Abstract
In the peptidoglycan of Mycobacterium leprae, l-alanine of the side chain is replaced by glycine. When expressed in Escherichia coli, MurC (UDP-N-acetyl-muramate:l-alanine ligase) of M. leprae showed Km and Vmax for l-alanine and glycine similar to those of Mycobacterium tuberculosis MurC, suggesting that another explanation should be sought for the presence of glycine.
Some chemical differences exist in the peptidoglycan of Mycobacterium spp. compared to other bacteria (3). Mycobacterial muramic acid is thought to be glycolylated instead of acetylated (14). In the case of Mycobacterium leprae, the first amino acid in the tetrapeptide side chain of the peptidoglycan is Gly instead of l-Ala (5), as found in Mycobacterium tuberculosis and many other bacteria, implying that the M. leprae genome may encode a unique UDP-N-acetylmuramate:l-Ala/Gly (UDP-MurNAc:l-Ala/Gly) ligase (MurC) specific for the addition of Gly to UDP-MurNAc. These special structural features of mycobacterial peptidoglycan suggest the presence of unique enzymes that could be exploited as drug targets.
The genes that encode MurC from several organisms have been sequenced (1, 6, 8, 11, 13), and the Escherichia coli MurC has been overexpressed and characterized (7, 11). However, the mycobacterial counterparts have not been studied. The availability of the genome sequences of M. tuberculosis (4) and M. leprae (ftp://ftp.sanger.ac.uk/pub/pathogens/leprae/) provides an opportunity to study the enzymes of these two pathogenic species, especially important in the case of M. leprae, which is not accessible to direct enzymatic study.
murC genes of Mycobacterium.
The complete sequence of the open reading frame of MLCB268.01c was revealed from the assembled genome sequence of M. leprae; it corresponds to bp 1084518 to 1086003. The resulting protein contains 595 amino acid residues with a theoretical molecular mass of 51 kDa, very similar to M. tuberculosis MurC (about 79% identity) but with only ∼34% identity to E. coli MurC. Both Rv2152c (M. tuberculosis) and MLCB268.01 (M. leprae) are found within the mra cluster and contain eight of nine invariant amino acids (2) that align perfectly with known MurCs from other organisms (Fig. 1). However, the MurC homologs found outside the mra clusters (Rv3712 and MLCB2407.24c) have only ∼22% identity with the putative MurCs found within the clusters, and four of the nine invariant amino acids either are absent or did not align.
FIG. 1.
Alignment of amino acid residues of MurC from M. tuberculosis, M. leprae, and E. coli, showing conserved residues (highlighted).
Cloning, expression, and purification of UDP-N-MurNAc:l-Ala ligase (MurC).
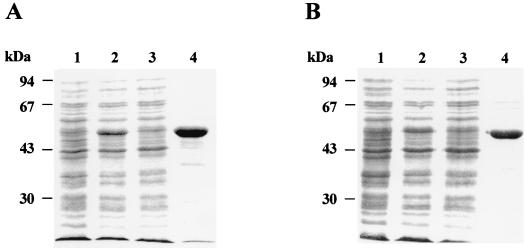
Rv2152c and Rv3712 were amplified from M. tuberculosis H37Rv genomic DNA and cloned into the pET29a+ vector (Novagen, Madison, Wis.) (16), yielding pSM201 and pSM203, respectively. The M. leprae MLCB268.01 and MLCB2407.24c genes were amplified from M. leprae genomic DNA and cloned into pET28a+ and pET29a+, respectively, yielding pSM206 and pSM208, respectively (16, 17). E. coli BL21(DE3) harboring plasmid pSM201, pSM203, pSM206, or pSM208 was grown in Luria-Bertani broth containing kanamycin, induced with isopropyl-β-d-thiogalactopyranoside, lysed by sonication on ice, and centrifuged at 30,000 × g for 30 min (17). The resulting supernatant containing the soluble His-tagged fusion proteins were loaded on a nickel-nitrilotriacetic acid (Ni-NTA) resin (Qiagen) column (18) which was washed with 20 mM Tris-HCl (pH 8.0)–10 mM MgCl2–2 mM β-mercaptoethanol–30 mM imidazole (pH 8.0) and 0.5 M NaCl, and the His-tagged proteins were eluted from the column with buffer containing 300 mM imidazole (pH 7.5) (18). Protein-containing fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10) to show a high degree of purification (Fig. 2).
FIG. 2.
SDS-PAGE gel showing the purification of Rv2152c (A) and MLCB268.01c (B). Whole-cell lysate of uninduced E. coli BL21(DE3) cells harboring pSM201 and pSM206 (lanes 1A and 1B, respectively), whole-cell lysates of isopropyl-β-d-thiogalactopyranoside-induced E. coli BL21(DE3) cells overproducing M. tuberculosis and M. leprae MurC (Lanes 2A and 2B, respectively), and clarified cell extracts (supernatant) obtained by centrifugation at 30,000 × g of the whole-cell lysate (lanes 3A and 3B). Note that most of the overexpressed protein was insoluble and hence gives the impression of lesser expression. Purified MurC proteins are shown in lanes 4A and 4B. In each panel, the positions of molecular size markers are shown on the left.
Assay for UDP-MurNAc:l-Ala ligase (MurC).
Purified fractions were pooled, and imidazole was removed by dialysis. The UDP-MurNAc:l-Ala and UDP-MurNAc:Gly ligase activities were assayed as described by Liger et al. (12). For this purpose, UDP-MurNAc was prepared by a two-step coupled enzymatic conversion of UDP-GlcNAc to UDP-MurNAc according to Jin et al. (9) and identified through negative-ion fast atom bombardment-mass spectroscopy (FAB-MS) (the expected mass of 679 was observed) and nuclear magnetic resonance (NMR) (300 MHz). The following signals were clearly identified by 1H NMR spectroscopy in heavy water at 300 MHz: d7.94 (doublet, j = 8.1HZ, H-6; uracyl), d 5.96 (doublet, j = 4.5HZ, H-1; ribosyl), d 5.96 (doublet, j = 8.1HZ, H-5; uracyl), d 5.60 broadened doublet (j = 4.2HZ, H-1; muramyl), d 2.03 (singlet, methyl; N-acetyl muramyl), and d 1.32 (doublet, j = 6.6HZ, methyl; lactyl-muramyl). Reaction mixtures contained 100 mM Tris-HCl (pH 8.6), 25 mM (NH4)2SO4, 20 mM MgCl2, 2 mM β-mercaptoethanol, 1 mM UDP-MurNAc, 2 mM ATP, 50 μM l-[14C]Ala (specific activity, 164 mCi/mmol) (ICN Radiochemicals, Irvine, Calif.) or [14C]Gly (46.87 mCi/mmol) (NEN Life Science Products, Boston, Mass.), and a predetermined amount of crude cell lysate or purified enzyme in a 25-μl reaction mix. Reactions were conducted under conditions in which product formation was linear with respect to both time and protein concentration. Reactions were stopped by the addition of 10 μl of glacial acetic acid and briefly centrifuged, and 3.5 μl of the supernatant was applied to a silica gel thin-layer chromatography plate which was developed in isobutyric acid–1 M ammonium hydroxide (5:3) to separate the reaction product from unreacted amino acids. Radioactivity was measured using Bioscan Imaging Scanner System 200-IBM (Bioscan Inc., Washington, D.C.). The proportion of counts of substrate and product compared to the total counts applied to the plate was used to calculate enzyme activity.
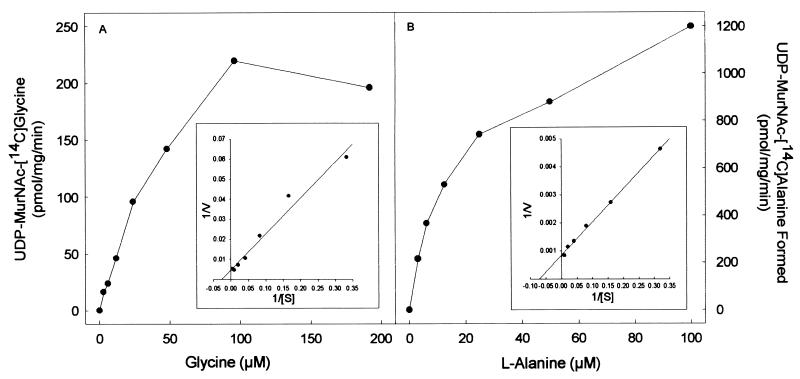
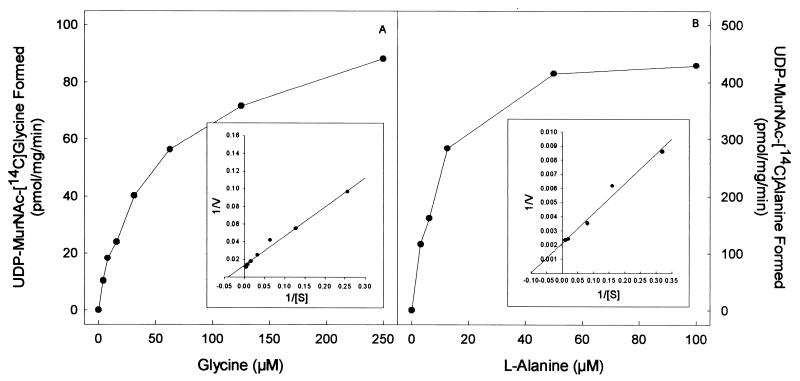
The purified proteins arising from cloned Rv2152c and MLCB268.01c showed good ligase activity using both l-Ala and Gly as substrates (Fig. 3 and 4). The products of the ligase reactions were also analyzed by MS; UDP-MurNAc-l-Ala gave the expected molecular weight of 750, and the UDP-MurNAc-Gly gave the expected molecular weight of 736. The Km and Vmax of both Rv2152c and MLCB268.01c were determined in the presence of either Gly or l-Ala (Fig. 3 and 4). Rv2152c showed an apparent Km of 38 μM and a Vmax of 220 pmol/mg/min for Gly. When assayed with various amounts of l-Ala, this enzyme showed an apparent Km of 14 μM and a Vmax of 1,200 pmol/mg/min. Even though the Km values for both of these substrates were similar, the Vmax for l-Ala was found to be much greater than that seen for Gly, indicating better catalysis with l-Ala as the substrate. Similar results were obtained with MLCB268.01c; this M. leprae enzyme had an apparent Km of 25 μM and a Vmax of 76 pmol/mg/min for Gly, and, when assayed with various amounts of l-Ala, this enzyme showed an apparent Km of 10 μM and a Vmax of 460 pmol/mg/min.
FIG. 3.
Effect of amino acid concentration on the rate of UDP-MurNAc-[14C]Gly (A) or UDP-MurNAc-l-[14C]Ala (B) biosynthesis by purified Rv2152c from M. tuberculosis. The apparent Km and Vmax values were derived from a double-reciprocal plot of these data (inset).
FIG. 4.
Effect of amino acid concentration on the rate of UDP-MurNAc:[14C]Gly (A) or UDP-MurNAc:l-[14C]Ala (B) biosynthesis by purified MLCB268.01c from M. leprae. The apparent Km and Vmax values were derived from a double-reciprocal plot of these data (inset).
Nonactive MurC homologs outside the mra cluster.
The MurC homologues Rv3712 and MLCB2407.24c were also expressed in E. coli as C-terminally His-tagged proteins using the T7 expression system, resulting in soluble proteins capable of being purified by Ni-NTA column chromatography. However, the purified proteins as well as the crude cell lysate from the overproducing E. coli cells showed no significant ligase activity when tested.
Molecular organization of the M. leprae, M. tuberculosis, and E. coli mra clusters.
In general, the basic genetic organization of the mra gene cluster responsible for peptidoglycan biosynthesis (19) of M. tuberculosis, M. leprae, and E. coli is similar except for four additional open reading frames in the M. tuberculosis mra cluster between pbpB and murE (Fig. 5). Clearly, Rv2152c from M. tuberculosis and MLCB268.01c from M. leprae encode the MurC enzymes of their respective species, in that the E. coli-overexpressed enzymes were enzymatically active, indicating that they underwent proper folding even when expressed in a nonhomologous system. The properties of these two ligases are very similar. The calculated apparent Km for Gly of both ligases was found to be much lower than the reported Km value of ∼2.5 to 10 mM for Gly of E. coli MurC (7, 12). The mycobacterial MurCs also had similar Km values for l-Ala, and in both cases, this value was slightly lower than the Km for Gly. However, the apparent Vmax for l-Ala is much higher than that for Gly in both cases, suggesting better catalysis of l-Ala ligase activity.
FIG. 5.
Comparison of the chromosomal organization of the mra clusters in M. tuberculosis, M. leprae, and E. coli.
The other two open reading frames (Rv3712 and MLCB2407.24c) that show homology to E. coli murC do not appear to encode any ligase activity, probably due to the absence of four of the nine invariant amino acids found in bona fide members of the MurC enzyme family. Therefore it can be concluded that M. tuberculosis and M. leprae, like other bacteria, have only one such ligase. Thus, the presence of a Gly-specific ligase can apparently be ruled out as the reason for the specific occurrence of Gly instead of l-Ala in the M. leprae peptidoglycan. M. leprae is always derived from host tissue because it is not possible to cultivate it in vitro, which may be due to the unusual peptidoglycan structure in this species. When E. coli and Salmonella cells are grown in human epithelial cells, changes in the chemical composition of the peptidoglycan are observed (15). From the data presented here, it can be hypothesized that, in M. leprae, the incorporation of Gly into peptidoglycan is due to a combination of the substrate specificity of the MurC and the nature of the intracellular environment.
Acknowledgments
We thank Philip Draper for his helpful discussions.
M. tuberculosis genomic DNA was obtained from J. T. Belisle through NIH, NIAID contract NO1 AI-75320. M. leprae genomic DNA was obtained through the resources of NIH, NIAID contract NO1 AI-55262. This work was supported by grant NIH, NIAID 18357 and contract NIH, NIAID NO1 AI-55262.
REFERENCES
- 1.Ansai T, Yamashita Y, Awano S, Shibata Y, Wachi M, Nagai K, Takehara T. A murC gene in Porphyromonas gingivalis 381. Microbiology. 1995;141:2047–2052. doi: 10.1099/13500872-141-9-2047. [DOI] [PubMed] [Google Scholar]
- 2.Bouhss A, Mengin-Lecreulx D, Blanot D, Van Heijenoort J, Parquet C. Invariant amino acids in the Mur peptide synthetases of bacterial peptidoglycan synthesis and their modification by site-directed mutagenesis in the UDP-MurNAc:l-alanine ligase from Escherichia coli. Biochemistry. 1997;36:11556–11563. doi: 10.1021/bi970797f. [DOI] [PubMed] [Google Scholar]
- 3.Brennan P J, Nikaido H. The envelope of mycobacteria. Annu Rev Biochem. 1995;64:29–63. doi: 10.1146/annurev.bi.64.070195.000333. [DOI] [PubMed] [Google Scholar]
- 4.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eigelmeier K, Gas S, Barry III C E, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 5.Draper P, Kandler O, Darbre A. Peptidoglycan and arabinogalactan of Mycobacterium. J Gen Microbiol. 1987;133:1187–1194. doi: 10.1099/00221287-133-5-1187. [DOI] [PubMed] [Google Scholar]
- 6.Duez C, Thamm I, Sapunaric F, Coyette J, Ghuysen J M. The division and cell wall gene cluster of Enterococcus hirae S185. DNA Seq. 1998;9:149–161. doi: 10.3109/10425179809072190. [DOI] [PubMed] [Google Scholar]
- 7.Emanuele J J, Jr, Jin H, Jacobson B L, Chang C Y, Einspahr H M, Villafranca J J. Kinetic and crystallographic studies of Escherichia coli UDP-N-acetylmuramate:l-alanine ligase. Protein Sci. 1996;5:2566–2574. doi: 10.1002/pro.5560051219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hishinuma F, Izaki K, Takahashi H. Inhibition of l-alanine adding enzyme by glycine. Agric Biol Chem. 1971;35:2050–2058. [Google Scholar]
- 9.Jin H, Emanuele J J, Jr, Fairman R, Robertson J G, Hail M E, Ho H T, Falk P J, Villafranca J J. Structural studies of Escherichia coli UDP-N-acetylmuramate:l-alanine ligase. Biochemistry. 1996;35:1423–1431. doi: 10.1021/bi952334k. [DOI] [PubMed] [Google Scholar]
- 10.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 11.Liger D, Masson A, Blanot D, Van Heijenoort J, Parquet C. Over-production, purification and properties of the uridine-diphosphate-N-acetylmuramate:l-alanine ligase from Escherichia coli. Eur J Biochem. 1995;230:80–87. doi: 10.1111/j.1432-1033.1995.0080i.x. [DOI] [PubMed] [Google Scholar]
- 12.Liger D, Masson A, Blanot D, Van Heijenoort J, Parquet C. Study of the overproduced uridine-diphosphate-N-acetylmuramate:l-alanine ligase from Escherichia coli. Microb Drug Resist. 1996;2:25–27. doi: 10.1089/mdr.1996.2.25. [DOI] [PubMed] [Google Scholar]
- 13.Lowe A M, Deresiewicz R L. Cloning and sequencing of Staphylococcus aureus murC, a gene essential for cell wall biosynthesis. DNA Seq. 1999;10:19–23. doi: 10.3109/10425179909033931. [DOI] [PubMed] [Google Scholar]
- 14.Petit J F, Adam A, Wietzerbin-Falszpan J, Lederer E, Ghuysen J M. Chemical structure of the cell wall of Mycobacterium smegmatis: isolation and partial characterization of the peptidoglycan. Biochem Biophys Res Commun. 1969;35:478–485. doi: 10.1016/0006-291x(69)90371-4. [DOI] [PubMed] [Google Scholar]
- 15.Quintela J C, de Pedro M A, Zollner P, Allmaier G, Garcia-del Portillo F. Peptidoglycan structure of Salmonella typhimurium growing within cultured mammalian cells. Mol Microbiol. 1997;23:693–704. doi: 10.1046/j.1365-2958.1997.2561621.x. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 17.Silbaq F S, Cho S-N, Cole S T, Brennan P J. Characterization of a 34-kilodalton protein of Mycobacterium leprae that is isologous to the immunodominant 34-kilodalton antigen of Mycobacterium paratuberculosis. Infect Immun. 1998;66:5576–5579. doi: 10.1128/iai.66.11.5576-5579.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takacs B T, Gordon M-F. Preparation of clinical grade proteins produced by recombinant DNA technologies. J Immunol Methods. 1981;143:231–240. doi: 10.1016/0022-1759(91)90048-k. [DOI] [PubMed] [Google Scholar]
- 19.Van Heijenoort J. Biosynthesis of the bacterial peptidoglycan unit. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. New York, N.Y: Elsevier Science Publishers; 1994. pp. 39–54. [Google Scholar]