Abstract
Background:
Histones have been associated with human diseases. However, the implication of extranuclear histone proteins and their potential mechanism in the pathophysiology of chronic rhinosinusitis (CRS) have not been thoroughly investigated. This study was designed to evaluate the role of histones in patients with CRS by comparing histone expression between patients and controls.
Methods:
Nasal polyp (NP) tissues were obtained, and their comprehensive gene expression profiles were investigated by microarray analysis. Differences in expression were verified by reverse transcriptase polymerase chain reaction and immunohistochemical staining. Cell culture and flow cytometry were used to evaluate the role of histones in the pathogenesis of polyps.
Results:
Significant differences in the microarray analysis were observed between the patient and control groups (P < 0.01). It was found by flow cytometry that the histone (H2BK) can promote cell apoptosis in NPs.
Conclusion:
Our results indicate that reduced expression of H2BK may contribute to the imbalance process of cell proliferation and apoptosis in CRS with NP.
Keywords: Apoptosis, histones, microarray analysis, nasal polyps, sinusitis
1. Introduction
Chronic rhinosinusitis (CRS) is a heterogeneous disorder characterized by inflammation of the nasal mucosa and paranasal sinuses, and it is generally classified into 2 subtypes based on the presence or absence of nasal polyps (NPs): CRS with NPs (CRSwNP) and CRS without NPs (CRSsNP) [1]. CRS affects approximately 5% to 15% of the general population, causes considerable impairment of performance and loss of quality of life, and is associated with high socioeconomic costs [2]. Although the pathogenic mechanisms underlying CRS pathogenesis have been extensively investigated, the etiology of NPs is still unknown.
Histones are evolutionarily conserved proteins that are major components of the nucleosome structure in eukaryotic cells. Posttranslational modifications (PTMs) of histone, such as acetylation, methylation, phosphorylation, and ubiquitination, play important roles in gene transcription regulation [3]. Histone PTMs can change the chromatin status to yield higher transcriptional activity or heterochromatin with lower transcriptional activity, resulting in gene repression or activation [4]. There is evidence pointing to the occurrence of histone PTMs in asthma [5]. Histone modifications have been shown to affect the differentiation of CD4+ and CD8+ cells, associated with asthma-related T cell and dendritic cell pathologies [6]. In addition to nuclear function, extranuclear histones released from activated immune cells have been demonstrated to be potential mediators of infection, sterile inflammation, antimicrobial activity, and cell apoptosis [7-9].
Histone H2BK(HIST1H2BK), also known as H2BC12, encodes a replication-dependent histone and is a member of the histone H2B family. Histones are the fundamental structural components of chromatin. Eukaryotic DNA has an octamer formed by 4 core histones (H2A, H2B, H3, and H4) tangled around the core histones. Histones H2A and H2B are located around the nucleosome, while their core regions include histones H3 and H4. HIST1H2BK(H2BK) is a member of histone H2B, encoding a replication-dependent histone that plays an indispensable role in processes related to transcription regulation, DNA repair, DNA replication, and chromosomal stability. Also, its functions involve the formation of a functional antibacterial barrier in the colon epithelium, as well as the bactericidal activity of amniotic fluid.
Although intranuclear and extranuclear histone functions have been identified in numerous diseases, their exact roles in various biological processes still remain to be defined. In particular, the implication of extranuclear histone proteins and their potential mechanism of action in the pathophysiology of CRS have not been well investigated.
In the present study, we studied the expression level and distribution pattern of histone in CRSwNP patients using DNA microarray, real-time quantitative polymerase chain reaction (qPCR), immunohistochemical (IHC) staining, and double immunofluorescent staining in order to assess histone function in NPs.
2. Materials and methods
2.1. Patients and tissue samples
Adult CRSwNP patients (n = 8) and healthy individuals without CRS (n = 7) were recruited from the Department of Otorhinolaryngology of the Ghent University Hospital. CRSwNP was diagnosed on the basis of symptoms, clinical examination, nasal endoscopy, and a computed tomography (CT) scan of the sinuses according to the European Position Paper on Rhinosinusitis and Nasal Polyps [10]. Tissue samples from CRSwNP patients were obtained during functional endoscopic sinus surgery. Inferior turbinate samples were collected from patients without sinus disease undergoing septoplasty or rhinoseptoplasty and used as the control samples. In the control group, none of the patients had a history of asthma, of which 2 had a positive skin prick test (SPT) result. In the CRSwNP group, 3 of the 8 patients had a history of asthma, of which 4 had a positive SPT result. Sinus disease was diagnosed on the basis of history, clinical examination, nasal endoscopy, and CT scan of the paranasal cavities according to the current European Position Paper on Rhinosinusitis and Nasal Polyps [1]. General exclusion criteria were based on the EP3OS definition for research (cystic fibrosis, gross immunodeficiency, congenital mucociliary problems, noninvasive fungal balls and invasive fungal disease, systemic vasculitis, and granulomatous diseases). The characteristics of the study subjects are shown in Table 1. This study was approved by the Ethics Committee of the Ghent University Hospital, and written informed consent was obtained from each subject before inclusion in the study. All patients stopped intranasal corticosteroids, antihistamines, antileukotrienes, oral and intranasal decongestants, or intranasal anticholinergics 1 week before surgery, and oral and/or intramuscular corticosteroid use was discontinued 4 weeks before the surgery.
Table 1.
Subjects’ characteristics
| Characteristics | Normal controls | CRSwNP patients |
|---|---|---|
| Number of subjects (n) | 7 | 8 |
| Gender (M/F) | 6/1 | 6/2 |
| Age range (yr) | 19–45 | 26–72 |
| Allergy (n) | 3 | 4 |
| Asthma (N) | 0 | 2 |
| Eosinophilic CRSwNP | 0 | 6 |
| Noneosinophilic CRSwNP | 0 | 2 |
CRSwNP, chronic rhinosinusitis with nasal polyps.
SPTs with a standard panel of 14 inhalant allergens were used to test the atopic status of all patients. A positive SPT was considered if the allergen caused a wheal >7 mm2 in area (diameter >3 mm). Each SPT contained negative and positive (10 mg/mL histamine solution) controls.
2.2. DNA microarray
Total RNA was isolated using an RNeasy MiniKit (Qiagen, Hilden, Germany), and the RNA was amplified and used to generate complementary DNA (cDNA) with an Ambion WT Expression Kit (Life technologies, Carlsbad, CA, USA) and fragmented and labeled with a GeneChip WT Terminal Labeling Kit (Affymetrix, Santa Clara, CA, USA). The labeled complementary RNA was then hybridized to the Affymetrix Human Gene 2.1 ST Array (Affymetrix, Santa Clara, CA, USA). Arrays were scanned, and data were generated with the GeneTitan Multi-Channel (MC) Instrument (Affymetrix, Santa Clara, CA, USA). The processed data were visualized using Affymetrix Expression Console Software (Affymetrix, Santa Clara, CA, USA). To identify the differential expression of genes between experimental groups, we performed an unpaired Student t test, and the obtained P values were adjusted for multiple testing using the Benjamini and Hochberg method [11].
2.3. Quantitative RT-PCR
Gene expression analysis was performed by reverse transcription quantitative polymerase chain reaction (RT-qPCR). Snap-frozen tissue samples (±30 mg) were disrupted using a mortar and pestle containing liquid nitrogen, directly thawed into lysis solution (Qiagen), and homogenized in a QIAshredder homogenizer (Qiagen). cDNA was synthesized from 1 µg of RNA using the iScript Advanced cDNA Synthesis Kit for RT-qPCR (Bio-Rad, Hercules, CA, USA). Amplification reactions were performed on a Light Cycler LC480 System (Roche, Basel, Switzerland) using a specific PrimePCR Assay (Bio-Rad). Transcription and amplification variations among samples were normalized to the expression levels of 2 reference genes, elongation factor 1 (EF-1) and succinate dehydrogenase complex flavoprotein subunit A (SDHA), which were used as the reference genes after validation with geNorm software (Biogazelle, Ghent, Belgium). Table 2 lists the primer sequences used in this study. The qPCR reaction mixture consisted of 5 ng of cDNA (total RNA equivalent), 250 nmol/L of each primer pair, and 2.5 µL of SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) to obtain a final volume of 5 µL. The normalized relative quantities (NRQs) were calculated using qBase+ software (Biogazelle, Belgium), and the results of the gene expression were expressed as the logarithm of NRQs per 5 ng of cDNA.
Table 2.
Primer sequences used for quantitative polymerase chain reaction
| Gene | Forward (5′–3′) | Reverse (5′–3′) | Amplicon size (bp) | Accession number |
|---|---|---|---|---|
| EF-1 | CTGAACCATCCAGGCCAAAT | GCCGTGTGGCAATCCAAT | 59 | NM_001402 |
| SDHA | TGGGAACAAGAGGGCATCTG | CCACCACTGCATCAAATTCATG | 86 | NM_004168 |
Sequences were obtained from the real-time polymerase chain reaction primer and probe database (http://www.rtprimerdb.org).
2.4. IHC staining
Tissue sections were embedded in paraffin and sliced into 4-µm sections for the IHC staining performed using the peroxidase-labeled streptavidin-biotin technique. Briefly, the 4-µm-thick tissue sections were deparaffinized by serial passages in an alcohol gradient. After blocking the endogenous peroxidase in 0.3% hydrogen peroxide and with 3% NaN3, the sections were incubated overnight at 4°C in the presence of primary antibodies (rabbit anti-H2BK antibody, Biorbyt, Brussels, Belgium, 1:500). Thereafter, each section was incubated with horseradish peroxidase-labeled streptavidin complex. Negative control studies were performed by replacing the primary antibodies with normal IgG in appropriate concentrations. The sections were examined and coded by an observer who was blinded to the sample and had no awareness of the clinical data using an Olympus CX40 Microscope (Olympus Optical Co, Hamburg, Germany).
2.5. Cell culture
A549 cells (accepted as a gift from Peking Union Medical College) were cultured in RPMI-1640 (Gibco, NY, USA) containing 10% fetal bovine serum (Gibco) in an incubator (Thermo, Waltham, MA, USA) at 37°C in the presence of air containing 5% CO2. The medium was changed every 2 days, and cells at a density of 80% to 90% were digested using 0.25% trypsin. The cells were then seeded into the wells of 24-well plates, cultured, and treated with recombinant H2BK (Abnova, Taiwan, China ) or phosphate buffered saline for 24 hours.
2.6. Flow cytometry
To investigate the role of H2BK in cell apoptosis, A549 cells were treated with or without 500 ng/mL H2BK recombinant protein and apoptosis was analyzed using an APC Annexin V Apoptosis Detection Kit with propidium iodide (PI; BioLegend, San Diego, CA, USA) according to the manufacturer’s protocol. Briefly, the cells were washed twice with cold BioLegend cell staining buffer, resuspended in 100 μL of Annexin V Binding Buffer, and 5 μL of APC Annexin V and 10 μL of PI solution were added to the cells. The cells were gently vortexed and incubated at room temperature (25°C) for 15 minutes in the dark. Then, 400 μL of Annexin V Binding Buffer was added. Flow cytometry was performed with an Attune NxT Flow Cytometer (Thermo Electron, San Jose, CA, USA).
2.7. Statistical analysis
An unpaired Student t test was performed to identify the genes that were differentially expressed between the experimental groups. The obtained P values were adjusted for multiple testing using the Benjamini and Hochberg method (Benjamini, Y., and Hochberg, Y., 1995). The nonparametric Mann-Whitney U test was used to analyze the qPCR data between groups. A P value <0.05 was considered statistically significant.
3. Results
3.1. Expression and cellular sources of H2BK in CRSwNP patients
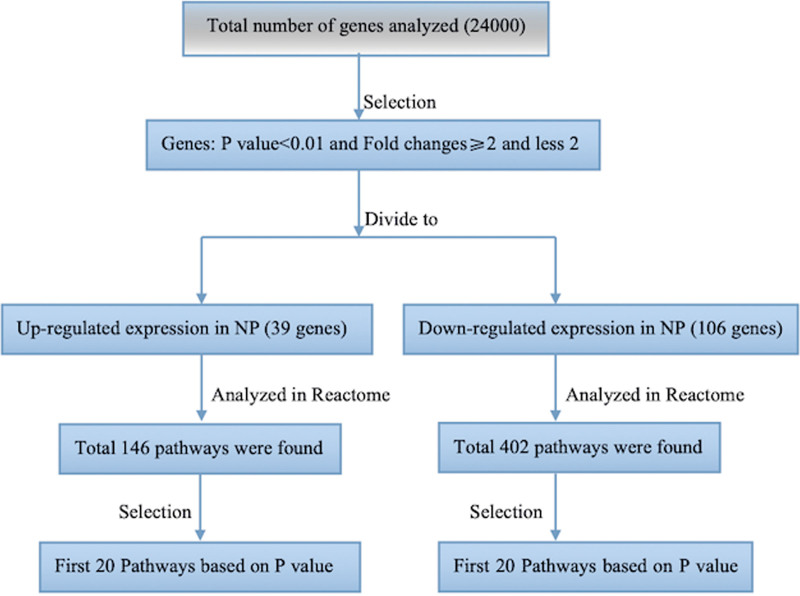
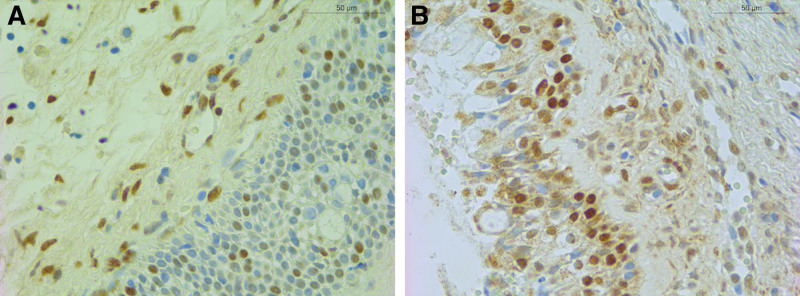
Based on the results of the DNA microarray analysis, there were 162 upregulated and 248 downregulated genes in NP tissues (P < 0.05). We narrowed down the genes from the microarray data with a P value cutoff of 0.01 and removing any gene with fold changes <2. The final list comprised 39 upregulated and 106 downregulated genes. This list was then submitted to the pathway database Reactome (Fig. 1, http://www.reactome.org). This software generates a list of the genes involved in pathways based on their P value (Tables 3 and 4). H2BK (P < 0.001, fold change = −3.7855) was found in each of the first 20 pathways (Table 4). However, H2BK messenger RNA (mRNA) levels did not significantly differ between the inferior turbinates from controls and NPs from CRSwNP patients. Interestingly, results of the IHC revealed that H2BK was less abundant in CRSwNP (Fig. 2A) tissues than in control tissue (Fig. 2B). H2BK was highly expressed in the epithelial cells and subepithelial inflammatory cells in both CRSwNP and control tissues, suggesting that nasal epithelial cells are one of the main cellular sources of H2BK production in patients with CRSwNP.
Figure 1.
Selection process of target genes through a Reactome software.
Table 3.
First 20 pathways based on P value for upregulated genes
| Symbol | Pathway |
|---|---|
| KDM4B | Histone demethylases demethylate histones |
| NOTCH4 | Pre-NOTCH transcription and translation |
| NOTCH4 | Pre-NOTCH expression and processing |
| IGHM | Classical antibody-mediated complement activation |
| IGHM | Fc-gamma receptor (FCGR) activation |
| IGHM | Creation of C4 and C2 activators |
| IGHM | Initial triggering of complement |
| IGHM | Role of phospholipids in phagocytosis |
| IGHM | Complement cascade |
| IGHM | Regulation of actin dynamics for phagocytic cup formation |
| KDM4B | Chromatin-modifying enzymes |
| KDM4B | Chromatin organization |
| PANX2 | Transmission across electrical synapses |
| PANX2 | Electric transmission across gap junctions |
| IGHM | FCGR-dependent phagocytosis |
|
COL27A1
COL18A1 |
Assembly of collagen fibrils and other multimeric structures |
| NOTCH4 | Signaling by NOTCH |
| PGF | Vascular endothelial growth factor(VEGF)ligand-receptor interactions |
| PGF | VEGF binds to VEGF receptor leading to receptor dimerization |
|
COL27A1
COL18A1 |
Collagen biosynthesis and modifying enzymes |
Table 4.
First 20 pathways based on P value for downregulated genes
| Genes | Pathways |
|---|---|
| HIST1H2BK POLR2K HIST2H2AB TWISTNB | RNA polymerase I chain elongation |
| RFC3 HIST1H2BK HIST2H2AB | Telomere maintenance |
| HIST1H2BK POLR2K HIST2H2AB TWISTNB | Nucleolar remodeling complex negatively regulates ribosomal RNA (rRNA) expression |
| HIST1H2BK HIST2H2AB | Packaging of telomere ends |
| HIST1H2BK HIST2H2AB | RNA polymerase I promoter opening |
| HIST1H2BK POLR2K HIST2H2AB TWISTNB | RNA polymerase I promoter clearance |
| HIST1H2BK POLR2K HIST2H2AB TWISTNB | RNA polymerase I transcription |
| HIST1H2BK HIST2H2AB | DNA methylation |
| HIST1H2BK POLR2K HIST2H2AB TWISTNB | Negative epigenetic regulation of rRNA expression |
| HIST1H2BK HIST2H2AB | Formation of the beta-catenin: tansactivation of T cell factor transactivating complex |
| HIST1H2BK HIST2H2AB | PRC2 methylates histones and DNA |
| HIST1H2BK HIST2H2AB ASF1A | DNA damage/telomere stress-induced senescence |
| HIST1H2BK HIST2H2AB RFC3 | Chromosome maintenance |
| HIST1H2BK HIST2H2AB | SIRT1 negatively regulates rRNA expression |
| HIST1H2BK HIST2H2AB | Activated PKN1 stimulates transcription of AR (androgen receptor)-regulated genes KLK2 and KLK3 |
| HIST1H2BK POLR2K HIST2H2AB | Transcriptional regulation by small RNAs |
| PAX3 ELP4 HIST2H2AB HIST1H2BK | Histone acetyltransferases acetylate histones |
| HIST1H2BK POLR2K HIST2H2AB TWISTNB | Epigenetic regulation of gene expression |
| HIST1H2BK HIST2H2AB | Condensation of prophase chromosomes |
| HIST1H2BK HIST2H2AB | Nucleosome assembly |
Figure 2.
H2BK was localized in subepithelial cells. Representative immunohistochemical staining of H2BK (A) in nasal polyp tissues and normal control (B) was shown (magnification, ×400).
3.2. Recombinant human H2BK-induced apoptosis in A549 cells in vitro
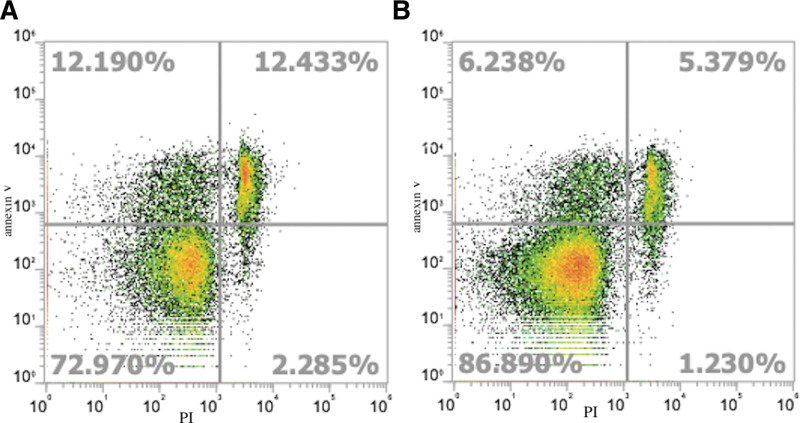
To define the role played by H2BK in apoptotic pathways in NP, A549 cells were treated with or without recombinant human H2BK and cells were counted by flow cytometry and apoptosis was detected using annexin V. The results revealed that the percentages of early (12.190% vs 6.238%) and late (12.433% vs 5.379%) apoptotic cells were higher in the H2BK-treated group than in the control group; furthermore, there were more necrotic cells in the H2BK-treated group than in the control group (Fig. 3).
Figure 3.
The percentage of apoptotic cells evaluated by Annexin V-APC/PI staining. Comparison of early and late apoptotic A549 cells after incubation for 24 hours with H2BK (A) and without H2BK (B). The results are visualized as representative dot plots.
4. Discussion
Histones are basic chromatin subunits and are important for the construction of the nucleosome within the nucleus. Histone PTMs are critical for the precise regulation of gene expression. There is growing interest in the contribution of histone modifications in regulating the development of allergic diseases such as allergic asthma, allergic rhinitis, and atopic dermatitis [12]. Histone is the main protein component of chromatin, which is widely modified after translation. More and more evidence show that the combination of epigenetic histone modifications can affect the whole chromatin structure and have clear functional consequences in cell processes including apoptosis. Histone modification regulates transcriptional activity through ubiquitination, methylation, and acetylation and thus participates in the regulation of cell cycle. More specific studies on histone family members have shown that the possible mechanisms involved in the nuclear event of cell apoptosis include phosphorylation of histones H2A, H2B, H3, and H4, dephosphorylation of histone H1, acetylation of histones H2B and H4, low acetylation of histone H4, methylation of components H3 and H4, and deubiquitination of histone H2A [13]. Researchers have reported that histone ubiquitination may play a role in transcriptional regulation, because ubiquitinated histones H2A and H2B have been proved to be related to the transcriptional activity of chromatin, which may be involved in the regulation of cell apoptosis [14]. Marushige and Marushige [15] have observed that histone H2A deubiquitination plays an important role in the apoptosis of rat glioma cells. Tanimoto et al. [16] also observed that histone H2A deubiquitination was accompanied by apoptotic chromatin condensation and DNA breakage. Histone modifications, especially phosphorylation and acetylation, have long been reported to affect the function and structure of chromatin during cell death. Lee et al. [17] used protein phosphatase inhibitors to confirm that histone phosphorylation may be involved in thymocyte apoptosis. This result was confirmed in the subsequent study of astrocyte apoptosis [18]. Ajiro [19] reported that the only apoptosis-related phosphorylation was observed on histone H2B in the process of cell line apoptosis triggered by a series of compounds. In addition, Cong et al. [20] have reported that histone methylation may be involved in macrophage apoptosis and unstable plaque formation of methionine-induced hyperhomocysteinemic ApoE −/− mice. Okubo et al. [21] and Li et al. [22] have reported that histone acetylation is involved in inducing apoptosis and have proposed that leptin may induce apoptosis by increasing the acetylation levels of histones H3 and H4 and inhibit apoptosis by reducing the acetylation levels of histones H3 and H4 [21, 22].
However, the role of extranuclear histone in CRS is not yet fully understood. This study is the first to report that H2BK may play a role in the pathogenesis of CRSwNP, highlighting its potential as a molecular target for therapeutic agents as it is involved in cell apoptosis.
Histone modification involves inflammatory cells such as T cells and macrophages, which contribute to remodeling of airways; furthermore, histone modification has been shown to directly regulate allergic phenotypes. In addition to altering nuclear function, histones can also act as damage-associated molecular pattern molecules when released into extracellular space. Histones have been detected at the cell surface or the cytoplasm of immune cells, and the levels of circulating histones in animals or patients with cancer, inflammation, and infection have been shown to increase [23]. As components of neutrophil extracellular traps (NETs), histones play a role in the innate immunity by capturing and degrading invading microorganisms [24]. Nonetheless, limited data have focused on extracellular histones and their influence on CRS. Our study was designed to investigate the role of extracellular histones in patients with CRS.
The presence of bacteria within the sinuses has been well documented. Staphylococcus aureus is an important infectious agent that triggers cytokine production in patients with upper respiratory tract inflammation or in individuals with NPs [25]. Although the antimicrobial activity of extracellular histones has been shown to occur by their binding to and blocking of both the core and lipid A moieties of Escherichia coli lipopolysaccharide [26], the present study did not find a similar binding or blocking for S. aureus (data not shown).
Toll-like receptors (TLRs) are essential to the innate immune system and have been demonstrated to play an important role in the development of CRS [27]. Extracellular histones can selectively bind to TLRs (eg, TLR9 and TLR4) and active TLR-dependent signaling pathways to produce cytokines, which in turn accelerates inflammatory responses of the airway [28]. However, in our study, we did not find any extracellular histone-mediated increase in TLR expression in NPs in vitro (data not shown).
Histologically, NPs consist of loose connective tissue, edematous myxoid stroma, and inflammatory cells. They are covered with different types of respiratory epithelium, which are characterized by hyperplasia and squamous metaplasia. Thus, the imbalance between apoptosis and proliferation could provide vital information for understanding its pathogenesis. Several studies have shown that apoptotic mechanisms are vital for the development and progression of NP [29]. Ours is the first study to investigate the expression of H2BK in CRSwNP patients. Here, results of the microarray analysis revealed that H2BK expression was downregulated in NP patients compared with controls. However, the level of H2BK mRNA expression did not differ between the inferior turbinates from controls and NP from CRSwNP patients. This could be attributed to the fact that H2BK is mainly distributed in epithelial cells, and the sample used for the polymerase chain reaction may contain a range of cells in addition to nasal mucosal epithelial cells. Results of the IHC analyses also verified that nasal epithelial cells were one of the main cellular sources of H2BK production in CRSwNP patients (Fig. 2).
To verify the role of H2BK in the apoptosis of NPs, we evaluated cells using flow cytometric Annexin V apoptosis detection with PI, which allowed us to differentiate between the contribution of necrosis and apoptosis on cell survival. The results show that the percentages of early and late apoptotic cells were higher in the H2BK-treated group than in the control group; moreover, there were more necrotic cells in the H2BK-treated group than in the control group (Fig. 3). Based on the results of this study, we can conclude that extracellular H2BK can promote apoptosis in NP, and the H2BK expression level may play an important role in the pathogenesis of NP. Histones were previously believed to play redundant roles in apoptosis. Saffarzadeh et al. [30] reported that recombinant histone H4 potently induces cell death, suggesting that NET-bound and free histone H4 exhibits considerable cytotoxicity. Silvestre-Roig et al. [31] found that extranuclear histone H4 synthesis is involved in a variety of atherosclerotic lesion processes, such as structural maintenance of chromosomes cell lysis, death, intimal neutrophil improvement, and atheromatous plaque development. This is the first study to investigate H2BK expression in CRSwNP patients. The reduced H2BK expression in CRSwNP patients might result in greater longevity of the cells, thus impairing proliferation control.
5. Conclusion
The process of cell death is an important phase for the perpetuation of cells and inflammatory processes in CRSwNP. Our results indicate that reduced expression of H2BK may contribute to the imbalances in cell proliferation and apoptosis in CRSwNP. The results of our study could provide critical information for the development of drugs aimed to increase the levels of apoptosis, facilitating the control and prevention of disease recurrence. Additional clinical studies are needed to investigate the dynamics of cell death in CRSwNP. However, due to the small number of noneosinophilic CRSwNP group (n = 2), the role of H2BK in the pathogenesis of sinusitis with different phenotypes has not been further explored. In the future, we will collect more samples to explore the role of histone and apoptosis regulation in the pathogenesis of different phenotypes of sinusitis.
Acknowledgements
This work was supported by Program for Changjiang Scholars and Innovative Research Team (IRT13082); National Science Fund for the Major International Joint Research Program (81420108009); National Natural Science Foundation of China (30973282, 81100706, 81570895, 81441031, 81441031, 81400470, and 81400447); Capital Health Research and Development of Special (2011-1017-06); Special Fund of Sanitation Elite Reconstruction of Beijing (2009-2-007); Beijing Health Bureau Program for High Level Talents (2011-3-043, 2014-3-015); Beijing Municipal Administration of Hospitals’ Mission Plan (SML20150203 to LZ, YZ); Priming Scientific Research Foundation for the Junior Researcher in Beijing Tongren Hospital, Capital Medical University (2018-YJJ-ZZL-010).
Conflicts of interest
The authors declare no conflicts of interest.
Author contributions
Yanming Zhao and Luo Zhang designed and supervised the study. Nan Zhang revised the study and the article. Claudina Perez Novo and Yang Wang performed data analysis, experiments of DNA sequencing, and flow cytometry. The final article was approved by all authors.
Footnotes
Published online 4 April 2024
References
- 1.Fokkens WJ, Lund VJ, Mullol J, Bachert C, Alobid I, Baroody F, Cohen N, Cervin A, Douglas R, Gevaert P, Georgalas C, Goossens H, Harvey R, Hellings P, Hopkins C, Jones N, Joos G, Kalogjera L, Kern B, Kowalski M, Price D, Riechelmann H, Schlosser R, Senior B, Thomas M, Toskala E, Voegels R, Wang DY, Wormald PJ. EPOS 2012: European Position Paper on Rhinosinusitis and Nasal Polyps 2012. A summary for otorhinolaryngologists. Rhinology. 2012;50:1-12. [DOI] [PubMed] [Google Scholar]
- 2.Kim YS, Kim NH, Seong SY, Kim KR, Lee GB, Kim KS. Prevalence and risk factors of chronic rhinosinusitis in Korea. Am J Rhinol Allergy. 2011;25:117-21. [DOI] [PubMed] [Google Scholar]
- 3.Zentner GE, Henikoff S. Regulation of nucleosome dynamics by histone modifications. Nat Struct Mol Biol. 2013;20:259-66. [DOI] [PubMed] [Google Scholar]
- 4.Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet. 2009;10:161-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kidd CD, Thompson PJ, Barrett L, Baltic S. Histone modifications and asthma. The interface of the epigenetic and genetic landscapes. Am J Respir Cell Mol Biol. 2016;54:3-12. [DOI] [PubMed] [Google Scholar]
- 6.Tumes DJ, Onodera A, Suzuki A, Shinoda K, Endo Y, Iwamura C, Hosokawa H, Koseki H, Tokoyoda K, Suzuki Y, Motohashi S, Nakayama T. The polycomb protein Ezh2 regulates differentiation and plasticity of CD4(+) T helper type 1 and type 2 cells. Immunity. 2013;39:819-32. [DOI] [PubMed] [Google Scholar]
- 7.Allam R, Darisipudi MN, Tschopp J, Anders HJ. Histones trigger sterile inflammation by activating the NLRP3 inflammasome. Eur J Immunol. 2013;43:3336-42. [DOI] [PubMed] [Google Scholar]
- 8.Barrero CA, Perez-Leal O, Aksoy M, Moncada C, Ji R, Lopez Y, Mallilankaraman K, Madesh M, Criner GJ, Kelsen SG, Merali S. Histone 3.3 participates in a self-sustaining cascade of apoptosis that contributes to the progression of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2013;188:673-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morita S, Tagai C, Shiraishi T, Miyaji K, Iwamuro S. Differential mode of antimicrobial actions of arginine-rich and lysine-rich histones against Gram-positive Staphylococcus aureus. Peptides. 2013;48:75-82. [DOI] [PubMed] [Google Scholar]
- 10.Fokkens W, Lund V, Bachert C, Clement P, Helllings P, Holmstrom M, Jones N, Kalogjera L, Kennedy D, Kowalski M, Malmberg H, Mullol J, Passali D, Stammberger H, Stierna P; EAACI. EAACI position paper on rhinosinusitis and nasal polyps executive summary. Allergy. 2005;60:583-601. [DOI] [PubMed] [Google Scholar]
- 11.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat. 1995;57:289-300. [Google Scholar]
- 12.Alaskhar Alhamwe B, Khalaila R, Wolf J, von Bülow V, Harb H, Alhamdan F, Hii CS, Prescott SL, Ferrante A, Renz H, Garn H, Potaczek DP. Histone modifications and their role in epigenetics of atopy and allergic diseases. Allergy Asthma Clin Immunol. 2018;14:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Th’ng JP. Histone modifications and apoptosis: cause or consequence? Biochem Cell Biol. 2001;79:305-11. [PubMed] [Google Scholar]
- 14.Nickel BE, Allis CD, Davie JR. Ubiquitinated histone H2B is preferentially located in transcriptionally active chromatin. Biochemistry. 1989;28:958-63. [DOI] [PubMed] [Google Scholar]
- 15.Marushige Y, Marushige K. Disappearance of ubiquitinated histone H2A during chromatin condensation in TGF beta 1-induced apoptosis. Anticancer Res. 1995;15:267-72. [PubMed] [Google Scholar]
- 16.Tanimoto Y, Onishi Y, Hashimoto S, Kizaki H. Peptidyl aldehyde inhibitors of proteasome induce apoptosis rapidly in mouse lymphoma RVC cells. J Biochem. 1997;121:542-9. [DOI] [PubMed] [Google Scholar]
- 17.Lee E, Nakatsuma A, Hiraoka R, Ishikawa E, Enomoto R, Yamauchi A. Involvement of histone phosphorylation in thymocyte apoptosis by protein phosphatase inhibitors. IUBMB Life. 1999;48:79-83. [DOI] [PubMed] [Google Scholar]
- 18.Enomoto R, Tatsuoka H, Komai T, Sugahara C, Takemura K, Yamauchi A, Nishimura M, Naito S, Matsuda T, Lee E. Involvement of histone phosphorylation in apoptosis of human astrocytes after exposure to saline solution. Neurochem Int. 2004;44:459-67. [DOI] [PubMed] [Google Scholar]
- 19.Ajiro K. Histone H2B phosphorylation in mammalian apoptotic cells. An association with DNA fragmentation. J Biol Chem. 2000;275:439-43. [DOI] [PubMed] [Google Scholar]
- 20.Cong G, Yan R, Huang H, Wang K, Yan N, Jin P, Zhang N, Hou J, Chen D, Jia S. Involvement of histone methylation in macrophage apoptosis and unstable plaque formation in methionine-induced hyperhomocysteinemic ApoE(-/-) mice. Life Sci. 2017;173:135-44. [DOI] [PubMed] [Google Scholar]
- 21.Okubo K, Isono M, Miyai K, Asano T, Sato A. Fluvastatin potentiates anticancer activity of vorinostat in renal cancer cells. Cancer Sci. 2020;111:112-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, Fu X, Li H, Gao Y, Wang W, Shen Y. Leptin differentially regulate cell apoptosis and cycle by histone acetylation in tibial and vertebral epiphyseal plates. Cell Biol Int. 2023;47:660-8. [DOI] [PubMed] [Google Scholar]
- 23.Allam R, Kumar SV, Darisipudi MN, Anders HJ. Extracellular histones in tissue injury and inflammation. J Mol Med (Berl). 2014;92:465-72. [DOI] [PubMed] [Google Scholar]
- 24.Saitoh T, Komano J, Saitoh Y, Misawa T, Takahama M, Kozaki T, Uehata T, Iwasaki H, Omori H, Yamaoka S, Yamamoto N, Akira S. Neutrophil extracellular traps mediate a host defense response to human immunodeficiency virus-1. Cell Host Microbe. 2012;12:109-16. [DOI] [PubMed] [Google Scholar]
- 25.Bachert C, Zhang N, Patou J, van Zele T, Gevaert P. Role of staphylococcal superantigens in upper airway disease. Curr Opin Allergy Clin Immunol. 2008;8:34-8. [DOI] [PubMed] [Google Scholar]
- 26.Kim HS, Cho JH, Park HW, Yoon H, Kim MS, Kim SC. Endotoxin-neutralizing antimicrobial proteins of the human placenta. J Immunol. 2002;168:2356-64. [DOI] [PubMed] [Google Scholar]
- 27.Liu JX, Liao B, Yu QH, Wang H, Liu Y-B, Guo C-L, Wang Z-C, Li Z-Y, Wang Z-Z, Ruan J-W, Pan Li, Yao Y, Chen C-L, Wang H, Liang Y, Zhen G, Liu Z. The IL-37-Mex3B-Toll-like receptor 3 axis in epithelial cells in patients with eosinophilic chronic rhinosinusitis with nasal polyps. J Allergy Clin Immunol. 2020;145:160-72. [DOI] [PubMed] [Google Scholar]
- 28.Xu J, Zhang X, Monestier M, Esmon NL, Esmon CT. Extracellular histones are mediators of death through TLR2 and TLR4 in mouse fatal liver injury. J Immunol. 2011;187:2626-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen D, Chen L, Xiao L. [The progress study of tumor suppressor gene and apoptosis gene in nasal polyps]. Lin Chuang Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2015;29:2099-102. [PubMed] [Google Scholar]
- 30.Saffarzadeh M, Juenemann C, Queisser MA, Lochnit G, Barreto G, Galuska SP, Lohmeyer J, Preissner KT. Neutrophil extracellular traps directly induce epithelial and endothelial cell death: a predominant role of histones. PLoS One. 2012;7:e32366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silvestre-Roig C, Braster Q, Wichapong K, Lee EY, Teulon JM, Berrebeh N, Winter J, Adrover JM, Santos GS, Froese A, Lemnitzer P, Ortega-Gómez A, Chevre R, Marschner J, Schumski A, Winter C, Perez-Olivares L, Pan C, Paulin N, Schoufour T, Hartwig H, González-Ramos S, Kamp F, Megens RTA, Mowen KA, Gunzer M, Maegdefessel L, Hackeng T, Lutgens E, Daemen M, von Blume J, Anders H-J, Nikolaev VO, Pellequer J-L, Weber C, Hidalgo A, Nicolaes GAF, Wong GCL, Soehnlein O. Externalized histone H4 orchestrates chronic inflammation by inducing lytic cell death. Nature. 2019;569:236-40. [DOI] [PMC free article] [PubMed] [Google Scholar]