyzing the first step of glycolysis, that regulates glucose metabolism. The HK1 deficiency in erythrocytes causes severe non-spherocytic hemolytic anemia. Previous reports also suggested that the HK1 locus could influence HbA1c levels via erythrocyte biology (Paré et al., 2008; Soranzo et al., 2010).
The only statistically significant discovery from our linkage-guided sequence analysis was the ARHGAP44-rs56340929 (p = 2e-6, MAF = 6%) for ΔHbA1c. We assessed all sequence elements under the linkage peak on 17p12 (within 1-LOD support interval from 12.7 Mb to 12.9 Mb) and found this lead variant accounted for nearly 30% of the linkage peak. Our RNAseq data showed that ARHGAP44 expression level was only marginally missed its association with the variant (p = 0.07) but significantly associated with ΔHbA1c (p = 0.02). Our lipidomics data did not reveal any metabolites that were significantly associated (p < 3e-4 after multiple testing correction) with ΔHbA1c or with the variant. This observation would suggest the ARHGAP44 gene variant does not directly regulate lipid metabolism. The ARHGAP44 encodes Rho GTPase activating protein 44 which is involved in the control of Rho-type GTPases (Xu et al., 2017). The ARHGAP44 gene has also been reportedly associated with cardiovascular diseases, serum creatinine and glycemic traits including HbA1c (Dornbos et al., 2022). Finally, we looked up the ARHGAP44-rs56340929 in the FOS and found encouraging validation evidence at a neighboring variant (rs14270267, 830 bp downstream, p = 0.0002, r2 = 0.002) and at the gene level (simpleM, p < 0.0003, Neff = 148, after correction for multiple testing) though not at the exact SNP site (p = 0.8).
Strengths of the current study included its extended family design, relatively large sample size, longitudinal data availability with multiple visits, well-defined phenotypic measures, and moreover availability of multi-omics data. However, there are also few limitations needed to be noted in this analysis. Sample heterogeneity and thus genetic heterogeneity may exist across the LLFS and FOS. This was evidenced by significant mean differences in HbA1c levels and key covariates between the two studies (see Table 1). The exceptional longevity of the LLFS sample compared to the general population may introduce selection bias that potentially rendered somewhat data heterogeneity. Additionally, incomplete access to the FOS data with missing covariate variables may impede the exact replication of phenotyping adjustment methods, which may further diminish the validity of the replication.
In conclusion, this integrated linkage-guided sequence analysis allowed for our identification of a novel gene the ARHGAP44 for ΔHbA1c in the LLFS with supportive evidence from our omics data as well as encouraging replication data using the FOS. Further independent replications from large cohorts are needed to confirm and extend our findings.
Funding
This work was supported by the National Institute on Aging (U01AG023746, U01AG023712, U01AG023749, U01AG023755, U01AG023744, and U19AG063893-01).
Footnotes
Statements of assistance
The investigators thank all the LLFS participants and staff for their valuable contributions. We are grateful to LeAnne Kniepkamp for her administrative help and effort. The authors thank Lihua Wang, M.D., M.S. for her assistance in statistical data analysis for this manuscript.
Prior published abstract of the study
Wang S, Thyagarajan B, Lee J, Zmuda J, Christensen K, Province M, An P. Novel gene variants associated with HbA1c changes over time among non-diabetic subjects in the Long Life Family Study. Innov Aging. 2023 Dec 21;7(Suppl 1):1088. doi: 10.1093/geroni/igad104.3496. PMCID: PMC10738390.
References
- Almasy L., & Blangero J. (1998). Multipoint Quantitative-Trait Linkage Analysis in General Pedigrees. The American Journal of Human Genetics, 62(5), 1198–1211. 10.1086/301844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An P., Miljkovic I., Thyagarajan B., Kraja A. T., Daw E. W., Pankow J. S., Selvin E., Kao W. H. L., Maruthur N. M., Nalls M. A., Liu Y., Harris T. B., Lee J. H., Borecki I. B., Christensen K., Eckfeldt J. H., Mayeux R., Perls T. T., Newman A. B., & Province M. A. (2014). Genome-wide association study identifies common loci influencing circulating glycated hemoglobin (HbA1c) levels in non-diabetic subjects: The Long Life Family Study (LLFS). Metabolism - Clinical and Experimental, 63(4), 461–468. 10.1016/j.metabol.2013.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Spracklen C. N., Marenne G., Varshney A., Corbin L. J., Luan J., Willems S. M., Wu Y., Zhang X., Horikoshi M., Boutin T. S., Mägi R., Waage J., Li-Gao R., Chan K. H. K., Yao J., Anasanti M. D., Chu A. Y., Claringbould A., … Lifelines Cohort Study. (2021). The trans-ancestral genomic architecture of glycemic traits. Nature Genetics, 53(6), 840–860. 10.1038/s41588-021-00852-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C., Layer R. M., Faust G. G., Lindberg M. R., Rose D. B., Garrison E. P., Marth G. T., Quinlan A. R., & Hall I. M. (2015). SpeedSeq: Ultra-fast personal genome analysis and interpretation. Nature Methods, 12(10), 966–968. 10.1038/nmeth.3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho K., Schwaiger-Haber M., Naser F. J., Stancliffe E., Sindelar M., & Patti G. J. (2021). Targeting unique biological signals on the fly to improve MS/MS coverage and identification efficiency in metabolomics. Analytica Chimica Acta, 1149, 338210. 10.1016/j.aca.2021.338210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornbos P., Singh P., Jang D.-K., Mahajan A., Biddinger S. B., Rotter J. I., McCarthy M. I., & Flannick J. (2022). Evaluating human genetic support for hypothesized metabolic disease genes. Cell Metabolism, 34(5), 661–666. 10.1016/j.cmet.2022.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust G. G., & Hall I. M. (2014). SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics, 30(17), 2503–2505. 10.1093/bioinformatics/btu314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Li L., Rao S., Shen G., Xi Q., Chen S., Zhang Z., Wang K., Ellis S. G., Chen Q., Topol E. J., & Wang Q. K. (2014). Genome-Wide Linkage Scan Identifies Two Novel Genetic Loci for Coronary Artery Disease: In GeneQuest Families. PLOS ONE, 9(12), e113935. 10.1371/journal.pone.0113935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissebah A. H., Sonnenberg G. E., Myklebust J., Goldstein M., Broman K., James R. G., Marks J. A., Krakower G. R., Jacob H. J., Weber J., Martin L., Blangero J., & Comuzzie A. G. (2000). Quantitative trait loci on chromosomes 3 and 17 influence phenotypes of the metabolic syndrome. Proceedings of the National Academy of Sciences, 97(26), 14478–14483. 10.1073/pnas.97.26.14478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren C. M., Mahtani M. M., Widén E., McCarthy M. I., Daly M. J., Kirby A., Reeve M. P., Kruglyak L., Parker A., Meyer J., Almgren P., Lehto M., Kanninen T., Tuomi T., Groop L. C., & Lander E. S. (2002). Genomewide Search for Type 2 Diabetes Mellitus Susceptibility Loci in Finnish Families: The Botnia Study. The American Journal of Human Genetics, 70(2), 509–516. 10.1086/338629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos R. J. F., Katzmarzyk P. T., Rao D. C., Rice T., Leon A. S., Skinner J. S., Wilmore J. H., Rankinen T., & Bouchard C. (2003). Genome-Wide Linkage Scan for the Metabolic Syndrome in the HERITAGE Family Study. The Journal of Clinical Endocrinology & Metabolism, 88(12), 5935–5943. 10.1210/jc.2003-030553 [DOI] [PubMed] [Google Scholar]
- Meigs J. B., Manning A. K., Fox C. S., Florez J. C., Liu C., Cupples L. A., & Dupuis J. (2007). Genome-wide association with diabetes-related traits in the Framingham Heart Study. BMC Medical Genetics, 8(1), S16. 10.1186/1471-2350-8-S1-S16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meigs J. B., Panhuysen C. I. M., Myers R. H., Wilson P. W. F., & Cupples L. A. (2002). A Genome-Wide Scan for Loci Linked to Plasma Levels of Glucose and HbA1c in a Community-Based Sample of Caucasian Pedigrees: The Framingham Offspring Study. Diabetes, 51(3), 833–840. 10.2337/diabetes.51.3.833 [DOI] [PubMed] [Google Scholar]
- Paré G., Chasman D. I., Parker A. N., Nathan D. M., Miletich J. P., Zee R. Y., & Ridker P. M. (2008). Novel Association of HK1 with Glycated Hemoglobin in a Non-Diabetic Population: A Genome-Wide Evaluation of 14,618 Participants in the Women’s Genome Health Study. PLOS Genetics, 4(12), e1000312. 10.1371/journal.pgen.1000312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen B. S., & Quinlan A. R. (2017). Who’s Who? Detecting and Resolving Sample Anomalies in Human DNA Sequencing Studies with Peddy. The American Journal of Human Genetics, 100(3), 406–413. 10.1016/j.ajhg.2017.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilia G., Chen W.-M., Scuteri A., Orrú M., Albai G., Dei M., Lai S., Usala G., Lai M., Loi P., Mameli C., Vacca L., Deiana M., Olla N., Masala M., Cao A., Najjar S. S., Terracciano A., Nedorezov T., … Schlessinger D. (2006). Heritability of Cardiovascular and Personality Traits in 6,148 Sardinians. PLOS Genetics, 2(8), e132. 10.1371/journal.pgen.0020132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnowski C., Leong A., Raffield L. M., Wu P., de Vries P. S., DiCorpo D., Guo X., Xu H., Liu Y., Zheng X., Hu Y., Brody J. A., Goodarzi M. O., Hidalgo B. A., Highland H. M., Jain D., Liu C.-T., Naik R. P., O’Connell J. R., … Meigs J. B. (2019). Impact of Rare and Common Genetic Variants on Diabetes Diagnosis by Hemoglobin A1c in Multi-Ancestry Cohorts: The Trans-Omics for Precision Medicine Program. The American Journal of Human Genetics, 105(4), 706–718. 10.1016/j.ajhg.2019.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwani S. I., Khan H. A., Ekhzaimy A., Masood A., & Sakharkar M. K. (2016). Significance of HbA1c Test in Diagnosis and Prognosis of Diabetic Patients. Biomarker Insights, 11, BMI.S38440. 10.4137/BMI.S38440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soranzo N. (2011). Genetic Determinants of Variability in Glycated Hemoglobin (HbA1c) in Humans: Review of Recent Progress and Prospects for Use in Diabetes Care. Current Diabetes Reports, 11(6), 562–569. 10.1007/s11892-011-0232-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soranzo N., Sanna S., Wheeler E., Gieger C., Radke D., Dupuis J., Bouatia-Naji N., Langenberg C., Prokopenko I., Stolerman E., Sandhu M. S., Heeney M. M., Devaney J. M., Reilly M. P., & Ricketts S. L. (2010). Common Variants at 10 Genomic Loci Influence Hemoglobin A1C Levels via Glycemic and Nonglycemic Pathways. Diabetes, 59(12), 3229–3239. 10.2337/db10-0502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojczynski M. K., Jiuan Lin S., Sebastiani P., Perls T. T., Lee J., Kulminski A., Newman A., Zmuda J. M., Christensen K., Province M. A., & on behalf of the Long Life Family Study. (2022). NIA Long Life Family Study: Objectives, Design, and Heritability of Cross-Sectional and Longitudinal Phenotypes. The Journals of Gerontology: Series A, 77(4), 717–727. 10.1093/gerona/glab333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Jiao J., Xu W., Ji L., Jiang D., Xie S., Kubra S., Li X., Fu J., Xiao J., & Zhang B. (2017). Mutant p53 promotes cell spreading and migration via ARHGAP44. Science China Life Sciences, 60(9), 1019–1029. 10.1007/s11427-016-9040-8 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Kent J. W. Jr., Olivier M., Ali O., Broeckel U., Abdou R. M., Dyer T. D., Comuzzie A., Curran J. E., Carless M. A., Rainwater D. L., Göring H. H. H., Blangero J., & Kissebah A. H. (2013). QTL-based association analyses reveal novel genes influencing pleiotropy of metabolic syndrome (MetS). Obesity, 21(10), 2099–2111. 10.1002/oby.20324 [DOI] [PMC free article] [PubMed] [Google Scholar]