Abstract
Background:
The diagnosis of allergic rhinitis (AR) primarily relies on symptoms and laboratory examinations. Due to limitations in outpatient settings, certain tests such as nasal provocation tests and nasal secretion smear examinations are not routinely conducted. Although there are clear diagnostic criteria, an accurate diagnosis still requires the expertise of an experienced doctor, considering the patient’s medical history and conducting examinations. However, differences in physician knowledge and limitations of examination methods can result in variations in diagnosis.
Objective:
Artificial intelligence is a significant outcome of the rapid advancement in computer technology today. This study aims to present an intelligent diagnosis and detection method based on ensemble learning for AR.
Method:
We conducted a study on AR cases and 7 other diseases exhibiting similar symptoms, including rhinosinusitis, chronic rhinitis, upper respiratory tract infection, etc. Clinical data, encompassing medical history, clinical symptoms, allergen detection, and imaging, was collected. To develop an effective classifier, multiple models were employed to train on the same batch of data. By utilizing ensemble learning algorithms, we obtained the final ensemble classifier known as adaptive random forest-out of bag-easy ensemble (ARF-OOBEE). In order to perform comparative experiments, we selected 5 commonly used machine learning classification algorithms: Naive Bayes, support vector machine, logistic regression, multilayer perceptron, deep forest (GC Forest), and extreme gradient boosting (XGBoost).To evaluate the prediction performance of AR samples, various parameters such as precision, sensitivity, specificity, G-mean, F1-score, and area under the curve (AUC) of the receiver operating characteristic curve were jointly employed as evaluation indicators.
Results:
We compared 7 classification models, including probability models, tree models, linear models, ensemble models, and neural network models. The ensemble classification algorithms, namely ARF-OOBEE and GC Forest, outperformed the other algorithms in terms of the comprehensive classification evaluation index. The accuracy of G-mean and AUC parameters improved by nearly 2% when compared to the other algorithms. Moreover, these ensemble classifiers exhibited excellent performance in handling large-scale data and unbalanced samples.
Conclusion:
The ARF-OOBEE ensemble learning model demonstrates strong generalization performance and comprehensive classification abilities, making it suitable for effective application in auxiliary AR diagnosis.
Keywords: Allergic rhinitis, artificial intelligence, deep learning, diagnosis, ensemble learning, machine learning
1. Introduction
Allergic rhinitis (AR) is a prevalent allergic inflammation affecting the upper respiratory tract, causing significant disruptions in people’s daily lives. It is considered a persistent condition with a global upward trend in prevalence. Approximately 500 million individuals worldwide are afflicted by AR, with the highest prevalence observed in developed regions such as Western Europe, Northern Europe, and North America, typically ranging from 12% to 30% [1]. An epidemiological survey conducted on Chinese adults revealed a rise in AR prevalence from 11.1% in 2005 to 17.6% in 2011 [2, 3].
The diagnosis of AR primarily relies on symptoms and laboratory examinations. However, due to limitations in outpatient settings, certain tests such as nasal provocation tests and nasal secretion smear examinations are not routinely conducted [4]. While the nasal provocation test is considered the gold standard for diagnosing AR, its usage as a routine clinical method is limited due to associated risks.
Typical symptoms of AR include paroxysmal sneezing, a runny nose, itchiness in the nose, and nasal congestion. These symptoms may be accompanied by eye-related symptoms such as itchy eyes, watery eyes, redness, and a burning sensation. The main signs of AR consist of paleness and swelling of the bilateral nasal mucosa, inferior turbinate edema, and a significant amount of watery discharge from the nasal cavity [5]. In some cases, patients with AR may also exhibit additional signs such as eczema and dermatitis. In addition to symptoms and signs, the diagnosis of AR relies on the detection of allergens through methods like the skin prick test, blood tIgE and sIgE tests [6]. Nasal secretion smears and sIgE analysis in nasal lavage fluid also aid in clinical diagnosis [7]. Endoscopy or computed tomography (CT) can be utilized to observe physical changes in patients, such as turbinate hypertrophy and mucosal swelling, which assist in diagnosing sinusitis and nasal polyps [8].
Although there are clear diagnostic criteria, an accurate diagnosis still requires the expertise of an experienced doctor, considering the patient’s medical history and conducting examinations. However, differences in physician knowledge and limitations of examination methods can result in variations in diagnosis. Artificial intelligence (AI) is an interdisciplinary field that encompasses theories, methods, technologies, and application systems aimed at simulating, extending, and expanding human intelligence. In recent years, AI has found widespread application across various industries [9]. AI has developed powerful algorithmic mathematical models such as decision trees, Naive Bayes (NB), and artificial neural networks (ANN), which have been utilized in intelligent control, pattern recognition, and prediction domains. Ensemble learning, a technique that has emerged in recent years, combines the predictions from multiple individual learning models to obtain more accurate, robust, and reliable results. Integrated learning models such as boosting, bagging, and random forest (RF) have been extensively employed in the analysis of various types of datasets. In this study, we aim to utilize a big data integrated learning model to analyze data from over 2,000 outpatient clinical cases and explore the application of AI-integrated learning in the clinical diagnosis of AR.
2. Materials and methods
2.1. Sample source
Data from a total of 2,231 cases were collected at Tongji Hospital and Anting Branch Hospital of Tongji University between April 1, 2019 and March 31, 2020. These cases comprised patients who were diagnosed with suspected AR. Among them, there were 1,335 males (59.84%) with an average age of (35.39 ± 19.71) years and 896 females (40.16%) with an average age of (37.69 ± 17.94) years. The clinical history of all patients was obtained, including information such as time of diagnosis, name, age, gender, duration of the disease, and 4 common symptoms: sneezing, runny nose, itchy nose, and nasal congestion. Additionally, 2 eye symptoms were recorded. Physical symptoms, such as the presence of nasal polyps and nasal discharge, were also noted. Blood tests were conducted, including routine blood tests, total IgE levels, allergen-specific IgE (SIgE) tests, and CT imaging.
This study primarily focused on collecting cases of AR and 6 other diseases that exhibit similar symptoms, including sinusitis (RS), chronic rhinitis (RS), upper respiratory tract infection (URI), nasal septum deviation (NSD), adenoid hypertrophy (AH), and others (OTH, which includes nasal tumors). The diagnosis of AR, in combination with medical history and clinical symptoms, can be categorized into 4 types: mild intermittent, mild persistent, moderate-severe intermittent, and moderate-severe persistent. The clinical symptom score was calculated using the Total Nasal Symptom Score and Total Ocular Symptom Score. These scores assessed the severity of symptoms related to stuffy nose, runny nose, itchy nose, and sneezing. The scores were assigned as follows: 0 for no symptoms, 1 for mild symptoms, 2 for moderate symptoms, and 3 for severe symptoms [10].
2.2. Experimental setup and algorithm structure design
The dataset comprises a total of 66 features, including 16 symptoms such as eye symptoms, nasal cavity examination, and runny nose. The presence or absence of symptoms is represented by 1 or 0. In the framework, the classification method based on association rules is compared with other classification methods, specifically the decision tree induction method (C4.5) as the former, and the probability classification method as the latter [11].
The classification of AR symptoms presents a unique challenge as it involves a multilabel learning problem, where patients may have other diseases concurrently. Additionally, certain tags are mutually exclusive. For instance, patients without AR should not be classified as having intermittent mild AR, and it is not possible to have both AR and intermittent AR simultaneously. To address this multilabel classification problem, conversion methods and adaptive algorithms are commonly employed.
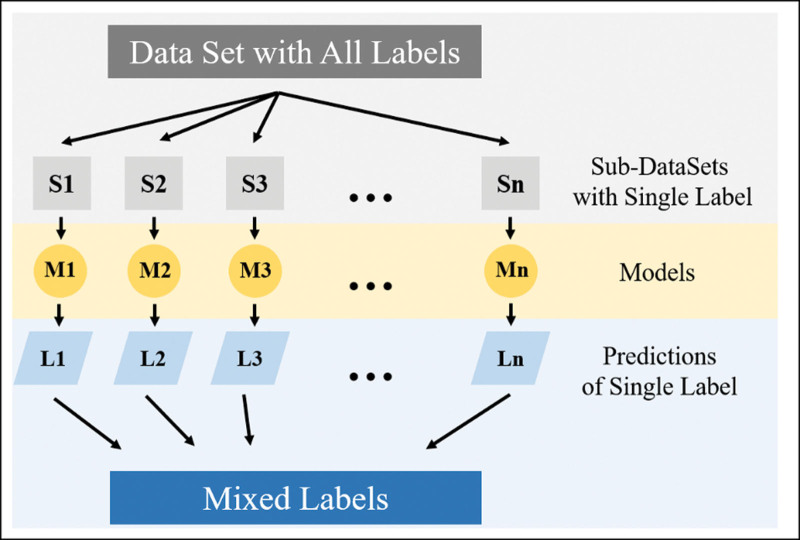
Traditionally, multilabel classification is converted into multiple binary classification problems with the same number of labels. Various basic machine learning algorithms are then utilized to train each model, creating an integrated classification model for multilabel classification, as depicted in Figure 1. Table 1 illustrates the different classification methods used for various rhinitis samples and types within the comprehensive classification model.
Figure 1.
Multilabel classification transformation.
Table 1.
Classification labels of diseases
| Rhinitis symbol | Rhinitis name | Classification criteria |
|---|---|---|
| AR | Allergic rhinitis | Binary classification |
| RS | Rhinosinusitis | Binary classification |
| NSD | Nasal septum deviation | Binary classification |
| CR | Chronic rhinitis | Binary classification |
| URI | Upper respiratory tract infection | Binary classification |
| AH | Adenoid hypertrophy | Binary classification |
| NAR | Nonallergic rhinitis | Binary classification |
| OTH | Others | Binary classification |
| Type | Types classification of AR | Multiclassification |
In the analysis, one-hot coding, also known as one-bit effective coding, is employed to encode all scenarios. This coding method represents N states using N binary bits (0s and 1s). Each state has its own binary bit, and only one-bit is active at any given time. One-hot encoding can handle discrete digital features and, to some extent, extend the features. For example, in case A, the patient exhibits clinical symptoms such as eye inflammation, turbinate hypertrophy, and clear discharge. Under these symptoms, case A has a value of 1, while symptoms like tearing, pale mucosa, and mucosal hyperemia have a value of 0. Eventually, the doctor diagnoses the patient with AR and NSD, assigning corresponding values of 1, while other nonexperienced symptoms have a value of 0, as shown in Table 2. All case data is processed as the symptom diagnosis input vector for the symptom classification model.
Table 2.
One-hot encoding
| Form of original data | ||||||||
|---|---|---|---|---|---|---|---|---|
| (*Property) | (*rhinitissymbol) | |||||||
| Property1 | Property2 | Property3 | … | Propertyn | AR | RS | … | Type |
| 1 | 0 | 0 | … | 1 | 1 | 0 | … | 1 |
| 0 | 1 | 0 | … | 1 | 0 | 1 | … | 0 |
| … | … | … | … | … | … | … | … | … |
| 1 | 0 | 1 | … | 0 | 1 | 0 | … | 0 |
AR, allergic rhinitis; RS, rhinosinusitis.
2.3. Unbalanced data processing
For multiclass classification, class imbalance methods such as SMOTE, ADASYN, and All-KNN are commonly used [12]. In the case of multilabel classification for rhinitis, AR patients accounted for 95.1% of the total patients, while a small number of patients with similar rhinitis symptoms had labels that constituted less than 10% of the total sample size. This leads to an imbalanced distribution of data across all categories, posing challenges for achieving balanced classification.
Applying oversampling techniques like SMOTE to the minority labels would increase the number of AR labels, further exacerbating the imbalance in the rhinitis symptom data and potentially reducing overall classification accuracy [13]. Actual clinical analysis of the collected data reveals that if the training set and test set are divided into a small number of labels, the limited number of samples in the test set can have a larger impact on the comprehensive classification, thus affecting the classification results and the balance between labels and sample size.
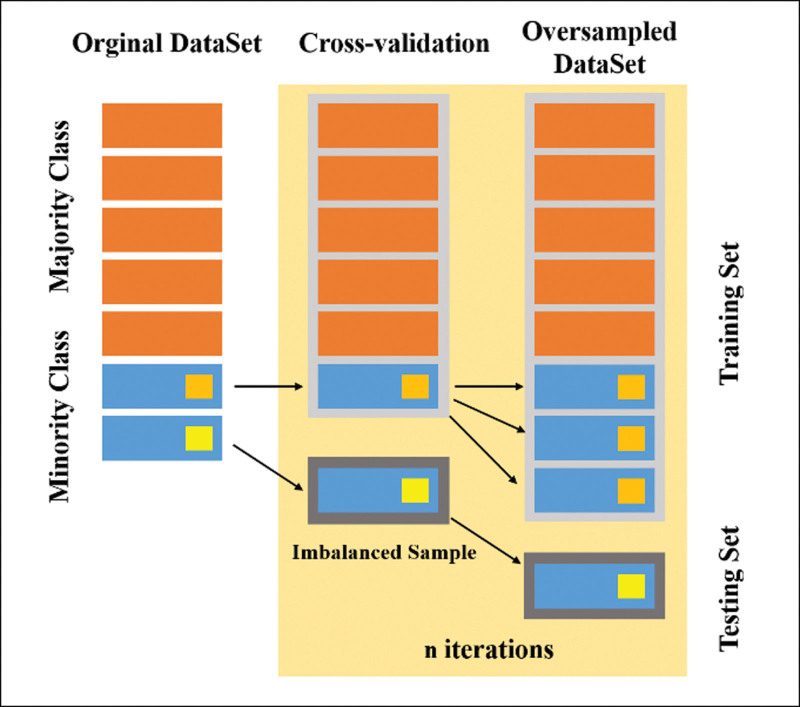
To address these issues, this study employs the ADASYN algorithm to process the imbalanced rhinitis sample data. This ensures a balanced strategy for AR and its labels, effectively improving the classification accuracy of AR and its labels for most similar rhinitis diseases, and enhancing overall classification accuracy. Additionally, some other cases of unbalanced rhinitis [14]. Figure 2 shows the unbalanced split of rhinitis sample data.
Figure 2.
Rhinitis sample set split and equalization.
2.4. Ensemble analysis of clinical data
To evaluate the prediction results of AR samples, various evaluation indicators are employed, including the confusion matrix comprehensive index: true positive, false negative, false positive, and true negative. Additionally, precision, sensitivity, specificity, G-means (calculated as the square root of sensitivity multiplied by specificity), F1-score, the area under the ROC curve (AUC), and other parameters are used as predictive evaluation indicators.
In this study, a heterogeneous ensemble rhinitis classifier model called adaptive random forest-out of bag simple orchestra (ARF-OOBEE) is proposed. This model is capable of identifying multiple diseases such as sinusitis (RS) (represented as a binary variable) and assessing the severity or persistence of symptoms. By converting heterogeneous multioutput classification problems into multilabel classification problems and multiple multiclass classification problems, this model effectively avoids interference between multilabel and multiclass symptom classification. It also allows for the use of multiple indexes or type labels for the same patient. The classifier is trained using multiple models on the same batch of data, and the ensemble learning algorithm is utilized to obtain the final ensemble classifier.
Furthermore, this study includes comparative experiments on 6 common machine learning classification algorithms: NB [15], support vector machine (SVM) [16, 17], logistic regression (LR) [18], multilayer perceptron [19], deep forest (GC Forest) [20], and extreme gradient boosting (XGBoost) [21].
3. Results
3.1. Clinical sample data analysis
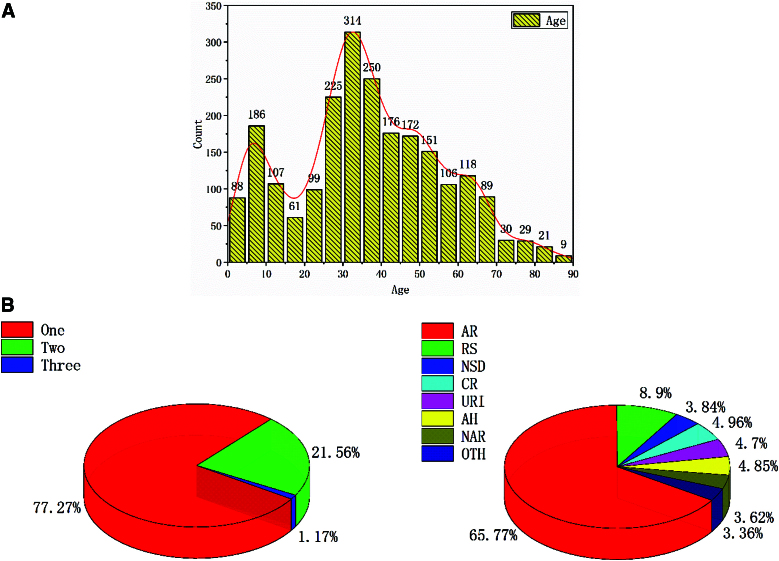
Based on the distribution of included data, 2 high incidence areas of rhinitis symptoms can be observed: one occurs in pediatric patients before the age of 10, and the other is seen between the ages of 30 and 40 (refer to Fig. 3). There was no statistically significant difference in morbidity between males and females. According to the statistics in this study, the most prevalent disease is AR, accounting for 65.77% (1,818 cases). The second most common disease is RS, accounting for 8.90% (246 cases). The remaining diseases, namely 137, 134, 130, 106, 100, and 93 cases, each account for less than 5% of the total. Additionally, the statistics on cumulative illnesses of patients indicate that the majority (77.27%, 1,724 cases) have only 1 disease, while 21.56% (481 cases) have 2 diseases, and a small portion (1.16%, 26 cases) have 3 diseases simultaneously.
Figure 3.
Age (A) and disease types (B) distribution in the samples: From the included data distribution, it can be found that there is a high incidence area of pediatric patients before the age of 10 years, and another high incidence area of rhinitis symptoms between 30 and 40 years old (A). According to statistics, among the 7 types of diseases studied in this article, the highest is AR accounted for 65.77% (1,818 cases), the second highest is RS accounted for 8.90% (246 cases), the rest are: 137, 134, 130, 106, 100, and 93 cases, accounting for less than 5%. Meanwhile, the statistics of the patients’ cumulative illnesses revealed that the patients had at most 3 diseases at the same time, which accounted for 1.16% (26 cases); patients with 2 diseases accounted for 21.56% (481 cases) and with 1 disease accounted for 77.27% (1,724 cases) (B). AR, allergic rhinitis.
3.2. Comprehensive evaluation index
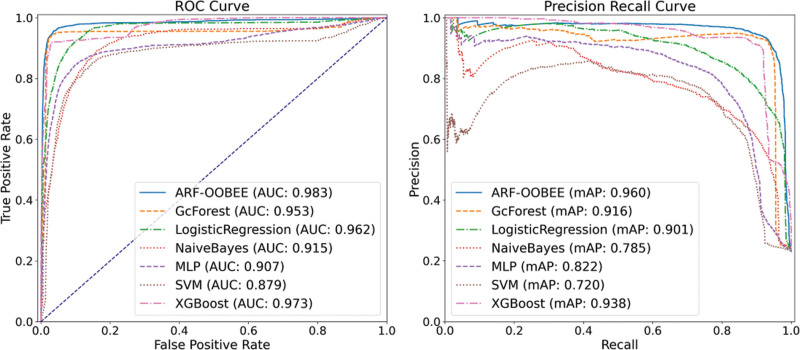
This article employs a random 10*2K-Folding cross-validation method to classify the samples based on the ARF-OOBEE ensemble model. After testing, it is found that the number of ensemble learning base classifiers is 70, with a depth of 12. The results are compared with the prediction outcomes of 5 common machine learning algorithms. According to the analysis of prediction indices in Table 3, the ARF-OOBEE algorithm has achieved an improvement of nearly 2% in the accuracy of G-mean and AUC parameters compared to the other 5 algorithms. This indicates that the ARF-OOBEE model exhibits good generalization performance and comprehensive classification ability for AR samples with clinical imbalance characteristics.
Table 3.
Comprehensive evaluation indicators of various machine learning algorithms
| Methods | F1-score | Sensitivity | Precision | Specificity | Hamming loss | Accuracy | G-mean | AUC |
|---|---|---|---|---|---|---|---|---|
| ARF-OOBEE | 0.9022 ± 0.0098 | 0.8949 ± 0.0118 | 0.9151 ± 0.0165 | 0.9805 ± 0.0338 | 0.0296 ± 0.0055 | 0.9704 ± 0.0168 | 0.9367 ± 0.0138 | 0.9830 ± 0.0202 |
| GcForest | 0.9140 ± 0.0145 | 0.8980 ± 0.0144 | 0.9420 ± 0.0169 | 0.9810 ± 0.0392 | 0.0252 ± 0.0078 | 0.9748 ± 0.0210 | 0.9386 ± 0.0236 | 0.9528 ± 0.0214 |
| LR | 0.8052 ± 0.0136 | 0.7905 ± 0.0110 | 0.8622 ± 0.0160 | 0.9581 ± 0.0300 | 0.0520 ± 0.0079 | 0.9480 ± 0.0196 | 0.8703 ± 0.0210 | 0.9616 ± 0.0225 |
| Naive Bayes | 0.7587 ± 0.0148 | 0.8085 ± 0.0106 | 0.7404 ± 0.0130 | 0.9113 ± 0.0380 | 0.0962 ± 0.038 | 0.9038 ± 0.0213 | 0.8584 ± 0.0220 | 0.9153 ± 0.0222 |
| MLP | 0.7673 ± 0.0152 | 0.7532 ± 0.0126 | 0.8327 ± 0.0165 | 0.9409 ± 0.0099 | 0.0745 ± 0.0380 | 0.9255 ± 0.0226 | 0.8418 ± 0.0232 | 0.9070 ± 0.0236 |
| SVM | 0.7411 ± 0.0133 | 0.7949 ± 0.0119 | 0.7137 ± 0.0135 | 0.8941 ± 0.0333 | 0.1090 ± 0.0083 | 0.8910 ± 0.0212 | 0.8430 ± 0.0231 | 0.8789 ± 0.0230 |
| XGBoost | 0.8804 ± 0.0116 | 0.8552 ± 0.0114 | 0.9435 ± 0.0176 | 0.9725 ± 0.0353 | 0.0335 ± 0.0079 | 0.9665 ± 0.0185 | 0.9120 ± 0.0227 | 0.9726 ± 0.0189 |
ARF-OOBEE, adaptive random forest-out of bag-easy ensemble; AUC, area under the curve; LR, logistic regression; MLP, multilayer perceptron; SVM, support vector machine; XGBoost, extreme gradient boosting.
Parameters such as precision, sensitivity, specificity, G-mean = sqrt (sensitivity × specificity), F1-score, and the area under the ROC curve (AUC) are used together as predictive evaluation metrics [22]. In Table 3 and Figure 4, 7 classification models are selected for comparison, including probability model, tree model, linear model, ensemble model, and neural network model. Among them, the ensemble model demonstrates the best and most stable performance in this article. The comprehensive classification evaluation index is lower than the ensemble classification algorithms ARF-OOBEE and GC Forest. The GC Forest algorithm consists of 2 RF and 2 extremely random trees (ERT) in a parallel structure, and it outperforms the single-structure RF algorithm in multiple comprehensive evaluation metrics, albeit with higher computational complexity.
Figure 4.
ROC curve for ensemble analysis: precision, sensitivity, specificity, G-mean = sqrt (sensitivity × specificity), F1-score, area under ROC curve AUC, and other parameters together were used as predictive evaluation indicators. ARF-OOBEE and 6 machine learning algorithms for comparative experiments, including Naive Bayes (NB), support vector machine (SVM), logistic regression (LR), multilayer perceptron (MLP), deep forest (GCForest), extreme gradient boosting (XGBoost). AUC, area under the curve; ROC, receiver operating characteristic.
Table 4 provides independent classification evaluation metrics for the 8 types of rhinitis symptoms in the original sample. Data analysis reveals that the prediction accuracy is higher for the binary classification of AR, RS, CS, SD, URI, AH, NAR, and OTH in rhinitis, while the classification of degree and type in multiclass rhinitis is lower. The ARF-OOBEE ensemble model transforms the compound label classification problem into a 4-label classification problem and 2 multiclass classification problems. Multilabel classification is employed for AR, RS, URI, and OTH, while multicategory classification is used for the degree and type of AR, thereby avoiding the presence of 2 or more AR classification labels in the same patient simultaneously.
Table 4.
Evaluation index of ARF-OOBEE in multiple label classification
| Classification | F1-score | Sensitivity | Precision | Specificity | Hamming loss | Accuracy | G-mean |
|---|---|---|---|---|---|---|---|
| AR | 0.9607 ± 0.0138 | 0.9472 ± 0.0115 | 0.9757 ± 0.0171 | 0.9884 ± 0.0225 | 0.0239 ± 0.0103 | 0.9761 ± 0.0363 | 0.9676 ± 0.0221 |
| RS | 0.9808 ± 0.0132 | 0.9733 ± 0.0122 | 0.9886 ± 0.0157 | 0.9984 ± 0.0165 | 0.0060 ± 0.0096 | 0.9940 ± 0.0321 | 0.9858 ± 0.0171 |
| NSD | 0.8687 ± 0.0122 | 0.8687 ± 0.0133 | 0.8687 ± 0.0202 | 0.9875 ± 0.0237 | 0.0243 ± 0.0088 | 0.9724 ± 0.0336 | 0.9262 ± 0.0226 |
| CR | 0.9085 ± 0.0136 | 0.9439 ± 0.0134 | 0.8791 ± 0.0241 | 0.9809 ± 0.0245 | 0.0239 ± 0.0087 | 0.9761 ± 0.0362 | 0.9622 ± 0.0251 |
| URI | 0.9142 ± 0.0123 | 0.9142 ± 0.0124 | 0.9142 ± 0.0173 | 0.9905 ± 0.0188 | 0.0179 ± 0.0083 | 0.9821 ± 0.0312 | 0.9516 ± 0.0213 |
| AH | 0.9706 ± 0.0131 | 0.9706 ± 0.0128 | 0.9706 ± 0.0219 | 0.9968 ± 0.0186 | 0.0060 ± 0.0079 | 0.9940 ± 0.0297 | 0.9837 ± 0.0228 |
| NAR | 0.7784 ± 0.0151 | 0.7258 ± 0.0116 | 0.8709 ± 0.0182 | 0.9921 ± 0.0193 | 0.0373 ± 0.0081 | 0.9627 ± 0.0312 | 0.8486 ± 0.0214 |
| OTH | 0.7974 ± 0.0142 | 0.7746 ± 0.0134 | 0.8249 ± 0.0193 | 0.9891 ± 0.0173 | 0.0269 ± 0.0099 | 0.9731 ± 0.0362 | 0.8753 ± 0.0184 |
| Degree of AR | 0.9270 ± 0.0097 | 0.9234 ± 0.0098 | 0.9311 ± 0.0211 | 0.9466 ± 0.0182 | 0.0597 ± 0.0096 | 0.9403 ± 0.0336 | 0.9349 ± 0.0204 |
| Types of AR | 0.9161 ± 0.0134 | 0.9075 ± 0.0108 | 0.9274 ± 0.0228 | 0.9349 ± 0.0207 | 0.0706 ± 0.0074 | 0.9294 ± 0.0321 | 0.9211 ± 0.0217 |
AH, adenoid hypertrophy; AR, allergic rhinitis; ARF-OOBEE, adaptive random forest-out of bag-easy ensemble; CR, chronic rhinitis; NAR, nonallergic rhinitis; NSD, nasal septum deviation; RS, rhinosinusitis; URI, upper respiratory tract infection.
4. Discussion
In recent years, there has been a significant increase in the prevalence of AR, and its diagnosis primarily relies on symptom evaluation and allergen detection. However, due to the lack of effective and reliable diagnostic tests, the final diagnosis often requires expert verification based on experience [23, 24]. To assist junior physicians and clinicians in diagnosing allergic diseases, this study utilizes AI methods to extract new insights from previous data for training [25, 26]. By dynamically verifying the rule base and employing rule inference methods, the clinical diagnosis support system becomes more adaptable. Additionally, the introduction of meta-heuristic data preprocessing technology and ensemble classification methods enhances the system’s efficiency. As a result, junior clinicians can enhance their clinical decision-making by accurately diagnosing allergic diseases, facilitating earlier detection and treatment of AR, and effectively managing patients’ symptoms to improve their quality of life.
The diagnosis of AR primarily relies on symptom assessment and allergen detection [27]. However, due to the complex and diverse nature of nasal inflammation, it is often associated with other conditions such as rhinosinusitis and nasal tumors. In such cases, imaging examinations can aid in the diagnosis of these additional diseases. Turbinate hypertrophy is also a characteristic change observed in AR. In our selected cases, coexisting conditions like rhinosinusitis and nasal polyps were identified. Therefore, the utilization of CT imaging can provide better assistance in diagnosing AR.
AI technology has the ability to learn tasks from a set of training examples without requiring human intervention. Furthermore, its objective is to generate output that is easily understandable by humans. In contrast, classical statistical methods typically involve a clear probability model and often require expert intervention in variable selection, problem transformation, and overall structure. The general process of data analysis typically consists of 4 stages: (1) collecting and encoding clinical data in an electronic format suitable for further processing; (2) utilizing feature extraction and dimensionality reduction techniques (such as principal component analysis) to process the data and select the most predictive parameters; (3) selecting an appropriate AI model through schema-model selection; and (4) extracting knowledge by evaluating accuracy, sensitivity, and specificity [28]. Currently, the most common computational models include ANN, SVM, Bayesian networks, fuzzy logic, and others.
In recent years, ensemble learning has emerged as a powerful technique for combining predictions from multiple individual learning models to obtain more accurate, stable, and robust results. Various ensemble learning models, such as boosting, bagging, and RF, have been proposed and applied to diverse datasets [29, 30]. In this study, we employed deep learning of ensemble learning models to compare 6 common machine learning classification algorithms, including RF, multilabel NB, multilabel SVM, multilabel LR, and GC Forest. The RF algorithm served as the base classification evaluation standard and formed the base classifier component of other algorithms. While RF exhibited good classification specificity, its comprehensive classification evaluation index was lower than that of ensemble classification algorithms such as ARF-OOBEE and GC Forest. The GC Forest algorithm, which consists of 2 RF and 2 ERT in a parallel structure, demonstrated superior performance across multiple comprehensive evaluation metrics compared to the single-structure RF algorithm, albeit at the cost of increased computational complexity [31].
There are 2 types of outputs for AR diseases, namely degree and types, which fall under the category of multiclass classification. In this study, the OOB (out-of-bag) EE ensemble classification algorithm is employed, using all samples as training data. The base classifier chosen is the extra-tree model, which helps balance the training data and enables predictions for unbalanced small samples. The OOBEE approach extracts data equal to the minority class from the majority class, combines the reused minority class data to construct a multigroup base classifier, and obtains the ensemble classifier through weighted voting. This technique reduces the impact of data imbalance on classification due to sample distribution.
The ARF-OOBEE model exhibits adaptive characteristics, allowing for dynamic adjustments in the number of ensemble RF and ERT base classifiers. Furthermore, it separately trains the parameters of the component classifiers. These adaptive features contribute to its strong comprehensive classification capabilities when dealing with massive datasets and unbalanced samples. The results indicate that the ARF-OOBEE algorithm has achieved an improvement of nearly 2% in accuracy for G-Mean and AUC parameters compared to the other 5 algorithms. This demonstrates the ARF-OOBEE model’s effectiveness in achieving good generalization performance and comprehensive classification ability for AR samples characterized by clinical imbalance.
There are certain limitations in this study. Firstly, the diagnosis of AR primarily relies on symptom scoring and allergen detection. However, some patients may still exhibit evident symptoms despite negative test results, requiring additional diagnostic methods such as nasal provocation tests. Unfortunately, these tests are not widely accessible in outpatient settings, leading to potential diagnostic errors in individual cases. While the AI system is designed to assist in diagnosis, it cannot completely replace the expertise of a rhinologist.
This study was conducted as a dual-center study at Tongji Hospital of Tongji University and Anting Branch Hospital, which may introduce a selection bias. To address this, future research should consider conducting a multicenter study to enhance the training database for AI systems and improve their diagnostic capabilities. Lastly, by means of self-learning, the system can assist junior doctors in completing AR diagnoses and enhancing their diagnostic skills. However, it is important to acknowledge that continuous professional development and collaboration with experienced clinicians remain crucial for the ongoing improvement of diagnostic accuracy.
Acknowledgements
This work was supported by National Key R&D Program of China (2022YFC2504100), the National Science Foundation of China (No.81873689), National Science Foundation of Shanghai (No. 23ZR1458000), Shanghai General Hospital Integrated Traditional Chinese and Western Medicine (No. ZHYY-ZXYJHZX-202118).
Conflicts of interest
The authors have no financial conflicts of interest.
Author contributions
Dai Fu, Zhao Chuanliang and Yang jingdong: Conception of the study, literature retrieval, and writing of article. Yu Shaoqing: Review and revise the article. All authors approved the final version of the article.
Footnotes
Published online 18 December 2023
Dai Fu, Zhao Chuanliang and Yang Jingdong contributed equally to this article as co-first authors.
The authors declare that no patient data appears in this article. Right to privacy and informed consent. Protection of human subjects and animals in research. The authors declare that no experiments are performed on humans or animals for this investigation.
References
- 1.Yorgancioğlu A, Kalayci O, Kalyoncu AF, Khaltaev N, Bousquet J. Allerjik rinit ve astim üzerine etkisi güncelleme (ARIA 2008) Türkiye deneyimi [Allergic rhinitis and its impact on asthma update (ARIA 2008) The Turkish perspective]. Tuberk Toraks. 2008;56:224-31. [PubMed] [Google Scholar]
- 2.Zhang Y, Zhang L. Prevalence of allergic rhinitis in china. Allergy Asthma Immunol Res. 2014;6:105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng M, Wang X, Bo M, Wang K, Zhao Y, He F, Cao F, Zhang L, Bachert C. Prevalence of allergic rhinitis among adults in urban and rural areas of china: a population-based cross-sectional survey. Allergy Asthma Immunol Res. 2015;7:148-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng L, Chen J, Fu Q, He S, Li H, Liu Z, Tan G, Tao Z, Wang D, Wen W, Xu R, Xu Y, Yang Q, Zhang C, Zhang G, Zhang R, Zhang Y, Zhou B, Zhu D, Chen L, Cui X, Deng Y, Guo Z, Huang Z, Huang Z, Li H, Li J, Li W, Li Y, Xi L, Lou H, Lu M, Ouyang Y, Shi W, Tao X, Tian H, Wang C, Wang M, Wang N, Wang X, Xie H, Yu S, Zhao R, Zheng M, Zhou H, Zhu L, Zhang L. Chinese society of allergy guidelines for diagnosis and treatment of allergic rhinitis. Allergy Asthma Immunol Res. 2018;10:300-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skoner DP. Allergic rhinitis: definition, epidemiology, pathophysiology, detection, and diagnosis. J Allergy Clin Immunol. 2001;108(1 Suppl):S2-S8. [DOI] [PubMed] [Google Scholar]
- 6.Dordal MT, Lluch-Bernal M, Sánchez MC, Rondón C, Navarro A, Montoro J, Matheu V, Ibáñez MD, Fernández-Parra B, Dávila I, Conde J, Antón E, Colás C, Valero A; SEAIC Rhinoconjunctivitis Committee. Allergen-specific nasal provocation testing: review by the rhinoconjunctivitis committee of the Spanish Society of Allergy and Clinical Immunology. J Investig Allergol Clin Immunol. 2011;21:1-12. [PubMed] [Google Scholar]
- 7.Chinoy B, Yee E, Bahna SL. Skin testing versus radioallergosorbent testing for indoor allergens. Clin Mol Allergy. 2005;3:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lloyd GA, Lund VJ, Scadding GK. CT of the paranasal sinuses and functional endoscopic surgery: a critical analysis of 100 symptomatic patients. J Laryngol Otol. 1991;105:181-85. [DOI] [PubMed] [Google Scholar]
- 9.Schwalbe N, Wahl B. Artificial intelligence and the future of global health. Lancet. 2020;395:1579-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaliner MA, Berger WE, Ratner PH, Siegel CJ. The efficacy of intranasal antihistamines in the treatment of allergic rhinitis. Ann Allergy Asthma Immunol. 2011;106(2 Suppl):S6-S11. [DOI] [PubMed] [Google Scholar]
- 11.Yan J, Zhang Z, Lin K, Yang F, Luo X. A hybrid scheme-based one-vs-all decision trees for multi-class classification tasks. Knowl-Based Syst. 2020;198:105922. [Google Scholar]
- 12.Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. SMOTE: synthetic minority over-sampling technique. J Artif Intell Res. 2002;16:321-57. [Google Scholar]
- 13.Zhao J, Jin J, Chen S, Zhang R, Yu B, Liu Q. A weighted hybrid ensemble method for classifying imbalanced data. Knowl-Based Syst. 2020;203:106087. [Google Scholar]
- 14.Zhang L, Shah SK, Kakadiaris IA. Hierarchical Multi-label Classification using Fully Associative Ensemble Learning. Pattern Recognit. 2017;70:89-103. [Google Scholar]
- 15.Zhang H, Liu CT, Mao J, Shen C, Xie R-L, Mu B. Development of novel in silico prediction model for drug-induced ototoxicity by using naïve Bayes classifier approach. Toxicol In Vitro. 2020;65:104812. [DOI] [PubMed] [Google Scholar]
- 16.Kumar S, Ong SH, Ranganath S, Ong TC, Chew FT. Classification of airspora using support vector machines (SVM). J Allergy Clin Immunol. 2003;111:S91. [Google Scholar]
- 17.Gao X, Hou J. An improved SVM integrated GS-PCA fault diagnosis approach of Tennessee Eastman process. Neurocomputing. 2016;174:906-11. [Google Scholar]
- 18.Mondal P, Dey D, Chandra Saha N, Moitra S, Saha GK, Bhattacharya S, Podder S. Investigation of house dust mite induced allergy using logistic regression in West Bengal, India. World Allergy Organ J. 2019;12:100088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heidari M, Shamsi H. Analog programmable neuron and case study on VLSI implementation of Multi-Layer Perceptron (MLP). Microelectron J. 2019;84:36-47. [Google Scholar]
- 20.Zhu G, Hu Q, Gu R, Yuan C, Huang Y. ForestLayer: efficient training of deep forests on distributed task-parallel platforms. J Parallel Distrib Comput. 2019;132:113-26. [Google Scholar]
- 21.Wang C, Deng C, Wang S. Imbalance-XGBoost: leveraging weighted and focal losses for binarylabel-imbalanced classification with XGBoost. Pattern Recognit Lett. 2020;136:190-97. [Google Scholar]
- 22.Izquierdo VE, Zurita MR. An evaluation of Guided Regularized Random Forest for classification and regression tasks in remote sensing. Int J Appl Earth Obs Geoinf. 2020;88:102051. [Google Scholar]
- 23.Greiner AN, Hellings PW, Rotiroti G, Scadding GK. Allergic rhinitis. Lancet. 2011;378:2112-122. [DOI] [PubMed] [Google Scholar]
- 24.Wang X, Du K, She W, Ouyang Y, Sima Y, Liu C, Zhang L. Recent advances in the diagnosis of allergic rhinitis. Expert Rev Clin Immunol. 2018;14:957-64. [DOI] [PubMed] [Google Scholar]
- 25.Ullah R, Khan S, Ali H, Chaudhary II, Bilal M, Ahmad I. A comparative study of machine learning classifiers for risk prediction of asthma disease. Photodiagn Photodyn Ther. 2019;28:292-96. [DOI] [PubMed] [Google Scholar]
- 26.Segura BI, Colón RC, Tejedor AM, Moro-Moro M. Predicting of anaphylaxis in big data EMR by exploring machine learning approaches. J Biomed Inform. 2018;87:50-59. [DOI] [PubMed] [Google Scholar]
- 27.Wide L, Bennich H, Johansson SG. Diagnosis of allergy by an in-vitro test for allergen antibodies. Lancet. 1967;2:1105-1107. [DOI] [PubMed] [Google Scholar]
- 28.Waring J, Lindvall C, Umeton R. Automated machine learning: review of the state-of-the-art and opportunities for healthcare. Artif Intell Med. 2020;104:101822. [DOI] [PubMed] [Google Scholar]
- 29.Kalaiselvi B, Thangamani M. An efficient Pearson correlation based improved random forest classification for protein structure prediction techniques. Measurement. 2020;162:107885. [Google Scholar]
- 30.Kadkhodaei HR, Moghadam AME, Dehghan M. HBoost: a heterogeneous ensemble classifier based on the Boosting method and entropy measurement. Expert Syst Appl. 2020;157:113482. [Google Scholar]
- 31.Bhardwaj R, Hooda N. Prediction of Pathological Complete Response after Neoadjuvant Chemotherapy for breast cancer using ensemble machine learning. Inf Med Unlocked. 2019;16:100219. [Google Scholar]