Abstract
The presence and sequence variation of the murM gene were studied in a large collection (814 strains) of genetically diverse Streptococcus pneumoniae isolates, which included 27 different serogroups and both penicillin-resistant (423 isolates, 67 pulsed-field gel electrophoretic [PFGE] types) and intermediately penicillin-resistant (165 isolates, 66 PFGE types) and penicillin-susceptible (226 isolates, 135 PFGE types) strains. Diversity of the murM sequences was tested by hybridization with mainly two kinds of probes: one derived from the amplification of the nucleotide sequence between nucleotides 201 and 624 in the penicillin-susceptible laboratory strain R36A (murMA probe) and a second probe that amplified the comparable, highly divergent sequence in the penicillin-resistant strain Pen6 (murMB probe). The great majority of the strains (761 of 814), including both penicillin-susceptible and penicillin-resistant isolates, reacted exclusively with the murMA probe. A smaller group of penicillin-resistant strains (48 of 814 isolates) reacted only with the murMB DNA probe, and an additional 5 isolates reacted with both probes. High-pressure liquid chromatography analysis of the peptidoglycan of strains hybridizing with murMB showed that they invariably contained an increased proportion of branched peptides. Complete sequencing of murM from a group of penicillin-resistant isolates allowed the identification of a number of different murMB alleles that differed in the length and exact position of the divergent (Pen6 type) sequences within the particular murM. The close similarity of these divergent sequences in the various murM alleles suggests a possible common heterologous origin.
Alteration of penicillin target proteins, the penicillin-binding proteins (PBPs), as a mechanism of penicillin resistance in Streptococcus pneumoniae, was first demonstrated by titration of the penicillin binding capacity of PBPs in several highly β-lactam-resistant isolates from South Africa (17). The PBPs of resistant strains were shown to have radically reduced affinities and/or binding capacities for the antibiotic molecule. Soon afterwards it was also noted that the cell wall peptidoglycan of the resistant strains from South Africa also had an abnormal chemical composition in which the proportion of branched muropeptides carrying an alanyl-serine or alanyl-alanine substituent on the lysine epsilon amino group of the stem peptide residues was significantly higher than the representation of these branched muropeptides in the cell wall of susceptible pneumococci. Using the South African isolates as DNA donors, both the abnormal low affinity of PBPs and the abnormal composition of the cell wall could be transferred to susceptible pneumococci in a multistep process of genetic transformation in which resistance to penicillin was the selective agent (7, 17).
An increased proportion of branched peptides was subsequently also demonstrated in several additional penicillin-resistant isolates from South Africa, Hungary, and the Czech Republic (6, 14). In contrast, penicillin-susceptible strains of different serotypes, isolation dates, and diverse genetic backgrounds appeared to produce a species-specific pneumococcal peptidoglycan in which the ratio of stem peptide units was preserved and in which branched peptides, while detectable, represented a relatively small proportion of all peptidoglycan building blocks (6, 14). However, the precise relationship between penicillin resistance level and cell wall composition has remained unclear, and examination of a large number of penicillin-resistant clinical isolates suggested that the abnormal cell wall stem peptide composition is related to the particular genetic lineage and is not necessarily a correlate of resistance itself (6, 13, 14).
Recent identification of the murM and murN genes encoding the enzymes involved with the biosynthesis of the branched structured muropeptide components in pneumococci has allowed a reexamination of this question. Inactivation of the murMN operon was shown to result in the formation of a peptidoglycan from which all branched muropeptide components were missing (5). In parallel, there was a complete loss of penicillin resistance in several of the resistant strains examined. The murM gene appears to be responsible for the addition of the first amino acid of the dipeptide bridge (4).
Comparison of the sequence of the murM genes carried by a penicillin-resistant South African isolate with that present in a susceptible S. pneumoniae laboratory strain demonstrated a considerable degree of divergence between the murM sequences of the resistant and susceptible pneumococci. We followed up this observation by examining a large number of penicillin-resistant and -susceptible clinical isolates of S. pneumoniae in order to obtain information about the frequency of mosaicism in the murM genes carried by these strains. The data suggest that the divergent sequences identified in various resistant strains of different genetic backgrounds may have originated from a common heterologous source.
MATERIALS AND METHODS
Strains and growth conditions.
The S. pneumoniae isolates and nonpneumococcal strains were obtained from the Rockefeller University collection. They were grown in a casein-based semisynthetic medium, C+Y, at 37°C without aeration, as previously described (6), or in tryptic soy agar (Difco, Detroit, Mich.) supplemented with 5% sterile sheep blood and incubated at 37°C.
Probes for the murM and murN genes.
DNA probes corresponding to internal fragments of the murM gene were amplified by using chromosomal DNA as a template either from the penicillin-susceptible strain R36A (MurMAI and MurMAII probes) or from the penicillin-resistant construct Pen6 (MurMBI probe). The MurMAI probe was generated by using primers 1 (5′-GCTGGATCCCATGAGAAGTTTGGTGTTTA-3′) and 2 (5′-GCTGAATTCCTGTTCGAATAGCCTGTT-3′) to amplify the R36A murM sequence between nucleotides 201 and 624. The MurMAII probe was generated by using primers 3 (5′-AAAATCATCATCCATACCAGC-3′) and 4 (5′-CAATATGGTGGACTGGAACT-3′) to amplify the R36A murM sequence between nucleotides 627 and 892. The DNA MurMBI probe was generated by using primers 5 (5′-TATGGATCCAGGGGAGAACTTACTGGCTGTGG-3′) and 6 (5′-GCTGAATTCCTTTGTTTCGTGCTGTTCGGATAG-3′) to amplify the Pen6 murM sequence between nucleotides 141 and 548.
A DNA probe corresponding to an internal fragment of the murN gene was generated with primers 7 (5′-TATGGATCCGGTTTCTTCTCGTTCCT-3′) and 8 (5′-GCCGAATTCACCTGTTGTTAAGCCATCA-3′) to amplify the R36A murN sequence between nucleotides 51 and 425.
The conditions for the PCR were 94°C for 5 min; 30 cycles of 94°C for 30 s, 53°C for 30 s, and 72°C for 2 min; and one final extension step of 72°C for 5 min. This PCR program was used for all amplifications, except for the extension time at 72°C, which was different depending on the size of the PCR fragment to be amplified. The 0.4-kb PCR fragments were purified with the Wizard PCR Preps DNA purification system (Promega, Madison, Wis.) and labeled with the ECL (enhanced chemiluminescence) direct labeling kit (Amersham, Little Chalfont, Buckinghamshire, United Kingdom).
PFGE.
Agarose discs for pulsed-field gel electrophoresis (PFGE) were prepared as previously described (15). The preparative procedure was changed for nonpneumococcal strains: lysozyme (final concentration, 100 μg/ml), mutanolysin (100 μg/ml), and affinity-purified pneumococcal amidase (10 μg/ml) were included as well as RNase (50 μg/ml) in the lysis solution, and the discs were incubated at 37°C for 5 h. Restriction of total DNA with SmaI (New England Biolabs) was performed as follows. One agarose disk was transferred into a microcentrifuge tube containing 500 μl of the commercially supplied buffer (50 mM potassium acetate, 20 mM Tris-acetate, 10 mM magnesium acetate, 1 mM dithiothreitol [pH 7.9]) and incubated for at least 1 h at 25°C. The buffer was then removed, and 50 μl of a solution of SmaI in a commercially supplied reaction buffer was added and the mixture was incubated at 25°C for a period of 14 to 16 h. The reaction was stopped by the addition of EDTA to a final concentration of 0.08 M, and the fragments were separated and analyzed as previously described (15).
Southern blot hybridization.
DNA fragments separated by PFGE were transferred to nylon membranes (Hybond N+; Amersham) with the Vacuum Gene System (Pharmacia LKB Biotech, Uppsala, Sweden), according to the manufacturer's instructions. Membranes were hybridized to the murM- and murN-specific DNA probes labeled with the ECL direct labeling system. The hybridization conditions were those recommended by the manufacturer, with a sodium chloride concentration of 0.25 M. The molecular sizes of the hybridization signal(s) and the corresponding SmaI fragments were then determined.
Amplification of the murMN genes.
Chromosomal DNA for PCR amplification of the murMN genes was prepared (12) with primers ZOO7 and ZOO8 described in a previous communication (5). The PCR-amplified fragments were purified with the Wizard PCR Preps DNA purification system, and DNA sequencing was done at the Rockefeller University Protein/DNA Technology Center with the Taq fluorescent dye terminator sequencing method by using a PE/ABI model 377 automated sequencer. Sequences were analyzed with DNASTAR software and the nonrandom distribution of the polymorphic sites by Maximum Chi-Squared (version 1.0) and MEGA software.
Cell wall preparation.
Pneumococcal cell walls were prepared by a previously published method (7, 14), except that breaking the cells was done by shaking the bacterial suspension with acid-washed glass beads with the help of the instrument FastPrep FP120 (Bio 101, La Jolla, Calif.).
Enzymatic digestion of cell walls.
Cell wall material (2 mg) was suspended in 25 mM sodium phosphate buffer (pH 7.4) and treated with affinity-purified pneumococcal amidase (5 μg) at 37°C for 18 to 24 h with constant stirring. The soluble wall material was washed with acetone, and the peptides were extracted with acetonitrile-isopropanol-water (25:25:50) containing 0.1% trifluoroacetic acid as described previously (7, 8, 14). After removal of the solvents by evaporation in a SpeedVac, the peptides were dissolved in 0.1% trifluoroacetic acid.
Separation and analysis of the cell wall stem peptides.
Peptides were separated with a Shimadzu LC-10AVP high-pressure liquid chromatography (HPLC) system on a Vydac 218TP54 column (The Separations Group, Hesperia, Calif.), as described previously (14). The peptides were eluted with an 80-min linear gradient from 0 to 15% acetonitrile (Fisher Scientific) in 0.1% trifluoroacetic acid (Pierce Chemical Co., Rockford, Ill.) pumped at a flow rate of 0.5 ml/min. The eluted fractions were detected and quantified by determination of their UV A210.
Nucleotide sequence accession number.
The sequences of the murM gene from several S. pneumoniae strains have been submitted to the GenBank database under the following accession numbers: Hun663, AF281135; DE1, AF281136; KY4, AF281137; KY17, AF281138; TX7, AF281139; and NE6, AF281140.
RESULTS
Distribution of the murM and murN genes in natural populations of S. pneumoniae.
A large number (814) of S. pneumoniae clinical isolates belonging to 27 different serogroups, including both penicillin-susceptible and -resistant strains, were screened with the DNA probes MurMAI and MurMBI. The collection included 226 penicillin-susceptible strains (MIC, ≤0.06 μg/ml), 165 penicillin intermediate-resistant strains (MIC, 0.12 to 1.0 μg/ml) and 423 penicillin-resistant strains (MIC, >2 μg/ml). The strains screened were genetically diverse: they included 261 different PFGE types and were recovered at a variety of geographic collection sites in the United States, Latin America, and Europe.
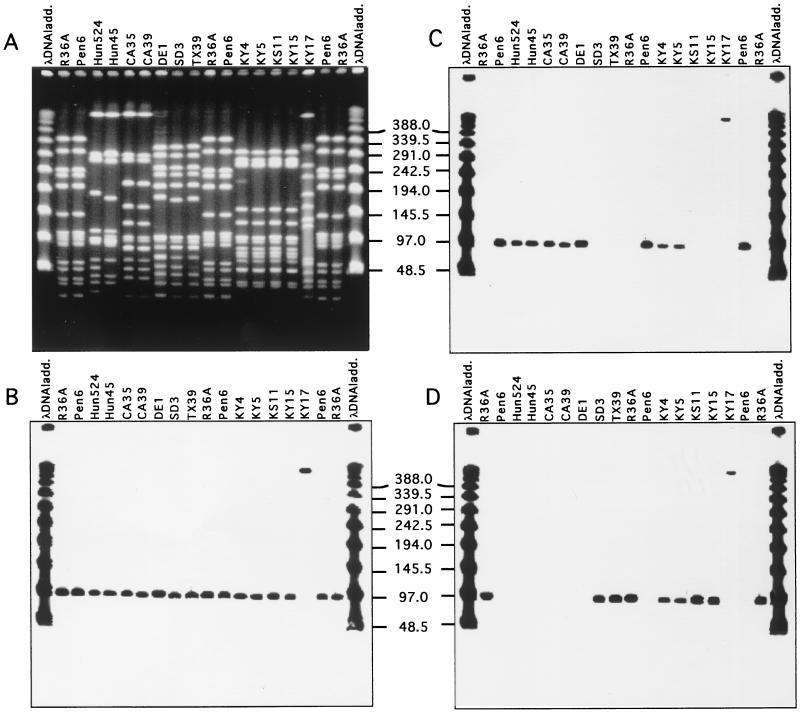
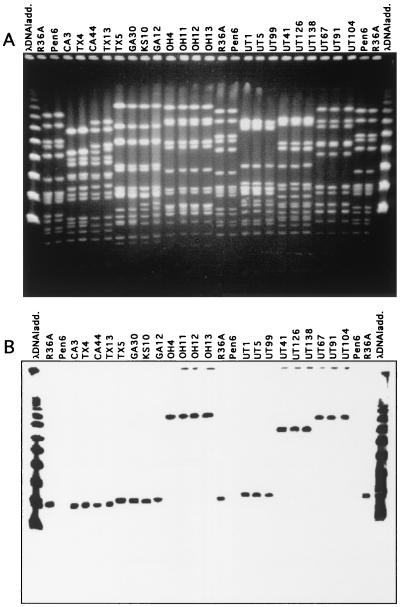
Screening of strains was done by hybridization of PFGE-separated chromosomal SmaI fragments with the MurMAI and/or MurMBI probes (Fig. 1 and 2). Each one of the 814 strains gave a positive hybridization signal with one or the other of the murM DNA probes. All strains gave strong hybridization signals with the murN DNA probe as well, and the murM and murN hybridization signals were always in the same SmaI fragment.
FIG. 1.
Ubiquitous distribution of murN and identification of different murM alleles in penicillin-resistant isolates of S. pneumoniae. DNA fragments separated by PFGE in panel A were transferred to membranes and tested with DNA probes specific for the murN gene (B), murMB1 (C), and murMA (D). A λ DNA ladder (ladd.) was used for size comparisons.
FIG. 2.
Variation in the molecular size of murM-hybridizing SmaI fragments in S. pneumoniae isolates belonging to different clonal types. DNA fragments separated by PFGE in panel A were transferred to membranes and tested with the R36A-specific murMA probe (B).
Screening S. pneumoniae isolates for different alleles of murM.
Comparison of the murM sequences from strains R36A and Pen6 (a genetic transformant constructed with the highly penicillin-resistant South African strain 8249 as a DNA donor) identified two regions in which the sequence of the Pen6 murM gene showed a high degree of divergence (higher than 20% at the DNA level and higher than 10% at the amino acid level) from that of the murM sequence in the penicillin-susceptible strain R36A (5). We constructed DNA probes MurMAI (amplifying the sequence between nucleotides 201 and 624 from strain R36A) and MurMBI (amplifying the sequence between nucleotides 141 and 548 from strain Pen6) to screen pneumococcal isolates by hybridization to one or the other of these DNA probes (Fig. 3). The sequence differences between the two probes were sufficient to distinguish strains carrying the murMA versus murMB type genes by DNA hybridization. The validation of the method was done by testing strains that were known to carry divergent murM alleles (Pen6 and Hun663) together with R36A (data not shown). The method was able to discern the different murM alleles: the MurMBI probe hybridized only with the penicillin-resistant strains Pen6 and Hun663, while the MurMAI probe hybridized only with the penicillin-susceptible strain R36A. Except for the cases of five strains which gave hybridization signals with both probes, no cross hybridization was observed in most of the pneumococcal isolates.
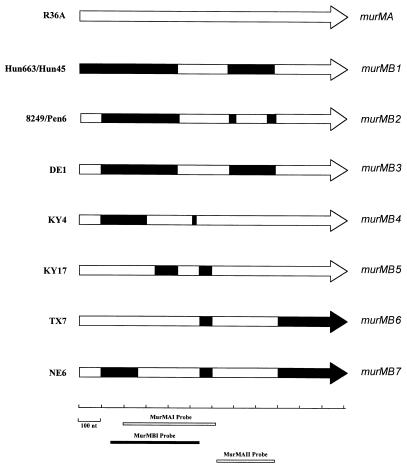
FIG. 3.
Genetic organization of the murM gene alleles in several S. pneumoniae strains and localization of probes used for ECL analysis. Regions in black represent mosaic sequences that are at least 20% divergent from the corresponding sequences of strain R36A.
Detection of several murM alleles among S. pneumoniae isolates.
The great majority of the S. pneumoniae isolates tested (761 of 814) hybridized only with the MurMAI probe. This large group had both penicillin-susceptible and penicillin-resistant isolates, including, among the latter, members of the geographically widely dispersed penicillin-resistant serotype 9 or 14 French/Spanish clone (penicillin MIC, 1 to 2 μg/ml); isolates belonging to the multidrug-resistant serotype 23F Spanish/USA clone, and also multidrug-resistant isolates for which penicillin MICs were extremely high, such as strains GA30 (MIC, 8.0 μg/ml) and BUL125 (MIC, 4.0 μg/ml) (Table 1).
TABLE 1.
Relevant properties of some of the strains analyzed
| Strain | Serotype | PFGE type | Penicillin MIC (μg/ml) | Susceptibility (S) or resistance (R)
|
No. of kb of SmaI hybridization pattern
|
murM allele | Cell wall composition | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Erythro mycin | Tetracycline | Chloramphen- icol | Sulfamethoxazole-trimethoprim | lytA |
murM probe
|
||||||||||
| MurMAI | MurMBI | ||||||||||||||
| Hun663 | 19A | Hun 1A | 2 | R | R | R | NDa | ND | 95 | murMB1 | Abnormally high Ala-Ala | ||||
| Hun524 | 19A | Hun 1A | 2 | R | R | R | R | 280/95 | 95 | murMB1 | Abnormally high Ala-Ala | ||||
| Hun45 | 19 | Hun 1A | 16 | R | R | R | R | 280/95 | 95 | murMB1 | Abnormally high Ala-Ala | ||||
| 8249 | 19A | SA1 | 6 | R | R | R | ND | ND | ND | ND | murMB2 | Abnormally high Ala-Ala/Ala-Ser | |||
| D20 | 19A | SA1 | 12 | R | R | R | ND | ND | ND | ND | murMB2 | Abnormally high Ala-Ala/Ala-Ser | |||
| CA 39 | 6 | 12 | 1.5 | S | S | S | I | >450/80 | 95 | murMB8 | Abnormally high Ala-Ala | ||||
| KY 4 | 23 | USA 5 | 2 | S | S | S | I | 260/85 | 95 | 95 | murMB4 | Abnormally high Ala-Ala/Ala-Ser | |||
| KY 5 | 23 | USA 5 | 3 | S | S | S | I | 260/85 | 95 | 95 | murMB4 | ND | |||
| KS 11 | 23 | USA 5 | 4 | S | S | S | R | 260/85 | 95 | murMA | ND | ||||
| WA2 | 23 | USA 5 | 0.023 | S | S | S | S | 85/70 | 95 | murMA | ND | ||||
| KY 17 | NTb | 30 | 1.5 | R | R | S | R | 450/160/60 | 450 | 450 | murMB5 | Abnormally high Ala-Ser | |||
| DE 1 | 14 | USA 22 | 3 | S | S | S | S | 85 | 95 | murMB3 | Abnormally high Ala-Ala | ||||
| SD 3 | 14 | USA 22 | 4 | R | S | S | R | 85 | 95 | murMA | ND | ||||
| MD1 | 14 | USA 22 | 0.05 | R | S | S | R | 85 | 95 | murMA | ND | ||||
| NE 6 | 6 | USA 10 | 6 | R | R | S | R | 360/100/50 | 95 | 95 | murMB7 | ND | |||
| FL10 | 6 | USA 10 | 2 | R | R | R | R | 360/85 | 95 | murMA | ND | ||||
| WI 3 | 6 | USA 10 | 4 | R | R | R | I | 360/100/50 | 95 | murMA | ND | ||||
| TX 6 | 6 | USA 17 | 6 | R | S | S | R | 105/60 | 95 | murMB6 | ND | ||||
| TX 7 | 6 | 14 | 4 | R | S | S | R | 105 | 95 | murMB6 | ND | ||||
| VP145 | 23 | A | 0.5 | ND | ND | ND | ND | 225/90/30 | 95 | murMA | Normal | ||||
| Sp2150 | 14 | B | 1 | S | S | S | ND | 260/95 | 50 | murMA | Normal, R6Hex | ||||
| TX 3 | 9 | B | 2 | S | S | S | I | 260/130 | 50 | murMA | ND | ||||
| Bul125 | 9 | B | 4 | S | S | S | R | 260/95 | 50 | murMA | Normal, R6Hex | ||||
| TX26 | 6 | USA 20 | 3 | S | S | S | S | 85 | 60 | murMA | ND | ||||
| CA3 | 19 | USA 3 | 3 | R | R | S | R | 115 | 90 | murMA | ND | ||||
| TX 4 | 19 | USA 3 | 2 | R | R | S | R | 115 | 90 | NSc | ND | ||||
| TX 5 | 14 | USA 16 | 6 | R | R | S | R | 95/40 | 95 | NS | ND | ||||
| GA 30 | NT | USA 16 | 8 | R | R | S | R | 95/40 | 95 | NS | ND | ||||
| GA12 | 14 | USA 16 | 4 | R | R | S | R | 95/70 | 95 | NS | ND | ||||
| OH 4 | 6 | USA 9 | 2 | R | S | S | R | 100/60 | 380 | NS | ND | ||||
| OH 11 | 6 | USA 9 | 2 | R | S | S | R | 100/60 | 380 | NS | ND | ||||
| OH 13 | 6 | USA 9 | 2 | R | S | S | R | 100/60 | 380 | NS | ND | ||||
| UT 1 | 19 | UT 20 | 0.008 | S | ND | ND | S | 280/95 | 100 | NS | ND | ||||
| UT 5 | 19 | UT 20 | 0.016 | S | ND | ND | S | 280/95 | 100 | NS | ND | ||||
| UT 41 | 11 | USA 27 | 0.016 | S | ND | ND | S | 85 | 300 | NS | ND | ||||
| UT 126 | 11 | USA 27 | 0.008 | S | ND | ND | S | 85 | 300 | NS | ND | ||||
| UT 67 | NT | USA 39 | 0.016 | S | ND | ND | S | 95 | 350 | NS | ND | ||||
| UT 91 | NT | USA 39 | 0.008 | S | ND | ND | S | 95 | 350 | NS | ND | ||||
ND, not done.
NT, nontypeable.
NS, not sequenced.
A smaller group of the isolates tested (55 of 814) showed different hybridization profiles: 48 of the 55 strains reacted with the MurMBI probe only, and PFGE analysis demonstrated that the great majority (45 of 48) of these strains belonged to the penicillin-resistant serotype 19 Hungarian clone of S. pneumoniae (11). On the other hand, isolates were also detected which showed variation in the murM allele in spite of the fact that they belonged to the same clonal type (as defined by PFGE and other shared properties). Examples of such variation are strains belonging to the PFGE type arbitrarily defined as USA 5. Two of the four such strains examined (KS11 and WA2) reacted only with the MurMAI probe, while two additional strains (KY4 and KY5) gave hybridization with both the MurMAI and MurBI probes. Similar reactions with both the MurMAI and MurMBI DNA probes were also observed in strains KY17 and NE6.
While most frequently the murM (and murN) gene hybridized with the SmaI fragment of 95 kb, variations in the size of the murM reactive fragment were also observed (Table 1).
A number of strains showing anomalous (i.e., murMB type) hybridization patterns as well others that only hybridized with the R36A murM probe (i.e., murMA type) were chosen for full sequencing of the murM alleles, and the positions of the polymorphic sites in the murM alleles of 14 such isolates are shown in Fig. 4. On the basis of sequence differences, the murMB alleles of these strains were assigned numbers, beginning with murMB1 (referring to the polymorphic murM sequence of the Hungarian clone, which was found to be the most divergent from the sequence found in the murMA gene) followed by the next most divergent sequence identified in South African strains 8249 and D20 (murMB2), among others. Some relevant properties of such strains are shown in Table 1.
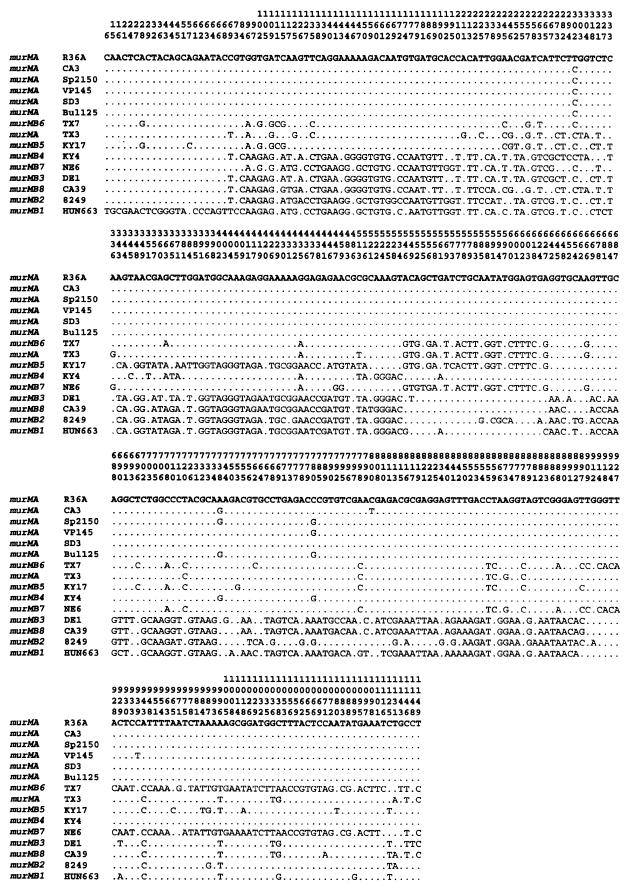
FIG. 4.
Distribution of polymorphic sites among the sequences of the murM alleles. The murM alleles were sequenced from the penicillin-susceptible laboratory strain R36A and from the penicillin-resistant strains listed. Numbers above the sequences identify positions at which a nucleotide alteration was detected in one of the alleles. Only positions with altered nucleotides are shown; residues identical to those in strain R36A are indicated by dots.
The murM genes show a mosaic structure.
The sequences from the different murM alleles were compared to corresponding parts of murMA from R36A. Comparison of the polymorphic sites within the murM gene from the sequenced alleles (Fig. 4) allowed the identification of regions with more than 20% divergence from R36A murMA. Based on these results, a mosaic structure for the murM divergent alleles is proposed. The limits for the divergent regions were defined, taking into account that the nonrandom distribution of the polymorphic sites should be significant (P < 0.01) in the maximum chi-square test (10). Divergent regions are shown pictorially in Fig. 3 and are also described in additional detail below.
Different alleles of murM identified in penicillin-resistant clinical isolates of S. pneumoniae.
The murMB1 allele (Hungarian clone) was very similar to murMB2 (South African clone) and to the alleles carried by strains DE1 (murMB3) and CA39 (murMB8): murMB1 was 97% identical to murMB2 (96% at the amino acid level) and 95% identical to murMB3 between nucleotides 95 and 447 but divergent from murMA (28 to 29% at the DNA level and 32% at the amino acid level). In the region upstream of nucleotide 95, the murMB2 and murMB3 alleles were very similar to murMA, but murMB1 showed extensive (29%) divergence. From nucleotide 447 up to nucleotide 677, there was considerable similarity between the different alleles: murMB1 was 96% identical to the corresponding sequences in murMB2 and murMB3, and all were 92% identical to the corresponding sequence in murMA. Using the MurMAII probe followed by sequencing, it was possible to show that between nucleotides 677 and 892, the alleles of murMB1 and murMB3 were again very divergent from murMA (42% at the DNA level and between 60 and 66% at the amino acid level) but were similar to one another (94% identity). This region had a different organization in the murMB2 allele: between nucleotides 711 and 850, the sequence was only 13% divergent (14% at the amino acid level) from that of murMA, whereas murMB1 and murMB3 showed 36% divergence (49 to 57% at the amino acid level).
Other examples for sequence divergence relative to the sequence of murMA were found in the murM alleles of strains KY4 and KY17. The divergent sequences in these alleles were located in different regions of the gene. The murM gene from KY4 (murMB4 allele) was very divergent between nucleotides 95 and 304 (27% divergence from murMA) but was very similar to other alleles like murMB1 and murMB3 (93 and 99% identity, respectively). The remaining sequence of murM in strain KY4 was very similar to that of murMA, except in the region between nucleotides 512 and 535, where the sequence was very similar to that in murMB1 and murMB3. The murM allele of KY17 (murMB5) was very similar to murMA, except in the regions between nucleotides 342 and 447 and between nucleotides 545 and 603. The first region showed 43% divergence from the sequence in murMA of R36A but was similar to the sequences characteristic of the murMB1 and murMB3 alleles (90 to 95% identity). The second region was divergent from all of the alleles described above (36 to 46% divergence for murMA and murMB1 through murMB4). This region was 96 to 98% identical to the sequence found in the murM alleles of strains TX7 (murMB6) and NE6 (murMB7).
The murMB6 allele was very similar to murMA, except in the region described above, and also in the region downstream of nucleotide 908, where the DNA sequence of murMB6 showed 18% divergence from all other murM alleles. The murMB7 allele (strain NE6) had a very similar sequence to that of murMB6, except between nucleotides 95 and 267, where it closely resembled the sequence of murMB1 through murMB4 (94 to 98% identity).
Strains with abnormal murM alleles show abnormalities in peptidoglycan composition.
Cell wall compositions of pneumococcal isolates carrying different murMB alleles and/or murMA alleles were analyzed by HPLC (Fig. 5). The peptidoglycans of the strains carrying various murMB alleles each had an increased percentage of branched peptides, but the particular branched peptide species showing the largest increase in abundance differed from one isolate to another (Table 2).
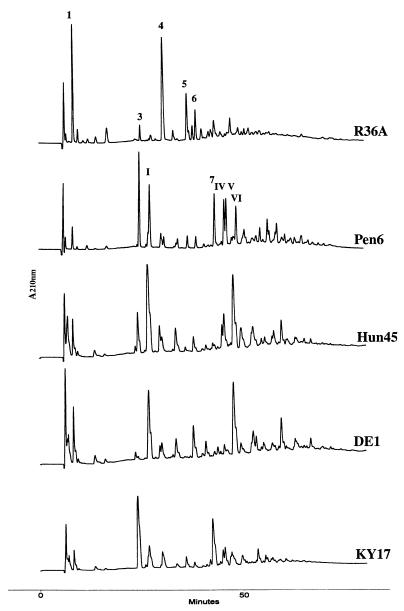
FIG. 5.
HPLC elution profiles of stem peptides of the peptidoglycan from the penicillin-susceptible strain R36A and several penicillin-resistant strains that carry different abnormal murM alleles. Preparation of the cell wall and stem peptide composition analysis were done as described in Materials and Methods.
TABLE 2.
Cell wall peptide composition of several strains of S. pneumoniae
| Peptide | % Peptide in:
|
||||
|---|---|---|---|---|---|
| R36A (murMA) | Pen6 (murMB2) | Hun45 (murMB1) | DE1 (murMB3) | KY17 (murMB5) | |
| 1 | 13.3 | 2.6 | 4.5 | 10.8 | 3.4 |
| 2 | 3.4 | 0.7 | 0.5 | 1.4 | 0.6 |
| 3 | 2.8 | 14.3 | 7.2 | 1.0 | 28.5 |
| I | 1.8 | 14.4 | 24.1 | 23.4 | 11.2 |
| II | 3.4 | 2.7 | 5.1 | 3.1 | 0.8 |
| 4 | 21.0 | 3.2 | 4.4 | 6.0 | 7.2 |
| III | 1.3 | 3.5 | 6.7 | 6.9 | 2.1 |
| 5 | 12.7 | 2.2 | 0.9 | 0.5 | 2.9 |
| 6 | 7.1 | 2.5 | 2.8 | 9.3 | 1.9 |
| 7 | 7.4 | 11.1 | 1.4 | 0.4 | 19.7 |
| IV | 2.6 | 6.5 | 4.7 | 0.6 | 4.1 |
| V | 2.8 | 12.3 | 8.1 | 3.7 | 5.2 |
| 8 | 7.8 | 2.5 | 1.4 | 0.6 | 5.8 |
| VI | 6.6 | 10.1 | 19.1 | 27.5 | 2.0 |
| 9 | 6.0 | 11.6 | 9.0 | 4.9 | 4.5 |
| Total | 100 | 100 | 100 | 100 | 100 |
| Monomers | 26 | 38 | 48 | 47 | 47 |
| Multimers | 74 | 62 | 52 | 53 | 53 |
| Linear | 50 | 12 | 14 | 23 | 17 |
| Branched | 50 | 88 | 86 | 77 | 83 |
| Branched/linear | 1.0 | 7.4 | 6.4 | 3.3 | 4.9 |
The composition of the peptidoglycan from strains belonging to the South African clone (strain 8249, murMB2) or to the Hungarian clone (strain Hun663 or Hun45, murMB1) has already been described (6, 14), but the HPLC profiles and cell wall composition of these strains are included here for comparison. The South African clone has a higher percentage of peptide 3 (relative to the percentage of peptide I), whereas the reverse is true for the Hungarian clone.
Peptidoglycan analyses were also carried out with strains carrying the murMA allele: in each one of the three cases tested (Table 1), the stem peptide compositions were similar to those of strain R36A or R6Hex (data not shown).
DISCUSSION
In a recent communication, we described the identification of two new genes—murM and murN—in the penicillin-susceptible laboratory strain R36A and in the penicillin-resistant South African strain 8249 of S. pneumoniae. Inactivation of the murMN operon caused the virtually complete disappearance of branched structured muropeptides from the cell wall and loss of resistance to penicillin, indicating that these two genes played important roles as determinants of stem peptide structure and in the expression of β-lactam resistance of this bacterium (5). Regions of the murM gene from the penicillin-resistant strain 8249 (or its genetic transformant Pen6) show sequences that were more than 10% divergent at the amino acid level from the corresponding sequence in the penicillin-susceptible strain R36A (5).
In this communication, we followed up these observations in several directions. Testing the large collection of genetically diverse clinical isolates of S. pneumoniae by hybridization with murM- and murN-specific probes demonstrated the ubiquity of these genes in the species.
The use of more selective DNA probes designed to amplify regions in which the murM sequences showed extensive divergence between the murM of the penicillin-susceptible strain R36A and the resistant strain Pen6 has allowed us to determine the frequency with which divergent murM genes are represented in the collection of clinical strains of S. pneumoniae. The great majority (761 of 814) of isolates carried the murMA allele, which was very similar or identical in sequence to that first identified in strain R36A (5). Only 55 strains (seven different PFGE patterns) had murM genes highly divergent from murMA. Strains with divergent murM alleles were associated with unique PFGE types or single isolates within clusters. A particularly noteworthy case is that of the penicillin-resistant Hungarian clone, in which 45 of the 48 strains examined shared the same divergent murM allele.
These findings raise questions about (i) the relationship between cell wall composition and the particular murM allele, (ii) the association between the penicillin resistance phenotype and the divergent murM sequences, and (iii) the evolutionary origin of the divergent murM sequences.
(i) Cell wall composition and the murM allele.
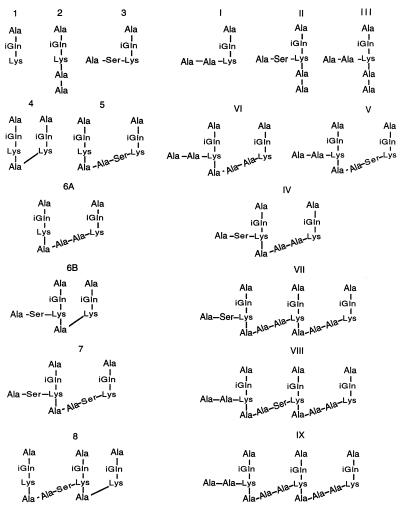
The striking shift towards increased percentages of branched peptides observed in the peptidoglycans of strains carrying a murMB allele suggests a possible correlation between a particular murMB allele and a matching—branched peptide-rich—peptidoglycan composition (Table 2). In strains HUN45 and DE1 carrying the closely related alleles murMB1 and murMB3, the peptidoglycan was enriched for peptide I, a branched peptide with an alanyl-alanine dipeptide substituent which increased from 1.8% (in strains carrying murMA) to 23 to 24% (Table 2). Also noticeable was the increased percentage (from 6.6% to 19 to 27%) of peptide VI, the dimeric species formed by transpeptidation of the monomeric peptide I. No major increase in peptide 3 (another branched peptide in which the bridge is composed of alanine and serine) was observed (Fig. 6). Compositional changes also included a modest decrease in the percentage of linear monomeric tripeptide (peptide 1) from 13.3 to 4.5 or 10.8% and in the linear dimeric peptide (peptide 4) from 21 to 4.4 or 6%. The level of cross-linking was similar to that in the penicillin-resistant strain Pen6, but the ratio of branched to linear peptides was intermediate between values found in Pen6 and R36A.
FIG. 6.
Structures of cell wall stem peptides identified in the pneumococcal peptidoglycan of penicillin-susceptible and -resistant strains of pneumococci. Structural assignments were based on methods described earlier (7, 14).
The peptidoglycan of KY17 (murBM5) was greatly enriched in branched peptide 3 (from 2.8 to 28%) and in the percentage of peptide 7, the dimeric species produced by transpeptidation of two monomeric peptides 3, was also increased (from 7.4 to over 19%). The important role that the murM allele plays as determinant of the unique stem peptide structure of the peptidoglycan is further supported by the results of recent experiments in which different alleles of murM were introduced into the same genetic background (4).
(ii) Penicillin resistance and the divergent murM alleles.
Abnormal murM was detected mainly in historically early isolates of resistant pneumococci (6) and was less frequently seen among more recent isolates, raising the question of whether there is any evolutionary advantage in losing this abnormal murM gene once penicillin resistance is established or at least in keeping it in some particular strains. The three S. pneumoniae isolates DE1, MD1, and SD3 differ in resistance to penicillin and erythromycin and they also carry different murM alleles (Table 1) in spite of the fact that they all belong to the same genetic lineage, called USA 22 (1). One may speculate that the penicillin-resistant (but erythromycin-susceptible) strain DE1 carrying a divergent murMB3 allele may have been the donor of penicillin resistance determinants to the penicillin-susceptible (erythromycin-resistant) strain MD1 that carries a murMA allele, thus generating the penicillin-resistant (and erythromycin-resistant) strain SD3, which has retained the murMA allele (Table 1). These results are reminiscent of the in vitro findings, which have demonstrated that abnormal cell wall composition (abnormal murM) may be separated from penicillin resistance during genetic transformation (13).
(iii) Origin of divergent murM sequences.
The DNA sequences of the several divergent murM alleles were compared with that of the penicillin-susceptible laboratory strain R36A. The upstream divergent regions of murM were very similar in all the various murMB alleles (except murMB6) identified so far (but were located within different regions of the murM gene), suggesting that this divergent region may have a common nonpneumococcal origin similar to the heterologous recombinational events responsible for the generation of mosaic pbp genes in penicillin-resistant pneumococci (2). We cannot rule out the possibility that the downstream divergent region from some murM alleles (murMB7 or murMB6) may have a different origin from the divergent upstream region (murMB1 through murMB5). We searched a group of isolates belonging to different streptococcal species for genes homologous to the murMB1 or murMB2 alleles by hybridization with the murMB-specific probe. No hybridization was obtained among the 16 particular streptococcal isolates that were representatives of the species Streptococcus oralis, Enterococcus faecalis, Enterococcus faecium, Streptococcus bovis, Streptococcus pyogenes, Streptococcus salivarius, Streptococcus mutans, and Streptococcus mitis. Recent findings about the variability within the S. mitis species (9, 16) suggest that a larger collection of these bacteria should be used to search for a putative murMB homologue.
The origin and mode of acquisition of the divergent murM sequences remains to be established. No pbp genes are present within at least a 6-kb distance in the upstream or downstream vicinity of the murMN operon in the preliminary sequence data from the unfinished genome of S. pneumoniae (obtained through The Institute for Genomic Research). This makes it unlikely that the divergent sequences in murM may have resulted from a hitchhiking event driven by a pbp gene, as was reported for the ddl gene (3).
ACKNOWLEDGMENTS
Partial support for these studies was provided by a grant from the National Institutes of Health (RO1 AI37275) and by the Irene Diamond Foundation. S.F. was supported by grant BD/9071/96 from PRAXIS XXI from Fundação para a Ciência e Tecnologia.
We are grateful to Mariana Pinho and Mario Ramirez for suggestions and discussions.
REFERENCES
- 1.Corso A, Severina E P, Petruk V F, Mauriz Y R, Tomasz A. Molecular characterization of penicillin resistant Streptococcus pneumoniae isolates causing respiratory disease in the United States. Microb Drug Resist. 1998;4:325–337. doi: 10.1089/mdr.1998.4.325. [DOI] [PubMed] [Google Scholar]
- 2.Dowson C G, Hutchison A, Brannigan J A, George R C, Hansman D, Liñares J, Tomasz A, Maynard Smith J, Spratt B G. Horizontal transfer of penicillin-binding protein genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1989;86:8842–8846. doi: 10.1073/pnas.86.22.8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Enright M C, Spratt B G. Extensive variation in the ddl gene of penicillin-resistant Streptococcus pneumoniae results from a hitchhiking effect driven by the penicillin-binding protein 2b gene. Mol Biol Evol. 1999;16:1687–1695. doi: 10.1093/oxfordjournals.molbev.a026082. [DOI] [PubMed] [Google Scholar]
- 4.Filipe S R, Pinho M G, Tomasz A. Characterization of the murMN operon involved in the synthesis of branched peptidoglycan peptides in Streptococcus pneumoniae. J Biol Chem. 2000;275:27768–27774. doi: 10.1074/jbc.M004675200. [DOI] [PubMed] [Google Scholar]
- 5.Filipe S R, Tomasz A. Inhibition of the expression of penicillin resistance in Streptococcus pneumoniae by inactivation of cell wall muropeptide branching genes. Proc Natl Acad Sci USA. 2000;97:4891–4896. doi: 10.1073/pnas.080067697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia-Bustos J, Tomasz A. A biological price of antibiotic resistance: major changes in the peptidoglycan structure of penicillin-resistant pneumococci. Proc Natl Acad Sci USA. 1990;87:5415–5419. doi: 10.1073/pnas.87.14.5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Bustos J F, Chait B T, Tomasz A. Altered peptidoglycan structure in a pneumococcal transformant resistant to penicillin. J Bacteriol. 1988;170:2143–2147. doi: 10.1128/jb.170.5.2143-2147.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Bustos J F, Tomasz A. Teichoic acid-containing muropeptides from Streptococcus pneumoniae as substrates for the pneumococcal autolysin. J Bacteriol. 1987;169:447–453. doi: 10.1128/jb.169.2.447-453.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawamura Y, Whiley R A, Shu S E, Ezaki T, Hardie J M. Genetic approaches to the identification of the mitis group within the genus Streptococcus. Microbiology. 1999;145:2605–2613. doi: 10.1099/00221287-145-9-2605. [DOI] [PubMed] [Google Scholar]
- 10.Maynard Smith J. Analysing the mosaic structure of genes. J Mol Evol. 1992;34:126–129. doi: 10.1007/BF00182389. [DOI] [PubMed] [Google Scholar]
- 11.Munoz R, Musser J M, Crain M, Briles D E, Marton A, Parkinson A J, Sorensen U, Tomasz A. Geographic distribution of penicillin-resistant clones of Streptococcus pneumoniae: characterization by penicillin-binding protein profile, surface protein A typing, and multilocus enzyme analysis. Clin Infect Dis. 1992;15:112–118. doi: 10.1093/clinids/15.1.112. [DOI] [PubMed] [Google Scholar]
- 12.Nesin M, Ramirez M, Tomasz A. Capsular transformation of a multidrug-resistant Streptococcus pneumoniae in vivo. J Infect Dis. 1998;177:707–713. doi: 10.1086/514242. [DOI] [PubMed] [Google Scholar]
- 13.Severin A, Figueiredo A M S, Tomasz A. Separation of abnormal cell wall composition from penicillin resistance through genetic transformation of Streptococcus pneumoniae. J Bacteriol. 1996;178:1788–1792. doi: 10.1128/jb.178.7.1788-1792.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Severin A, Tomasz A. Naturally occurring peptidoglycan variants of Streptococcus pneumoniae. J Bacteriol. 1996;178:168–174. doi: 10.1128/jb.178.1.168-174.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soares S, Kristinsson K G, Musser J M, Tomasz A. Evidence for the introduction of a multiresistant clone of serotype 6B Streptococcus pneumoniae from Spain to Iceland in the late 1980s. J Infect Dis. 1993;168:158–163. doi: 10.1093/infdis/168.1.158. [DOI] [PubMed] [Google Scholar]
- 16.Whatmore A M, Efstratiou A, Pickerill A P, Broughton K, Woodard G, Sturgeon D, George R, Dowson C G. Genetic relationships between clinical isolates of Streptococcus pneumoniae, Streptococcus oralis, and Streptococcus mitis: characterization of “atypical” pneumococci and organisms allied to S. mitis harboring S. pneumoniae virulence factor-encoding genes. Infect Immun. 2000;68:1374–1382. doi: 10.1128/iai.68.3.1374-1382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zighelboim S, Tomasz A. Penicillin-binding proteins of multiply antibiotic-resistant South African strains of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1980;17:434–442. doi: 10.1128/aac.17.3.434. [DOI] [PMC free article] [PubMed] [Google Scholar]