Abstract
A full-length heme oxygenase gene from the gram-negative pathogen Neisseria meningitidis was cloned and expressed in Escherichia coli. Expression of the enzyme yielded soluble catalytically active protein and caused accumulation of biliverdin within the E. coli cells. The purified HemO forms a 1:1 complex with heme and has a heme protein spectrum similar to that previously reported for the purified heme oxygenase (HmuO) from the gram-positive pathogen Corynebacterium diphtheriae and for eukaryotic heme oxygenases. The overall sequence identity between HemO and these heme oxygenases is, however, low. In the presence of ascorbate or the human NADPH cytochrome P450 reductase system, the heme-HemO complex is converted to ferric-biliverdin IXα and carbon monoxide as the final products. Homologs of the hemO gene were identified and characterized in six commensal Neisseria isolates, Neisseria lactamica, Neisseria subflava, Neisseria flava, Neisseria polysacchareae, Neisseria kochii, and Neisseria cinerea. All HemO orthologs shared between 95 and 98% identity in amino acid sequences with functionally important residues being completely conserved. This is the first heme oxygenase identified in a gram-negative pathogen. The identification of HemO as a heme oxygenase provides further evidence that oxidative cleavage of the heme is the mechanism by which some bacteria acquire iron for further use.
Survival of microorganisms in any biological niche depends on a constant supply of iron, which participates in numerous metabolic pathways either alone or in the form of heme (1, 18). Besides being able to assimilate iron via siderophore-dependent and -independent pathways, bacteria can also use hemoglobin (Hb), heme, and other heme-containing proteins as sources of iron (23, 32). Active transport of the whole heme molecule into the cytoplasm is the common theme of all heme-iron assimilation strategies in bacteria studied so far (6, 26, 27). In gram-negative bacteria, heme is extracted from heme proteins at the bacterial surface and transported into the periplasm via specific heme receptors. Once in the periplasm, heme is subjected to another active transport process that requires ATP and the presence of a heme-specific ATP-binding cassette transporter (27). These types of systems, initially described in Yersinia enterocolitica, Vibrio cholerae, and Haemophilus influenzae, were subsequently found in many other gram-negative bacteria including Pseudomonas aeruginosa, Neisseria meningitidis, Serratia marcescens, and other enterobacteria (4, 11–13, 17, 26–28, 38).
In contrast to the breadth of knowledge about heme transport processes in microorganisms, relatively little is known about the fate of heme in the cellular interior. It is assumed that heme must be degraded to provide cells with iron since more than 90% of total cell iron is not associated with heme. The process of heme degradation has been well studied in eukaryotes, in which a family of enzymes—heme oxygenases—catalyze oxidative cleavage of heme to biliverdin (16, 31). Detection of carbon monoxide, a product of the heme oxygenase reaction, in gram-positive organisms such as Bacillus cereus and Streptomyces mitis, suggested the existence of heme oxygenase activity (8). A heme oxygenase (HmuO), essential for utilization of heme and Hb-iron, was recently identified and characterized from the gram-positive pathogen Corynebacterium diphtheriae (22, 34). A high degree of identity (33%) with the eukaryotic heme oxygenases suggested a role for HmuO in the oxidative cleavage of heme and subsequent release of iron (22). Recently, another heme oxygenase-like gene product (HemO) was identified in the gram-negative organisms N. meningitidis and Neisseria gonorrhoeae. Although it shares limited homology to known heme oxygenases, phenotypic studies implicated the hemO gene product in assimilation of heme- and Hb-iron and protection against heme toxicity (43).
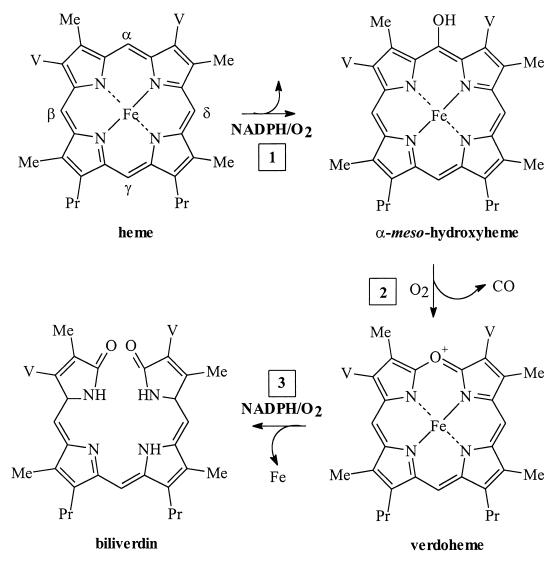
Heme oxygenases are enzymatically unique in that they use heme both as a substrate and cofactor by binding oxygen for intramolecular degradation of the porphyrin macrocycle. The intermediates in the oxidative cleavage of heme as determined from studies of the human heme oxygenases and the HmuO enzyme are shown in Fig. 1 (34, 39–42). The initial step in heme degradation is its reduction to the ferrous form (Fe2+) by an NADPH-dependent reductase. Oxygen is then bound to give the ferrous-dioxygen complex (Fe2+-O2), which is reduced further by a two-electron reduction of oxygen to yield a species formally equivalent to H2O2. The terminal oxygen of the protonated peroxide intermediate [Fe3+-O-OH] reacts specifically with the α-meso carbon of the heme to give the first intermediate in heme degradation, α-meso-hydroxyheme (Fig. 1 [step 1]). The α-meso-hydroxyheme in the presence of oxygen is converted directly to verdoheme with the release of the α-meso carbon as CO (Fig. 1 [step 2]). An additional two-electron reduction of the ferric (Fe3+) verdoheme-HO complex and subsequent binding of oxygen yields ferric (Fe3+)-biliverdin (Fig. 1 [step 3]). In mammalian systems, reduction of ferric (Fe3+)-biliverdin to ferrous (Fe2+)-biliverdin releases the iron and biliverdin as the final products of the reaction (15). The conversion of heme to biliverdin, iron, and CO by heme oxygenase requires five electrons provided by the reductase system and three molecules of oxygen (15).
FIG. 1.
Chemical steps in heme degradation as defined by the studies of eukaryotic heme oxygenases and C. diphtheriae HmuO. Degradation of heme by heme oxygenase yields iron, biliverdin, and CO. Me, methyl side chain; V, vinyl side chain; Pr, propionic side chain.
In this report, we describe purification of the hemO gene product and characterization of its enzymatic activity. These studies unequivocally identify HemO as a heme oxygenase and represent the first characterization of a heme oxygenase activity in a gram-negative pathogenic bacterium.
MATERIALS AND METHODS
General methods.
Plasmid purification, subcloning, and bacterial transformations were carried out as previously described (21). Deionized, double distilled water was used for all experiments. Oligonucleotides were purchased from GIBCO-BRL. All absorption spectra of the heme-HemO complex were recorded on a Cary Varian Bio 100 UV spectrophotometer.
Bacterial strains.
E. coli strain DH5α-9CR [F− mcrA (mrr-hsdRMS-mcrBC) φ80dlacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 deoR thi-1 phoA supE44 8− gyrA96 relA1] was used for DNA manipulation and E. coli strain BL21(DE3)/pLysS [F− ompT hsdSR(rB− mB−) gal dcm(DE3)] was used for expression of the heme oxygenase. Commensal and pathogenic Neisseria strains used in this study have been described previously (19). HemO genes from commensal Neisseria isolates were cloned by PCR amplification using procedures and primers previously described (44). DNA sequence analysis was done using dye terminator cycle sequencing on an ABI model 377 automated sequencer at the Emory Core Sequencing Facility.
Construction of HemO expression plasmid pWMZ1651.
The pWMZ1651 construct which encodes the wild-type HemO was constructed as described below. PCRs were performed with Pfx DNA polymerase (Promega) according to the instructions of the manufacturer. The hemO gene was amplified by PCR from the plasmid pDJH1550 (44) using oligonucleotides OLIGO-1 (5′-CGCGCATATGAGTGAAACCGAAAATCAAGCC-3′) and HEMO-XHO (5′-CGCTCGAGTTTTTAGTGCCTGTGCGGCATCATTCC-3′), encoding the stop codon (TAA) followed by an XhoI site at the 3′ end of the hemO gene. The 0.65-kb PCR product was cloned into the pCR2.1-TOPO vector (Invitrogen) to generate pWMZ1643. Following sequence verification, the gene was subcloned into the pET21a expression vector utilizing the NdeI and XhoI restriction sites to create pWMZ1651.
Expression and purification of HemO.
The E. coli BL21(DE3) strain carrying pWMZ1651 was grown in Luria-Bertani medium containing 100 mg of ampicillin per liter, overnight at 37°C. The cells were subsequently subcultured into fresh LB-ampicillin medium (100 ml) and grown at 37°C to mid-log phase. The cells were then subcultured (10 ml) into Luria Bertani LB-ampicillin medium (1 liter) and, on reaching mid-log phase, their expression was induced by addition of isopropyl-1-thiol-(d)-galactopyranoside to a final concentration of 1 mM. Cell growth was continued for 4 to 5 h at 30°C, and cells were harvested by centrifugation (10,000 × g for 20 min). Cells were lysed by sonication in 50 mM Tris-HCl (pH 7.8) containing 1 mM EDTA and 1 mM phenylmethysulfony fluoride. The cell suspension was then centrifuged at 27,000 × g for 40 min.
The soluble fraction was applied to a Sepharose-Q Fast Flow column (1.5 by 10 cm) previously equilibrated with 20 mM Tris-HCl (pH 7.5). The column was washed with 3 volumes of 20 mM Tris-HCl (pH 7.5) containing 50 mM NaCl. The protein was then eluted with the same buffer with a linear gradient of NaCl from 50 to 500 mM. The protein eluted at a concentration of 150 mM NaCl, and the peak fractions were pooled and dialyzed against 10 mM potassium phosphate (pH 7.4) (2 × 4 liters) at 4°C. The HemO protein was then stored at −80°C or reconstituted with heme as described below.
Reconstitution of HemO with heme.
The HemO-heme complex was prepared as described previously (34). Hemin was added to the purified HemO to a final 2:1 heme-protein ratio. The sample was then applied to a Bio-Gel HTP column (1.5 by 6 cm) preequilibrated with 10 mM potassium phosphate (pH 7.4). The column was then washed with the same buffer (5 volumes), and the protein was eluted in 150 mM potassium phosphate. The protein was concentrated by an Amicon filtration unit and stored at −80°C. For preparations of the heme-His-HemO complex. His-HemO had hemin added as described above, and the unbound heme was removed by applying the complex to a Sepharose-Q Fast Flow column pre-equilibrated with 20 mM Tris-HCl (pH 7.8). The column was washed with 2 to 3 volumes of 20 mM Tris-HCl (pH 7.8), and the heme-His-HemO complex was eluted in the same buffer with a linear gradient of 50 to 500 mM NaCl. The protein fractions were pooled and dialyzed (2 × 4 liters) against 20 mM Tris-HCl (pH 7.8) at 4°C.
Determination of the extinction coefficient for the HemO-heme complex.
The millimolar extinction coefficient at 405 nm (ɛ405) for the HemO-heme complex was determined as previously described (10). The absorbance of a purified heme-HemO sample at 405 nm was measured. The solution was diluted with alkaline pyridine, and the spectrum of the oxidized pyridine hemochrome was recorded. An excess of dithionite was added, and the spectrum of the reduced ferrous pyridine hemochrome was then recorded. The concentration was calculated from the ΔAox-red at 557 nm using an ɛ405 value of 34.53.
Reaction of heme-HemO with NADPH cytochrome P450 reductase.
The reaction of heme-HemO in presence of NADPH reductase was similar to that described previously (34). Purified human cytochrome P450 reductase was added to the heme-HemO complex (10 μM) at a ratio of reductase/HemO equal to 3:1 in a final volume of 1 ml of 20 mM Tris-HCl (pH 7.5). The reaction was initiated by the addition of NADPH in 10-μM increments to a final concentration of 100 μM. The spectral changes between 300 and 750 nm were monitored over a 30-min time period. Following completion of the reaction, the product was extracted for analysis by high-pressure liquid chromatography (HPLC) as described below.
The ascorbic-acid-dependent conversion of heme to biliverdin was also monitored. Ascorbic acid at a final concentration of 5 mM was added directly to the heme-HemO complex (10 μM) in 20 mM Tris-HCl buffer (pH 7.5). The spectral changes between 300 and 750 nm were recorded. The products of the reaction were extracted and subjected to HPLC analysis as described below.
Reaction of the heme-HemO complex with H2O2.
Five equivalents of 10 mM H2O2 (5 μl) in Tris-HCl (pH 7.5) were added to the heme-HemO complex (10 μM) in the same buffer. The reaction was monitored spectroscopically. Following maximum decrease in the Soret band and maximum increase in the 680-nm-absorption, 20% (final concentration) pyridine was added to the reaction mixture. The products were extracted into chloroform, and the solvent was removed under a stream of nitrogen. The verdoheme product formed in the H2O2-dependent reaction was hydrolytically converted to biliverdin by the method of Saito and Itano (20). The biliverdin was extracted into chloroform and reduced to dryness. The residue was resuspended in 5% sulfuric acid in methanol, esterified, and analyzed by HPLC as described below.
Detection of carbon monoxide as a reaction product.
Detection of CO as a product of the reaction was carried out as previously described (34). Purified heme-HemO, purified NADPH cytochrome P450 reductase (1.5 μM), and NADPH (100 μM) in a final volume of 1 ml were placed in both the reference and reaction cuvettes and zeroed immediately. Myoglobin (50 μl, 125 μM) was added to the reaction cuvette, and the same volume of buffer was added to the reference cuvette. The spectrum was recorded at 2-min intervals between 350 and 650 nm, and the transition from 410 to 420 nm was monitored. The transition from the ferrous-dioxygen myoglobin complex, which has a characteristic Soret band at 408 nm, to the ferrous-CO myoglobin complex with a Soret band at 420 nm is indicative of CO production as a consequence of oxidative cleavage of the heme. Control reactions in the absence of HemO did not induce a transition in the Soret band from 408 to 420 nm.
HPLC analysis of heme-HemO reaction products.
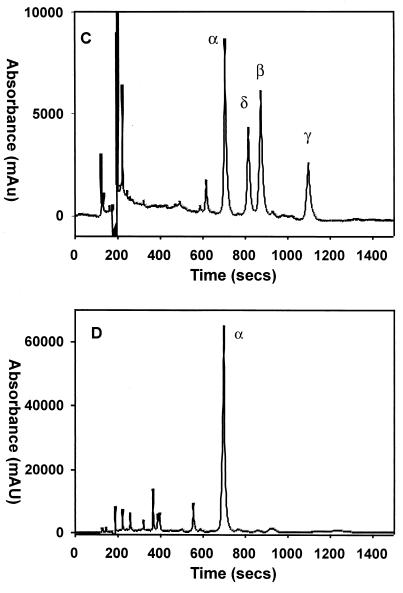
Following the reaction of the heme-HemO complex with NADPH reductase or ascorbate, glacial acetic acid (200 μl) and 3 M HCl (200 μl) were added to the reaction mixture (1 ml) before extracting into chloroform. The organic layer was washed with distilled water (3 × 1 ml), and the chloroform layer was removed under a stream of argon. The resultant residue was dissolved in 1 ml of 4% sulfuric acid in methanol and esterified for 12 h at room temperature. The esters were diluted (fourfold) with distilled water and extracted into chloroform. The organic layer was washed further with distilled water and dried over sulfate. The chloroform was again removed under a stream of argon. The residue was dissolved in HPLC solvent prior to HPLC analysis. The samples were analyzed on reverse-phase HPLC on an ODS-AQ C18 (S-5:) (YMC, Inc., Wilmington, N.C.) column (3.0 by 250 mm) eluted with 85:15 (vol/vol) methanol/water at a flow rate of 0.4 ml/min. The elutant was monitored at 380 nm, and the biliverdin standards were eluted in the order α (11.9 min), β (13.9 min), γ (14.8 min), and δ (18.5 min) (7).
RESULTS
Identification and cloning of hemO genes from commensal neisseriae.
In a previous study, we identified the product of the hemO gene as essential for heme utilization as an iron source by pathogenic neisseriae (43). Nucleotide sequences homologous to the hemO gene were identified in different pathogenic isolates of N. meningitidis and N. gonorrhoeae as well as in some commensal neisseriae. To confirm these findings, hemO homologues from six members of commensal nisseriae were PCR amplified, and their nucleotide sequences were determined. All six amplified open reading frames were highly homologous to the N. meningitidis hemO, sharing between 95 and 98% identical amino acid residues. Comparison of HemO homologues and the human heme oxygenase 1 (HO-1) is shown in Fig. 2. The proximal ligand of the HO-1, His-25, is conserved in all neisserial HemO proteins (His-23), suggesting involvement of this histidine in heme binding (24). The HO-1 residues Thr-21, Glu-29, and Phe-207 are positioned in close proximity to the heme molecule (24). The same residues are highly conserved in neisserial HemO proteins with only one conservative change of aspartic acid for glutamic acid at position 29 (position 26 in HemO) (Fig. 2). Furthermore, HO-1 residues Gly-139, Gly-143, and Leu-147, which provide the flexibility of the distal helix in the opening and closing of the active site, are conserved in the HemO sequences. Finally, residues Met-34 and Phe-37, which interact with the α-meso edge of heme in HO-1, are found conserved in the HemO sequences. Although the overall homology between HemO and HO-1 does not exceed 20%, the high degree of conservation of functionally or structurally important residues suggests that HemO catalyzes oxidative degradation of heme (Fig. 2). Based on this limited but significant homology between neisserial HemO proteins and human HO-1, it was expected that the HemO-dependent heme degradation proceeds via the same steps and intermediates as previously described for HO-1 (Fig. 1). In order to probe the enzymatic activity, the HemO protein was purified in its native form, and its ability to bind heme and catalyze oxidative degradation of heme in vitro was explored.
FIG. 2.
Amino acid sequence alignment of neisserial HemO proteins and HO-1. Comparison was done with the ClustalW 1.8 program on the Baylor College of Medicine Search Launcher. H1, human heme oxygenase 1; NB, N. meningitidis MC58; NA, N. meningitidis A 2855, AF133695; NP, N. polysacchareae AF216858; NF, N. flava; NS, N. subflava AF216745; NK, N. kochii AF216856; NL, N. lactamica AF216855; NG, N. gonorrhoeae F1090; and NC, N. cinerea AF216744. “.” indicates similar, while “∗” indicates identical amino acid residues between human heme oxygenase and at least one neisserial protein.
Expression and purification of HemO.
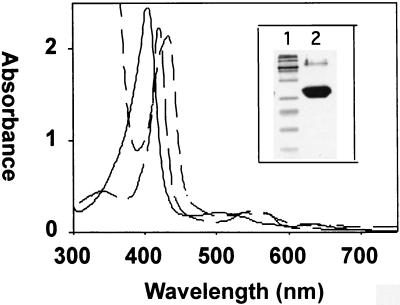
The wild-type HemO was expressed as a soluble, catalytically active protein. As previously reported for the HmuO from C. diphtheriae, expression of HemO in Escherichia coli BL21(DE3) turned the cells green owing to the accumulation of biliverdin (35). Purification of HemO by ion exchange and gel filtration yielded a protein that formed a single band at 26 kDa on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 3, inset, lane 2). The yield of purified protein from a liter of cells ranged from 20 to 30 mg.
FIG. 3.
Absorption spectra of the heme-HemO complexes and SDS-PAGE of the purified HemO proteins (inset). The spectra are ferric (—), ferrous deoxy (—..—), and ferrous CO-bound (— —) heme-HemO. Inset: SDS-PAGE of the purified HemO. Lane 1, markers; lane 2, purified native HemO.
Properties of the heme-HemO complex.
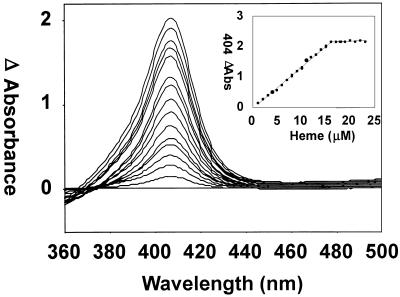
Reconstitution of the protein with the substrate heme provided information about the nature of the proximal ligand of the heme. The maximum absorbance (Soret band) of the heme-HemO complex following removal of the excess heme was at 406 nm (Fig. 3). Reduction of the heme-HemO complex with dithionite under an atmosphere of CO gave a spectrum typical of a reduced ferrous-CO complex with a Soret band at 421 nm and α and β bands at 568 and 538 nm, respectively (Fig. 3). The ferrous-deoxy heme-HemO was generated by the addition of dithionite under an atmosphere of argon. The Soret peak dropped in intensity and shifted to 434 nm with the appearance of a band in the visible region at 550 nm (Fig. 3). The spectral properties of the ferrous-deoxy heme-HemO complex are comparable to those previously reported for the eukaryotic heme oxygenases and the bacterial HmuO and suggest that the proximal ligand is a histidine (33, 34, 36). Unlike HmuO and the human HO-1, passage of the ferrous-CO heme-HemO through Sephadex G-25 to remove the excess reductant did not yield the ferrous-O2 heme-HemO complex. Instead, the heme-HemO complex spontaneously oxidized back to the resting-state ferric heme-HemO complex (data not shown). The millimolar extinction coefficient at 406 nm (ɛ406) was calculated from the pyridine hemochrome method to be 179 mM−1 cm−1 (10). The ratio of heme bound to HemO was calculated by measuring the difference spectrum at various concentrations of free heme versus the heme-HemO complex by UV-visible spectroscopy (Fig. 4). The HemO protein was saturated at a ratio of 1:1 heme to protein.
FIG. 4.
Absorption difference spectra of heme binding to HemO. Increasing amounts of heme (0.5 to 25 μM) were added to both the sample (15 μM) and reference cuvettes. The inset shows the saturation at a 1:1 ratio of heme to protein (15 μM).
Catalytic activity of the heme-HemO complex.
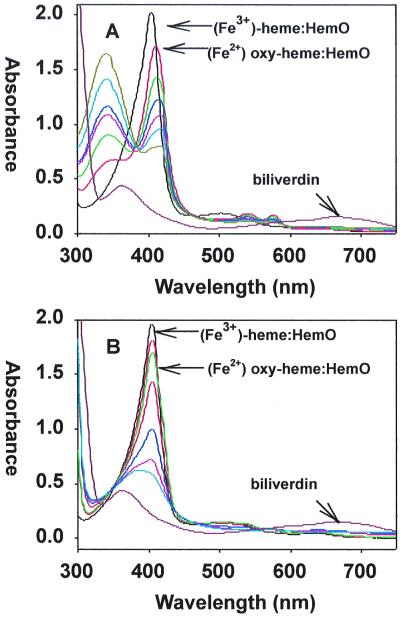
In order to demonstrate HemO catalytic activity, the heme-HemO complex was reacted in the presence of either cytochrome P450 reductase plus NADPH or ascorbate, and the reaction was monitored by UV-visible spectroscopy. The products were determined by HPLC analysis following their extraction and methylation. The ferric heme-HemO complex was quantitatively converted to ferric biliverdin in the presence of the human cytochrome P450 reductase system (Fig. 5A). Reaction of the heme-HemO complex with NADPH initiated the formation of a ferrous-dioxygen complex, indicated by the drop and shift of the Soret band from 406 to 410 nm and the appearance of α and β bands at 570 and 540 nm. Over a period of 30 min, the reaction proceeded with a decrease in the Soret and α and β bands. The subsequent decrease in intensity of these bands occurred as the ferrous-dioxygen complex was converted to the ferric biliverdin-HemO complex and the Soret band eventually shifted back toward 400 nm with loss of any absorbance in the α/β region of the spectrum (Fig. 5A). Acidification of the product yielded a spectrum with broad maxima at 380 nm and 680 nm, indicative of the iron-free biliverdin (Fig. 5A).
FIG. 5.
Conversion of the heme-HemO to biliverdin in the presence of NADPH cytochrome P450 reductase or ascorbate. (A) Ferric heme-HemO complex (black). After the addition of NADPH in 10-μM increments, spectra were taken at 10-min intervals (curves of different colors); the final biliverdin product after acidification (brown). (B) The ferric heme-HemO complex (black). After the addition of ascorbate (5 mM), spectra were taken at 10-min intervals (curves of different colors); the final biliverdin product after acidification (brown). (C) HPLC chromatogram of a mixture of all four biliverdin dimethyl esters as standards. (D) HPLC chromatogram of the product of the heme-HemO reaction with ascorbate following extraction and methylation. The UV-visible spectra and HPLC product analysis lead to the conclusion that oxidative cleavage of heme by HemO is regiospecific, with only the biliverdin IXα isomer being formed in both the NADPH- and ascorbate-dependent reactions.
The reaction in the presence of ascorbate also yielded ferric biliverdin as the final product (Fig. 5B). Acidification and extraction of the product followed by HPLC analysis gave a single peak with a retention time and spectrum identical to those of biliverdin IXα for both NADPH and ascorbate-dependent reactions (Fig. 5D). The HPLC chromatogram of all four possible biliverdin isomers is shown for comparison (Fig. 5C). Coinjection of the product of both reactions with known standards verified biliverdin IXα as the final product (data not shown).
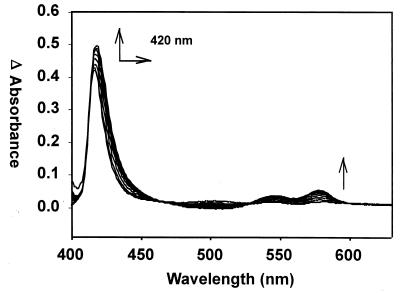
Difference absorption spectroscopy in the presence of myoglobin confirmed carbon monoxide as a product of oxidative cleavage of heme by HemO. The myoglobin absorption spectrum was recorded at 2-min intervals in order to monitor for characteristic spectral changes of a myoglobin-CO complex (Fig. 6). The shift in the Soret band from 408 to 420 nm as well as the appearance of the α and β bands at 568 and 538 nm occurred on the transition of the ferrous-dioxygen myoglobin to the ferrous-CO myoglobin complex. The increased affinity of myoglobin for CO over oxygen (200-fold) allows for the detection of any CO produced as a consequence of oxidative cleavage of the heme. Control reactions in the absence of the heme-HemO complex showed no shift in Soret band throughout the experiment (data not shown). The complete conversion of ferrous dioxygen myoglobin to the ferrous carbon monoxide complex indicated that carbon monoxide was generated as a product of oxidative heme cleavage (Step 2 in Fig. 1).
FIG. 6.
Difference absorption spectra of the heme-HemO and NADPH cytochrome P450 reductase reaction in the presence of myoglobin. The reference and sample cuvette contained the heme-HemO complex, NADPH cytochrome P450 reductase, and NADPH. The reaction was zeroed immediately after the addition of NADPH, after which myoglobin was added to the sample cuvette. The shift in the Soret band from 408 to 420 nm and the appearance of α and β bands at 568 and 538 nm are indicative of the ferrous-CO myoglobin complex formation, confirming the release of CO on conversion of heme to iron-biliverdin (arrows).
Reaction of the heme-HemO complex with H2O2.
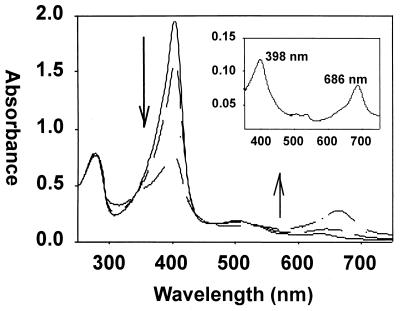
The first step in the reaction of heme to mesohydroxyheme (see Fig. 1) involved reduction of the ferric heme-HemO complex to the ferrous dioxygen heme-HemO, which is formally equivalent to the ferric peroxy heme-HemO intermediate. The ability of H2O2 to substitute for activated oxygen in forming the ferric peroxy heme-HemO intermediate was examined. The heme-HemO complex was reacted with 5 equivalents of H2O2. A decrease in the Soret band over a 10-min period was accompanied by an increase in the absorbance at 660 nm (Fig. 7). The product of the reaction was extracted into chloroform after the addition of pyridine to a final concentration of 20%. The resulting spectrum was typical of a ferric verdoheme product with a Soret peak at 398 nm and a strong visible band at 686 nm (Fig. 7 [inset]) (36, 39–42). Hydrolytic conversion of the product to biliverdin and subsequent HPLC analysis verified that the initial H2O2-dependent hydroxylation occurred solely at the α-meso carbon (data not shown).
FIG. 7.
Conversion of the ferric heme-HemO complex to verdoheme in the presence of H2O2. Ferric heme-HemO (—), 10, 20, and 30 min after the addition of H2O2 (— —). Inset: addition of 20% pyridine yields the bis-pyridine complex of ferric verdoheme with the characteristic absorption band at 686 nm.
DISCUSSION
HemO is a heme oxygenase.
The critical step in heme utilization is the release of iron from the heme. Although the heme receptor and transport proteins responsible for the internalization of heme have been identified, the mechanism of iron release has only recently been elucidated (32). The recent identification and characterization of a soluble heme oxygenase (HmuO) from the gram-positive C. diphtheriae provided the first evidence that iron release involved the oxidative cleavage of the heme to biliverdin, with the release of carbon monoxide and iron (22, 34). Subsequent identification in the gram-negative pathogen N. meningitidis of the hemO gene, which is required for the utilization of heme as an iron source, suggested that the product of the hemO gene may be a heme oxygenase (43). Besides being essential in iron assimilation, the product of the hemO gene also protected neisseriae against heme toxicity. In this report, we have described the expression and purification of HemO and have shown that HemO oxidatively cleaves heme in vitro to yield ferric iron-biliverdin and CO as final products. The heme oxygenase-dependent cleavage of the porphyrin macrocycle is the critical step in the ability to further utilize heme iron. The identification of HemO as a heme oxygenase provides the first conclusive evidence that oxidative cleavage of the porphyrin is required for heme utilization in neisseriae.
The UV-visible spectra of the heme-HemO complexes are similar to those previously reported for HO-1 and HmuO, in which the proximal ligand to the heme is a histidine with a water molecule bound in the six-ligand position of heme (2, 3, 33–36). In the presence of the human NADPH cytochrome P450 reductase system or of ascorbate as reductant, HemO-bound heme was quantitatively converted to ferric (Fe3+) biliverdin. It is interesting that both reactions yielded ferric biliverdin as the final product, which inhibited any subsequent enzyme turnover because the product remained bound to the protein. The reaction catalyzed by the C. diphtheriae HmuO has been reported to yield biliverdin as the product in both the ascorbate-driven and NADPH-dependent reactions (2, 34). However, in the ascorbate-driven reaction, the free-biliverdin spectrum as indicated by the broad absorbance band at 680 nm was observed only after a 2-h period (34). It is clear that in vitro iron is not displaced from the biliverdin complex easily. Indeed, the human heme oxygenase in the presence of ascorbate yields ferric biliverdin, which yields the iron-free biliverdin complex only on the addition of iron chelators (15). In contrast, the NADPH cytochrome P450 reductase-dependent reaction reduces the ferric biliverdin to the ferrous complex with the subsequent release of iron. The inability of the human cytochrome P450 reductase to reduce the ferric-biliverdin HemO complex further may be due to inefficient coupling of the enzymes. The sequence identity of HemO orthologs to that of the eukaryotic heme oxygenases is relatively low and may partly explain the inability to reduce the ferric-biliverdin HemO complex to the ferrous form with the release of the ferrous iron (43). The reductase partner in vivo has not yet been identified; however, we are currently attempting to isolate the protein by both conventional biochemical and molecular approaches.
As previously observed for the eukaryotic heme oxygenases (36, 37), H2O2 was able to support the first step in the reaction from heme to mesohydroxyheme, which in the presence of oxygen was converted to verdoheme with the release of CO. In agreement with previous studies, the initial step in heme oxidation most probably occurs via electrophilic addition of the protonated complex (Fe3+-O-OH) to the α-meso edge of heme (37). However, similar to results previously described with HO-1, the reaction of HemO with H2O2 does not support subsequent steps in converting verdoheme to biliverdin.
The model of heme utilization in neisseriae.
This study has initiated the characterization of the heme utilization cycle in a gram-negative pathogen. The cycle begins at the surface of the bacterium, in this case N. meningitidis, where highly specific receptors, HmbR and HpuAB, bind heme-containing compounds, Hb, haptoglobin-Hb, or heme (14, 23, 28). Through a not yet fully understood mechanism, neisserial proteins TonB, ExbB, and ExbD energize the active transport process of heme through the pores in specific receptors (29). Currently, the fate of heme after it enters the periplasm of neisseriae is not clear. From analogies with other gram-negative bacteria, we conclude that heme is most likely transported by an ABC-type heme-specific system into the cytoplasm (27). Studies in this communication clearly implicate HemO in the oxidative degradation of heme, with ferric iron-biliverdin and CO as products of the reaction.
While iron is either released to a storage protein such as bacterioferritin, utilized in different iron-dependent metabolic reactions, or stored, the fate of CO and biliverdin in neisseriae is not clear. In eukaryotes, biliverdin is transformed to bilirubin, which protects cells against oxidative damage (25). Enzymatic conversion of biliverdin to bilirubin has another important role; as mentioned above, the rate-limiting step in heme degradation is the release of biliverdin from heme oxygenase (39). Eukaryotes accomplish this by employing biliverdin reductase that directly interacts with heme oxygenase. Inhibition of heme oxygenase activity by ferric-biliverdin was observed in HemO-catalyzed heme degradation in vitro. Presumably, facilitation of both iron and biliverdin release from the protein must take place in vivo. Furthermore, the reductase or reducing equivalents that support the HemO reaction are not known. The identification of proteins involved in electron transfer to heme-HemO, as well as those involved in biliverdin release and further degradation, is the subject of current investigation.
It is unclear whether heme degradation products confer any advantage to neisseriae during the colonization of the human host, apart from supplying iron and protecting against heme toxicity. However, both heme degradation products are potentially biologically active; biliverdin can be reduced to bilirubin, which is a powerful antioxidant, and CO binds to eukaryotic guanylyl cyclase and increases intracellular levels of cGMP (16). These secondary effects of HemO-dependent heme degradation might be particulary pronounced in infections of the female genital tract with N. gonorrhoeae. It is interesting that oviduct infections usually occur within the first week after onset of menstruation (9, 30). Therefore, it seems that large amounts of heme in the menstrual flow assist in the dissemination of infection from an uncomplicated and local cervicitis to more serious inflammation of adnexal tissues. The release of heme iron by HemO is surely an important contributor to infection since iron is rate limiting for growth within the human host (5). However, it remains to be seen whether CO released from heme degradation might further contribute to the infection process by affecting the function of polymorphonuclear leukocytes and vasculature through an increase in intracellular cGMP levels.
In conclusion, HemO is the first heme oxygenase identified in a gram-negative pathogen. The identification of HemO as a heme oxygenase provides further evidence that oxidative cleavage of heme is the mechanism by which some bacteria acquire iron for further use.
ACKNOWLEDGMENTS
We thank the Gonococcal Genome Sequencing Project, supported by USPHS/NIH grant #AI38399, and B. A. Roe, S. P. Lin, L. Song, X. Yuan, S. Clifton, T. Ducey, L. Lewis, and D. W. Dyer for gonococcal hemO nucleotide sequence data. We thank Donna Balding-Perkins, Melanie Ratliff, Heather Alexander, and Veena Kumar for reading the manuscript and for helpful suggestions.
This work is supported by Public Health Service grant AI472870-01A1 to I.S.
REFERENCES
- 1.Braun V, Hantke K, Koster W. Bacterial iron transport: mechanisms, genetics, and regulation. Met Ions Biol Syst. 1998;35:67–145. [PubMed] [Google Scholar]
- 2.Chu G C, Katakuya K, Zhang X, Yoshida T, Keda-Saito M I. Heme degradation as catalyzed by a recombinant bacterial heme oxygenase (HmuO) from Corynebacterium diphtheriae. J Biol Chem. 1999;274:21319–21325. doi: 10.1074/jbc.274.30.21319. [DOI] [PubMed] [Google Scholar]
- 3.Chu G C, Tomita T, Sonnichsen F D, Yoshida T, Keda-Saito M I. The heme complex of Hmu O, a bacterial heme degradation enzyme from Corynebacterium diphtheriae: structure of the catalytic site. J Biol Chem. 1999;274:24490–24496. doi: 10.1074/jbc.274.35.24490. [DOI] [PubMed] [Google Scholar]
- 4.Cope L D, Yogev R, Muller-Eberhard U, Hansen E J. A gene cluster involved in the utilization of both free heme and heme: hemopexin by Haemophilus influenzae type b. J Bacteriol. 1995;177:2644–2653. doi: 10.1128/jb.177.10.2644-2653.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornelissen C N, Kelley M, Hobbs M M, Anderson J E, Cannon J G, Cohen M S, Sparling P F. The transferrin receptor expressed by gonococcal strain FA1090 is required for the experimental infection of human male volunteers. Mol Microbiol. 1998;27:611–616. doi: 10.1046/j.1365-2958.1998.00710.x. [DOI] [PubMed] [Google Scholar]
- 6.Coulton J W, Pang J C S. Transport of haemin by Haemophilus influenzae type b. Curr Microbiol. 1983;9:93–98. [Google Scholar]
- 7.Crusats J, Suzuki A, Mizutani T, Ogoshi H. Regioselective porphyrin bridge cleavage controlled by electronic effects: coupled oxidation of 3-demethyl-3-(trifluoromethyl)mesohemin IX and identification of its four biliverdin derivatives. J Org Chem. 1998;63:602–607. doi: 10.1021/jo9714728. [DOI] [PubMed] [Google Scholar]
- 8.Engel R R, Matsen J M, Chapman S S, Swartz S. Carbon monoxide production from heme compounds by bacteria. J Bacteriol. 1972;112:1310–1315. doi: 10.1128/jb.112.3.1310-1315.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eschenbach D A, Harnisch J P, Holmes K K. Pathogenesis of acute pelvic inflammatory diseases: role of contraception and other risk factors. Am J Obstet Gynecol. 1977;128:838–850. doi: 10.1016/0002-9378(77)90051-5. [DOI] [PubMed] [Google Scholar]
- 10.Falk J E. Part A: pyrrole pigments: chemistry and biochemistry of porphyrins and metalloporphyrins. In: Florkin M, Stotz E H, editors. Comprehensive biochemistry. Vol. 9. Amsterdam: Elsevier Publishing Company; 1963. pp. 3–33. [Google Scholar]
- 11.Henderson D P, Payne S M. Characterization of the Vibrio cholerae outer membrane heme transport protein HutA: sequence of the gene, regulation of expression, and homology to the family of TonB-dependent proteins. J Bacteriol. 1994;176:3269–3277. doi: 10.1128/jb.176.11.3269-3277.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Letoffe S, Redeker V, Wandersman C. Isolation and characterization of an extracellular haem-binding protein from Pseudomonas aeruginosa that shares function and sequence similarities with Serratia marcescens HasA haemophore. Mol Microbiol. 1998;28:1223–1234. doi: 10.1046/j.1365-2958.1998.00885.x. [DOI] [PubMed] [Google Scholar]
- 13.Letoffe S, Ghigo J M, Wandersman C. Iron acquisition from heme and hemoglobin by a Serratia marcescens extracellular protein. Proc Natl Acad Sci USA. 1994;91:9876–9880. doi: 10.1073/pnas.91.21.9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis L A, Gray E, Wang Y P, Roe B A, Dyer D W. Molecular characterization of hpuAB, the haemoglobin-haptoglobin-utilization operon of Neisseria meningitidis. Mol Microbiol. 1997;23:737–749. doi: 10.1046/j.1365-2958.1997.2501619.x. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Ortiz de Montellano P R. Reaction intermediates and single turnover rate constants for the oxidation of heme by human heme oxygenase-1. J Biol Chem. 2000;275:5297–5307. doi: 10.1074/jbc.275.8.5297. [DOI] [PubMed] [Google Scholar]
- 16.Maines M D. Heme oxygenase: function, multiplicity, regulatory mechanism, and clinical applications. FASEB J. 1988;2:2557–2568. [PubMed] [Google Scholar]
- 17.Ochsner U A, Johnson Z, Vasil M L. Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology. 2000;146:185–198. doi: 10.1099/00221287-146-1-185. [DOI] [PubMed] [Google Scholar]
- 18.Payne S M. Iron acquisition in microbial pathogenesis. Trends Microbiol. 1993;1:66–69. doi: 10.1016/0966-842x(93)90036-q. [DOI] [PubMed] [Google Scholar]
- 19.Richardson A R, Stojiljkovic I. HmbR, a hemoglobin-binding outer membrane protein of Neisseria meningitidis, undergoes phase variation. J Bacteriol. 1999;181:2067–2074. doi: 10.1128/jb.181.7.2067-2074.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saito S, Itano H A. Verdohemochrome IX alpha: preparation and oxidoreductive cleavage to biliverdin IX alpha. Proc Natl Acad Sci USA. 1982;79:1393–1397. doi: 10.1073/pnas.79.5.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 22.Schmitt M P. Utilization of host iron sources by Corynebacterium diphtheriae: identification of a gene whose product is homologous to eukaryotic heme oxygenases and is required for acquisition of iron from heme and hemoglobin. J Bacteriol. 1997;179:838–845. doi: 10.1128/jb.179.3.838-845.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schryvers A B, Stojiljkovic I. Iron acquisition systems in the pathogenic neisseria. Mol Microbiol. 1999;32:1117–1123. doi: 10.1046/j.1365-2958.1999.01411.x. [DOI] [PubMed] [Google Scholar]
- 24.Schuller D J, Wilks A, Ortiz de Montellano P R, Poulos T L. Crystal structure of human heme oxygenase-1. Nat Struct Biol. 1999;6:860–867. doi: 10.1038/12319. [DOI] [PubMed] [Google Scholar]
- 25.Stocker R, Yamamoto Y, McDonagh A F, Glazer A N, Ames B N. Bilirubin is antioxidant of possible physiological importance. Science. 1987;235:1043–1046. doi: 10.1126/science.3029864. [DOI] [PubMed] [Google Scholar]
- 26.Stojiljkovic I, Hantke K. Haemin uptake system of Yersinia enterocolitica: similarities with other TonB-dependent systems in gram-negative bacteria. EMBO J. 1992;11:4359–4367. doi: 10.1002/j.1460-2075.1992.tb05535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stojiljkovic I, Hantke K. Transport of haemin across the cytoplasmic membrane through a haemin-specific periplasmic binding-protein-dependent transport system in Yersinia enterocolitica. Mol Microbiol. 1994;13:719–732. doi: 10.1111/j.1365-2958.1994.tb00465.x. [DOI] [PubMed] [Google Scholar]
- 28.Stojiljkovic I, Hwa B, de Saint Martin L, O'Gaora P, Nassif X, Hefferon F, So M. The Neisseria meningitidis haemoglobin receptor: its role in iron utilization and virulence. Mol Microbiol. 1995;15:531–541. doi: 10.1111/j.1365-2958.1995.tb02266.x. [DOI] [PubMed] [Google Scholar]
- 29.Stojiljkovic I, Srinivasan N. Neisseria meningitidis tonB, exbB, and exbD genes: Ton-dependent utilization of protein-bound iron in Neisseriae. J Bacteriol. 1997;179:805–812. doi: 10.1128/jb.179.3.805-812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sweet R L, Blankfort-Doyle M, Robbie M O, Schacter J. The occurrence of chlamydial and gonococcal salpingitis during the menstrual cycle. JAMA. 1986;255:2062–2064. [PubMed] [Google Scholar]
- 31.Tenhunen R, Marver H S, Schmid R. The enzymatic conversion of heme to bilirubin by microsomal heme oxygenase. Proc Natl Acad Sci USA. 1968;61:748–755. doi: 10.1073/pnas.61.2.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wandersman C, Stojiljkovic I. Bacterial heme sources: the role of heme, hemoprotein receptors and hemophores. Curr Opin Microbiol. 2000;3:215–220. doi: 10.1016/s1369-5274(00)00078-3. [DOI] [PubMed] [Google Scholar]
- 33.Wilks A, Black S M, Miller W L, Ortiz de Montellano P R. Expression and characterization of truncated human heme oxygenase (hHO-1) and a fusion protein of hHO-1 with human cytochrome P450 reductase. Biochemistry. 1995;34:4421–4427. doi: 10.1021/bi00013a034. [DOI] [PubMed] [Google Scholar]
- 34.Wilks A, Schmitt M P. Expression and characterization of a heme oxygenase (HmuO) from Corynebacterium diphtheriae. J Biol Chem. 1998;273:837–841. doi: 10.1074/jbc.273.2.837. [DOI] [PubMed] [Google Scholar]
- 35.Wilks A, Moenne-Loccoz P. Identification of the proximal ligand His-20 in heme oxygenase (Hmu O) from Corynebacterium diphtheriae: oxidative cleavage of the heme macrocycle does not require the proximal histidine. J Biol Chem. 2000;275:11686–11692. doi: 10.1074/jbc.275.16.11686. [DOI] [PubMed] [Google Scholar]
- 36.Wilks A, Ortiz de Montellano P R. Rat liver heme oxygenase: high level expression of a truncated soluble form and nature of the meso-hydroxylating species. J Biol Chem. 1993;268:22357–22362. [PubMed] [Google Scholar]
- 37.Wilks A, Torpey J, Ortiz de Montellano P R. Heme oxygenase (HO-1): evidence for electrophilic oxygen addition to the porphyrin ring in the formation of alpha-meso-hydroxyheme. J Biol Chem. 1994;269:29553–29556. [PubMed] [Google Scholar]
- 38.Wyckoff E E, Duncan D, Torres A G, Mills M, Maase K, Payne S M. Structure of the Shigella dysenteriae haem transport locus and its phylogenetic distribution in enteric bacteria. Mol Microbiol. 1998;28:1139–1152. doi: 10.1046/j.1365-2958.1998.00873.x. [DOI] [PubMed] [Google Scholar]
- 39.Yoshida T, Noguchi M, Kikuchi G. A new intermediate of heme degradation catalyzed by the heme oxygenase system. J Biochem (Tokyo) 1980;88:557–563. doi: 10.1093/oxfordjournals.jbchem.a133003. [DOI] [PubMed] [Google Scholar]
- 40.Yoshida T, Noguchi M, Kikuchi G, Sano S. Degradation of mesoheme and hydroxymesoheme catalyzed by the heme oxygenase system: involvement of hydroxyheme in the sequence of heme catabolism. J Biochem (Tokyo) 1981;90:125–131. doi: 10.1093/oxfordjournals.jbchem.a133441. [DOI] [PubMed] [Google Scholar]
- 41.Yoshida T, Noguchi M, Kikuchi G. The step of carbon monoxide liberation in the sequence of heme degradation catalyzed by the reconstituted microsomal heme oxygenase system. J Biol Chem. 1982;257:9345–9348. [PubMed] [Google Scholar]
- 42.Yoshida T, Noguchi M. Features of intermediary steps around the 688-nm substance in the heme oxygenase reaction. J Biochem (Tokyo) 1984;96:563–570. doi: 10.1093/oxfordjournals.jbchem.a134868. [DOI] [PubMed] [Google Scholar]
- 43.Zhu W, Hunt D J, Richardson A R, Stojiljkovic I. Use of heme compounds as iron sources by pathogenic neisseriae requires the product of the hemO gene. J Bacteriol. 2000;182:439–447. doi: 10.1128/jb.182.2.439-447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]