Abstract
YsxC is a member of a family of GTP-binding proteins carried by a diverse range of organisms from bacteria to yeasts, plants, and humans. To resolve the issue of whether ysxC of Bacillus subtilis is essential for growth, we attempted to construct mutants in which ysxC was either inactivated or placed under the control of an inducible promoter. Viable mutants were obtained only in the latter case, and these were inducer dependent, demonstrating unambiguously that ysxC is an essential gene.
GTP-binding proteins are frequently involved in regulatory pathways as ubiquitous molecular switches (6), operating at various stages of growth and life cycles. The majority of these proteins are guanine nucleotide-binding proteins, while others use GTP as a substrate for phosphorylation and/or guanylation (10). In Bacillus subtilis, GTP-binding proteins are involved in translation initiation and elongation; cell division; protein secretion via the signal recognition particle pathway; biosynthesis of flagella; the synthesis of adenylosuccinate, pyrimidine, folic acid, and riboflavin; and the oxidation of thiophene (7). The function of several small putative GTP-binding proteins (e.g., Obg, Bex, EngA, YloQ, YsxC, and YyaF) is, as yet, unknown. YloQ is essential for the growth of B. subtilis (2), while Bex has been shown to complement the Escherichia coli essential gene era (EMBL accession no. U18532, available at http://www.embl-heidelberg.de/srs5/). Obg is essential for both growth and sporulation of B. subtilis (18, 21) and is required for the activation of ςB by stress (17).
The putative GTP-binding protein YsxC (also called OrfX [13]) is encoded by a bicistronic operon that includes lonA (Fig. 1A), which encodes a cytosolic ATP-dependent serine endopeptidase (22). ysxC is likely to be transcribed together with lonA, since its start codon overlaps the lonA coding sequence and no ysxC-specific promoter or transcriptional initiation site has been detected. Furthermore, lonA and ysxC showed similar transcription patterns, including induction by heat and other stresses (13).
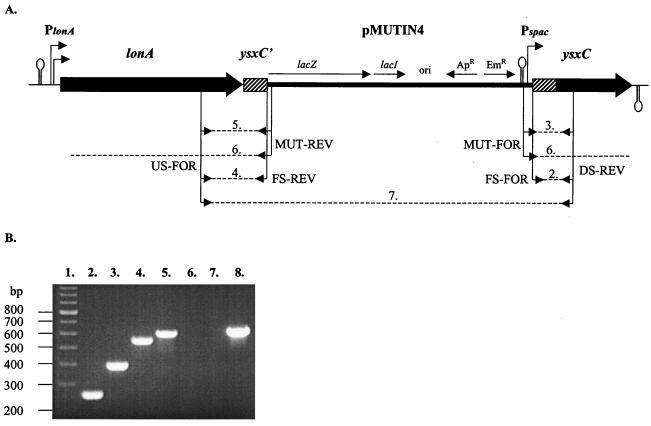
FIG. 1.
Construction of a fusion B. subtilis mutant of ysxC. (A) Schematic representation of the ysxC region of BFA2414 after integration of pYSXCF, which is a pMUTIN4-based integration plasmid. Filled thick arrows indicate structural genes, and putative ρ-independent terminators are shown as stem-loop structures. Two promoters upstream of lonA (PlonA) are marked with fine broken arrows. Striped boxes show the tandem duplication of the RBS and 5′ end of ysxC. ysxC′ is the 5′ end of ysxC. Plasmid pMUTIN4 is shown as a thick line. The lacZ reporter gene, lacI, and ampicillin resistance (Apr) and Emr genes are marked with fine arrows, and promoter Pspac is marked with a fine broken arrow above that of pMUTIN4. The region in pMUTIN4 used for replication in E. coli is labeled “ori,” and three terminators (t1t2t0) upstream of Pspac are indicated as a stem-loop structure. The arrows below the genes indicate the location and orientation of the primers, while the dashed lines indicate their expected PCR products. The positions of the primers specific for B. subtilis 168 in respect to the entire genome (7, 11) are as follows: DS-REV, 2879355 to 2879372; FS-REV, 2879433 to 2879450; FS-FOR, 2879596 to 2879580; and US-FOR, 2879970 to 2879954. The 5′ ends of the forward (FOR) primers included a 10-bp linker with a HindIII restriction site, while the reverse (REV) primers included a 9-bp linker with a BamHI site. Positions of the primers specific for pMUTIN4 were as follows: MUT-FOR, 147 to 165, and MUT-REV, 361 to 379. The numbers above the dashed lines correspond to the lanes of the agarose electrophoresis gel shown in panel B. (B) Diagnostic PCR confirming the correct integration of pYSXCF into ysxC in mutant BFA2414. Lane 1, 100-bp ladder (Amersham Pharmacia Biotech Inc., Little Chalfont, United Kingdom). PCR was performed with BFA2414 (lanes 2 through 7) or B. subtilis 168 (lane 8) chromosomal DNA. The primers (and expected product lengths) were as follows: lane 2, FS-FOR and DS-REV (261 bp); lane 3, MUT-FOR and DS-REV (399 bp); lane 4, US-FOR and FS-REV (557 bp); lane 5, US-FOR and MUT-REV (611 bp); lane 6, MUT-FOR and MUT-REV (no PCR product expected since these primers are oriented away from each other); lane 7, US-FOR and DS-REV (no PCR product expected since the polymerization reaction time of 50 s is too short for the synthesis of the 9,409-bp fragment); and lane 8, US-FOR and DS-REV (635 bp).
The insertional inactivation of ysxC was previously reported to have no effect on growth, nor did it result in any demonstrable phenotype (16). However, in a more recent genome-based approach designed to identify essential genes in E. coli, B. subtilis, and Saccharomyces cerevisiae, ysxC of B. subtilis was reported to be essential (2). In that study, which lacked experimental data, of six genes of unknown function that were essential for the growth of E. coli, five orthologs were essential for B. subtilis and one was essential for S. cerevisiae. Three of the five essential B. subtilis genes, namely, obg, yloQ, and ysxC, encode putative GTP-binding proteins, while yrrA (now called trmU, SWISS-PROT accession no. O35020) encodes a putative tRNA, (5-methylaminomethyl-2-thiouridylate)-methyltransferase, and ydiE encodes a putative metalloprotease (probably o-sialoglycoprotein endopeptidase). These conserved essential bacterial genes with nonessential orthologs in yeast represent potential targets for novel broad-spectrum antimicrobial agents (6).
Construction of integrational mutations in ysxC.
To determine whether ysxC is essential for growth of B. subtilis 168, integrational mutants were constructed in which ysxC was either inactivated (knockout mutant) or placed under the control of a tightly regulated Pspac promoter (23) (fusion mutant). In the case of the knockout mutant, a 303-bp internal fragment of ysxC (bp 2879356 to 2879054 [7]) was cloned into the integrational plasmid pMUTIN4 (20), resulting in plasmid pYSXCK (Table 1). For the construction of the fusion mutant, a 164-bp fragment from the 5′ end of ysxC, incorporating the ribosome binding site (RBS) and start codon of ysxC, was also cloned into pMUTIN4, resulting in plasmid pYSXCF (Table 1).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| B. subtilis | ||
| 168 | trpC2 | 1 |
| BFA2414 | trpC2 ysxC::pYSXCF Emr | This study (Fig. 1) |
| BFA2414(p65) | trpC2 ysxC::pYSXCF p65 Emr Kmr | This study |
| BFA2414SUP(p65) | trpC2 ysxC::pYSXCF p65 Emr Kmr, constitutive Pspac promoter | This study |
| Plasmids | ||
| pMUTIN4 | Apr EmrspoVG-lacZ Pspac (8.61 kb) | 20 |
| pYSXCK | pMUTIN4 containing a 303-bp internal fragment of ysxC (8.913 kb) | This study |
| pYSXCF | pMUTIN4 containing a 164-bp fragment with RBS and 5′ end of ysxC (8.774 kb) | This study |
| p65 | pUB110-based, Kmr PpenP::lacI (4.962 kb) | S. D. Ehrlich |
Kmr, kanamycin resistance.
If ysxC is essential for the growth of B. subtilis 168, it should not be possible to isolate an integrational mutant with pYSXCK, while integration of pYSXCF should lead to isopropyl-β-d-thiogalactopyranoside (IPTG)-dependent growth. Repeated but unsuccessful attempts were made to generate a pYSXCK-based knockout mutant, irrespective of the presence of IPTG. In the case of the pYSXCF-based fusion mutant, about 100 erythromycin-resistant (Emr) and lincomycin-resistant (Lmr) transformants per μg of pYSXCF DNA were isolated in the presence of IPTG, while none were isolated in its absence. These data indicate that ysxC is essential for growth. The authenticity of the integration event in this mutant (BFA2414) (Fig. 1A) was confirmed by PCR (Fig. 1B). To ensure tight regulation of the Pspac promoter, BFA2414 was transformed with plasmid p65, which provides multiple copies of the E. coli lacI gene.
Expression of ysxC is essential for growth.
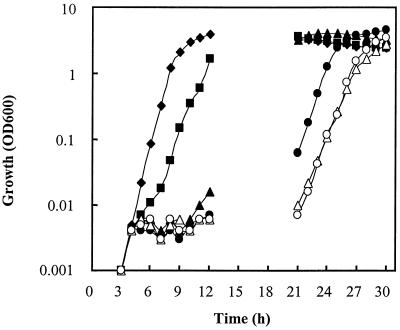
The IPTG dependence of BFA2414(p65) was confirmed by growing the organism to exponential phase (optical density at 600 nm [OD600] = ∼0.3) in Luria-Bertani (LB) medium (15) containing 1 mM IPTG and 0.3 μg of erythromycin and 10 μg of kanamycin per ml. The cells were washed twice with prewarmed LB medium and diluted 10−5-fold into prewarmed LB media containing erythromycin, kanamycin, and a range of concentrations of IPTG from 0 to 1 mM. As shown in Fig. 2, growth was eventually observed in each of the cultures. However, the time at which growth was first observed was increasingly delayed with decreasing IPTG concentrations. In the case of the culture with 0.1 mM IPTG, growth was delayed by approximately 2 h with respect to the culture containing 1 mM IPTG, while growth of the culture with no added IPTG was delayed by more than 20 h. During exponential phase, the mean generation times of cultures with smaller amounts (≤0.1 mM) or no IPTG was increased to ∼50 min, compared to ∼30 min for the culture with 1 mM IPTG.
FIG. 2.
OD600 of BFA2414(p65) in LB media with the following concentrations of IPTG: 1 mM (⧫), 0.1 mM (■), 0.01 mM (▴), 0.001 mM (●), 0.0001 mM (▵), and 0 mM (○).
The transcription of ysxC was monitored (9) by fusing the spoVG-lacZ reporter gene of pMUTIN4 to its native promoter (Fig. 1A). Irrespective of the IPTG concentration, β-galactosidase production increased during exponential phase, reaching a peak of ca. 22 nmol of ONP/min/OD600 unit at or about the transition between exponential and stationary growth phase; thereafter, the values declined.
To determine whether the delayed growth and decreased growth rate of BFA2414(p65) at the lower (i.e., ≤0.1 mM) IPTG concentrations (Fig. 2) were due to overgrowth by suppressor mutants, stationary-phase samples were plated to determine the ratio of the IPTG-independent colonies (putative suppressor mutants) to total CFU. There was a marked difference in the plating efficiencies of the various cultures. In the case of the culture with 1 mM IPTG, the plating efficiency was (4.9 ± 1.2) × 10−7. The number of the IPTG-independent colonies increased as the IPTG concentration decreased, and cultures with 0.001 mM IPTG or less exhibited a plating efficiency close to 1. These data suggest that the growth in the latter cultures was due to the accumulation of derivatives with suppressor mutations.
To determine the location of putative suppressor mutation(s), chromosomal DNA from IPTG-independent colonies [BFA2414SUP(p65)] was used to transform B. subtilis 168. In each case, similar numbers (ca. 103/μg of DNA) of Emr Lmr transformants were observed, irrespective of the presence of IPTG. In contrast, when chromosomal DNA from the IPTG-dependent BFA2414 was used, transformants (ca. 9 × 102/μg of DNA) were obtained only in the presence of IPTG. These results suggested that the observed suppression was linked to the integrated pYSXCF.
One possibility was that the suppressor mutation(s) occurred in the “oid” lac operator (14) of the Pspac promoter, leading to its constitutive expression. The oid, or ideal, lac operator has perfect symmetry and a 10-fold-higher affinity for the Lac repressor than the native lac operator. Consequently, the native lac operator associated with the Pspac promoter of integrational vector pMUTIN2 was replaced by the oid operator in pMUTIN4 to reduce the noninduced level of expression of this promoter (20). The Pspac promoter regions from several IPTG-independent BFA2414SUP(p65) mutants were PCR amplified using primers MUT-FOR and DS-REV (Fig. 1A). Sequencing of the PCR products revealed that all of the BFA2414SUP(p65) mutants contained a single C→T transition at nucleotide 10 of the oid lac operator. An identical base pair substitution at the same nucleotide of the native lac operator has been shown to decrease its affinity for the Lac repressor by 96% and to generate a constitutive phenotype (3). We therefore concluded that the observed IPTG-independent growth is due to the selection of clones with a mutation in the integrated pYSXCF that severely reduces the capacity of the lac operator upstream of the functional ysxC gene to bind the lactose repressor. Since a single spontaneous mutation in the lac operator of pMUTIN4 can result in the loss of the IPTG dependence of target gene expression, this may lead to an underestimation of the number of essential genes when a gene fusion rather than a gene knockout strategy is initially used to isolate such mutants. Our results indicated that appropriate care needs to be taken when selective pressure is applied to this controllable promoter system.
YsxC protein family.
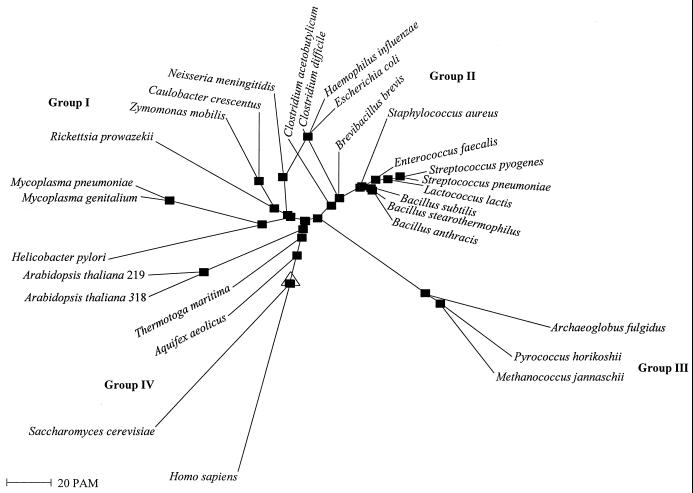
YsxC is a member of a family of small GTP-binding proteins that are carried by a diverse range of organisms from bacteria to yeast, plants, and humans. An analysis of 29 members of the YsxC protein family (Fig. 3) shows that they can be classified into four distinct phylogenetic groups. Group I includes YsxC orthologs from gram-negative bacteria and Mycoplasma species, group II includes orthologs from gram-positive bacteria (e.g., Bacillus, Clostridium, and Staphylococcus), group III includes orthologs from the Archaea, and group IV includes orthologs from S. cerevisiae, Homo sapiens, Arabidopsis thaliana, and, interestingly, two hyperthermophilic bacteria, Aquifex aeolicus and Thermotoga maritima.
FIG. 3.
Radial phylogenetic tree of the YsxC protein family. Multiple-sequence alignment and phylogenetic analysis were performed using the web site of the Institut National de la Recherche Agronomique (http://www.toulouse.inra.fr/multalin.html; see reference 4). The symbol comparison table was Blosum62, the gap weight was 12, and the gap length weight was 2. The root of the tree is marked with a triangle. Proteins with SWISS-PROT accession numbers (in parentheses) are as follows: B. subtilis YsxC (P38424), E. coli YihA (P24253), Haemophilus influenzae HI1118 (P46453), Helicobacter pylori HP1567 (O26087), Mycoplasma genitalium MG335 (P47577), M. pneumoniae MP359 (P75303), Methanococcus jannaschii MJ0320 (Q57768), Rickettsia prowazekii RP102 (Q9ZE46), A. aeolicus AQ_1815 (O67679), and Caulobacter crescentus CgpA (Q9ZG89). Proteins with GenBank accession number are as follows: Archaeoglobus fulgidus AF1326 (AAB89919), P. horikoshii PH0200 (BAA29269), T. maritima TM1466 (AAD36534), L. lactis (AAF63739), Neisseria meningitidis NMB1806 (AAF42143), Zymomonas mobilis CgpA (AAD56911), S. cerevisiae Ydr336wp (AAB64772), H. sapiens HSPC135 (AAF29099), A. thaliana 219 is At2g22870 (AAC32434) with a length of 219 amino acid residues, and A. thaliana 318 is F15N18_70 (CAB87708) with a length of 318 amino acid residues. YsxC orthologs of B. anthracis, B. stearothermophilus, C. acetobutylicum, C. difficile, E. faecalis, S. aureus, S. pneumoniae, and S. pyogenes were obtained from unfinished genome sequencing projects at TIGR, the Sanger Centre, Genome Therapeutics Corporation, and the University of Oklahoma. In B. stearothermophilus, the 130 amino acids at the N terminus of the YsxC homolog were deduced from sequence contig 552. In B. brevis, the 162 amino acids at the N terminus were deduced from the GenBank sequence (D00863) after inserting one nucleotide between positions 2745 and 2746 (16). PAM, percent accepted mutation.
In group II organisms, ysxC is located downstream of lonA or clpX, both of which code for class III ATP-dependent heat shock proteases. In the case of Bacillus and Brevibacillus, ysxC is located downstream of lonA, and its start codon overlaps the 3′ end of the lonA coding sequence by one nucleotide. In Clostridium difficile and Clostridium acetobutylicum, the coding sequence of ysxC overlaps the 3′ end of lonA by eight nucleotides. In Streptococcus spp., Enterococcus faecalis, Staphylococcus aureus, and Lactococcus lactis, ysxC is in a putative operon with clpX. In L. lactis the start codon of ysxC overlaps the 3′ end of the clpX coding sequence by one nucleotide. In the other three phylogenetic groups (I, III, and IV), lonA or clpX is located at a distal site on the chromosome with respect to ysxC. Linkage between an ATP-dependent protease and ysxC was also observed in Pyrococcus horikoshii from group III, in which the ysxC ortholog, PH0200, is located 59 bp downstream of a gene encoding a putative regulatory subunit of the ATP-dependent 26S protease. In addition to YsxC orthologs in eukaryotes, two bacterial homologs from A. aeolicus and T. maritima belong to group IV. A. aeolicus is one of the earliest diverging and most thermophilic bacteria known, and as a chemolithoautotroph, it can grow on hydrogen, oxygen, carbon dioxide, and mineral salts (5). T. maritima is one of the deepest and most slowly evolving lineages in the Eubacteria. Although the core of T. maritima may be eubacterial, almost one quarter of the genome is archaeal in nature (12). A. thaliana has at least two homologs of YsxC, one 219 and the other 318 amino acid residues in length, encoded on chromosomes II and V, respectively (8). The length of the smaller homolog is in the range of prokaryotic YsxC proteins (190 to 219 amino acids). The length of the longer YsxC homolog is similar to that of the YsxC orthologs from S. cerevisiae and H. sapiens. With respect to YsxC of B. subtilis, these proteins have a 100- to 110-amino-acid extension at their amino terminus. Their N termini, which are highly conserved in S. cerevisiae and H. sapiens but not in A. thaliana, showed no homology to any other bacterial and archaeal proteins. In the case of A. thaliana, the N terminus contained a putative transmembrane helix (19) between amino acid residues 11 and 29 and the protein is currently the only putative membrane-bound member of the YsxC family.
No homologs of ysxC were observed on the complete genome sequences of Borrelia burgdorferi, Chlamydia trachomatis, Chlamydia pneumoniae, Deinococcus radiodurans, Mycobacterium tubercolosis, and Treponema pallidum using databases at The Institute for Genomic Research (TIGR; http://www.tigr.org/tdb) and the Pasteur Institute (http://genolist.pasteur.fr).
An alignment of YsxC and 28 homologs (data not shown) revealed four regions of conservation: (i) (G/R)X(S/T)N(V/A)GKS(S/T), a putative GTP-binding motif located toward the amino terminus; (ii) PGXTXXX(N/I), located 15 to 23 residues downstream of the first region; (iii) DXPG(Y/F)G(Y/F), a second putative GTP-binding motif located 10 to 20 residues downstream of the second region; and (iv) KXDK, located 56 to 74 residues downstream of the third motif. The second motif is shorter in S. cerevisiae (GXTXXXN), and only the threonine residue is conserved in the case of H. sapiens.
We were not able to generate a viable mutant of ysxC with pYSXCK, which, after integration into the B. subtilis chromosome, generates a strain carrying a YsxC protein that is truncated at its C terminus by just 23 amino acids. Since this protein includes the four conserved motifs described above, this indicates that the highly charged (5 K residues, 2 E residues, 1 D residue, 1 R residue, and a serine dyad) C terminus is essential for function.
Acknowledgments
We thank S. D. Ehrlich for the gift of plasmids pMUTIN4 and p65. We acknowledge the release of preliminary sequence data for the YsxC orthologs in Bacillus anthracis, Bacillus stearothermophilus, C. acetobutylicum, C. difficile, E. faecalis, S. aureus, Streptococcus pneumoniae, and Streptococcus pyogenes, which were obtained using the TIGR website (http://www.tigr.org) from unfinished genome sequencing projects at TIGR, the Sanger Centre, Genome Therapeutics Corporation, and the University of Oklahoma.
The work was funded by the European Commission (BIO4-CT95-0278).
REFERENCES
- 1.Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arigoni F, Talabot F, Peitsch M, Edgerton M D, Meldrum E, Allet E, Fish R, Jamotte T, Curchod M L, Loferer H. A genome-based approach for the identification of essential bacterial genes. Nat Biotechnol. 1998;16:851–856. doi: 10.1038/nbt0998-851. [DOI] [PubMed] [Google Scholar]
- 3.Barkley M D, Bourgeois S. Repressor recognition of operator and effectors. In: Miller J H, Reznikoff W S, editors. The operon. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1978. pp. 177–220. [Google Scholar]
- 4.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olsen G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 6.Koonin E V. Genomic microbiology: right on target? Nat Biotechnol. 1998;16:821–822. doi: 10.1038/nbt0998-821. [DOI] [PubMed] [Google Scholar]
- 7.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Cummings N J, Daniel R A, Denizot F, Devine K M, Dusterhoft A, Ehrlich S D, Emmerson P T, Entian K D, Errington J, Fabret C, Ferrari E, Foulger D, Fritz C, Fujita M, Fujita Y, Fuma S, Galizzi A, Galleron N, Ghim S Y, Glaser P, Goffeau A, Golightly E J, Grandi G, Guiseppi G, Guy B J, Haga K, Haiech J, Harwood C R, Henaut A, Hilbert H, Holsappel S, Hosono S, Hullo M F, Itaya M, Jones L, Joris B, Karamata D, Kasahara Y, Klaerr-Blanchard M, Klein C, Kobayashi Y, Koetter P, Koningstein G, Krogh S, Kumano M, Kurita K, Lapidus A, Lardinois S, Lauber J, Lazarevic V, Lee S M, Levine A, Liu H, Masuda S, Mauel C, Medigue C, Medina N, Mellado R P, Mizuno M, Moestl D, Nakai S, Noback M, Noone D, O'Reilly M, Ogawa K, Ogiwara A, Oudega B, Park S H, Parro V, Pohl T M, Portetelle D, Porwollik S, Prescott A M, Presecan E, Pujic P, Purnelle B, Rapoport G, Rey M, Reynolds S, Rieger M, Rivolta C, Rocha E, Roche B, Rose M, Sadaie Y, Sato T, Scanlan E, Schleich S, Schroeter R, Scoffone F, Sekiguchi J, Sekowska A, Seror S J, Serror P, Shin B S, Soldo B, Sorokin A, Tacconi E, Takagi T, Takahashi H, Takemaru K, Takeuchi M, Tamakoshi A, Tanaka T, Terpstra P, Tognoni A, Tosato V, Uchiyama S, Vandenbol M, Vannier F, Vassarotti A, Viari A, Wambutt R, Wedler E, Wedler H, Weitzenegger T, Winters P, Wipat A, Yamamoto H, Yamane K, Yasumoto K, Yata K, Yoshida K, Yoshikawa H F, Zumstein E, Yoshikawa H, Danchin A. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 8.Lin X, Kaul S, Rounsley S, Shea T P, Benito M I, Town C D, Fujii C Y, Mason T, Bowman C L, Barnstead M, Feldblyum T V, Buell C R, Ketchum K A, Lee J, Ronning C M, Koo H L, Moffat K S, Cronin L A, Shen M, Pai G, Van Aken S, Umayam L, Tallon L J, Gill J E, Venter J C, et al. Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature. 1999;402:761–768. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- 9.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 10.Mitchell C, Vary J C. Proteins that interact with GTP during sporulation of Bacillus subtilis. J Bacteriol. 1989;171:2915–2918. doi: 10.1128/jb.171.6.2915-2918.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moszer I, Kunst F, Danchin A. The European Bacillus subtilisgenome sequencing project: current status and accessibility of the data from a new World Wide Web site. Microbiology. 1996;142:2987–2991. doi: 10.1099/13500872-142-11-2987. [DOI] [PubMed] [Google Scholar]
- 12.Nelson K E, Clayton R A, Gill S R, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Nelson W C, Ketchum K A, McDonald L, Utterback T R, Malek J A, Linher K D, Garrett M M, Stewart A M, Cotton M D, Pratt M S, Phillips C A, Richardson D, Heidelberg J, Sutton G G, Fleischmann R D, Eisen J A, Fraser C M, et al. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 13.Riethdorf S, Völker U, Gerth U, Winkler A, Engelmann S, Hecker M. Cloning, nucleotide sequence, and expression of the Bacillus subtilis longene. J Bacteriol. 1994;176:6518–6527. doi: 10.1128/jb.176.21.6518-6527.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sadler J R, Sasmor H, Betz J L. A perfectly symmetric lac operator binds the lacrepressor very tightly. Proc Natl Acad Sci USA. 1983;80:6785–6789. doi: 10.1073/pnas.80.22.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 16.Schmidt R, Decatur A L, Rather P N, Moran C P, Jr, Losick R. Bacillus subtilis Lon protease prevents inappropriate transcription of genes under the control of the sporulation transcription factor ςG. J Bacteriol. 1994;176:6528–6537. doi: 10.1128/jb.176.21.6528-6537.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scott J M, Haldenwang W G. Obg, an essential GTP binding protein of Bacillus subtilis, is necessary for stress activation of transcription factor ςB. J Bacteriol. 1999;181:4653–4660. doi: 10.1128/jb.181.15.4653-4660.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trach K, Hoch J A. The Bacillus subtilis spo0Bstage 0 sporulation operon encodes an essential GTP-binding protein. J Bacteriol. 1989;171:1362–1371. doi: 10.1128/jb.171.3.1362-1371.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tusnády G E, Simon I. Principles governing amino acid composition of integral membrane proteins: applications to topology prediction. J Mol Biol. 1998;283:489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- 20.Vagner V, Dervyn E, Ehrlich S D. A vector for systematic inactivation in Bacillus subtilis. Microbiology. 1998;144:3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 21.Vidwans S J, Ireton K, Grossman A D. Possible role for the essential GTP-binding protein Obg in regulating the initiation of sporulation in Bacillus subtilis. J Bacteriol. 1995;177:3308–3311. doi: 10.1128/jb.177.11.3308-3311.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wipat A, Carter N, Brignell S C, Guy B J, Piper K, Sanders J, Emmerson P T, Harwood C R. The dnaB-pheA (256°-240°) region of the Bacillus subtilischromosome containing genes responsible for stress responses, the utilization of plant cell walls and primary metabolism. Microbiology. 1996;142:3067–3078. doi: 10.1099/13500872-142-11-3067. [DOI] [PubMed] [Google Scholar]
- 23.Yansura D G, Henner D J. Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc Natl Acad Sci USA. 1984;81:439–443. doi: 10.1073/pnas.81.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]