Abstract
Osmotic stress was found to induce biofilm formation in a Staphylococcus aureus mucosal isolate. Inactivation of a global regulator of the bacterial stress response, the alternative transcription factor ςB, resulted in a biofilm-negative phenotype and loss of salt-induced biofilm production. Complementation of the mutant strain with an expression plasmid encoding ςB completely restored the wild-type phenotype. The combined data suggest a critical role of ςB in S. aureus biofilm regulation under environmental stress conditions.
For numerous pathogenic bacteria, biofilms represent a source of persisting and relapsing infections and thus contribute significantly to pathogenesis (4). Biofilms seem to protect bacteria from unfavorable external conditions, and, in some bacterial ecosystems, the conversion of planktonic cells into a biofilm-producing community is triggered by environmental stress factors (6, 12, 13). In the human pathogen Staphylococcus aureus, biofilm formation is mediated by the production of the extracellular polysaccharide adhesin PIA, whose synthesis depends on the expression of the icaADBC-encoded enzymes (5, 15). The regulation of biofilm expression in this organism is poorly understood. All S. aureus strains analyzed so far contain the entire ica gene cluster, but only a few express the operon and produce biofilms in vitro (5). In this study, we investigated whether the alternative transcription factor ςB is involved in the regulation of ica expression. ςB is known to be a global regulator of the stress response in S. aureus and also influences various virulence-associated genes (7, 11, 16, 20). To elucidate the possible role of ςB in biofilm formation, we used a genetic approach and constructed an S. aureus sigB::ermB insertion mutant of the biofilm-forming, methicillin-sensitive mucosal isolate S. aureus MA12 (18). Biofilm formation and ica expression of the mutant were compared with the phenotypes of the corresponding wild-type strain and a complemented strain that carried a sigB copy on an expression vector.
Construction of a sigB insertion mutant and complementation of the mutation.
The inactivation of sigB was done by insertion of an erythromycin resistance cassette into the sigB gene of S. aureus MA12 by double-crossover integration. For this purpose, the temperature-sensitive shuttle vector pSK8, which carries a sigB::ermB mutation, was constructed. A 937-bp fragment containing the entire sigB gene was amplified by PCR from S. aureus MA12 by using the primers 5′ CGG GAT CCG GTG TGA CAA TCA GTA TGA C 3′ and 5′ CGG AAT TCG CGA CAT TTA TGT GGA TAC AC 3′. The DNA fragment was inserted into the shuttle vector pBT1 (17), resulting in pSK7. Then the ermB cassette of pEC1 (1) was excised by XbaI-HindIII digestion, treated with the Klenow fragment of Escherichia coli DNA polymerase, and ligated with EcoRV-digested pSK7, resulting in plasmid pSK8. Following passage through the restriction-negative strain S. aureus RN4220, pSK8 was reisolated and transformed into S. aureus MA12 by electroporation (19). Replacement of the chromosomal S. aureus MA12 sigB wild-type gene was achieved by double-crossover integration of the sigB::ermB insert of pSK8 following a temperature shift to the nonpermissive temperature of the shuttle vector (42°C). Erythromycin-resistant and chloramphenicol-sensitive colonies were isolated, and the sigB::ermB integrations were confirmed by Southern hybridization, PCR, and nucleotide sequencing (data not shown). From these experiments, the sigB::ermB insertion mutant S. aureus MA12.2 was selected for further analysis. To restore the ςB function, the S. aureus MA12.2 sigB mutant strain was complemented with plasmid pSK9. Plasmid pSK9 was constructed by inserting the sigB PCR fragment into the expression shuttle vector pHPS9 (8). Prior to transformation into S. aureus MA12.2, the vector was transformed into the restriction-negative cloning host S. aureus RN4220. The plasmid was reisolated and transferred into S. aureus MA12.2, resulting in the complemented strain S. aureus MA12.2-1(pSK9).
Analysis of the ςB function.
Inactivation of the sigB gene in S. aureus MA12.2 and restoration of its function in the complemented strain S. aureus MA12.2-1(pSK9) was investigated by Northern analysis of the asp23-specific gene expression. asp23 encodes an alkaline shock protein, and the gene was shown to be preceded by a ςB-dependent promoter (7, 11). Recent studies have shown that asp23 transcription is absent in S. aureus sigB mutants. It has therefore been concluded that asp23 transcription reliably reflects the activity of the ςB factor (7, 11, 16).
The Northern hybridization experiments revealed a strong asp23-specific signal in both the wild-type and the complemented strains. In contrast, no asp23-specific transcription was detected in the mutant strain S. aureus MA12.2 (data not shown). Furthermore, as expected for a ςB-negative strain (2, 11), and in contrast to both the wild-type and complemented strains, the mutant strain had lost its yellow pigmentation and showed enhanced hemolysis on blood agar plates (data not shown). Taken together, these data indicate that S. aureus MA12.2 is a sigB mutant and that the sigB mutation is restored in the complemented strain S. aureus MA12.2-1(pSK9).
Effect of the sigB mutation on biofilm production and ica expression.
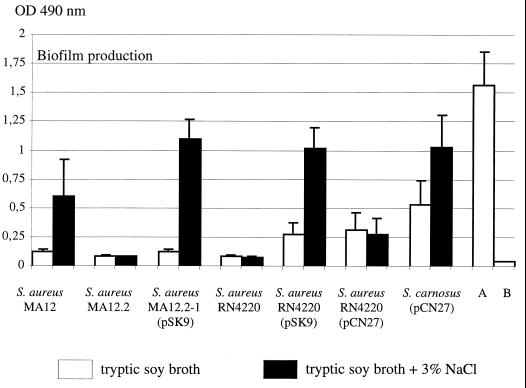
Quantitative biofilm measurement was done in a microtiter assay as described previously (3, 22). Bacteria were grown overnight in 96-well, flat-bottomed tissue culture plates (Greiner, Nürtingen, Germany) at 37°C using either tryptic soy broth (TSB) (Difco, BBL, Detroit, Mich.) or TSB supplemented with 3% sodium chloride as growth medium. Based upon the optical densities (OD) of the biofilms, the strains were classified as nonadherent strains (OD ≤ 0.120), weak biofilm producers (0.120 < OD ≤ 0.240), or strongly adherent strains (OD > 0.240) according to the scheme introduced by Christensen et al. (3). As shown in Fig. 1, the biofilm formation of S. aureus MA12 was weak when the strain was grown in TSB, but it could be significantly stimulated under osmotic stress conditions (that is, TSB containing 3% sodium chloride). In contrast, the S. aureus MA12.2 sigB insertion mutant failed to produce any detectable biofilm, irrespective of whether it was grown in TSB alone or in TSB containing 3% sodium chloride. In the case of S. aureus MA12.2(pSK9), in which the inactivated chromosomal copy of sigB was complemented by an expression plasmid carrying sigB, both basal and the osmotic stress-stimulated biofilm formation were restored.
FIG. 1.
Biofilm formation on polystyrene tissue culture plates of the wild-type strain (S. aureus MA12), the sigB::ermB insertion mutant (S. aureus MA12.2), the complemented strain [S. aureus MA12.2-1(pSK9)], the cloning host S. aureus RN4220, S. aureus RN4220(pSK9), and S. aureus RN4220(pCN27) carrying the icaADBC operon of S. epidermidis on plasmid pCN27 (9), and S. carnosus TM300(pCN27) (9) after growth in unsupplemented TSB and TSB supplemented with 3% sodium chloride, respectively. A, S. epidermidis RP62A (positive control); and B, S. carnosus TM300 (negative control).
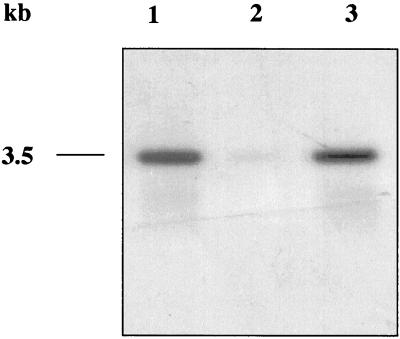
Next, we examined the effect of the sigB inactivation on the transcription of the ica gene cluster by Northern blotting experiments. To this end, RNA was isolated from mid-log cultures of the wild-type strain, the sigB mutant, and the complemented strain, which were grown in TSB containing 3% sodium chloride. Hybridization with a 32P-radiolabeled icaA-specific gene probe revealed a strong signal in the S. aureus MA12 wild-type strain (Fig. 2, lane 1). In contrast, ica transcription was drastically diminished in the sigB mutant (Fig. 2, lane 2) and restored in the complemented strain S. aureus MA12.2SK(pSK9) (Fig. 2, lane 3). These data are consistent with the observed differences in biofilm production under high-salt conditions between the wild-type and mutant strains and again support the conclusion that sigB is involved in the control of biofilm formation in S. aureus MA12.
FIG. 2.
Northern blot analysis of ica transcription in the S. aureus MA12 wild-type strain (lane 1), the sigB::ermB insertion mutant S. aureus MA12.2 (lane 2), and the complemented strain S. aureus MA12.2-1(pSK9) (lane 3) after growth in TSB supplemented with 3% sodium chloride.
We then extended the study to another S. aureus strain, RN4220. S. aureus RN4220 is a restriction-negative strain that is commonly used as a cloning host for plasmids prior to their transformation into the staphylococcal strain of interest (10). S. aureus RN4220 had been derived from S. aureus 8325, which was described recently as a spontaneous sigB-negative mutant with an 11-bp deletion in the rsbU regulatory gene (11, 20). Normally, S. aureus RN4220 does not produce biofilms, under either low-salt or high-salt conditions (Fig. 1). We found that, when transformed with plasmid pSK9, this strain formed biofilms and, moreover, that the biofilm production was induced under high-salt conditions. These results led us to propose that the biofilm-negative phenotype of S. aureus RN4220 is caused by the absence of ςB. Obviously, this conclusion appears to contradict previous results which demonstrated that S. aureus RN4220 exhibits a biofilm-positive phenotype when the icaADBC genes are provided on a plasmid (5, 14). Therefore, we have done an additional set of experiments. We have transformed plasmid pCN27 (9), which carries the entire ica operon of Staphylococcus epidermidis, into S. aureus RN4220. Consistent with previous data (14), the strain formed a biofilm when grown in TSB (Fig. 1). However, in obvious contrast to the pSK9 complementation experiments described above, no induction of biofilm production was observed under osmotic stress conditions. Finally, we found that propagation of pCN27 in Staphylococcus carnosus TM300 (9) increased the biofilm production of this strain under high-salt conditions. The combined data led us to hypothesize that the biofilm-forming phenotype of S. aureus RN4220(pCN27) may result from a basal, vector-driven ica expression, which is enhanced by the copy effect of the plasmid. The absence of biofilm induction, however, suggests that ςB is required for the activation of biofilm formation in response to osmotic stress.
Conclusions.
In this study, we provide evidence that the biofilm-forming phenotype observed in a mucosal isolate of S. aureus can be induced by changing environmental conditions (in this case, osmotic stress). The results are consistent with our recent data obtained for S. epidermidis (S. Rachid and W. Ziebuhr, unpublished data), which demonstrated that the expression of the biofilm-mediating ica operon is enhanced by high osmolarity, high temperature, and subinhibitory concentrations of certain antibiotics. In contrast to S. epidermidis, all S. aureus strains analyzed so far carry the ica gene cluster, but only a few spontaneously express biofilms in vitro (5). It is thus tempting to speculate that, in these biofilm-negative strains, the ica expression may be tightly controlled. The data presented in this study strongly support the idea that this suppression might be overcome by activation of ςB in response to external stress. Another explanation for the biofilm-negative phenotype in the majority of the S. aureus strains would be the presence of mutations in the sigB gene itself or in its adjacent rsbU, rsbV, and rsbW regulatory genes (11, 20). It is also conceivable that S. aureus biofilm production may be influenced by mechanisms that have been shown to be used by S. epidermidis in the control of the ica expression (e.g., phase variation and gene rearrangements) (21–23). All these hypotheses still need careful experimental evaluation. It should also be noted that, in this study, we have characterized only the mucosal isolate S. aureus MA12 in detail. Even though we have obtained conclusive evidence for the critical involvement of the ςB factor in biofilm formation in this isolate, the actual role of ςB in ica expression remains to be determined. The nucleotide sequence immediately upstream of the ica operon does not resemble any of the known ςB-dependent promoter structures, suggesting an indirect activation of the ica expression by additional unknown regulatory factors. This idea is also supported by the identification of S. aureus strains which express the sigB gene but, nevertheless, are biofilm negative (Rachid and Ziebuhr, unpublished). To answer the question of whether ςB is directly or indirectly involved in S. aureus ica expression, more experimental work, including primer extension analyses under different growth conditions, is needed. Finally, a broad range of S. aureus strains has to be analyzed to identify possible variations in biofilm regulation among different strains.
Acknowledgments
This work was supported by the BMBF (grant no. 01KI9608), the Deutsche Forschungsgemeinschaft (Graduiertenkolleg Infektiologie), and the Fond der Chemischen Industrie.
We are grateful to Jürgen Kreft, Lehrstuhl für Mikrobiologie, Universität Würzburg, for providing plasmid pHSP9, and to Friedrich Götz, Mikrobielle Genetik, Universität Tübingen, for plasmid pCN27.
REFERENCES
- 1.Brückner R. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol Lett. 1997;151:1–8. doi: 10.1111/j.1574-6968.1997.tb10387.x. [DOI] [PubMed] [Google Scholar]
- 2.Cheung A L, Chien Y-T, Bayer A S. Hyperproduction of alpha-hemolysin in a sigB mutant is associated with elevated SarA expression in Staphylococcus aureus. Infect Immun. 1999;67:1331–1337. doi: 10.1128/iai.67.3.1331-1337.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christensen G D, Simpson W A, Younger J J, Baddour L M, Barrett F F, Melton D M, Beachey E H. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J Clin Microbiol. 1985;22:996–1006. doi: 10.1128/jcm.22.6.996-1006.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costerton J W, Stewart P S, Greenberg E P. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 5.Cramton S E, Gerke C, Schnell N F, Nichols W W, Götz F. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect Immun. 1999;67:5427–5433. doi: 10.1128/iai.67.10.5427-5433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deretic V, Schurr M J, Boucher J C, Martin D W. Conversion of Pseudomonas aeruginosa to mucoidy in cystic fibrosis: environmental stress and regulation of bacterial virulence by alternative sigma factors. J Bacteriol. 1994;176:2773–2780. doi: 10.1128/jb.176.10.2773-2780.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gertz S, Engelmann S, Schmid R, Ohlsen K, Hacker J, Hecker M. Regulation of sigmaB-dependent transcription of sigB and asp23 in two different Staphylococcus aureus strains. Mol Gen Genet. 1999;261:558–566. doi: 10.1007/s004380051001. [DOI] [PubMed] [Google Scholar]
- 8.Haima P, van Sinderen D, Bron S, Venema G. An improved beta-galactosidase alpha-complementation system for molecular cloning in Bacillus subtilis. Gene. 1990;93:41–47. doi: 10.1016/0378-1119(90)90133-c. [DOI] [PubMed] [Google Scholar]
- 9.Heilmann C, Schweitzer O, Gerke C, Vanittanakom N, Mack D, Götz F. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol. 1996;20:1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x. [DOI] [PubMed] [Google Scholar]
- 10.Kreiswirth B N, Lofdahl S, Betley M J, O'Reilly M, Schlievert P M, Bergdoll M S, Novick R P. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 11.Kullik I, Giachino P, Fuchs T. Deletion of the alternative sigma factor ςB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J Bacteriol. 1998;180:4814–4820. doi: 10.1128/jb.180.18.4814-4820.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LaPaglia C, Hartzell P L. Stress-induced production of biofilm in the hyperthermophile Archaeoglobus fulgidus. Appl Environ Microbiol. 1997;63:3158–3163. doi: 10.1128/aem.63.8.3158-3163.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathee K, Ciofu O, Sternberg C, Lindum P W, Campbell J I, Jensen P, Johnsen A H, Givskov M, Ohman D E, Molin S, Hoiby N, Kharazmi A. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology. 1999;145:1349–1357. doi: 10.1099/13500872-145-6-1349. [DOI] [PubMed] [Google Scholar]
- 14.McKenney D, Hübner J, Muller E, Wang Y, Goldmann D A, Pier G B. The ica locus of Staphylococcus epidermidis encodes production of the capsular polysaccharide/adhesin. Infect Immun. 1998;66:4711–4720. doi: 10.1128/iai.66.10.4711-4720.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKenney D, Pouliot K L, Wang Y, Murthy V, Ulrich M, Doring G, Lee J C, Goldmann D A, Pier G B. Broadly protective vaccine for Staphylococcus aureus based on an in vivo-expressed antigen. Science. 1999;284:1523–1527. doi: 10.1126/science.284.5419.1523. [DOI] [PubMed] [Google Scholar]
- 16.Miyazaki E, Chen J-M, Ko C, Bishai W R. The Staphylococcus aureus rsbW (orf159) gene encodes an anti-sigma factor of SigB. J Bacteriol. 1999;181:2846–2851. [Google Scholar]
- 17.Ohlsen K, Koller K-P, Hacker J. Analysis of expression of the alpha-toxin gene (hla) of Staphylococcus aureus by using a chromosomally encoded hla::lacZ gene fusion. Infect Immun. 1997;65:3606–3614. doi: 10.1128/iai.65.9.3606-3614.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohlsen K, Ziebuhr W, Koller K-P, Hell W, Wichelhaus T A, Hacker J. Effects of subinhibitory concentrations of antibiotics on alpha-toxin (hla) gene expression of methicillin-sensitive and methicillin-resistant Staphylococcus aureus isolates. Antimicrob Agents Chemother. 1998;42:2817–2823. doi: 10.1128/aac.42.11.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schenk S, Laddaga R A. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol Lett. 1992;73:133–138. doi: 10.1016/0378-1097(92)90596-g. [DOI] [PubMed] [Google Scholar]
- 20.Wu S, de Lencastre H, Tomasz A. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J Bacteriol. 1996;178:6036–6042. doi: 10.1128/jb.178.20.6036-6042.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ziebuhr W, Dietrich K, Trautmann M, Wilhelm M. Chromosomal rearrangements affecting biofilm production and antibiotic resistance in a S. epidermidis strain causing shunt-associated ventriculitis. Int J Med Microbiol. 2000;290:115–120. doi: 10.1016/S1438-4221(00)80115-0. [DOI] [PubMed] [Google Scholar]
- 22.Ziebuhr W, Heilmann C, Götz F, Meyer P, Wilms K, Straube E, Hacker J. Detection of the intercellular adhesion gene cluster (ica) and phase variation in Staphylococcus epidermidis blood culture strains and mucosal isolates. Infect Immun. 1997;65:890–896. doi: 10.1128/iai.65.3.890-896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ziebuhr W, Krimmer V, Rachid S, Lößner I, Götz F, Hacker J. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol Microbiol. 1999;32:345–356. doi: 10.1046/j.1365-2958.1999.01353.x. [DOI] [PubMed] [Google Scholar]