Abstract
Background:
Volumetry of subregions in the medial temporal lobe (MTL) computed from automatic segmentation in MRI can track neurodegeneration in Alzheimer’s disease. However, image quality may vary in MRI. Poor quality MR images can lead to unreliable segmentation of MTL subregions. Considering that different MRI contrast mechanisms and field strengths (jointly referred to as “modalities” here) offer distinct advantages in imaging different parts of the MTL, we developed a muti-modality segmentation model using both 7 tesla (7T) and 3 tesla (3T) structural MRI to obtain robust segmentation in poor-quality images.
Method:
MRI modalities including 3T T1-weighted, 3T T2-weighted, 7T T1-weighted and 7T T2-weighted (7T-T2w) of 197 participants were collected from a longitudinal aging study at the Penn Alzheimer’s Disease Research Center. Among them, 7T-T2w was used as the primary modality, and all other modalities were rigidly registered to the 7T-T2w. A model derived from nnU-Net took these registered modalities as input and outputted subregion segmentation in 7T-T2w space. 7T-T2w images most of which had high quality from 25 selected training participants were manually segmented to train the multi-modality model. Modality augmentation, which randomly replaced certain modalities with Gaussian noise, was applied during training to guide the model to extract information from all modalities. To compare our proposed model with a baseline single-modality model in the full dataset with mixed high/poor image quality, we evaluated the ability of derived volume/thickness measures to discriminate Amyloid+ mild cognitive impairment (A+MCI) and Amyloid− cognitively unimpaired (A–CU) groups, as well as the stability of these measurements in longitudinal data.
Results:
The multi-modality model delivered good performance regardless of 7T-T2w quality, while the single-modality model under-segmented subregions in poor-quality images. The multi-modality model generally demonstrated stronger discrimination of A+MCI versus A–CU. Intra-class correlation and Bland-Altman plots demonstrate that the multi-modality model had higher longitudinal segmentation consistency in all subregions while the single-modality model had low consistency in poor-quality images.
Conclusion:
The multi-modality MRI segmentation model provides an improved biomarker for neurodegeneration in the MTL that is robust to image quality. It also provides a framework for other studies which may benefit from multimodal imaging.
Keywords: multi-modality, medial temporal lobe, subregion segmentation
1. Introduction
Different MRI contrast mechanisms and field strengths (jointly referred to as “modalities”) offer distinct advantages in imaging different parts of the brain. Multi-modality analysis has been widely used in many brain disease-related tasks, such as brain tumor segmentation (Menze et al., 2015), ischemic stroke lesion segmentation (Lin & Liebeskind, 2016), brain tissue segmentation (Išgum et al., 2015) as well as subregion segmentation in medial temporal lobe (MTL) (Xie et al., 2023).
The MTL, which includes the hippocampus and parahippocampal gyrus, is a frequent focus of research given its role in episodic memory, healthy aging (Maillet & Rajah, 2013; Raz et al., 2004) and brain disorders including neurodegenerative diseases, such as Alzheimer’s disease (AD), (Barkhof et al., 2007; Burton et al., 2009; Korf et al., 2004). The MTL is the earliest cortical region affected by tau proteinopathy, a hallmark pathology of AD (Bilgel, n.d.; Braak and Braak, 1995, 1991; Nelson et al., 2012). Its morphometric abnormalities are not only visible in magnetic resonance imaging (MRI) acquired from patients with AD, but also subtle loss in its subregions can be detected in patients with mild cognitive impairment (MCI) and preclinical AD (Duara et al., 2008; Visser et al., 2002; Xie et al., 2020).
The MTL can be anatomically divided into hippocampal subfields and MTL cortical subregions (called MTL subregions together hereinafter). MRI-based MTL subregional volumetry and morphometry can provide highly sensitive measures for analyzing patterns of neurodegeneration in AD and related disorders (Barkhof et al., 2007; Burton et al., 2009) as well as for mapping of cognition and memory (Squire et al., 2004).
T1-weighted (T1w) 3 tesla (3T) (combined as 3T-T1w) MRI at approximately 1×1×1mm3 resolution is the most commonly used MRI contrast for quantifying neurodegeneration. In most large AD neuroimaging studies, such as the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (Petersen et al., 2010) or A4 (Sperling et al., 2020), 3T-T1w modality is the primary structural sequence. However, the appearance of the hippocampus in 3T-T1w MRI lacks sufficient parenchymal contrast, to reliably identify and label the subfields in the hippocampus (Wisse et al., 2021; Yushkevich et al., 2015). On the other hand, a “dedicated” 3T T2-weighted (T2w) (combined as 3T-T2w) sequence offers higher resolution in an oblique coronal plane perpendicular to the hippocampus’s main axis with signal contrast allowing for visualization of subfields (Bonnici et al., 2012; Winterburn et al., 2013). Such dedicated 3T-T2w MRI is being collected in some research protocols, including ADNI, but is less common than 3T-T1w.
The 7 tesla (7T) MRI provides higher resolution and contrast than lower magnetic field scanners for imaging the MTL (Cho et al., 2010; Derix et al., 2014; Prudent et al., 2010; Wisse et al., 2014), and therefore can trace the intricate anatomy of complex regions such as the hippocampal head. It has the potential to identify structural changes in brain disorders with greater accuracy than 3T MRI. Many recent studies utilize the MP2RAGE sequence which combines two volumes acquired at different inversion times, resulting in an image with more inhomogeneity but outstanding T1w tissue contrast in the brain (Forstmann et al., 2014). Although 7T-T2w MRI has a high contrast and resolution and can help us observe the subregion structure of MTL clearly (Kollia et al., 2009; Zwanenburg et al., 2011), it is also susceptible to the lateral signal dropout, making the image look like blurry, especially for the BA36 subregion (Grande et al., 2023).
More advantages and disadvantages of these modalities are shown in Table 1.
Table 1.
The advantages and disadvantages of different MRI sequences for MTL subregional morphometry.
| Sequence | Advantages | Disadvantages |
|---|---|---|
| 3T-T2w 0.4 × 0.4 × 1.2 mm3 | • Clear dark band (The stratum radiatum, lacunosum and moleculare, SRLM) crucial for hippocampal subfields • Sharper visualization of sulci • Easier to separate dura from cortex (Xie et al., 2016) |
• More prone to motion • Only covers a part of the brain |
| 7T-T2w 0.42 × 0.42 × 1 mm3 | • Clear SRLM • Higher resolution and contrast • Whole-brain coverage |
• Lateral signal dropout affecting MTL cortex |
| 3T-T1w 0.8 × 0.8 × 0.8 mm3 and 7T-T1w 0.69 × 0.69 × 0.69 mm3 | • Isotropic voxels which are better for thickness measurements • Whole-brain coverage but less prone to signal dropout |
• Difficult to separate dura from cortex • SRLM not clearly visible on 3T, affecting hippocampal subfield segmentation (Wisse et al., 2021) |
Segmenting the MTL subregions manually is a challenging and time-consuming task, which is prone to inter-rater differences and requires a lot of anatomical knowledge (Yushkevich et al., 2015). Automatic segmentation of the MTL subregions is less laborious and more reproducible.
Atlas-based techniques are a subclass of supervised automatic medical image segmentation algorithms that rely on deformable registration to match expert-segmented example images (atlases) to target images, providing an implicit shape, intensity and context prior for segmentation (Iglesias & Sabuncu, 2015). Such complex priors are essential for MTL subregion segmentation, where there is high intersubject anatomical variability; and until the advent of deep learning, multi-atlas segmentation has been the leading approach for MTL subregion segmentation in MRI. Yushkevich et al. proposed the model named ‘Automatic Segmentation of Hippocampal Subfields (ASHS)’ in 3T-T2w, and the corresponding atlas set is open source (Yushkevich et al., 2015). It was also then developed for 3T-T1w sequence where hippocampal subfields combined into a single hippocampus label that is then split into anterior and posterior portions (Xie et al., 2016, 2019) and 7T-T2w sequence with a new 7T-T2w segmentation protocol (Wisse et al., 2016). Xie et al. combined a deep-learning method and an atlas-based segmentation pipeline to create the model named ‘deep label fusion’, which achieved a higher segmentation accuracy than the conventional ASHS (Xie et al., 2023), while also demonstrating better generalization ability from 3T to 7T than nnU-Net, a leading deep learning 3D image segmentation pipeline (Isensee et al., 2021).
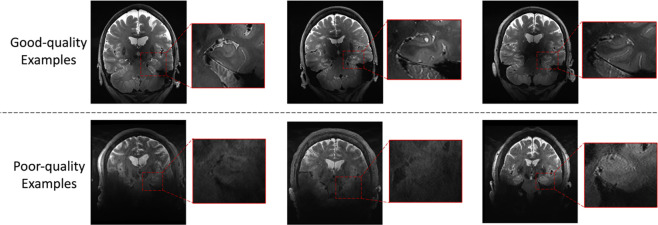
While 7T MRI offers distinct avantages over 3T MRI in terms of resolution and contrast, image quality of 7T MRI is more variable. In particular, higher magnetic field strength is associated with more severe susceptibility artifacts and therefore some kinds of MRI artifacts are more pronounced at 7T than at 3T. Based on its anatomical location relative to the sinuses, the MTL is particularly vulnerable, especially in the most inferior portion of the MTL with low contrast and low sharpness (see Figure 1). The existing 7T-T2w ASHS-based model performs poorly on such poor-quality images. This issue cannot be addressed by improving the algorithm’s robustness or designing a specific model architecture to adapt to this particular type of image because most of the MRI scans in the ASHS atlas set have high image quality where manual labeling is possible, whereas manually labeling subfields and subregions is challenging even for experienced annotators in poor-quality images. Therefore, there is no training data with poor image quality available to train a segmentation model. On the other hand, the segmentation difficulties encountered in low-quality images are due to the lack of sufficient anatomical information for segmentation, so that neither experienced experts nor segmentation models can complete accurate segmentation. To overcome this issue, it is necessary to consider additional information outside of 7T-T2w.
Figure 1.
Examples of good image quality and poor image quality in 7T-T2w modality
The current study presents an automated MTL subregion segmentation deep learning model for multi-scanner multi-modality MRI. The model combines all available modalities (3T-T1w, 3T-T2w, 7T-T1w, 7T-T2w) to fuse information and extract complementary information from other modalities when information from one or more modalities is missing or corrupted. This approach addresses the problem of poor segmentation driven by poor quality of the 7T-T2w modality.
Our study is carried out in a large dataset (n=197) of 3T and 7T MRI in the same set of research study participants at the University of Pennsylvania Alzheimer’s Disease Research Center (ADRC). The input for the model consists of multi-modality images co-registered and resliced to the space of the 7T-T2w image. A modality augmentation scheme is proposed to force the model to extract useful features from all modalities. The nnU-Net, one of the most sophisticated segmentation models, is used as the segmentation backbone. The model’s performance is first evaluated through cross-validation on the available set of manually-labeled atlases. However, recognizing that evaluation in this high-quality image dataset is likely not representative of real-world performance on individuals whose 7T-T2w scans are of poorer quality, we perform three additional indirect evaluations, including (1) volume comparison, (2) longitudinal comparison and (3) a task-based evaluation comparing the ability of different models to distinguish Amyloid− cognitively unimpaired (A–CU) and Amyloid+ MCI (A+MCI) individuals. Together, these indirect evaluations demonstrate that the proposed multi-modality segmentation model outperforms the baseline single-modality model that only considers the 7T-T2w modality, especially for poor quality 7T-T2w images.
This study is an application of using multi-modality images to reduce the impact of poor quality images on segmentation performance. Our evaluation focuses on coping with differences in image quality between the images for which expert annotations are available and on which models are trained on the one hand, and the images to which they are eventually applied on the other. The challenges addressed in this paper are relevant to other medical image analysis applications in which multimodality data is available, including other MRI contrasts such as diffusion tensor imaging (DTI) and susceptibility weighted imaging (SWI).
2. Materials and Methods
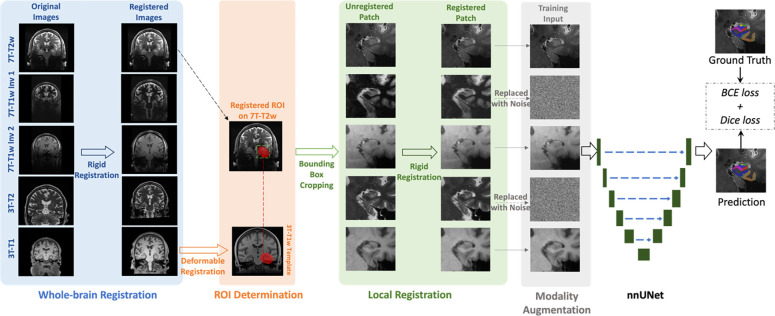
The flowchart of the training process of the segmentation model is shown in Figure 2. It includes whole-brain registration, ROI determination, local registration, modality augmentation, and nnU-Net training. In the model inference, the modality augmentation will be skipped and the model will use full modality as input. Details of each step are explained below.
Figure 2.
The flowchart of the training process of the segmentation model. All modalities are rigidly registered to 7T-T2w in the whole-brain registration step. Then, the MTL ROI (red region in ROI determination step) is mapped from 3T-T1w template to 7T-T2w space by deformable registration. MTL patches are cropped along the bounding box of the ROIs in all registered modalities and local registration step refines the misalignment between 7T-T2w and other modalities specifically on MTL. Before feeding into the nnU-Net segmentation model, two to four modalities (two modalities in this figure as examples) are randomly replaced by noise in modality augmentation step. Finally, the nnU-Net is trained using combined BCE and Dice as loss function
2.1. Dataset
2.1.1. Participants and MRI protocol
MRI scans were acquired from the ADRC at the University of Pennsylvania. Both 7T and 3T MRI were considered in this study. The 7T protocol included both T1w (MP2RAGE) and T2w (TSE) scans. The 7T-T1w scan contained two inversion contrasts with different T1 weightings, INV1 and INV2, which had a resolution of 0.69 × 0.69 × 0.69 mm3. The 7T-T2w scan covered the whole brain and had in-plane resolution of 0.42 × 0.42 mm2 and slice thickness of 1 mm.
The 3T protocol included a ‘routine’ whole-brain T1w (MPRAGE) scan and a ‘dedicated’ T2w (TSE) scan with partial brain coverage. The resolution of the 3T-T1w scan was 0.8 × 0.8 × 0.8mm3. The in-plane resolution and slice thickness of the 3T-T2w scan were 0.4 × 0.4 mm2 and 1.2 mm, respectively, with the slice orientation approximately orthogonal to the main axis of the hippocampi.
The inclusion and exclusion criteria used in this study are shown in Supplementary Section S1. We finally obtained 25 training participants and 172 test participants. The most recent diagnosis result and amyloid PET to the date of 7T scan of each participant were collected. Of these participants, 40 were diagnosed with MCI, while the remaining 157 were cognitively unimpaired (CU) individuals, 122 of them had Amyloid PET with 40 diagnosed as positive and 82 negative scans based on visual read. Table 2 shows specific cohort characteristics in this study.
Table 2.
Characteristics of the cohort in this study
| Training Set | Test Set | |||
|---|---|---|---|---|
| Number of participants | 25 | 172 | ||
| Age at the time of 7T scan | 59 – 97 (72.2 ± 7.5) | 23 – 94 (63.0 ± 16.7) | ||
| Sex (female/male) | 16/9 | 103/69 | ||
| High-quality 7T-T2w ROIs* | 47 | 231 | ||
| Low-quality 7T-T2w ROIs | 3 | 113 | ||
| CU participants | 15 | (Amyloid-: 9) (Amyloid+: 3) | 142 | (Amyloid-: 65) (Amyloid+: 15) |
| MCI participants | 10 | (Amyloid-: 2) (Amyloid+: 4) | 30 | (Amyloid-: 6) (Amyloid+: 18) |
| Amyloid PET | 18 | 104 | ||
| Longitudinal scan | - | 29 | ||
Because each participant has two ROIs, left and right, the total number of ROIs is twice the total number of participants. The quality assessment is described in section 2.1.3
Among 172 test participants, some of them had more than one 7T scan. Using the inclusion and exclusion criteria described in Supplementary Section 1, longitudinal scans of 29 participants were collected.
The distributions of scanning date interval between 7T and 3T MRI of 172 test participants and 25 training set, respectively, scanning interval between two longitudinal 7T scans in 29 participants with longitudinal data are shown in Supplementary Figure S2.
2.1.2. Ground-truth Annotation for Training Set
A total of 25 participants were previously selected as the ASHS “atlas set” and underwent manual segmentation (Xie et al., 2023). As shown in Table 2, most ROIs in atlas set had high quality (the quality assessment process is described in Section 2.1.3). This subset also serves as the training set for the proposed segmentation model.
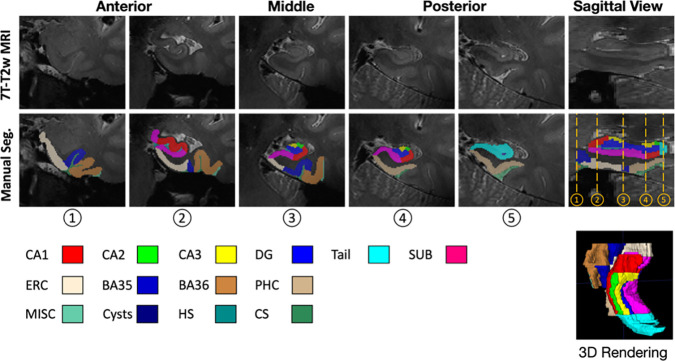
The “Penn Aging Brain Cohort (ABC)” protocol, originally described in (Berron et al., 2017), and modified in (Xie et al., 2023) was used to segment the hippocampal subfields and cortical MTL subregions manually on both left and right sides in 7T-T2w scans of training set. The specific subregions segmented are as follows and shown in Figure 3.
Figure 3.
Examples of ground-truth annotation in the training set
Hippocampus subfields included cornu ammonis (CA) 1, CA2, CA3, dentate gyrus (DG), subiculum (SUB), and the tail of hippocampus. The tail is not partitioned into subfields because of its complex anatomy and curvature of the hippocampal axis relative to the imaging plane (Berron et al., 2017).
MTL cortical subregions included entorhinal cortex (ERC), Brodmann areas 35 and 36 (BA35/36) and parahippocampal cortex (PHC).
Four non-gray-matter labels including hippocampal sulcus (HS), collateral sulcus (CS), cysts and miscellaneous (MISC) were also annotated.
The remaining 172 participants without ground-truth (manual segmentation) were used as the test set to evaluate the model. The test set was not selected according to image quality, and therefore contained participants with poor 7T-T2w image quality, which is common in real-world 7T-T2w dataset.
2.1.3. Quality Assessment
To evaluate segmentation performance across different image qualities, the image quality of 7T-T2w modality for both training and test sets was assessed and labeled by author YL. The participants in the training and test sets were mixed together and randomly shuffled for assessment. The assessment process was blinded to the rater. The specific steps are as follows:
The MTL region of each participant was cropped from the 7T-T2w MRI.
The participants were inspected in random order.
The rater navigated through the ROIs in 3D in ITK-SNAP (Yushkevich et al., 2006) and rated them on a scale of 1 to 9 based on the visibility and sharpness of hippocampus and cortical regions, with higher numbers indicating better image quality. When assessing, rater used the number 5 as a threshold, and all images considered to be of high quality would have a score greater than or equal to 5.
The assessments were conducted three times and the average rating of each ROI was calculated to minimize bias.
After rounding, the rating will be used as the final rating for each participant.
2.2. Pre-processing
2.2.1. Whole-brain Rigid Registration
The different modalities had different resolutions (as displayed in Figure 2), voxel spacing and slice thicknesses. All modalities’ images were registered to primary modality (7T-T2w) space rigidly using the registration tool greedy (Venet et al., 2021) in each participant.
This process in each participant can be divided into three steps.
7T-T1w to 7T-T2w: The 7T-T1w INV1 and 7T-T1w INV2 had same resolution and voxel spacing. They were registered together to 7T-T2w with normalized mutual information (NMI) as image similarity metric, center alignment by translation as initial registration, and 100, 50 and 10 as number of iterations at coarsest level (4x), intermediate (2x) and full (1x) resolution, respectively.
3T-T1w to 7T-T2w: The 3T-T1w modality was registered to 7T-T2w rigidly with NMI as image similarity metric, 165 center alignment by translation as initial registration, and 100, 50 and 10 as number of iterations at coarsest level (4x), intermediate (2x) and full (1x) resolution, respectively.
3T-T2w to 7T-T2w: The 3T-T2w image was “dedicated” scan with partial brain coverage that targets the hippocampal region specifically. Performing registration from 3T-T2w to 7T-T2w directly could result in misalignment because the uncovered region (zero-value voxels) were involved in the calculation of registration similarity. Since 3T-T1w and 3T-T2w were acquired in the same scan, they were roughly aligned. The rigid registration matrix calculated from 3T-T1w to 7T-T2w was used to map 3T-T2w image to 7T-T2w space.
Registration accuracy was visually checked. The examples of successful and failed registrations are shown in Supplementary Figure S3.
2.2.2. Region of Interest Determination
The current study focuses on MTL subregions. Thus, the regions surrounding the left and right MTL were selected as regions of interest (ROI), respectively. An unbiased population template (Joshi et al., 2004) constructed using 29 3T-T1w MRI scans which were from a different population with this study, and template-space ROIs for the left and right MTL, generated as part of the 3T-T1w ASHS package (Xie et al., 2019), were used in this process.
The ROI determination for each participant includes the following steps.
First, the neck was trimmed in the 3T-T1w image.
Then, the template was registered to the neck-trimmed 3T-T1w image using deformable registration (using greedy (Venet et al., 2021)) with the similarity metric of normalized cross-correlation (NCC) calculated with the neighborhoods of 5×5×5 voxels. The deformation field was saved and the left and right ROIs in template were mapped to the 3T-T1w image using the deformation field.
Since the 3T-T1w image had been registered to 7T-T2w space in step 2.2.1, these ROIs were further mapped to the 7T-T2w space using registration matrix obtained in 2.2.1.
Left and right MTL patches along the bounding box of the registered ROIs were cropped from 7T-T2w image and other modalities’ images in 7T-T2w space according to the ROIs in 7T-T2w.
Registration accuracy was visually checked. The examples of successful and failed registrations are shown in Supplementary Figure S4.
2.2.3. Local Rigid Registration
The whole-brain affine registration was not sufficient to align the local MTL region well due to modality-related non-linear deformations, e.g., MRI gradient distortion. This motived the need for further local registration. Therefore, a more reliable rigid registration that focused only on left and right ROIs, respectively, was performed by greedy (Venet et al., 2021).
The local registration can be divided into three steps.
7T-T1w to 7T-T2w: 7T-T1w inv1 had similar intensity distribution to 7T-T2w, so the NCC calculated in neighborhoods of 5×5×5 was chosen as similarity metric for the rigid registration from 7T-T1w INV1 patch to 7T-T2w patch. Then, the 7T-T1w INV2 patch was mapped to 7T-T2w using the same registration matrix as 7T-T1w INV1.
3T-T1w to 7T-T2w: the 3T-T1w patch was rigidly registered to the 7T-T2w with NMI as similarity metric, identical alignment as initialization, and 100 and 50 as number of iterations at the coarsest level and intermediate resolution, respectively.
3T-T2w to 7T-T2w: 3T-T2w image had similary intensity distribution as 7T-T2w. 3T-T2w patch was rigidly registered to 7T-T2w with weighted-NCC as similarity metric, identical alignment as initialization, and 100 and 50 as number of iterations at the coarsest level and intermediate resolution, respectively.
This resulted in two ROIs for each participant with each ROI containing five well-registered modalities. Registration accuracy was visually checked. The examples of successful and failed registrations are shown in Supplementary Figure S5.
2.3. Segmentation Model
The current study focuses on leveraging multiple MRI field strengths and modalities in MTL subregion segmentation rather than on developing a novel segmentation backbone. We conducted experiments using the extensively validated nnU-Net framework (Isensee et al., 2021) as the segmentation model. Specifically, the 3D full-resolution version of nnU-Net was selected.
The input of the model was a five-channel 3D input (as per the order, the five channels were 7T-T2w, 7T-T1w inv1, 7T-T1w inv2, 3T-T2w, and 3T-T1w) which was formed by stacking all modalities in the same ROI with 7T-T2w. All ROIs on the right were flipped to the left side before feeding into the segmentation model.
Due to the high quality of the 7T-T2w modality in the training set and the fact that manual segmentation was based on the 7T-T2w, we would expect the nnU-Net model to primarily rely on the 7T-T2w to minimize its loss during training and largely ignore information from other modalities. However, when applied to the larger testing set, where the average quality of the 7T-T2w is lower, we hypothesize that a model that primarily relies on 7T-T2w would underperform compared to a model that effectively synthesizes information from all available modalities. To encourage nnU-Net to use all available modalities during training, we incorporate an additional augmentation scheme, called modality augmentation (ModAug), which we employ together with the standard data augmentation schemes in nnU-Net.
As shown in Figure 4, in each iteration of training process, there was a 50% chance that input data was fed into an augmentation branch. Otherwise, all five modalities were fed into the segmentation model. In the augmentation branch, there was an equal chance that data were processed by each of four different sub-branches, where one, two, three or four modalities in the input data would be replaced by random noise. This step was designed to force the model to extract critical information from all modalities, rather than focusing only on the information associated with the primary modality.
Figure 4.
The pipeline of modality augmentation. When five channels were fed into the network, they would be processed by augmentation branch with probability of 50%. Othewise, the full modalities were used in the segmentation. In augmentation branch, there was equal probability that one, two, three and four modalities were replaced by noise randomly.
Combined Dice and binary cross-entropy (BCE) loss function was used to supervise the training process using deep-supervision scheme. The model was trained in a five-fold cross-validation setting. In each fold, the training process was terminated after 400 epochs, where the validation loss of the validation set plateaued to constant. All other hyper-parameters remained at their default settings. When performing inference on the test set without manual segmentations (n=172), the five per-fold models were ensembled to generate the final prediction (Isensee et al., 2021).
2.4. Model Evaluation
2.4.1. Baseline Model
To verify the advantage of using multiple modalities, another nnU-Net model which only used the primary modality (7T-T2w modality) as input was trained as baseline model. It is referred to as the single-modality model.
2.4.2. Cross-validation in the Annotated Training Set
As a metric to measure the consistency between predicted segmentation and ground-truth, Dice similarity coefficient (Dice, 1945) was used to evaluate segmentation accuracy in five-fold cross-validation in the training process. For each subregion, the average of Dice scores from all validation participants in five folds were calculated.
2.4.2. Indirect Validation in the Test Set
Due to the variable quality of the 7T-T2w, and the expert effort it would entail, we did not attempt to generate manual segmentations for the test set, which made direct quantitative evaluation, such as Dice coefficient, impossible. Three indirect evaluation methods were used to compare the segmentation performance of proposed model and baseline model.
Inter-model consistency: volume comparison. In order to discover the difference between multi-modality and single-modality models, the volume of each subregion in each participant was calculated and was compared between models. Outliers (subregions with low consistency) were identified and analyzed to determine how they relate to image quality.
Longitudinal consistency. A good segmentation model should perform well regardless image quality. When presented with two longitudinal scans for the same participant in our study, a good model should provide largely consistent MTL subregion volume measurements, considering that the loss of hippocampal MTL volume in healthy aging and MCI in the range of 0.2–2.55% a year (Fjell et al., 2009; Jack et al., 2000; Kurth et al., 2017). Intraclass correlation coefficients (ICC) was used to evaluate longitudinal consistency for each model in the subset of 29 participants who had longitudinal 3T and 7T MRI.
Classification between NC and MCI. Previous studies showed that regional measurements such as cortical thickness and subfield volumes had the ability to discriminate between A+MCI participants and A–CU participants (Whitwell et al., 2007; Wolf et al., 2004). Correct computation of measurements relies on accurate segmentation of subregions, so the ability to separate A–CU and A+MCI was used as a metric for evaluating segmentation models.
To make the volumes of subregions comparable across participants, volumes of hippocampus subfields were adjusted by the volume of hippocampal tail (for each subfield, the proportion of the tail volume was determined by proportion of the rest of the hippocampus) and the volumes of MTL cortical subregions were normalized by the length of their segmentation in the 7T-T2w slicing direction (number of slices spanned times slice thickness) (Yushkevich et al., 2015).
3. Results
3.1. Quality Assessment
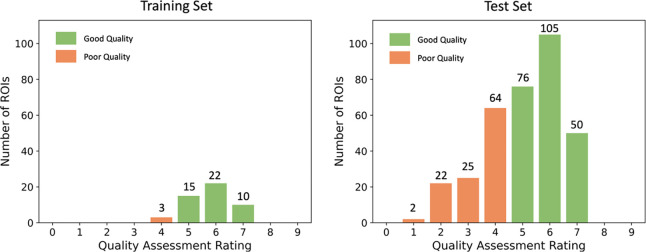
Image quality was first assessed so that we could evaluate the performance of the models in images of different quality. Left and right ROIs of all participants in the whole dataset were assessed. Figure 5 shows the distribution of image quality in the training set and the test set. The rating greater or equal to 5 represents the good quality and less than 5 represents the poor quality.
Figure 5.
The distribution of image quality in the training and test set respectively.
The average quality of the training set was 5.79, while the average quality of the test set was 5.04. The majority of the ROIs in the training set had good image quality. Only three ROIs in the training set (6%) had quality rating 4 and none were rated below 4. As expected, the test set had a substantially larger proportion of poor quality ROIs (32.85% with rating lower than 5). However, the distribution of the quality ratings in the [5,9] range was similar for the test set and the training set (the ratio of samples rated 5, 6 and 7 in both sets was approximately 3:4:2).
3.2. Cross-validation in the Training Set
Dice coefficient between automated and manual segmentation was calculated for each subregion in the cross-validation experiments. Table 3 compares the average Dice coefficient for each subregion among the single-modality (7T-T2w only) model, the multi-modality model trained without ModAug, and the multi-modality model with ModAug (proposed model).
Table 3.
Dice score (average Dice ± standard deviation) comparison of single-modality and multi-modality models in five-fold cross-validation (Bold numbers represent the highest values among all models)
| Subregions | Single-modality model | Multi-modality model without ModAug | Multi-modality model with ModAug |
|---|---|---|---|
|
| |||
| Dice | Dice | ||
|
| |||
| CA1 | 0.814 ± 0.036 | 0.817 ± 0.034 | 0.817 ± 0.036 |
| CA2 | 0.735 ± 0.059 | 0.747 ± 0.054 | 0.741 ± 0.055 |
| CA3 | 0.705 ± 0.064 ** | 0.728 ± 0.058 | 0.724 ± 0.057 |
| DG | 0.870 ± 0.026 ** | 0.871 ± 0.026 ** | 0.866 ± 0.023 |
| SUB | 0.861 ± 0.026 | 0.862 ± 0.024 | 0.862 ± 0.026 |
| Tail | 0.845 ± 0.053 *** | 0.859 ± 0.053 | 0.861 ± 0.052 |
| ERC | 0.846 ± 0.043 *** | 0.845 ± 0.042 *** | 0.855 ± 0.040 |
| BA35 | 0.730 ± 0.080 *** | 0.727 ± 0.072 *** | 0.751 ± 0.073 |
| BA36 | 0.812 ± 0.057 *** | 0.802 ± 0.058 *** | 0.825 ± 0.052 |
| PHC | 0.815 ± 0.065 | 0.818 ± 0.064 | 0.822 ± 0.062 |
p < 0.01;
p < 0.001 in paired t-test comparison between corresponding model and multi-modality model with ModAug
For each subregion, the highest Dice value was obtained from multi-modality model, either with ModAug or without ModAug. It shows the advantage of involving other modalities in a segmentation model, which indeed provides more useful information than 7T-T2w alone. However, the difference between single-modality model and multi-modality model was not particularly large. The subregion Dice difference between single-modality model the best multi-modality model did not surpass 0.023, which was quite close, and some subregions even did not have statistical significance. This was due to the fact that most ROIs in training set had high image quality, allowing the single-modality model to segment subregions effectively without assistance from multi-modality data.
The comparison between two mutli-modality models with and without ModAug, repectively, shows the model without ModAug performed better in hippocampus subfields and the model with ModAug performed better in extrahippocampal subregions. The p-value shows the segmentation on hippocampus was comparable between these two models in all subfields except for DG, while the model with ModAug significantly performed better in ERC, BA35 and BA36. This indicates the ModAug encouraged the model to learn multi-modality information better.
3.3. Inter-model consistency: volume comparison
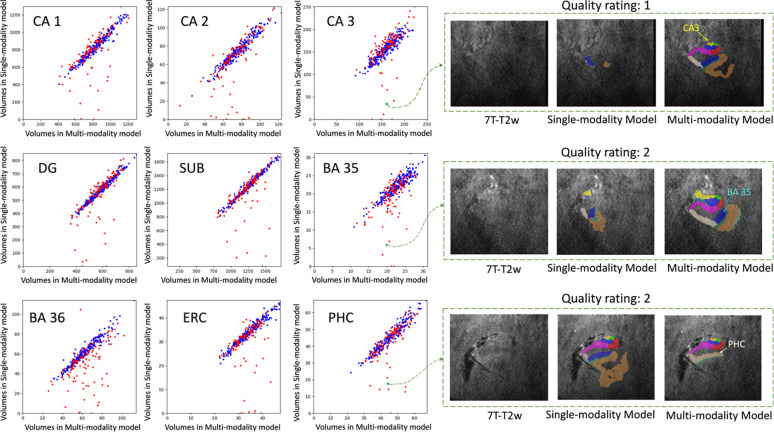
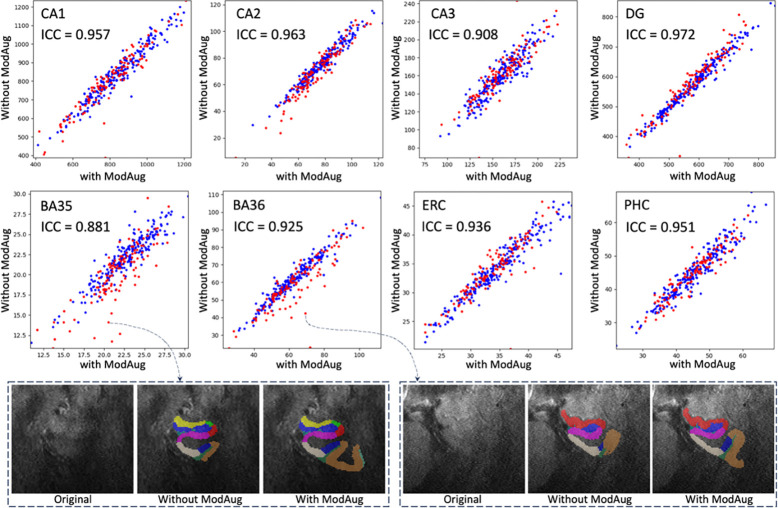
The inference of the single-modality model and the inference of the multi-modality model with ModAug were carried out on the test set. Figure 6 shows the subregion volume comparisons between these two models.
Figure 6.
The volume comparison for different subregions on the left side. X-axis: subregion volume by the multi-modality model with ModAug. Y-axis: subregion volume by the single-modality model. Blue dots: participants with good image quality. Red dots: participants with poor image quality.
The blue dots representing high quality ROIs (quality rating > 5) in each subplot of Figure 6 are distributed largely along the diagonal line. This demonstrates the consistency of segmentation results between single-modality and multi-modality models with ModAug in high-quality images. However, the red dots, which represent poor-quality ROIs, are not always distributed along the diagonal line in each subplot in Figure 6. Most outliers fall below the diagonal line, indicating that the segmentation results of the single-modality model produce smaller subregions than those of the multi-modality model. As can be seen from the examples in Figure 6, these outliers were poor quality images and segmentations. The single-modality model had severe under-segmentation, while the segmentation results from multi-modality model had typical segmentation size and morphology.
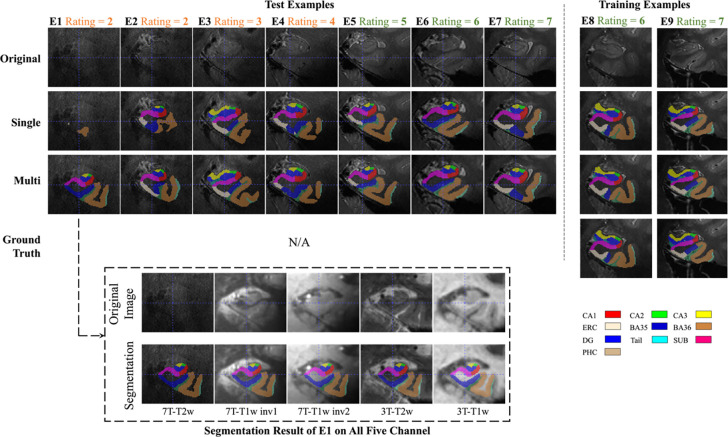
To show how the segmentation results varied with the quality of the image, seven examples with different image qualities in the test set and two examples with high quality in the training set were randomly selected. Segmentation results of the single-modality and multi-modality models on these nine examples are shown in Figure 7. When the image quality was poor, the single-modality model exhibited under-segmentation while the multi-modality model could still segment all subregions resonably. As the image quality improved, the under-segmentation of single-modality model became less and its segmentation results approached the multi-modality model. In the high-quality images, these two models performed similarly. Further, the segmentation results of the two models were both similar to the ground-truth in the two training examples in Figure 7. This explains the high Dice score of each model in cross-validation.
Figure 7. Subregion segmentation in examples with imaging quality from low to high.
The segmentation in training examples are obtained by cross-validation. The higher the image quality, the more consistent between single-modality and multi-modality models. For the first example (E1), other modalities are shown in dashed box and the segmentation by multi-modality model aligns well in these modalities. (CA = cornu ammonis; DG = dentate gyrus; SUB = subiculum; ERC = entorhinal cortex; BA = Brodmann area; CS = collateral sulcus; HS = hippo sulcus; MISC = miscellaneous label)
3.4. Longitudinal Comparison
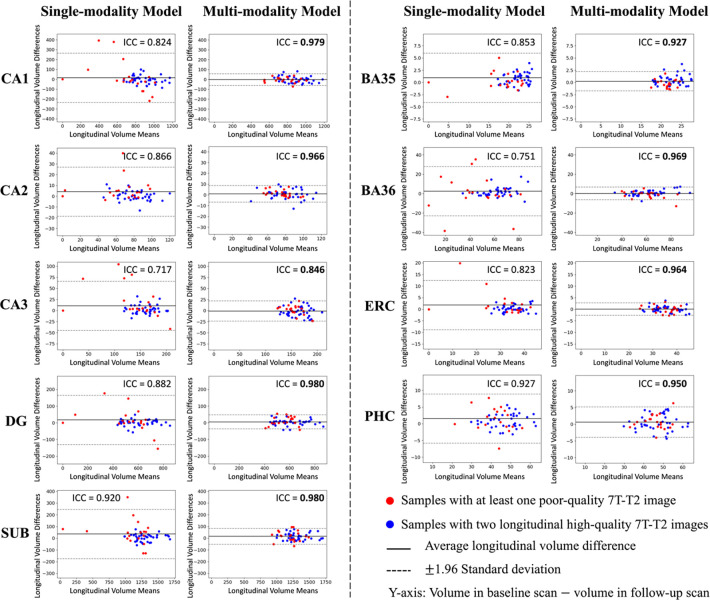
For the 29 test participants with longitudinal MRI scans, the ICC of volumes obtained from different timepoints of each subregion were computed. Additionally, the Bland-Altman plot for each subregion was plotted in order to visually compare the differences between the models. As shown in Figure 8, the multi-modality model had higher ICC values on each subregion compared to single-modality model, suggesting that multi-modality segmentation was more consistent across longitudinal scans.
Figure 8. Longitudinal segmentation consistency comparison.
For the 29 participants with two longitudinal scans, each scan was segmented using both the multi-modality model and the single-modality model, and the volume of each subregion was calculated from the segmentation. In this figure, the volume consistency between the two scans was represented by Bland-Altman plot and intra-class correlation (ICC) for each model. The multi-modality model had smaller volume mean values and smaller standard deviations of longitudinal volume difference in all subregions. For the single-modality model, the distribution of results from poor-quality data (red dots) was more scattered. The multi-modality model provided much higher ICC values in all subregions. Higher ICC values are highlighted in bold
As can be seen from the Bland-Altman plot in Figure 8, the segmentation results of multi-modality model in the longitudinal scan pairs had smaller standard deviation, regardless of the quality of the image. The segmentation results of the single-modality model were less consistent across the two scans than those of the multi-modality model. In terms of the distribution of image quality, inconsistency was mainly in the low-quality images.
3.5. Distinguishing CU and MCI
To indirectly assess the segmentation accuracy of the model, a general linear model (GLM) was fitted in each subregion with the regional volume or thickness measure as the dependent variable and diagnostic group (A+MCI and A–CU) as the independent variable. For the subfields of the hippocampus, the dependent variable was volume adjusted by the volume of hippocampal tail and the GLM also included age and intracranial volume (ICV) as nuissance covariants; for the cortical subregions, the dependent variable was median thickness calculated by computing the pruned Voronoi skeleton (Ogniewicz & Kübler, 1995) of the MTL cortex and integrating the radius field over each MTL subregion, as implemented in the software CM-Rep (Pouch et al., 2015), and only age was included as the nuissance covariant.
The p-value, AUC, and Cohen’s D for the fitted GLMs are shown in Table 4. The multi-modality model was associated with a greater difference and effect size between the two diagnostic groups, in absolute terms, compared to single-modality model.
Table 4. Amyloid+ MCI and Amyloid— CU comparison in the test set.
The volumes of hippocampus subfields and median thicknesses of cortical subregions of 65 Amyloid— CU and 18 Amyloid+ MCI participants were calculated separately based on the segmentation results of single-modality and multi-modality models. The volumes of subfields were adjusted for the hippocampal tail. p-value, AUC and Cohen's D of multi-modality model and single-modality model were compared. For each subregion, if the p-value of one of the two models was less than 0.05, the smaller p-value as well as the larger AUC and Cohen's D were marked bold. (MCI: mild cognitive impairment; CU: cognitively unimpaired; AUC: area under the ROC curve)
| Single-modality Model | Multi-modality Model | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Side | Measure | Subregion | p-value | AUC | Cohen’D | p-value | AUC | Cohen’s D |
|
| ||||||||
| Left | Adjusted Volume | CA1 | 0.047 | 0.73 | 0.48 | 0.00040 | 0.75 | 0.86 |
| CA2 | 0.0098 | 0.69 | 0.78 | 3.0E-05 | 0.78 | 1.25 | ||
| CA3 | 0.38 | 0.58 | 0.23 | 0.00029 | 0.76 | 0.93 | ||
| DG | 0.057 | 0.71 | 0.49 | 7.4E-05 | 0.78 | 0.99 | ||
| SUB | 0.080 | 0.73 | 0.48 | 0.00010 | 0.76 | 0.89 | ||
|
| ||||||||
| Median Thickness | ERC | 0.43 | 0.50 | 0.24 | 0.033 | 0.65 | 0.67 | |
| BA35 | 0.61 | 0.49 | 0.09 | 0.39 | 0.51 | 0.30 | ||
| BA36 | 0.28 | 0.41 | 0.26 | 0.71 | 0.46 | 0.01 | ||
| PHC | 0.37 | 0.39 | 0.28 | 0.66 | 0.50 | 0.16 | ||
|
| ||||||||
| Right | Adjusted Volume | CA1 | 0.033 | 0.66 | 0.46 | 0.0012 | 0.72 | 0.66 |
| CA2 | 0.043 | 0.69 | 0.60 | 0.00062 | 0.77 | 0.94 | ||
| CA3 | 0.45 | 0.62 | 0.15 | 0.026 | 0.67 | 0.48 | ||
| DG | 0.013 | 0.71 | 0.63 | 0.0010 | 0.73 | 0.78 | ||
| SUB | 0.012 | 0.68 | 0.56 | 0.0036 | 0.69 | 0.64 | ||
|
| ||||||||
| Median Thickness | ERC | 0.45 | 0.47 | 0.16 | 0.21 | 0.58 | 0.42 | |
| BA35 | 0.71 | 0.48 | 0.04 | 0.21 | 0.58 | 0.44 | ||
| BA36 | 0.54 | 0.45 | 0.10 | 0.91 | 0.52 | 0.14 | ||
| PHC | 0.38 | 0.43 | 0.24 | 0.68 | 0.52 | 0.14 | ||
4. Discussion
We developed a multi-modality segmentation model for MTL subregions in structural MRI using 7T-T2w, 7T-T1w (INV1 and INV2), 3T-T2w and 3T-T1w modalities and a modality augmentation scheme to guide the model to learn features from all available modalities. The multi-modality model showed stability in segmentation even when the quality of the primary imaging modality was poor, which is common in real-world MRI data but was not captured by the training set due to the demands of manual segmentation. The proposed multi-modality model also had higher consistency in longitudinal scans and better ability to discriminate A+MCI and A–CU groups when compared with the single-modality model, which only considers the 7T-T2w modality.
4.1. Considerations for segmentation evaluation metric
When evaluating the accuracy of a segmentation model, it is common to report average metrics such as Dice coefficient in cross-validation experiments conducted on all available annotated data. However, there may be an inherent bias in image quality between images that are annotated for training and images on which the trained model is ultimately used. This was indeed the case in our dataset, where the proportion of low-quality images was much higher (32.85% vs 6%) for the non-annotated test images. If we only considered this type of evaluation, we would have concluded that there is no benefit from using multiple modalities in MTL subregion segmentation. The Dice coefficient computed in cross-validation experiments did not reflect the superior performance of multi-modality model because the data in the training set were selected from high quality images, and as demonstrated in Table 3 and Figure 7. The single-modality model also obtained good segmentation results from high-quality images.
However, our indirect evaluation on the non-annotated test set clearly points to greater robustness of the multi-modality model. In order to compare the models’ performance in images with different qualities, we evaluated them in a test dataset that included low quality images. However, manual annotation of these data was not practical, or outright impossible, precisely because the image quality of the modality used for manual segmentation was low. Hence, quantitative analyses, including measuring Dice coefficient, could not be performed. Instead, we measured the robustness of the segmentations generated by the two models in images with different qualities through a series of indirect tasks, where two aspects were verified: stability and accuracy. Specifically, by comparing the segmentation results of the two models and by comparing the segmentation results of the same model at different longitudinal time points, we verified that the multi-modality model likely had better segmentation stability, particularly in low-quality images. By demonstating greater separation between disease and control groups (higher AUCs) on a classification task, we also got strong evidence in the higher segmentation accuracy of the multimodality model.
4.2. MTL Cortical Thickness Findings in A+MCI in the Context of Prior Work.
When distinguishing between A+MCI and A–CU, difference in median thickness for BA35 was not significant and in ERC thickness was only significantly smaller on the left. This was inconsistent with our experience and with existing research (Duara et al., 2008; Hata et al., 2019; Ogawa et al., 2019; Wolk et al., 2017; Xie et al., 2020; Yushkevich et al., 2015), especially given that the volumes of hippocampus subfields were significantly smaller in A+MCI group. Since BA35 approximates the transentorhinal region, the site of earliest cortical tau tangle pathology and neurodegeneration in AD, we expect this region to show differences as pronounced as the hippocampus and hippocampal subfields. Earlier MTL morphometry studies using ASHS also showed results consistent with these expectations (Wolk et al., 2017; Yushkevich et al., 2015).
We identified three possible reasons for this discrepancy. First, the definition of BA35 boundaries in the current protocol may not completely align with prior versions of procotol (Yushkevich et al., 2015) used in prior studies. Second, the segmentation by nnU-Net may have been less accurate (oversegmentation or undersegmentation) than ASHS. Third, this lack of significant effect could have been due to the small sample size (18 A+MCI) and potentially idiosyncratic differences from prior populations studied. We conducted two additional experiments to better understand the source of the discrepancy. A 3T-T1w atlas, labeled based on the “Penn Memory Center (PMC)” protocol (Xie et al., 2019), was used in these experiments. First, a 3T-T1w ASHS was trained on 3T-T1w atlas and run on the 3T-T1w images of the 83 test participants diagnosed as A+MCI and A–CU (same participants in Section 3.5), and the relationship between each cortical median thickness and diagnosis group was fitted using a GLM with age as covariate. Second, an nnU-Net was trained on 3T-T1w atlas and applied to the 3T-T1w images in the same dataset, and the same analysis was performed. The p-values of GLM in these models are shown in Table 5.
Table 5.
p-value of GLM with dependent variable of cortical subregion median thickness and independent variable of diagnosis group for different models in 3T-T1w
| Side | Subregion | ASHS | nnU-Net |
|---|---|---|---|
|
| |||
| Left | ERC | 0.012 | 0.0031 |
| BA35 | 0.030 | 0.00036 | |
| BA36 | 0.28 | 0.18 | |
| PHC | 0.32 | 0.40 | |
|
| |||
| Right | ERC | 0.58 | 0.051 |
| BA35 | 0.97 | 0.13 | |
| BA36 | 0.79 | 0.13 | |
| PHC | 0.15 | 0.31 | |
Median thickness of BA35 segmented by 3T-T1w ASHS and 3T-T1w nnUNet was significant on the left (p-values: 0.030 and 0.00036, repectively) although not so on the right. Since the same dataset was used in the analysis, the dataset was not the primary reason for the lack of effect using the proposed model, nor was the use of nnU-Net specifically. This leads us to conclude that the main difference between proposed mutli-modality model and 3T-T1w nnU-Net was probably the segmentation protocol. The 3T-T1w atlas is based on the “PMC” protocol described in (Yushkevich et al., 2015) and the 7T-T2w atlas is based on the ABC protocol by (Berron et al., 2017). These protocols employ slightly different rules to label MTL subregions and these differences might explain the lack of group effects in BA35 and right ERC, even though they were developed in collaboration with the same neuronatomist (Ding & Van Hoesen, 2010).
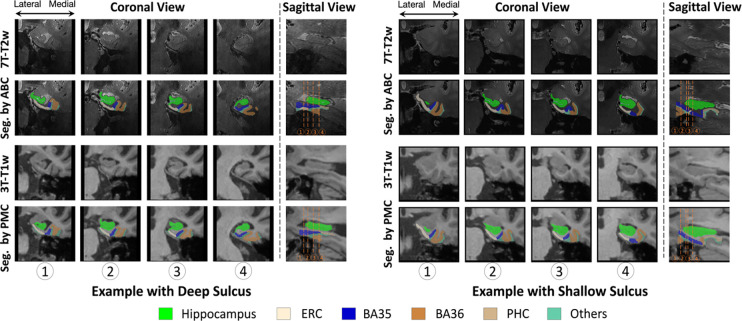
To demonstrate these differences, two participants’ segmentation of BA35 in 7T-T2w by multi-modality model trained by ABC protocol and 3T-T1w by nnU-Net trained by PMC protocol are shown in Figure 9. For the first participant in Figure 9, BA35 was located on the medial side (i.e., left) of the collateral sulcus in both 3T-T1w and 7T-T2w. For the second individual, the BA35 in 3T-T1w was mainly located on the medial side of the collateral sulcus and BA35 in 7T-T2w was on medial side in regions when the the sulcus was deep (the first slice in coronal view) and on both sides when more shallow (the second and third slices in coronal view).
Figure 9.
Comparison of BA35 (dark blue) segmentation by nnU-Net trained in PMC protocol (3T-T1w) and multi-modality model trained in ABC protocol (7T-T2w). The first example had deep sulcus and segmentations for BA35 by both methods were at the medial side of collateral sulcus. The second example had shallow sulcus in slice 2 and 3 in coronal view and segmentations of BA35 by multi-modality model were on both sides of collateral sulcus. (The 3T-T1w images and segmentations have been registered to 7T-T2w here, making it possible to compare them with segmentations in 7T-T2w.)
However, we have only verified that the difference in protocols was a possible reason for the difference in significance in the experiments, but have not found a specific way of how exactly the ABC protocol affects the thickness of the subregion. Further experiments are necessary in the future.
4.3. The role of modality augmentation
In cross-validation, the multi-modality model with and without ModAug did not show significant difference according to Dice scores in the training set, which had high-quality images. We expected that ModAug would have greater advantage in the test set. The subregion volume comparison of the multi-modality model trained with and without ModAug is shown in Figure 10.
Figure 10.
Subregion volumes comparison between multi-modality model with and without ModAug. (Red dots: low-quality images; blue dots: high-quality images)
In Figure 10, except for BA35 and BA36, high-quality dots and low-quality dots have similar distribution in the plots. In BA35 and BA36, there were more poor-quality images as outliers. Two outliers were picked and shown at the bottom of Figure 10. In these two examples, the model trained without ModAug might have undersegmentation in BA35 and BA36. The model trained with ModAug output the results with more reasonable shape of MTL. This is in line with a common issue of signal drop out in more lateral regions in the primary modality 7T-T2w which mostly affects segmentation accuracy of BA35 and BA36. Relying more on the other modalities, which are less prone to signal drop out in these areas, should indeed provide a more complete segmentation of BA35 and BA36.
4.4. Importance of each modality
ModAug appeared to allow extraction of useful information from alternative modalities when image quality was poor in the primary one. It allowed the model to be robust to missing modalities, but we did not know how much information provided by each modality.
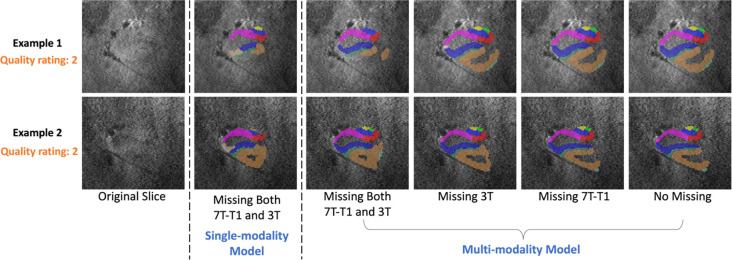
In order to further verifiy the importance of each modality, we ran the inference of the multi-modality model with certain modalities discarded. Specificly, 3T, 7T-T1w and both 3T and 7T-T1w were replaced by noise images in the inference, respectively, and we observed the changes of segmentations after removing these modalities.
The subregion volume consistency between these results and the model with complete input (no modality missing) was calculated by ICC. As shown in Table 6, when both 3T and 7T-T1w were missing, there was the most significant loss of information resulting in lower ICC values compared with input in which only one of these modalities was missing. The input missing 7T-T1w had higher ICC values than the input missing 3T, indicating that the model was more dependent on the 3T modality than the 7T-T1w. This conclusion can be also reflected in Figure 11, which shows two poor-quality examples segmented by the multi-modality model on input with different modalities missing. The results on input only missing 7T-T1w were closest to the complete input.
Table 6.
ICC values of subregion volume comparison between multi-modality model with certain modalities missing and model without modality missing
| Model | Missing modality | CA1 | CA2 | CA3 | DG | SUB | BA35 | BA36 | ERC | PHC |
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Multi-modality Model with ModAug | 3T | 0.995 | 0.989 | 0.981 | 0.972 | 0.992 | 0.971 | 0.98 | 0.988 | 0.986 |
| 7T-T1w | 0.998 | 0.994 | 0.99 | 0.997 | 0.998 | 0.986 | 0.998 | 0.993 | 0.992 | |
| 3T and 7T-T1w | 0.871 | 0.811 | 0.793 | 0.869 | 0.854 | 0.758 | 0.767 | 0.851 | 0.804 | |
|
| ||||||||||
| Single-modality Model | 3T and 7T-T1w * | 0.795 | 0.747 | 0.662 | 0.783 | 0.75 | 0.696 | 0.675 | 0.595 | 0.786 |
The single-modality model only used 7T-T2w as input. Therefore, there was no 3T or 7T-T1w fed into the model. We also call this situation as “missing 3T and 7T-T1w” here.
Figure 11.
Two examples of poor-quality images segmented by multi-modality models with input missing 7T-T1w and 3T, missing 3T, missing 7T-T1w and input without modality missing. The segmentations by single-modality for these two examples are also compared here.
Besides different situations of missing modality, the segmentation by single-modality model was also compared with other scenarios. The ICC values in Table 6 show that single-modality model had lowest consistency with multi-modality model with full modalities as input. The examples in Figure 11 also reflect the low consistency between the single-modality model and multi-modality model. Although we input the same amount of information into single-modality model and multi-modality model with both 3T and 7T-T1w input missing, the single-modality model had more undersegmentation. The reason might be in the training process. For multi-modality model, the information of different modalities from training set was stored in the network parameters of nnU-Net. Therefore, when encountering missing modality, these embedded information can help the model make relatively better predictions.
4.5. Limitations and future work
One limitation of the present study is that it only considered the quality of the 7T-T2w in evaluation. The impact of poor quality of other modalities was not evaluated. The quality assessment for other modalities will be done in the future and we will evaluate how other modalities’ quality influences the segmentation.
Another limitation is that the proposed multi-modality model required one modality to be selected as “primary”, in our case this was 7T-T2w. This modality is used as a target for registration and all other modalities are resampled in its space, causing loss of resolution, such as the isotropic resolution and finer plane thickness in 3T-T1w, and aliasing. In the future it may be possible to develop a model that allows all images to be analyzed in their own space. With the 3T-T2w ASHS atlas, which was developed using the same ABC protocol as 7T-T2w ASHS atlas, it is possible to develop a segmentation model which considers both 3T and 7T modalities as target spaces of segmentation. This can also solve the problem that other centers do not have mixed 3T and 7T MRI. In this case, for the participants with only 3T modality, the model should output the segmentation in 3T space; for the participants with both 7T and 3T modalities, the model should output consistent segmentation in the spaces of both modalities.
5. Conclusion
This study developed a segmentation model for MTL subregions using 7T-T2w, 7T-T1w, 3T-T2w and 3T-T1w MR images. Incorporating these modalities during training with the help of together with modality augmentation led to a model that is more resilient to low image quality, resulting in more accurate segmentation. When the primary modality’s image quality was low, proposed multi-modality model still could generate stable segmentaitons by extracting useful information from other modalities.
While the current study focused on a very unique dataset with four structural modalities collected at two MRI field strengths, the challenges and the insights gained in the evaluation regarding the utility of multi-modality models in the context of poor/variable image quality and the danger of only using cross-validation performance to select the best segmentation model when manually annotated data exhibit selection bias relative to the real-world data distribution are likely relevant to other image segmentation contexts with multiple modalities available, such as DTI, SWI.
Supplementary Material
Funding
This work was supported by NIH (grant numbers R01-AG069474, RF1-AG056014, P30-AG072979, R01-AG070592)
Footnotes
Declaration of Competing Interests
D.A.W. has served as a paid consultant to Eli Lilly, GE Healthcare, Merck, Janssen and Qynapse. He serves on a DSMB for Functional Neuromodulation. He received grants from Eli Lilly/Avid Radiopharmaceuticals, Merck and Biogen. He is also now on a DSMB for GSK.
S.R.D. received consultation fees from Rancho Biosciences and Nia Therapeutics.
L.X. is a paid employee of Siemens Healthineers. He received personal consulting fees from Galileo CDS, Inc.
L.E.M.W. was supported by MultiPark, a strategic research area at Lund University.
Ethics approval and consent to participate
Written informed consent was obtained for all participants under a protocol approved by the University of Pennsylvania Institutional Review Board.
Consent for publication
All authors have reviewed the contents of the manuscript being submitted, approved of its contents and validated the accuracy of the data and consented to publication.
Data and Code Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- Barkhof F., Polvikoski T. M., Van Straaten E. C. W., Kalaria R. N., Sulkava R., Aronen H. J., Niinistö L., Rastas S., Oinas M., Scheltens P., & Erkinjuntti T. (2007). The significance of medial temporal lobe atrophy: A postmortem MRI study in the very old. Neurology, 69(15), 1521–1527. 10.1212/01.wnl.0000277459.83543.99 [DOI] [PubMed] [Google Scholar]
- Berron D., Vieweg P., Hochkeppler A., Pluta J. B., Ding S.-L., Maass A., Luther A., Xie L., Das S. R., Wolk D. A., Wolbers T., Yushkevich P. A., Düzel E., & Wisse L. E. M. (2017). A protocol for manual segmentation of medial temporal lobe subregions in 7 Tesla MRI. NeuroImage: Clinical, 15, 466–482. 10.1016/j.nicl.2017.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilgel M. (n.d.). Causal links among amyloid, tau, and neurodegeneration. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnici H. M., Chadwick M. J., Kumaran D., Hassabis D., Weiskopf N., & Maguire E. A. (2012). Multi-voxel pattern analysis in human hippocampal subfields. Frontiers in Human Neuroscience, 6. 10.3389/fnhum.2012.00290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton E. J., Barber R., Mukaetova-Ladinska E. B., Robson J., Perry R. H., Jaros E., Kalaria R. N., & O’Brien J. T. (2009). Medial temporal lobe atrophy on MRI differentiates Alzheimer’s disease from dementia with Lewy bodies and vascular cognitive impairment: A prospective study with pathological verification of diagnosis. Brain, 132(1), 195–203. 10.1093/brain/awn298 [DOI] [PubMed] [Google Scholar]
- Cho Z.-H., Han J.-Y., Hwang S.-I., Kim D., Kim K.-N., Kim N.-B., Kim S. J., Chi J.-G., Park C.-W., & Kim Y.-B. (2010). Quantitative analysis of the hippocampus using images obtained from 7.0 T MRI. NeuroImage, 49(3), 2134–2140. 10.1016/j.neuroimage.2009.11.002 [DOI] [PubMed] [Google Scholar]
- Derix J., Yang S., Lüsebrink F., Fiederer L. D. J., Schulze-Bonhage A., Aertsen A., Speck O., & Ball T. (2014). Visualization of the amygdalo–hippocampal border and its structural variability by 7T and 3T magnetic resonance imaging. Human Brain Mapping, 35(9), 4316–4329. 10.1002/hbm.22477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dice L. R. (1945). Measures of the Amount of Ecologic Association Between Species. Ecology, 26(3), 297–302. 10.2307/1932409 [DOI] [Google Scholar]
- Ding S., & Van Hoesen G. W. (2010). Borders, extent, and topography of human perirhinal cortex as revealed using multiple modern neuroanatomical and pathological markers. Human Brain Mapping, 31(9), 1359–1379. 10.1002/hbm.20940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duara R., Loewenstein D. A., Potter E., Appel J., Greig M. T., Urs R., Shen Q., Raj A., Small B., Barker W., Schofield E., Wu Y., & Potter H. (2008). Medial temporal lobe atrophy on MRI scans and the diagnosis of Alzheimer disease. Neurology, 71(24), 1986–1992. 10.1212/01.wnl.0000336925.79704.9f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjell A. M., Walhovd K. B., Fennema-Notestine C., McEvoy L. K., Hagler D. J., Holland D., Brewer J. B., & Dale A. M. (2009). One-Year Brain Atrophy Evident in Healthy Aging. The Journal of Neuroscience, 29(48), 15223–15231. 10.1523/JNEUROSCI.3252-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forstmann B. U., Keuken M. C., Schafer A., Bazin P.-L., Alkemade A., & Turner R. (2014). Multi-modal ultra-high resolution structural 7-Tesla MRI data repository. Scientific Data, 1(1), 140050. 10.1038/sdata.2014.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande X., Wisse L., & Berron D. (2023). Ultra-high field imaging of the human medial temporal lobe. In Advances in Magnetic Resonance Technology and Applications (Vol. 10, pp. 259–272). Elsevier. 10.1016/B978-0-323-99898-7.00031-6 [DOI] [Google Scholar]
- Hata K., Nakamoto K., Nunomura A., Sone D., Maikusa N., Ogawa M., Sato N., & Matsuda H. (2019). Automated Volumetry of Medial Temporal Lobe Subregions in Mild Cognitive Impairment and Alzheimer Disease. Alzheimer Disease & Associated Disorders, 33(3), 206–211. 10.1097/WAD.0000000000000318 [DOI] [PubMed] [Google Scholar]
- Iglesias J. E., & Sabuncu M. R. (2015). Multi-atlas segmentation of biomedical images: A survey. Medical Image Analysis, 24(1), 205–219. 10.1016/j.media.2015.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isensee F., Jaeger P. F., Kohl S. A. A., Petersen J., & Maier-Hein K. H. (2021). nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nature Methods, 18(2), 203–211. 10.1038/s41592-020-01008-z [DOI] [PubMed] [Google Scholar]
- Išgum I., Benders M. J. N. L., Avants B., Cardoso M. J., Counsell S. J., Gomez E. F., Gui L., Hűppi P. S., Kersbergen K. J., Makropoulos A., Melbourne A., Moeskops P., Mol C. P., Kuklisova-Murgasova M., Rueckert D., Schnabel J. A., Srhoj-Egekher V., Wu J., Wang S., … Viergever M. A. (2015). Evaluation of automatic neonatal brain segmentation algorithms: The NeoBrainS12 challenge. Medical Image Analysis, 20(1), 135–151. 10.1016/j.media.2014.11.001 [DOI] [PubMed] [Google Scholar]
- Jack C. R., Petersen R. C., Xu Y., O’Brien P. C., Smith G. E., Ivnik R. J., Boeve B. F., Tangalos E. G., & Kokmen E. (2000). Rates of hippocampal atrophy correlate with change in clinical status in aging and AD. Neurology, 55(4), 484–490. 10.1212/WNL.55.4.484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S., Davis B., Jomier M., & Gerig G. (2004). Unbiased diffeomorphic atlas construction for computational anatomy. NeuroImage, 23, S151–S160. 10.1016/j.neuroimage.2004.07.068 [DOI] [PubMed] [Google Scholar]
- Kollia K., Maderwald S., Putzki N., Schlamann M., Theysohn J. M., Kraff O., Ladd M. E., Forsting M., & Wanke I. (2009). First Clinical Study on Ultra-High-Field MR Imaging in Patients with Multiple Sclerosis: Comparison of 1.5T and 7T. American Journal of Neuroradiology, 30(4), 699–702. 10.3174/ajnr.A1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf E. S. C., Wahlund L.-O., Visser P. J., & Scheltens P. (2004). Medial temporal lobe atrophy on MRI predicts dementia in patients with mild cognitive impairment. Neurology, 63(1), 94–100. 10.1212/01.WNL.0000133114.92694.93 [DOI] [PubMed] [Google Scholar]
- Kurth F., Cherbuin N., & Luders E. (2017). The impact of aging on subregions of the hippocampal complex in healthy adults. NeuroImage, 163, 296–300. 10.1016/j.neuroimage.2017.09.016 [DOI] [PubMed] [Google Scholar]
- Lin M. P., & Liebeskind D. S. (2016). Imaging of Ischemic Stroke. CONTINUUM: Lifelong Learning in Neurology, 22(5), 1399–1423. 10.1212/CON.0000000000000376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet D., & Rajah M. N. (2013). Association between prefrontal activity and volume change in prefrontal and medial temporal lobes in aging and dementia: A review. Ageing Research Reviews, 12(2), 479–489. 10.1016/j.arr.2012.11.001 [DOI] [PubMed] [Google Scholar]
- Menze B. H., Jakab A., Bauer S., Kalpathy-Cramer J., Farahani K., Kirby J., Burren Y., Porz N., Slotboom J., Wiest R., Lanczi L., Gerstner E., Weber M.-A., Arbel T., Avants B. B., Ayache N., Buendia P., Collins D. L., Cordier N., … Van Leemput K. (2015). The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Transactions on Medical Imaging, 34(10), 1993–2024. 10.1109/TMI.2014.2377694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson P. T., Alafuzoff I., Bigio E. H., Bouras C., Braak H., Cairns N. J., Castellani R. J., Crain B. J., Davies P., Tredici K. D., Duyckaerts C., Frosch M. P., Haroutunian V., Hof P. R., Hulette C. M., Hyman B. T., Iwatsubo T., Jellinger K. A., Jicha G. A., … Beach T. G. (2012). Correlation of Alzheimer Disease Neuropathologic Changes With Cognitive Status: A Review of the Literature. Journal of Neuropathology & Experimental Neurology, 71(5), 362–381. 10.1097/NEN.0b013e31825018f7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M., Sone D., Beheshti I., Maikusa N., Okita K., Takano H., & Matsuda H. (2019). Association between subfield volumes of the medial temporal lobe and cognitive assessments. Heliyon, 5(6), e01828. 10.1016/j.heliyon.2019.e01828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogniewicz R. L., & Kübler O. (1995). Hierarchic Voronoi Skeletons. Pattern Recognition, 28(3), 343–359. 10.1016/0031-3203(94)00105-U [DOI] [Google Scholar]
- Petersen R. C., Aisen P. S., Beckett L. A., Donohue M. C., Gamst A. C., Harvey D. J., Jack C. R., Jagust W. J., Shaw L. M., Toga A. W., Trojanowski J. Q., & Weiner M. W. (2010). Alzheimer’s Disease Neuroimaging Initiative (ADNI): Clinical characterization. Neurology, 74(3), 201–209. 10.1212/WNL.0b013e3181cb3e25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouch A. M., Tian S., Takebe M., Yuan J., Gorman R., Cheung A. T., Wang H., Jackson B. M., Gorman J. H., Gorman R. C., & Yushkevich P. A. (2015). Medially constrained deformable modeling for segmentation of branching medial structures: Application to aortic valve segmentation and morphometry. Medical Image Analysis, 26(1), 217–231. 10.1016/j.media.2015.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudent V., Kumar A., Liu S., Wiggins G., Malaspina D., & Gonen O. (2010). Human Hippocampal Subfields in Young Adults at 7.0 T: Feasibility of Imaging. Radiology, 254(3), 900–906. 10.1148/radiol.09090897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz N., Rodrigue K. M., Head D., Kennedy K. M., & Acker J. D. (2004). Differential aging of the medial temporal lobe: A study of a five-year change. Neurology, 62(3), 433–438. 10.1212/01.WNL.0000106466.09835.46 [DOI] [PubMed] [Google Scholar]
- Sperling R. A., Donohue M. C., Raman R., Sun C.-K., Yaari R., Holdridge K., Siemers E., Johnson K. A., Aisen P. S., & for the A4 Study Team. (2020). Association of Factors With Elevated Amyloid Burden in Clinically Normal Older Individuals. JAMA Neurology, 77(6), 735. 10.1001/jamaneurol.2020.0387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squire L. R., Stark C. E. L., & Clark R. E. (2004). THE MEDIAL TEMPORAL LOBE. Annual Review of Neuroscience, 27(1), 279–306. 10.1146/annurev.neuro.27.070203.144130 [DOI] [PubMed] [Google Scholar]
- Venet L., Pati S., Feldman M. D., Nasrallah M. P., Yushkevich P., & Bakas S. (2021). Accurate and Robust Alignment of Differently Stained Histologic Images Based on Greedy Diffeomorphic Registration. Applied Sciences, 11(4), 1892. 10.3390/app11041892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser P. J., Verhey F. R. J., Hofman P. A. M., Scheltens P., & Jolles J. (2002). Medial temporal lobe atrophy predicts Alzheimer’s disease in patients with minor cognitive impairment. Journal of Neurology, Neurosurgery and Psychiatry, 72(4), 491–497. 10.1136/jnnp.72.4.491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitwell J. L., Petersen R. C., Negash S., Weigand S. D., Kantarci K., Ivnik R. J., Knopman D. S., Boeve B. F., Smith G. E., & Jack C. R. (2007). Patterns of Atrophy Differ Among Specific Subtypes of Mild Cognitive Impairment. Archives of Neurology, 64(8), 1130. 10.1001/archneur.64.8.1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterburn J. L., Pruessner J. C., Chavez S., Schira M. M., Lobaugh N. J., Voineskos A. N., & Chakravarty M. M. (2013). A novel in vivo atlas of human hippocampal subfields using high-resolution 3T magnetic resonance imaging. NeuroImage, 74, 254–265. 10.1016/j.neuroimage.2013.02.003 [DOI] [PubMed] [Google Scholar]
- Wisse L. E. M., Biessels G. J., Heringa S. M., Kuijf H. J., Koek D. (H.) L., Luijten P. R., & Geerlings M. I. (2014). Hippocampal subfield volumes at 7T in early Alzheimer’s disease and normal aging. Neurobiology of Aging, 35(9), 2039–2045. 10.1016/j.neurobiolaging.2014.02.021 [DOI] [PubMed] [Google Scholar]
- Wisse L. E. M., Chételat G., Daugherty A. M., De Flores R., La Joie R., Mueller S. G., Stark C. E. L., Wang L., Yushkevich P. A., Berron D., Raz N., Bakker A., Olsen R. K., & Carr V. A. (2021). Hippocampal subfield volumetry from structural isotropic 1 mm 3 MRI scans: A note of caution. Human Brain Mapping, 42(2), 539–550. 10.1002/hbm.25234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisse L. E. M., Kuijf H. J., Honingh A. M., Wang H., Pluta J. B., Das S. R., Wolk D. A., Zwanenburg J. J. M., Yushkevich P. A., & Geerlings M. I. (2016). Automated Hippocampal Subfield Segmentation at 7T MRI. American Journal of Neuroradiology, 37(6), 1050–1057. 10.3174/ajnr.A4659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf H., Hensel A., Kruggel F., Riedel-Heller S. G., Arendt T., Wahlund L.-O., & Gertz H.-J. (2004). Structural correlates of mild cognitive impairment. Neurobiology of Aging, 25(7), 913–924. 10.1016/j.neurobiolaging.2003.08.006 [DOI] [PubMed] [Google Scholar]
- Wolk D. A., Das S. R., Mueller S. G., Weiner M. W., & Yushkevich P. A. (2017). Medial temporal lobe subregional morphometry using high resolution MRI in Alzheimer’s disease. Neurobiology of Aging, 49, 204–213. 10.1016/j.neurobiolaging.2016.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L., Wisse L. E. M., Das S. R., Vergnet N., Dong M., Ittyerah R., De Flores R., Yushkevich P. A., Wolk D. A., & for the Alzheimer’s Disease Neuroimaging Initiative. (2020). Longitudinal atrophy in early Braak regions in preclinical Alzheimer’s disease. Human Brain Mapping, 41(16), 4704–4717. 10.1002/hbm.25151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L., Wisse L. E. M., Das S. R., Wang H., Wolk D. A., Manjón J. V., & Yushkevich P. A. (2016). Accounting for the Confound of Meninges in Segmenting Entorhinal and Perirhinal Cortices in T1-Weighted MRI. In Ourselin S., Joskowicz L., Sabuncu M. R., Unal G., & Wells W. (Eds.), Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016 (Vol. 9901, pp. 564–571). Springer International Publishing. 10.1007/978-3-319-46723-8_65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L., Wisse L. E. M., Pluta J., De Flores R., Piskin V., Manjón J. V., Wang H., Das S. R., Ding S., Wolk D. A., Yushkevich P. A., & for the Alzheimer’s Disease Neuroimaging Initiative. (2019). Automated segmentation of medial temporal lobe subregions on in vivo T1-weighted MRI in early stages of Alzheimer’s disease. Human Brain Mapping, 40(12), 3431–3451. 10.1002/hbm.24607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L., Wisse L. E. M., Wang J., Ravikumar S., Khandelwal P., Glenn T., Luther A., Lim S., Wolk D. A., & Yushkevich P. A. (2023). Deep label fusion: A generalizable hybrid multi-atlas and deep convolutional neural network for medical image segmentation. Medical Image Analysis, 83, 102683. 10.1016/j.media.2022.102683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich P. A., Piven J., Hazlett H. C., Smith R. G., Ho S., Gee J. C., & Gerig G. (2006). User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. NeuroImage, 31(3), 1116–1128. 10.1016/j.neuroimage.2006.01.015 [DOI] [PubMed] [Google Scholar]
- Yushkevich P. A., Pluta J. B., Wang H., Xie L., Ding S.-L., Gertje E. C., Mancuso L., Kliot D., Das S. R., & Wolk D. A. (2015). Automated volumetry and regional thickness analysis of hippocampal subfields and medial temporal cortical structures in mild cognitive impairment: Automatic Morphometry of MTL Subfields in MCI. Human Brain Mapping, 36(1), 258–287. 10.1002/hbm.22627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwanenburg J. J. M., Versluis M. J., Luijten P. R., & Petridou N. (2011). Fast high resolution whole brain T2* weighted imaging using echo planar imaging at 7T. NeuroImage, 56(4), 1902–1907. 10.1016/j.neuroimage.2011.03.046 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.