Abstract
Ferrous iron (Fe2+) is required for the growth and virulence of many pathogenic bacteria, including Vibrio cholerae (Vc), the causative agent of the disease cholera. For this bacterium, Feo is the primary system that transports Fe2+ into the cytosol. FeoB, the main component of this system, is regulated by a soluble cytosolic domain termed NFeoB. Recent reanalysis has shown that NFeoBs can be classified as either GTP-specific or NTP-promiscuous, but the structural and mechanistic bases for these differences were not known. To explore this intriguing property of FeoB, we solved the X-ray crystal structures of VcNFeoB in both the apo and GDP-bound forms. Surprisingly, this promiscuous NTPase displayed a canonical NFeoB G-protein fold like GTP-specific NFeoBs. Using structural bioinformatics, we hypothesized that residues surrounding the nucleobase could be important for both nucleotide affinity and specificity. We then solved the X-ray crystal structures of N150T VcNFeoB in the apo and GDP-bound forms to reveal H-bonding differences surround the guanine nucleobase. Interestingly, isothermal titration calorimetry revealed similar binding thermodynamics of the WT and N150T proteins to guanine nucleotides, while the behavior in the presence of adenine nucleotides was dramatically different. AlphaFold models of VcNFeoB in the presence of ADP and ATP showed important conformational changes that contribute to nucleotide specificity among FeoBs. Combined, these results provide a structural framework for understanding FeoB nucleotide promiscuity, which could be an adaptive measure utilized by pathogens to ensure adequate levels of intracellular iron across multiple metabolic landscapes.
Keywords: Feo, iron, NTPase, X-ray crystallography, isothermal titration calorimetry
INTRODUCTION
Iron (Fe) is an essential nutrient for nearly all lifeforms due to its use as a cofactor in numerous biochemical processes, including oxidative phosphorylation, de novo DNA synthesis, and nitrogen fixation, among others(1–5). To harness the power of this element, iron must first be acquired from the environment before it can be biologically incorporated into proteins and enzymes. For many organisms, including most bacteria, the prevalent environmental oxidation state of iron dictates its mode of acquisition. For example, the highly insoluble ferric iron (Fe3+) is prevalent in oxic environments, and bacteria will commonly deploy siderophores to solubilize and to capture this form of iron. Membrane receptors then translocate the siderophore-chelated iron into the cytosol, where the iron is either removed by reductive dissociation or by cleavage of the siderophore(6, 7). Additionally, some pathogenic bacteria can sequester either free heme (iron protoporphyrin IX) or utilize hemophores to remove heme from host hemoproteins (e.g., hemoglobin and myoglobin). Like siderophoremediated uptake, membrane receptors then translocate the heme into the cytosol where the heme is either recycled or destroyed to remove the iron contained within(8–11). In contrast, when living within anoxic or acidic environments, bacteria commonly encounter the more labile, but also more reactive, ferrous iron (Fe2+)(12–15). Unfortunately, despite its strong contribution to bacterial metal homeostasis and pathogenesis, the mechanisms of bacterial Fe2+ acquisition are poorly understood compared to the mechanisms of Fe3+ uptake and heme acquisition.
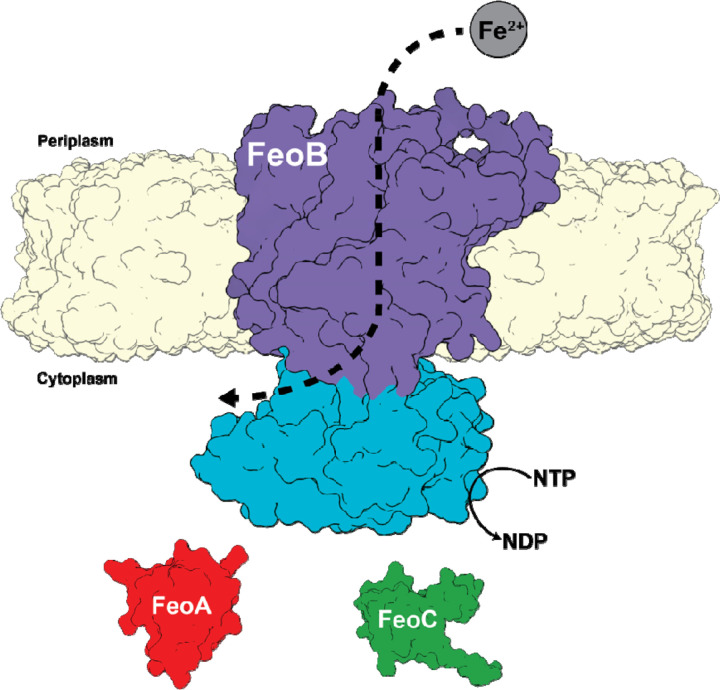
While some auxiliary Fe2+ transport systems have been identified, the ferrous iron transport (Feo) system is the most widely distributed and conserved Fe2+ acquisition system across the prokaryotic domain(12–15), although Feo’s precise mechanism of function remains unclear. Canonically, the feo operon encodes for three proteins, FeoA, FeoB, and FeoC(16), although FeoC is the least conserved of these proteins(13–15, 17) (Fig. 1). FeoA and FeoC are known to be small (ca. 7–10 kDa) cytosolic proteins, while FeoB is a large (ca. 80–100 kDa) polytopic transmembrane protein that contains an N-terminal soluble G-protein-like domain termed NFeoB(12, 14, 18). The roles of FeoA and FeoC remain somewhat enigmatic; however, these proteins have been shown to interact with NFeoB in vitro(19), FeoA appears to regulate GTP hydrolysis in vitro(17), and some FeoCs bind oxygen-sensitive [Fe-S] clusters, presumably for regulatory purposes(20). In vivo, several observations indicate that both proteins interact with FeoB and are required for Feo-dependent iron uptake in Vibrio cholerae (Vc)(21, 22), the pathogenic bacterium responsible for the diarrheal disease cholera. Interestingly, bacterial two hybrid (BACTH) systems demonstrated that both VcFeoA and VcFeoC were found to interact with intact VcFeoB in the cell(23), although the precise nature of this complex and its mechanism are still unclear.
Fig. 1.
Cartoon depiction of the tripartite Vibrio cholerae ferrous iron transport (Feo) system. In V. cholerae the Feo system consists of three proteins: FeoA (colored in red) and FeoC (colored in green), both of which are cytosolic, and a polytopic transmembrane protein, FeoB (colored in purple), with soluble N-terminal domain termed NFeoB (colored in teal). The NFeoB domain of V. cholerae FeoB wa recently discovered to be nucleotide promiscuous and is best classified as an NTPase rather than a strict GTPase.
FeoB from V. cholerae was recently shown to hydrolyze ATP, GTP, and other NTPs in vitro, and this function could serve to supply V. cholerae with Fe2+ in vivo, indicating that this FeoB may be better classified as an NTPase rather than a strict GTPase(24, 25). This promiscuity for NTP consumption was then shown to occur in vitro for other FeoBs from a handful of infectious bacterial species such as Helicobacter pylori, Streptococcus mutans, Staphylococcus aureus, and Bacillus cereus, which could suggest that NTP promiscuity may be a common theme used by some pathogenic bacteria to acquire iron and to establish infection. NFeoB proteins contain generally conserved G-motifs that are common amongst G-proteins and are responsible for binding to different segments of the guanine nucleotide, with G1, G2, and G3 binding to the α-, β-, and γ-phosphates, while the G4 and G5 motifs interact with the nucleobase(26). Sequence analyses suggested that differentially conserved residues within the G4 and G5 motifs might be important for guanine recognition and for NTPase activity in FeoB(27, 28). Specifically, the G5 motif residues Ser148 and Asn150, when altered, displayed significant effects on ATPase activity, but minimal effects on GTPase activity, indicating that these G5 residues may play a critical role in nucleotide discrimination(24). However, the structural basis of nucleotide promiscuity in FeoB remained unknown, precluding a more comprehensive understanding of this unique aspect of the Feo system.
In this work, we have structurally and biophysically characterized the VcNFeoB domain in order to understand its nucleotide promiscuity. Using X-ray crystallography, we determined the structures of apo and GDP-bound VcNFeoB, which reveals a conserved NFeoB fold composed of a G-protein like domain tethered to a GDI domain, despite the nucleotide promiscuous nature of VcFeoB. Additionally, we determined the X-ray crystal structures of apo and GDP-bound N150T VcNFeoB, and we show how differences in residues at the G5 motif alter the hydrogen-bonding interactions surrounding the guanine nucleobase. Isothermal titration calorimetry (ITC) was used to determine substrate affinities and stoichiometries of different nucleotides to both the wild-type and variant VcNFeoBs. Intriguingly, we demonstrate dramatic differences in the behavior of the VcNFeoB towards GDP/GMP-PNP binding compared to ADP/AMP-PNP binding, which could be rationalized using AlphaFold modeling. Taken together, these findings provide a structural framework for understanding the nucleotide promiscuity of VcFeoB, which could be leveraged for future developments of targeted therapeutics to tackle issues of V. cholerae pathogenesis, as recently demonstrated(29).
RESULTS
The VcNFeoB NTPase Domain Purifies as a Nucleotide-Free Monomer that Displays Broad NTPase Activity
To prepare the VcNFeoB NTPase domain for crystallization trials, we overproduced in E. coli a previously designed construct that encodes for a non-cleavable (His)6-tagged version of the protein (i.e., VcNFeoB(His)6))(24). As this protein was not initially suitable for crystallization from immobilized metal-affinity chromatography (IMAC), we added two additional purification steps: anion exchange chromatography (AEX) and size-exclusion chromatography (SEC), both of which revealed interesting biophysical properties of VcNFeoB(His)6. First, regarding AEX, we monitored the 260 nm / 280 nm ratio through the entire chromatography process, and noted a normal value of ≈0.6, indicating that VcNFeoB(His)6 does not co-purify with nucleotide, unlike previous reports of E. coli NFeoB overproduced in E. coli(30). Second, SEC of either crudely purified VcNFeoB(His)6 (IMAC only) or polished VcNFeoB(His)6 (after SEC) showed only the presence of a dominant monomeric species in solution (Fig. S1), consistent with our previous in vitro studies on Feo proteins (from V. cholerae and others) recombinantly produced in E. coli. It is possible that FeoA and/or FeoC may be necessary to induce FeoB oligomerization, and in vivo studies have suggested this to be the case for Vibrio cholerae(23). However, other Feo systems appear to be functional monomers in vitro, and this highly pure, monomeric VcNFeoB(His)6 domain was active against multiple nucleotide triphosphates, most notably ATP and GTP, as previously described(24), thus the precise oligomeric state of FeoB remains controversial and requires further exploration. For all subsequent constructs (vide infra), a similar overproduction and purification process was employed, producing highly pure, homogeneous, and monomeric protein (Fig. S1).
The Structure of the Apo VcNFeoB NTPase Domain Reveals a Typical NFeoB Fold
In order to characterize the three dimensional structure of the VcNFeoB NTPase domain, we sought to crystallize the VcNFeoB(His)6 protein. Crystals of the apo domain were generated and ultimately diffracted to a modest 3.7 Å resolution in the P1 space group consistent with 8 molecules (dimer of tetramers) in the asymmetric unit (ASU) (Table S1). This oligomerization is likely crystallization induced based on our in-solution studies (Fig. S1). We solved this structure by using molecular replacement coupled with model building and restrained refinement approaches (Rw/Rf = 0.210/0.268) (Table S1), and an analysis of the crystal contacts suggested that the (His)6 tag may have impacted the crystal quality. To overcome this issue, VcNFeoB was recloned into a vector encoding for N-terminal (His)6 tag fused to a cleavable SUMO moiety. Expression and purification followed the same procedures as previously described (vide supra), and the SUMO tag was cleaved prior to crystallization. Crystals of the SUMO-cleaved apo domain were generated and ultimately diffracted to 2.3 Å resolution in the P121 space group consistent with 2 molecules in the asymmetric unit (ASU) (Table S1). This oligomerization is likely crystallization induced based on our in-solution studies (Fig. S1). Using the 3.7 Å resolution model, we were able to solve the 2.3 Å resolution structure of the tagless protein (Rw/Rf = 0.203/0.266) (Table S1).
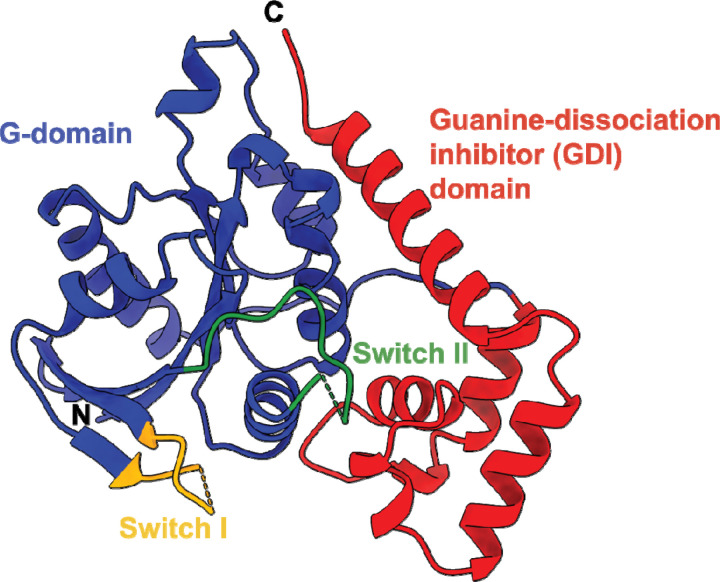
The X-ray crystal structure of the apo VcNFeoB NTPase domain reveals the presence of a typical NFeoB fold (Fig. 2). Distinctly present are the two common NFeoB subdomains: the globular G-protein subdomain that is responsible for binding and hydrolyzing nucleotides (31) (Fig. 2, blue) and the hammer-shaped guanine-dissociation inhibitor (GDI) subdomain that regulates nucleotide dissociation (32) and connects directly to the FeoB transmembrane region (Fig. 2, red). Within the G-protein domain, two switch regions (known as Switch I and Switch II) that regulate nucleotide hydrolysis and communicate nucleotide status to the GDI domain (31, 32) are present, albeit Switch I is mostly disordered, while Switch II is mostly ordered in the apo form. Surprisingly, comparison of the VcNFeoB NTPase domain to other structurally characterized NFeoB domains displays strong structural conservation in both subdomains (Cα RMSD < 1Å on average) even though the NFeoB is not a strict GTPase (Fig. S2). These observations indicate that gross structural changes in the NFeoB region do not account for the observed nucleotide promiscuity of V. cholerae FeoB per se.
Fig. 2.
X-ray crystal structure of the SUMO-cleaved Vibrio cholerae NFeoB NTPase domain in the apo state (PDB ID 9BA6). The overall structure of the VcNFeoB has a typical NFeoB fold and comprises two major domains: the guanine-dissociation inhibitor (GDI) domain (labeled in red) that regulates GDP release and connects to the transmembrane region, and the G-protein domain (labeled in blue) that binds and catalyzes nucleotide hydrolysis. Within the G-protein domain are two key switch regions (Switch I and Switch II, labeled yellow and green respectively) that regulate nucleotide hydrolysis and transmit information to the GDI domain. In the absence of nucleotide, Switch I is mostly disordered, while Switch II is mostly ordered. ‘N’ and ‘C’ represent the N- and C-termini in the structure, respectively.
The GDP-bound Structure of the VcNFeoB NTPase Domain Reveals Important Nucleotide-Binding Residues
To determine whether structural properties within the nucleotide-binding pocket could contribute to nucleotide promiscuity, we sought to determine the structure of VcNFeoB in the presence of various nucleotides. To do so, we co-crystallized apo VcNFeoB (both SUMO-cleaved and (His)6 tagged) in the presence of hydrolyzed nucleotides (e.g., ADP and GDP) and the presence of non- or slowly-hydrolyzable triphosphate mimics (e.g., AMP-PNP, AMP-PCP, and GMP-PNP). Despite extensive fine screening and testing of multiple conditions, we were only able to generate datasets of GDP-bound VcNFeoB(His)6 that diffracted modestly, but completely, to 4.2 Å resolution in the C121 space group consistent with 4 molecules in the asymmetric unit (ASU) (Table S1). This oligomerization is likely crystallization induced based on our in-solution studies (Fig. S1). Using molecular replacement with our 2.3 Å apo VcNFeoB structure coupled with strongly restrained refinement approaches, we built a model that clearly displayed GDP in the nucleotide-binding pocket of all molecules in the ASU based on omit maps (Fig. S3). Using the X-ray structure of GDP-bound E. coli NFeoB as a guide (PDB ID 3I8X; 2.3 Å resolution), we were able to place GDP in all VcNFeoB molecules in the ASU and to solve this structure (Rw/Rf = 0.223/0.273) (Table S1).
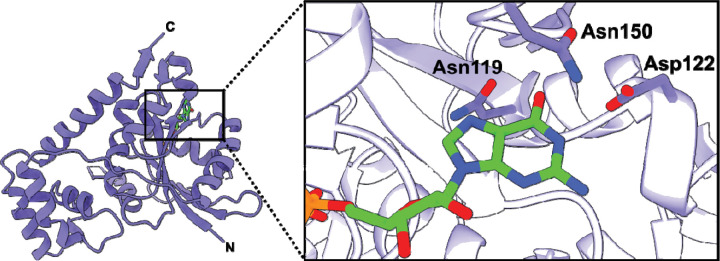
The crystal structure of VcNFeoB(His)6 bound to GDP reveals multiple amino acids that contribute to nucleotide binding (Fig. 3). In the absence of nucleotide, the binding pocket is fairly open, while binding of GDP elicits a contraction surrounding the nucleobase with the associated amino acids responsible for nucleotide recognition coming together (Fig. S4). In this structure, the Switch I loop that has been shown to be important for GTP hydrolysis (31) is mostly disordered, which may be attributed to the flexibility of this region when GDP is bound. As the nucleobase enters the binding pocket, a region of random coil from Asn119 to Asp122 tightens (Fig. S4), and both residues (conserved amongst NFeoBs) become within hydrogen-bonding distance (3.5 and 2.8 Å, respectively) of the guanine purine (Figs. 3,4a). Underneath the guanine purine is Lys120, also conserved in many NFeoBs, that appears to prop up the hydrolyzed nucleotide in the binding pocket; electrostatic contributions from this residue are not observed in our structure, and in some NFeoBs (like those in S. thermophilus, K. pneumoniae, E. coli and Gallionella capsiferriformans, and Thermotoga maritima) this residue corresponds to a non-polar amino acid, either a Met or an Ala(31, 33–36). Interestingly, Asn150 is positioned along a region of random coil (known as the G5 motif) above the guanine purine but turned towards, and tightly hydrogen bonded with Asp122 on the G4 motif (2.3 Å distance) (Figs. 3,4a). Because of the high variability within the G5 region, we sought to use bioinformatics to gain a better understanding of whether Asn150 might have an important role in nucleotide discrimination.
Fig. 3.
X-ray crystal structure of the Vibrio cholerae NFeoB NTPase domain in the GDP-bound state. The right panel represents a zoomed-in view of the nucleotide-binding pocket bound to GDP. Three residues make important contacts with the purine ring: Asn119, Asp122, and Asn150. Of these residues, only Asn150 is located on a variable loop region (G5) and lacks conservation. In the presence of GDP, the Switch I region is nearly fully disordered, while the Switch II region is only partially disordered. ‘N’ and ‘C’ represent the N- and C-termini in the structure, respectively.
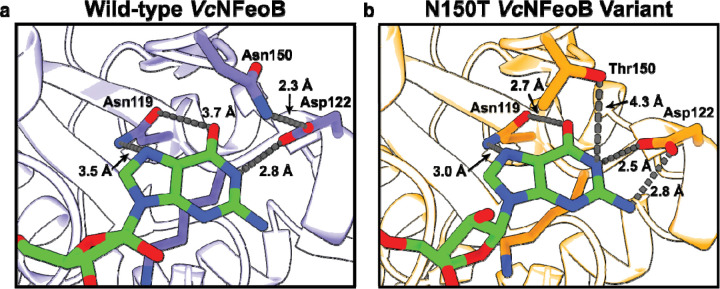
Fig. 4.
Comparisons of the nucleotide-binding pockets of the WT (a) and N150T (b) VcNFeoB NTPase domain structures in their GDP-bound forms. Fewer hydrogen-bonding interactions are observed in the WT GDP-bound structure compared to the N150T variant of the NTPase domain. We hypothesize that the fewer hydrogen-bonding interactions in the WT structure allow for greater plasticity in NTP/NDP binding, unlike the strict GTPases that do not bind and hydrolyze other NTPs.
Alterations in Hydrogen-Bonding Surrounding the Nucleotide-Binding Pocket Are Likely Linked To the Nucleotide Promiscuity of VcNFeoB
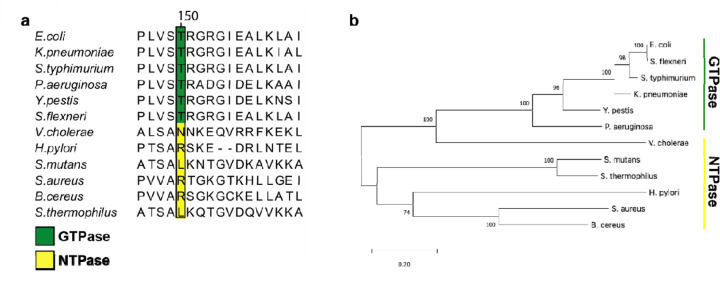
To gain a better understanding of residues that are either conserved or variable amongst NFeoB NTPases and GTPases, and to understand whether these sequence differences might contribute to functional differences, we used multiple sequence analysis (MSA) and phylogenetics. To do so, we limited our approach only to the sequences that have been previously tested in vitro to have either NTPase or strictly GTPase activities(28). Partial sequence alignments revealed that the position analogous to Asn150 within the G5 motif of the VcNFeoB NTPase domain is highly variable for NFeoBs known to be NTPases, while this same position is an invariant Thr residue for NFeoBs known to be strict GTPases (Fig. 5A). The flanking regions of the G5 motif are also highly variable in NFeoB NTPases, while the same regions are highly conserved in NFeoB GTPases (Fig. 5A), suggesting that a degree of flexibility near the nucleobase may be important for nucleotide promiscuity. Intriguingly, phylogenetic analyses using the full-length sequences of bona fide FeoB NTPases and GTPases reveal a differential clustering among GTPase and NTPase proteins (Fig. 5B). While the number of FeoB proteins with verified nucleotide activity is low, this analysis could suggest that convergent evolution (and/or horizontal gene transfer) may have occurred amongst these organisms, similar to a previous hypothesis for the Feo system(13).
Fig. 5.
Partial multiple sequence alignment (MSA) and phylogenetic analysis of nucleotide-specific and nucleotide-promiscuous FeoBs. a. The partial MSA compares residues in the variable G5 region of experimentally determined GTPases (green) and experimentally determined NTPases (yellow). At position 150 in V. cholerae FeoB (NTPase) is an Asn residue, while the analogous position 150 in the strict GTPase FeoBs is a Thr residue. The numbering above the MSA is based on the V. cholerae FeoB sequence. b. Phylogenetic analysis of the FeoB sequences from the respective organisms in panel ‘a’ shows distinct clustering of the NTPase FeoBs from those of the GTPase FeoBs. The 0.20 scalebar indicates the amount of genetic change at a certain length. The numbers above the nodes represent the bootstrap values above 50% from 500 iterations.
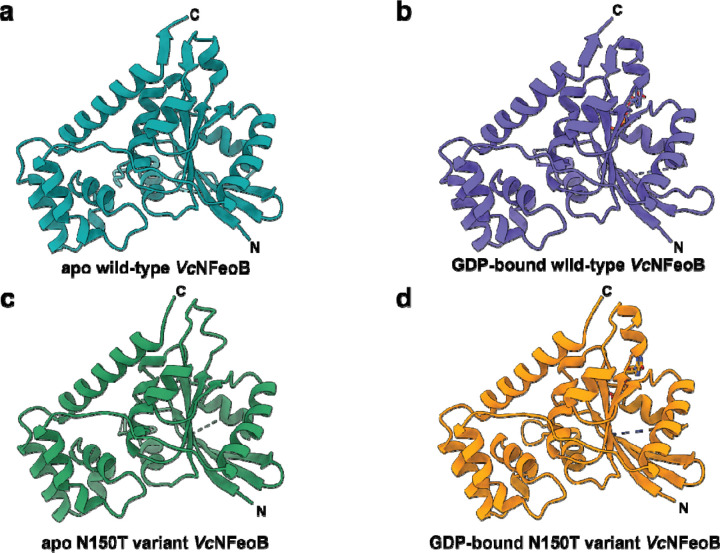
To test whether alteration of Asn150 to a Thr residue would inform on structural changes that may take place within the nucleotide-binding pocket, we expressed, purified, and crystallized the N150T variant of VcNFeoB(His)6 in the presence of GDP. The crystals diffracted to 2.9 Å resolution (Table S1), and initial analysis revealed one apo VcNFeoB(His)6 NTPase domain and one GDP-bound VcNFeoB(His)6 NTPase domain both within the asymmetric unit. This hetero-oligomerization is likely crystallization induced based on our in-solution studies (Fig. S1). Omit maps confirmed that substantial density was present and consistent with GDP in one, but not both, molecules within the asymmetric unit (Fig. S3), allowing us to visualize a direct comparison between the apo and the GDP-bound N150T variant at the same resolution (Fig. 6). Interestingly, the presence of Thr in position 150 affects the hydrogen bonding pattern surrounding the guanine nucleobase both directly and indirectly. First, the Thr hydroxyl moiety now makes a new hydrogen bond with position N1 along the purine ring (Fig. 4b). Second, removal of Asn150 releases Asp122 to tighten and to extend its hydrogen bonds with positions N1 and the H2N-C1 group (Fig. 4b). Third, Asn119 appears to pull the guanine further into the binding pocket, although uncertainty due to modest resolution of the GDP-bound WT VcNFeoB NTPase domain structure prevents a definitive statement regarding Asn119 hydrogen bonding strength. However, by comparison to the WT protein, the N150T variant appears to have two key additional hydrogen bonds surrounding the guanine purine that could affect nucleotide stability and may afford the discrimination of GTP relative to other NTPs. Finally, like the WT VcNFeoB NTPase domain, the N150T variant displays a mostly disordered Switch I region and a partially disordered Switch II region in both the GDP-bound and apo forms.
Fig. 6.
Comparisons of the WT and N150T VcNFeoB NTPase domain structures in their apo and GDP-bound forms. a. Apo WT VcNFeoB(His)6 NTPase domain (PDB ID 8VWL). b. GDP-bound WT VcNFeoB(His)6 NTPase domain (PDB ID 8VWN). c. Apo N150T VcNFeoB(His)6 NTPase domain (PDB ID 9BA7). d. GDP-bound N150T VcNFeoB(His)6 NTPase domain (PDB ID 9BA7). In general, the structural similarities among the WT and variant VcNFeoB(His)6 NTPase domains are very high: 1.36 Å Cα RMSD (apo WT and apo N150T) and 0.88 Å Cα RMSD (GDP-bound WT and GDP-bound N150T). ‘N’ and ‘C’ represent the N- and C-termini in the structure, respectively.
Isothermal Titration Calorimetry Coupled with AlphaFold Modeling Reveal Key Differences in GTP/GDP and ATP/ADP Binding
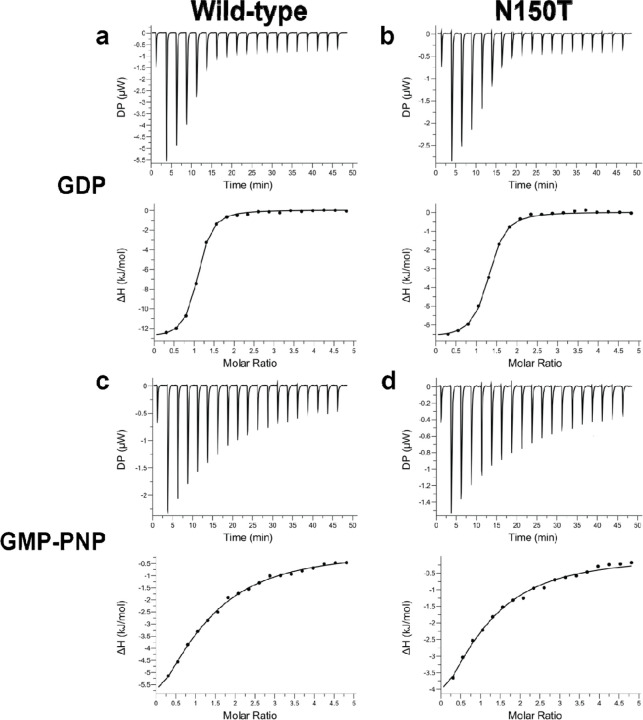
To test the binding strength and binding stoichiometry of various nucleotides to the WT VcNFeoB NTPase domain and its N150T variant, we used isothermal titration calorimetry (ITC) (Fig. 7). Despite the hydrogen-bonding differences in our X-ray crystal structures, the WT and N150T VcNFeoB proteins displayed nearly identical binding strengths to GDP: WT Kd of 3.18 μM ± 0.23 μM; N150T Kd of 3.60 μM ± 0.36 μM (Fig. 7a,b) with a single binding site (N ≈ 1). By comparison to the binding of GMP-PNP (a non-hydrolyzable GTP analog), the WT and N150T VcNFeoB constructs displayed slightly different binding strengths, consistent with our observed increase in hydrogen bonding in the N150T variant: WT Kd of 121.23 μM ± 34.13 μM; N150T Kd of 93.95 μM ± 15.63 μM (Fig. 7c,d) with a single binding site (N ≈ 1). GDP is known to have a stronger affinity to NFeoBs than GTP/GMP-PNP due to the presence and function of the GDI domain(32, 37), and we observed a similar trend as our structural work also reveals the presence of a GDI domain in VcNFeoB. However, based on these data, the presence/absence of Asn150 alone does not dramatically change the binding strength of GTP/GDP to the VcNFeoB NTPase domain. We then tested the ability of the VcNFeoB NTPase domain to bind adenosine containing nucleotides. While we attempted multiple different concentrations and stoichiometries using both hydrolyzed (ADP) and non-hydrolyzable analogs (AMP-PNP), we did not observe any appreciable saturation that could be fitted to any logical binding isotherm for both the WT and N150T variant proteins (Fig. S5). However, titrations of ADP/AMP-PNP showed strong amounts of heat evolution (up to 12 μW per injection) even when corrected for dilutions of nucleotide in the absence of protein (Fig. S5). These observations suggest either a rapid kinetic reversibility (i.e., fast kon and fast koff), and/or a potential conformational change that may be occurring as the domain samples ADP/AMP-PNP in the binding pocket.
Fig. 7.
The VcNFeoB NTPase domain binds GDP and GMP-PNP similarly to GTP-specific NFeoB domains. Representative ITC thermograms (top) and ΔH vs. molar ratio traces (bottom) of WT (left) and N150T (right) VcNFeoB titrated with either GDP (a, b) or GMP-PNP (c, d). All datasets have been corrected for nucleotide dilution into buffer in the absence of protein. The strength of binding for GDP to both WT (3.18 μM ± 0.23 μM) and N150T VcNFeoB (3.60 μM ± 0.36 μM) are nearly identical, with both having a single binding site (N ≈ 1). The strength of binding for GMP-PNP to WT VcNFeoB (121.23 μM ± 34.13 μM) is slightly weaker than binding to N150T VcNFeoB (93.95 μM ± 15.63 μM), but both reveal a single binding site (N ≈ 1). All values were determined in triplicate and represent the mean ± one standard deviation of the mean.
Finally, to gain a better understanding of what may be occurring within the binding pocket, we used the AlphaFold3 server to predict the WT and N150T VcNFeoB structures bound to ADP and ATP (Fig. S6). Interestingly, in the predicted WT structures in the presence of adenine-containing nucleotides, Asn150 makes a very weak interaction with the adenine purine while Asp122 and Ser148 (part of the G4 and G5 motifs, respectively) turn completely away from the binding pocket and make tight hydrogen-bonds (≈ 2.7 Å) with one another (Fig. S6a). A similar result was observed in the N150T variant with Thr150, making even weaker interactions with the adenine nucleotide (Fig. S6b). In both cases, strong hydrogen-bond interactions (≤ 2.8 Å) with Ser148 cause the NTPase domain to adopt an “Asp off” conformation, i.e., Asp122 makes no interactions with the adenine nucleobase. These results differ from the GDP-bound structures, as both adopt a “Asp on” conformation in which Asp122 makes strong interactions (≈ 2.4 Å – 2.8 Å) with the guanine nucleobase. When we compare the ADP/ATP-bound behavior to the apo structure of VcNFeoB (Fig. S4), we note that the G4 and G5 regions would need to undergo structural rearrangements upon nucleotide binding, which could explain the strong heat evolution upon ADP and AMP-PNP titrations, but fewer hydrogen bonding interactions likely preclude stable or prolonged binding within the pocket, explaining the ITC results.
DISCUSSION
Whether FeoB, the primary prokaryotic ferrous iron transporter, is nucleotide promiscuous or nucleotide specific has vacillated for some time. Before the structure of NFeoB was known, early studies of FeoB predicted that the protein might hydrolyze ATP due to its sequence similarity to other ATPases, and decreased FeoB-dependent iron uptake in Helicobacter pylori was observed when ATP synthesis was disrupted by proton uncouplers(38–40). However, subsequent studies of FeoB showed that the NFeoB domain from E. coli was GTP-specific(41), and structures of NFeoB from Methanocaldococcus jannaschii and E. coli revealed the presence of a G-protein like domain(34, 42), strongly implying that NFeoB bound and hydrolyzed only guanine nucleotides. This presumption continued for nearly two additional decades, as additional NFeoB structures were determined and FeoB was further explored in an almost GTP-exclusive manner(12, 14, 18). However, despite this assumption, Shin et al reexamined NTPase activity in the context of V. cholerae FeoB and found this protein to be nucleotide promiscuous both in vitro and in vivo(24). These observations were further expanded to show that several bacterial FeoBs could be differentially classified as GTP-specific while others could be classified as nucleotide promiscuous(24, 25). However, the structural basis for this functional divergence of FeoB was not known.
In this work, we provided the first X-ray crystal structure of V. cholerae NFeoB, a notably promiscuous NTPase, in its WT and variant forms, both in the presence and absence of nucleotides. While the general NFeoB G-protein like fold is conserved in our structures, the GDP-bound WT VcNFeoB(His)6 structure revealed interactions between Asn150 (G5 motif) and Asp122 (G4 motif) that caused Asp122 to decrease the number of interactions it makes with the guanine nucleobase. As Asn150 is highly variable among NFeoB NTPases but conserved as a Thr among NFeoB GTPases, we wondered whether alteration of this residue alone could change nucleotide binding strength and/or specificity. Our structure of N150T VcNFeoB(His)6 revealed increased H-bonding to the nucleobase due to the presence of Thr150 in the G5 motif, but this modification alone did not dramatically alter the binding affinity between NFeoB and GDP and only modestly increased the binding affinity between NFeoB and GMP-PNP. Instead, we hypothesized that the observed altered interactions might contribute more to nucleotide promiscuity.
While we were unable to determine experimental structures of adenine-based nucleotides bound to VcNFeoB, ITC data and AlphaFold modeling provided insight into nucleotide promiscuity when taken in context of our other experimentally-determined structures. For example, ITC analyses of ADP- or AMP-PNP titrated into VcNFeoB revealed an isotherm that failed to saturate but displayed strong heat evolution even after correction for appropriate dilutions, distinct from the response of this domain in the presence of GDP and GMP-PNP. This unusual behavior could be interpreted to mean weak binding but also that conformational changes may accompany the interactions of ADP/AMP-PNP within the nucleotide binding pocket, perhaps explaining why we failed to produce crystals of adenosine nucleotides bound to VcNFeoB despite exhaustive trials. Structural modeling using AlphaFold supports this notion, as predicted structures of both ADP- and ATP-bound VcNFeoB revealed movement of the G4 Asp122 residue and G5 Ser148 residue to lock the two amino acids into a strong H-bond preventing Asp122 from interacting with the nucleobase (i.e., “Asp off”). This conformational change opens up the binding pocket and results in only weak interactions as ADP/ATP enters but then exits the domain, likely rapidly. However, the rate of this conformational change must be slower than the rate of ATP hydrolysis, as we note that VcNFeoB still hydrolyzes ATP robustly under these conditions(24, 25). In fact, all three residues (Asp122, Ser148, and Asn150) combined displayed an important role for ATP hydrolysis(24), suggesting the G4 and G5 motif are working in concert to contribute to nucleotide specificity.
Interestingly, structural analyses of other bacterial NTPases reveal a similar amino acid pattern that may contribute to a more general nucleotide promiscuity (Fig. S7), which could be leveraged for a functional advantage by select organisms. For example, among these NTPases in the G5 motif position analogous to Asn150 in VcNFeoB are a diverse set of amino acids that only interact with the nucleotide base weakly at best(43), but this weak interaction may be important for plasticity within the binding pocket. In contrast, the positions analogous to Asp122 and Ser148 in VcNFeoB are conserved in these other bacterial NTPases. Conservation of Asp in this region is unsurprising, as the G4 NxxD motif is required for GTP hydrolysis(44), but Ser is not; it may be possible that the Ser residue is needed to stabilize the “Asp off” conformation for NTPases to facilitate promiscuity in general. Based on these observations, we propose that a combination of these three amino acids in G4 and G5 provide conformational flexibility to allow the utilization of both GTP and ATP for protein function. In the case of FeoB, it is possible that this nucleotide promiscuity may provide an advantage to the several bacteria pathogens that are NTP agnostic. For these organisms, we propose that FeoB could leverage ATP hydrolysis when GTP levels in the cell are low, perhaps as a virulence factor or another adaptive mechanism to ensure Fe2+ uptake across a wide array of conditions, given iron’s essential nature to bacterial function. However, under homeostatic conditions, NTP promiscuous FeoBs likely rely on GTP and are likely still regulated by GDP based on the stable interactions observed in this study. Moreover, given the highly reactive nature of its translocated substrate, regulation of FeoB-mediated Fe2+ uptake by the status of GDP/GTP is presumably an important protective mechanism to prevent iron overload within the cell. It is possible that this intriguing mechanism of FeoB function could be leveraged as a means to combat bacterial virulence in the future.
EXPERIMENTAL PROCEDURES
Cloning of NFeoB Constructs
The VcNFeoB(His)6 WT and N150T variant constructs were cloned into the pET-21a(+) plasmid as described previously (24) based on the sequence of WT VcFeoB (Uniprot ID C3LP27). To create the N-terminal (His)6-SUMO-VcNFeoB fusion, the gene encoding for VcNFeoB was subcloned, and a synthetically added sequence for the Small Ubiquitin-like Modifier (SUMO) protein (Uniprot ID Q12306) was commercially appended (GenScript). The entire sequence was then subcloned into the pET-45b(+) plasmid between the PmlI and PacI restriction sites, which allows for the translation of the N-terminal (His)6-SUMO-VcNFeoB fusion when read in frame.
Expression of NFeoB Constructs
The VcNFeoB(His)6 WT and N150T variant plasmids were separately transformed into BL21(DE3) electrocompetent cells via electroporation, plated on Luria-Bertani (LB) plates supplemented with ampicillin (100 μg/mL), and incubated at 30 ºC overnight. The next day, starter flasks containing 100 mL LB broth and ampicillin (100 μg/mL) were inoculated with a single colony each (WT and N150T) and allowed to grow overnight at 30 ºC with 200 RPM shaking. The next day, 25 mL of the overnight cultures were inoculated into 1 L flasks charged with 1 L LB broth and ampicillin (100 μg/mL, final), and these cells were grown at 37 ºC with shaking of 200 RPM. When OD600 reached 0.4–0.8, the cells in the flasks were then cold shocked at 4 ºC for 2 h before induction with isopropyl β-D-1-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. Cells were then grown for ca. 20 h overnight and harvested the next day by spinning at 5000 xg, resuspended in resuspension buffer (50 mM Tris pH 8.0, 300 mM NaCl, and 10 % (v/v) glycerol) before being flash frozen on N2(l) and stored at - 80 ºC.
All purification steps were conducted at 4 ºC unless otherwise stated. Frozen cells were thawed and diluted to 100 mL with resuspension buffer, and 1 mM (final) phenylmethylsulfonyl fluoride (PMSF) was added prior to sonication at 80 % amplitude, 30 s on pulse, 30 s rest pulse, 12 min total. Lysed cells were clarified by spinning at 163,000 xg for 1 h. The supernatant was then applied to a 5 mL HisTrap HP column (Cytiva) that was pre-charged with Ni2+ and equilibrated with 5 column volumes (CVs) of wash buffer (50 mM Tris pH 8.0, 300 mM NaCl, 10 % (v/v) glycerol and 1 mM TCEP). After the sample was applied, the column was washed with 10 CVs of wash buffer, then with wash buffer containing 50 mM imidazole, and eluted with wash buffer containing 150 mM imidazole. Eluted protein fractions were then pooled, and buffer exchanged into ion exchange wash buffer (50 mM Tris pH 8, 10 % (v/v) glycerol) using a 50 mL HiPrep 26/10 desalting column to remove all salt content before anion exchange chromatography. After desalting, the protein was then applied to a 5 mL HiTrap Q HP anion exchange column (Cytiva) that was then washed extensively with 10 CVs of the ion exchange buffer. The protein was purified via a linear elution gradient from 0 M to 1 M NaCl. Eluted protein fractions were pooled, concentrated via a 10 kDa molecular weight cutoff (MWCO) filter, and injected onto a 120 mL Superdex 75 preparative grade gel filtration column (Cytiva) after equilibration with 1.5 CVs of size exclusion buffer (25 mM Tris pH 8.0, 100 mM NaCl, 5 % (v/v) glycerol and 1 mM TCEP). Fractions corresponding to pure, monomeric VcNFeoB(His)6 were pooled and concentrated via a 10 kDa MWCO filter to ca. 12 mg/mL, aliquoted, flash frozen on N2(l) and stored at −80 ºC. An identical procedure was followed for the VcNFeoB(His)6 N150T variant.
The cellular transformation, expression, cellular harvesting, cellular lysis, and initial Ni2+-based purification of the (His)6-SUMO-VcNFeoB mirrored that of VcNFeoB(His)6. After fractions were eluted from the 5 mL HisTrap HP column, the (His)6-SUMO-VcNFeoB protein was buffer exchanged into SUMO cleavage buffer (50 mM Tris pH 8.0, 300 mM NaCl, 10 % (v/v) glycerol, and 10 mM β-mercaptoethanol (BME)) and concentrated via a 10 kDa MWCO filter. House-made SUMO protease Ulp1 was then added at a 1:100 (mg/mg) ratio and allowed to cleave overnight with gentle rocking at 4 ºC. The next day, the solution was applied again to a 5 mL HisTrap HP column to separate the now cleaved VcNFeoB from any uncleaved protein. Eluted fractions containing cleaved VcNFeoB were concentrated via a 10 kDa MWCO filter injected onto a 120 mL preparative Superdex 75 column, eluted isocratically, pooled, and stored identically to VcNFeoB(His)6 (vide supra).
Crystallization of NFeoB Constructs
Apo VcNFeoB(His)6 was initially thawed and diluted to 10 mg/mL with size exclusion buffer prior to crystallization trials. Several commercial sparse-matrix screens were used to test for crystallization using vapor diffusion in sitting drop format. After incubation at 25 ºC for ca. 4 months, crystals appeared in a condition containing 25 % (w/v) PEG 3350, 0.1 M bis-Tris pH 5.5, and 0.2 M MgCl2. The crystals were then looped, cryo-protected, and frozen in N2(l). Unfortunately, fine screens failed to replicate crystallization for further optimization.
To crystallize GDP-bound VcNFeoB(His)6, protein at 10 mg/mL was incubated with 3 mM GDP for 2 h at room temperature before sparse-matrix screens were used to test for crystallization using vapor diffusion in sitting drop format at 25 ºC. After 2 weeks, small, cubic-shaped crystals appeared in a condition containing 25 % (w/v) PEG 3350, 0.1 M bis-Tris pH 5.5, 0.2 M MgCl2, and 0.1 M LiCl. Crystals were then looped, cryo-protected, and frozen in N2(l).
The preparation of GDP-bound VcNFeoB(His)6 N150T was identical to that of the WT protein. Sparse-matrix screens were used to test for crystallization using vapor diffusion in sitting drop format at 25 ºC. Crystals initially appeared in conditions containing 24 % (w/v) PEG 3350, 0.1 M bis-Tris pH 5.5, 0.03 M MgCl2, and 0.2 M (NH4)2SO4. Crystals matured after two weeks and were then looped, cryo-protected, and frozen in N2(l).
To crystallize SUMO-cleaved VcNFeoB, the protein was initially thawed and incubated with 3 mM ADP prior to dilution to 10 mg/mL. Sparse-matrix screens were used to test for crystallization using vapor diffusion in sitting drop format at 20 ºC. After two weeks of incubation, clustered crystals appeared in a condition containing 30 % (w/v) PEG 2000, 0.1 M Tris pH 8.0. Single crystals were separated manually using crystallization tools after the clusters were transferred to a drop containing cryo-protectant. Separated single crystals were then looped and frozen in N2(l).
X-ray Diffraction, Data Reduction, and Structural Determination
Diffraction data were collected at the Advanced Photon Source (APS), Argonne National Laboratory on LS-CAT beamline 21-ID-D and at Brookhaven National Laboratory beamline 17-ID-2 (FMX). Data were automatically processed using Xia2 (45) and/or AutoProc(46). The initial phases of all datasets were determined by molecular replacement (MR) using Phenix Phaser (47) with an AlphaFold-generated model as an initial search input(48). After an initial MR solution was identified, further model building was accomplished using Phenix AutoBuild(47). The unambiguous presence of GDP in the nucleotide-binding site was confirmed by the generation of Polder maps in Phenix for GDP-bound VcNFeoB(His)6 WT and N150T datasets. The initial placement of GDP was determined based on the structure of E. coli NFeoB bound to GDP (PDB ID 3I8X) that was then further refined. Iterative rounds of manual model building and refinement were accomplished in Coot (49) and Phenix Refine(47), respectively, until model convergence and the final placement of any visible solvent molecules. Ramachandran statistics and clash values were determined from the MolProbity program (50) within the Phenix software suite. The following structures have been deposited in the Protein Data Bank: WT apo VcNFeoB(His)6 (PDB ID 8VWL); WT GDP-bound VcNFeoB(His)6 (PDB ID 8VWN); N150T GDP-bound VcNFeoB(His)6 (PDB ID 9BA7); SUMO-cleaved apo VcNFeoB (PDB ID 9BA6). Data collection and refinement statistics are provided for all structures in SI Table 1.
Isothermal Titration Calorimetry
Purified WT or N150T VcNFeoB(His)6 was diluted to 0.1 mM (3.2 mg/mL) in SEC buffer for all isothermal titration calorimetry (ITC) experiments. Experiments were conducted using the MicroCal PEAQ-ITC Automated (Malvern Panalytical) to probe nucleotide binding. All titrations with nucleotide diphosphates (GDP and ADP) were performed in 25 mM Tris pH 8.0, 100 mM NaCl, 5% (v/v) glycerol and 1 mM TCEP, while experiments involving the triphosphate mimics (GMP-PNP and AMP-PNP) were conducted using the same buffer conditions except with added MgCl2 to 10 mM final concentration. The calorimetry cell was loaded with 200 μL of WT or N150T VcNFeoB(His)6, and 40 μL of nucleotide (GDP, GMP-PNP, ADP, or AMP-PNP) at 2.5 mM concentration (GDP and GMP-PNP) or 5.0 mM concentration (ADP and AMP-PNP) was loaded into the injection syringe. Thermal equilibrium was reached at 25 ºC after an initial 60 s delay followed by 19X 2 μL serial injections into the cell with 150 s interval delays between injection points with high spinning. Data were analyzed using the Malvern MicroCal PEAQ ITC analysis tool and fitted to a binding isotherm that has a single site using the following equations:
| Equation 1 |
Where the heat released, , from the ith injection is represented by .
| Equation 2 |
Where the total heat (Q) is related to the number of sites (n), the fractional occupation (Θ) the total free concentration of the macromolecule (Mt), the molar heat of ligand binding (ΔH), and the volume determined relative to zero for the unbound species (V0).
Bioinformatic Analyses
Based on previous studies in which nucleotide promiscuity of NFeoB from multiple organisms was initially uncovered(28), sequences were obtained from the Uniprot database of intact FeoBs. A multiple sequence alignment was constructed through the EMBL MUSCLE program using default parameters(51). The resultant alignment was visualized via Jalview (52–54) and was then entered into the MEGAX software (55–57) for phylogenetic analysis using the maximum likelihood method and 500 bootstrap iterations with a minimum coverage of 95%. The final phylogenetic results were also visualized using MEGAX.
Structural prediction using AlphaFold3
Predicted VcNFeoB structures with adenosine nucleotides (ATP and ADP) were generated using the AlphaFold3 (58) server by submitting amino acids 1–261 from VcFeoB (Uniprot ID C3LP27) with either ADP and Mg2+ or ATP and Mg2+ and utilizing the default parameters. In all cases, the lowest energy calculated structure is displayed as being representative, but the resulting five calculated structures for each prediction reveal very similar results. The calculated structures were both visualized and analyzed using ChimeraX(59).
Supplementary Material
Acknowledgements:
This work was primarily supported by NSF CAREER grant CHE1844624 (A. T. S.), and in part by NIH-NIGMS grant R35 GM133497 (A.T.S), NIH-NIGMS grant T32 GM066706 (A. T. S. and M.L.), and by NIH-NIAID grants R01 AI091957 and R37 AI016935 (S.M.P.). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). NSLS2 is a U.S.DOE Office of Science User Facility operated under Contract No. DE-SC0012704. This publication resulted from the data collected using the beamtime obtained through NECAT BAG proposal # 311950. Sequence searches utilized both database and analysis functions of the Universal Protein Resource (UniProt) Knowledgebase and Reference Clusters (http://www.uniprot.org) and the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/).
Abbreviations:
- GDP
guanosine diphosphate
- GMP-PNP
5’guanylyl-imidodiphosphate
- GTP
guanosine triphosphate
- ADP
adenosine diphosphate
- AMP-PNP
5’adenylyl-imidodiphosphate
- ATP
adenosine triphosphate
- IMAC
immobilized metal affinity chromatography
- AIEX
anion exchange chromatography
- IPTG
isopropyl β-D-l-thiogalactopyranoside
- NFeoB
soluble N-terminal GTP-binding domain of FeoB
- SUMO
small ubiquitin-like modifier
- Ulp1
Ubl-specific protease
- PMSF
phenylmethylsulfonyl fluoride
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SEC
size-exclusion chromatography
- R.M.S.D.
root-mean-square deviation
- Tris
tris(hydroxymethyl)aminomethane
- TCEP
tris(2-carboxyethyl)phosphine
- ITC
isothermal titration calorimetry
Footnotes
Supporting Information: Supporting information is available online, including:
Data collection and refinement statistics for apo WTVcNFeoB(His)6 (P1 space group), GDP-bound WT VcNFeoB(His)6, cleaved WTVcNFeoB (P121 space group), and apo and GDP-bound N150T VcNFeoB(His)6 (Table S1)
Size-exclusion chromatograms and 15 % SDS-PAGE analyses of purified VcNFeoB constructs (Fig. S1)
Superpositioning of NFeoB domains (Fig. S2)
Quality of the electron density and omit maps surrounding the GDP molecule in WT VcNFeoB(His)6 and N150T VcNFeoB(His)6 (Fig. S3)
Comparison of the GTP-binding pocket of WT VcNFeoB(His)6 in the apo state and the GDP-bound state (Fig. S4)
ITC data of adenine-containing nucleotides titrated into VcNFeoB (Fig. S5)
AlphaFold3 predictions of VcNFeoB bound to adenine nucleotides (Fig. S6)
Altered hydrogen bonding interactions among various bacterial NTPases (Fig. S7)
Conflict of interest: The authors declare no competing financial interests.
Data Availability Statement:
All data are contained within the manuscript, either in the main body or in the Supplemental data submitted with the manuscript, and/or deposited in repositories such as the Protein Data Bank (PDB).
References
- 1.Steimle S., van Eeuwen T., Ozturk Y., Kim H. J., Braitbard M., Selamoglu N. et al. (2021) Cryo-EM structures of engineered active bc1-cbb1 type CIII2CIV super-complexes and electronic communication between the complex Nat Commun 12, 929 10.1038/s41467-021-21051-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Booker S., Broderick J., and Stubbe J. (1993) Ribonucleotide reductases: radical enzymes with suicidal tendencies Biochem Soc Trans 21 (Pt 3), 727–730 10.1042/bst0210727 [DOI] [PubMed] [Google Scholar]
- 3.Glorieux C., and Calderon P. B. (2017) Catalase, a remarkable enzyme: targeting the oldest antioxidant enzyme to find a new cancer treatment approach Biol Chem 398, 1095–1108 10.1515/hsz-2017-0131 [DOI] [PubMed] [Google Scholar]
- 4.Einsle O., and Rees D. C. (2020) Structural Enzymology of Nitrogenase Enzymes Chem Rev 120, 4969–5004 10.1021/acs.chemrev.0c00067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu Y., and Ribbe M. W. (2015) Nitrogenase and homologs J Biol Inorg Chem 20, 435–445 10.1007/s00775-014-1225-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schalk I. J., and Mislin G. L. A. (2017) Bacterial Iron Uptake Pathways: Gates for the Import of Bactericide Compounds J Med Chem 60, 4573–4576 10.1021/acs.jmedchem.7b00554 [DOI] [PubMed] [Google Scholar]
- 7.Cain T. J., and Smith A. T. (2021) Ferric iron reductases and their contribution to unicellular ferrous iron uptake J Inorg Biochem 218, 111407 10.1016/j.jinorgbio.2021.111407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choby J. E., and Skaar E. P. (2016) Heme Synthesis and Acquisition in Bacterial Pathogens J Mol Biol 428, 3408–3428 10.1016/j.jmb.2016.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skaar E. P. (2010) The Battle for Iron between Bacterial Pathogens and Their Vertebrate Hosts PloS Pathogens 6, 1–4 10.1371/journal.ppat.1000949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith A. D., and Wilks A. (2015) Differential contributions of the outer membrane receptors PhuR and HasR to heme acquisition in Pseudomonas aeruginosa J Biol Chem 290, 7756–7766 10.1074/jbc.M114.633495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cescau S., Cwerman H., Létoffé S., Delepelaire P., Wandersman C., and Biville F. (2007) Heme acquisition by hemophores Biometals 20, 603–613 10.1007/s10534-006-9050-y [DOI] [PubMed] [Google Scholar]
- 12.Brown J. B., Lee M. A., and Smith A. T. (2021) Ins and Outs: Recent Advancements in Membrane Protein-Mediated Prokaryotic Ferrous Iron Transport Biochemistry 10.1021/acs.biochem.1c00586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gómez-Garzón C., Barrick J. E., and Payne S. M. (2022) Disentangling the Evolutionary History of Feo, the Major Ferrous Iron Transport System in Bacteria mBio 13, e0351221 10.1128/mbio.03512-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sestok A. E., Linkous R. O., and Smith A. T. (2018) Toward a mechanistic understanding of Feo-mediated ferrous iron uptake Metallomics 10, 887–898 10.1039/c8mt00097b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lau C. K., Krewulak K. D., and Vogel H. J. (2016) Bacterial ferrous iron transport: the Feo system FEMS Microbiol Rev 40, 273–298 10.1093/femsre/fuv049 [DOI] [PubMed] [Google Scholar]
- 16.Hantke K. (1987) Ferrous iron transport mutants in Escherichia coli K12 FEMS Microbiology Letters 44, 53–57 10.1111/j.1574-6968.1987.tb02241.x [DOI] [PubMed] [Google Scholar]
- 17.Sestok A. E., Brown J. B., Obi J. O., O’Sullivan S. M., Garcin E. D., Deredge D. J. et al. (2022) A fusion of the Bacteroides fragilis ferrous iron import proteins reveals a role for FeoA in stabilizing GTP-bound FeoB J Biol Chem 298, 101808 10.1016/j.jbc.2022.101808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sestok A. E., Lee M. A., and Smith A. T. (2020) Prokaryotic ferrous iron uptake: exploiting pools of reduced iron across multiple microbial environments. In Advances in Environmental Microbiology: Microbial Metabolism of Metals and Metalloids, Hurst CJ, ed. [Google Scholar]
- 19.Brown J. B., Lee M. A., and Smith A. T. (2022) The structure of Vibrio cholerae FeoC reveals conservation of the helix-turn-helix motif but not the cluster-binding domain J Biol Inorg Chem 27, 485–495 10.1007/s00775-022-01945-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith A. T., Linkous R. O., Max N. J., Sestok A. E., Szalai V. A., and Chacón K. N. (2019) The FeoC [4Fe-4S] Cluster Is Redox-Active and Rapidly Oxygen-Sensitive Biochemistry 58, 4935–4949 10.1021/acs.biochem.9b00745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weaver E. A., Wyckoff E. E., Mey A. R., Morrison R., and Payne S. M. (2013) FeoA and FeoC are essential components of the Vibrio cholerae ferrous iron uptake system, and FeoC interacts with FeoB J Bacteriol 195, 4826–4835 10.1128/JB.00738-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faruque S. M., Albert M. J., and Mekalanos J. J. (1998) Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae Microbiol Mol Biol Rev 62, 1301–1314 10.1128/MMBR.62.4.1301-1314.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stevenson B., Wyckoff E. E., and Payne S. M. (2016) Vibrio cholerae FeoA, FeoB, and FeoC Interact To Form a Complex J Bacteriol 198, 1160–1170 10.1128/JB.00930-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shin M., Mey A. R., and Payne S. M. (2019) FeoB contains a dual nucleotide-specific NTPase domain essential for ferrous iron uptake Proc Natl Acad Sci U S A 116, 4599–4604 10.1073/pnas.1817964116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gómez-Garzón C., and Payne S. M. (2020) FeoB hydrolyzes ATP and GTP Metallomics 12, 2065–2074 10.1039/d0mt00195c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo M., Han Z., Huang G., Li R., Liu Y., Lu J. et al. (2022) Structural comparison of unconventional G protein YchF with heterotrimeric G protein and small G protein Plant Signal Behav 17, 2024405 10.1080/15592324.2021.2024405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Osaka N., Hirota Y., Ito D., Ikeda Y., Kamata R., Fujii Y. et al. (2021) Divergent Mechanisms Activating RAS and Small GTPases Through Post-translational Modification Front Mol Biosci 8, 707439 10.3389/fmolb.2021.707439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin M., Park J., Jin Y., Kim I. J., Payne S. M., and Kim K. H. (2020) Biochemical characterization of bacterial FeoBs: A perspective on nucleotide specificity Arch Biochem Biophys 685, 108350 10.1016/j.abb.2020.108350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shin M., Jin Y., Park J., Mun D., Kim S. R., Payne S. M. et al. (2021) Characterization of an antibacterial agent targeting ferrous iron transport protein FeoB against Staphylococcus aureus and Gram-positive bacteria ACS Chem Biol 16, 136–149, [DOI] [PubMed] [Google Scholar]
- 30.Hagelueken G., Hoffmann J., Schubert E., Duthie F. G., Florin N., Konrad L. et al. (2016) Studies on the X-Ray and Solution Structure of FeoB from Escherichia coli BL21 Biophys J 110, 2642–2650 10.1016/j.bpj.2016.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ash M. R., Maher M. J., Guss J. M., and Jormakka M. (2011) A suite of Switch I and Switch II mutant structures from the G-protein domain of FeoB Acta Crystallogr D Biol Crystallogr 67, 973–980 10.1107/S0907444911039461 [DOI] [PubMed] [Google Scholar]
- 32.Eng E. T., Jalilian A. R., Spasov K. A., and Unger V. M. (2008) Characterization of a novel prokaryotic GDP dissociation inhibitor domain from the G protein coupled membrane protein FeoB J Mol Biol 375, 1086–1097 10.1016/j.jmb.2007.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hung K. W., Chang Y. W., Eng E. T., Chen J. H., Chen Y. C., Sun Y. J. et al. (2010) Structural fold, conservation and Fe(II) binding of the intracellular domain of prokaryote FeoB J Struct Biol 170, 501–512 10.1016/j.jsb.2010.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guilfoyle A., Maher M. J., Rapp M., Clarke R., Harrop S., and Jormakka M. (2009) Structural basis of GDP release and gating in G protein coupled Fe2+ transport EMBO J 28, 2677–2685 10.1038/emboj.2009.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deshpande C. N., McGrath A. P., Font J., Guilfoyle A. P., Maher M. J., and Jormakka M. (2013) Structure of an atypical FeoB G-domain reveals a putative domain-swapped dimer Acta Crystallogr Sect F Struct Biol Cryst Commun 69, 399–404 10.1107/S1744309113005939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hattori M., Jin Y., Nishimasu H., Tanaka Y., Mochizuki M., Uchiumi T. et al. (2009) Structural basis of novel interactions between the small-GTPase and GDI-like domains in prokaryotic FeoB iron transporter Structure 17, 1345–1355 10.1016/j.str.2009.08.007 [DOI] [PubMed] [Google Scholar]
- 37.Petermann N., Hansen G., Schmidt C. L., and Hilgenfeld R. (2010) Structure of the GTPase and GDI domains of FeoB, the ferrous iron transporter of Legionella pneumophila FEBS Lett 584, 733–738 10.1016/j.febslet.2009.12.045 [DOI] [PubMed] [Google Scholar]
- 38.Hantke K. (1987) Ferrous iron transport mutants in Escherichia coli K12 FEMS Microbiol Lett 44, 53–57 10.1111/j.1574-6968.1987.tb02241.x [DOI] [Google Scholar]
- 39.Kammler M., Schön C., and Hantke K. (1993) Characterization of the ferrous iron uptake system of Escherichia coli J Bacteriol 175, 6212–6219 10.1128/jb.175.19.6212-6219.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Velayudhan J., Hughes N. J., McColm A. A., Bagshaw J., Clayton C. L., Andrews S. C. et al. (2000) Iron acquisition and virulence in Helicobacter pylori: a major role for FeoB, a high-affinity ferrous iron transporter Mol Microbiol 37, 274–286 10.1046/j.1365-2958.2000.01987.x [DOI] [PubMed] [Google Scholar]
- 41.Marlovits T. C., Haase W., Herrmann C., Aller S. G., and Unger V. M. (2002) The membrane protein FeoB contains an intramolecular G protein essential for Fe(II) uptake in bacteria Proc Natl Acad Sci U S A 99, 16243–16248 10.1073/pnas.242338299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Köster S., Wehner M., Herrmann C., Kühlbrandt W., andYildiz O. (2009) Structure and function of the FeoB G-domain from Methanococcus jannaschii J Mol Biol 392, 405–419 10.1016/j.jmb.2009.07.020 [DOI] [PubMed] [Google Scholar]
- 43.Vaccaro F. A., and Drennan C. L. (2022) The role of nucleoside triphosphate hydrolase metallochaperones in making metalloenzymes Metallomics 14, 10.1093/mtomcs/mfac030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peterson Y. K., Hazard S., Graber S. G., and Lanier S. M. (2002) Identification of structural features in the G-protein regulatory motif required for regulation of heterotrimeric G-proteins J Biol Chem 277, 6767–6770 10.1074/jbc.C100699200 [DOI] [PubMed] [Google Scholar]
- 45.Winter G. (2010) Xia2: an expert system for macromolecular crystallography data reduction J Appl Crystallogr 43, 186–190 doi: 10.1107/S0021889809045701 [DOI] [Google Scholar]
- 46.Vonrhein C., Flensburg C., Keller P., Sharff A., Smart O., Paciorek W. et al. (2011) Data processing and analysis with the autoPROC toolbox Acta Crystallogr D Biol Crystallogr 67, 293–302 10.1107/S0907444911007773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N. et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution Acta Crystallogr D Biol Crystallogr 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O. et al. (2021) Highly accurate protein structure prediction with AlphaFold Nature 596, 583–589 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics Acta Crystallogr D, Biol Crystallogr 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 50.Williams C. J., Headd J. J., Moriarty N. W., Prisant M. G., Videau L. L., Deis L. N. et al. (2018) MolProbity: More and better reference data for improved all-atom structure validation Protein Sci 27, 293–315 10.1002/pro.3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edgar R. C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput Nucleic Acids Res 32, 1792–1797 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Troshin P. V., Procter J. B., and Barton G. J. (2011) Java bioinformatics analysis web services for multiple sequence alignment--JABAWS:MSA Bioinformatics 27, 2001–2002 10.1093/bioinformatics/btr304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Troshin P. V., Procter J. B., Sherstnev A., Barton D. L., Madeira F., and Barton G. J. (2018) JABAWS 2.2 distributed web services for Bioinformatics: protein disorder, conservation and RNA secondary structure Bioinformatics 34, 1939–1940 10.1093/bioinformatics/bty045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., and Barton G. J. (2009) Jalview Version 2--a multiple sequence alignment editor and analysis workbench Bioinformatics 25, 1189–1191 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumar S., Stecher G., Li M., Knyaz C., and Tamura K. (2018) MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms Mol Biol Evol 35, 1547–1549 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stecher G., Tamura K., and Kumar S. (2020) Molecular Evolutionary Genetics Analysis (MEGA) for macOS Mol Biol Evol 37, 1237–1239 10.1093/molbev/msz312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tamura K., Stecher G., and Kumar S. (2021) MEGA11: Molecular Evolutionary Genetics Analysis Version 11 Mol Biol Evol 38, 3022–3027 10.1093/molbev/msab120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abramson J., Adler J., Dunger J., Evans R., Green T., Pritzel A. et al. (2024) Accurate structure prediction of biomolecular interactions with AlphaFold 3 Nature 10.1038/s41586-024-07487-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meng E. C., Goddard T. D., Pettersen E. F., Couch G. S., Pearson Z. J., Morris J. H. et al. (2023) UCSF ChimeraX: Tools for structure building and analysis Protein Sci 32, e4792 10.1002/pro.4792 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the manuscript, either in the main body or in the Supplemental data submitted with the manuscript, and/or deposited in repositories such as the Protein Data Bank (PDB).