Abstract
Autism is associated with genomic instability, which is regulated by telomere length (TL) and index of global methylation (LINE-1). This study will determine relative TL (RTL) and LINE-1 methylation percentage for 69 patients and 33 control subjects to evaluate their potential role as biomarkers for autism. The results displayed a significant decrease of both RTL and LINE-1 methylation in autistic cases relative to controls (P < 0.001). Analysis of receiver operating characteristics curve revealed that both of RTL and LINE-1 methylation percentage have the ability to serve as autism biomarkers (area under the curve = 0.817 and 0.889, respectively). The statistical analysis revealed positive correlation between the two biomarkers (correlation coefficient = 0.439 and P < 0.001).
Keywords: Autism, Relative Telomere Length, LINE-1, Methylation
Introduction
Autism spectrum disorder (ASD) is a group of early-onset and lifelong neurodevelopmental disorders with distinct phenotype of behavioural characteristics (ranging from mild to severe), including impairments in social communication and restricted repetitive behaviours, interests, or activities. Co-occurrence with other disorders, such as motor delays, depression, intellectual disability, anxiety disorder, gastrointestinal disturbances, attention deficit hyperactivity disorder, and seizure disorder, is a common event in ASD patients (Fyke & Velinov, 2021). ASD affects one in every 54 children aged 8 years and is 4.3 times more prevalent in boys than in girls (Maenner et al., 2020). A recent study in Egypt determined the prevalence of ASD to be 5.4/1000 (Yousef et al., 2021).
Complexity and risk factors heterogeneity, including environmental and genetic factors of ASD have been well documented. For example, although numerous single genes, copy-number, and double hit models have been identified in autism, approximately 70% of autistic subjects have unknown causes (Homs et al., 2016). In addition, epigenetic factors such as DNA methylation (Kimura et al., 2019), histone modifications (Sun et al., 2016), and non-coding RNAs (Tonacci et al., 2019) have been implicated in ASD incidence.
A telomere is an evolutionarily conserved tract of repetitive DNA sequence located at the end of a chromosome and bound by a series of specialized proteins to function as protective cap for the chromosome. Mammalian telomeric DNA comprises ~ 5–15 kb of double-stranded tandem repeats (5ʹTTAGGG repeats for the G-rich strand and 5ʹCCCTAA repeats for the complementary C-rich strand) ended in a 3’ single-stranded overhang of ~ 12–300 bases. During cell division, the human telomere loses approximately 50–200 base pairs (Stewart et al., 2012); consequently, telomere length (TL) is a hallmark of cellular senescence and organismal aging, and its shortening is a biomarker for numerous diseases (Zhao et al., 2009).
One of best-characterized epigenetic mechanisms is cytosine residues methylation at CpG sites. 60–90% of CpGs throughout the genome were found to be methylated (Nguyen et al., 2010). Long interspersed element-1 (LINE-1) is a ~ 6,000 bp-long transposable mobile element that exists in over 500,000 copies to represent approximately 17% of the human genome. LINE-1 contains a 5ʹ untranslated region (UTR) with an internal promoter, two non-overlapping open reading frames (ORF1 and ORF2), and a 3ʹ UTR (Tangsuwansri et al., 2018). The proteins encoded by LINE-1 (ORF1p and ORF2p) are responsible for its retrotransposition after their assembly into a functional ribonucleoprotein (RNP) particle. LINE-1 contributes to genomic instability and altered gene expression via a “copy-and-paste” mechanism of amplification (Belancio, 2015). DNA methylation and histone modifications are suggested to play a role in regulating transcription and retrotransposition of LINE-1. Hypomethylation of LINE-1 leads to its reverse transcription into DNA sequence and transposition throughout the genome, contributing to gene disruption and genomic instability (Swets et al., 2016). Altered methylation pattern of LINE-1 is associated with numerous diseases, including cancer (Swets et al., 2016), heart diseases (Guarrera et al., 2015), and neurodevelopmental disorders (Shpyleva et al., 2018). Due to the extensive genomic distribution of LINE-1 and its high methylation level in most normal tissues, LINE-1 methylation regarded as an index and surrogate marker for global DNA methylation (Guarrera et al., 2015).
Both TL and LINE-1 are involved in regulating genomic stability, which is implicated in autism (Williams et al., 2013); therefore, this study will evaluate relative TL (RTL) and LINE-1 methylation as biomarkers for autism, as well as their correlation to each other, disease severity, and other clinical characteristics.
Subjects and Methods
Subjects
The research was conducted in 2019–2020 with 69 Egyptian participants (55 males and 14 females) who met the DSM-V diagnostic criteria for ASD. Diagnosis and severity rating of autism were carried out by Childhood Autism Rating Scale (CARS). Any syndromic ASD was ruled out. The ages of patients ranged from 2 to 20 years (mean age: 6.8 years) and they are classified as children (2–10 year) and adolescents (11–20 year). The ages of the subjects in each group are distributed in small rang, except for three cases that were considered as outliers. All patients were recruited from the outpatient clinic of Clinical Genetics Department, Centre of Excellence of Human Genetics, National Research Centre (NRC), Egypt. They were subjected to detailed medical history assessment (personal, pregnancy, delivery, perinatal and neonatal). Subsequently, 33 age-matched healthy subjects were analyzed as the control group. The onset of symptoms, their progress, the presence of a sleep disorder and gastrointestinal (GIT) manifestation were meticulously documented. EEG was performed on all patients at the Neurophysiological unit, Faculty of Medicine, Kaser El-Ainy, Cairo University (patient’s characteristics are listed in Table 1). It was done under sedation by chloral hydrate for 2 h. Patients provided informed and written consent to participate and the study protocol was approved by the ethical research committee of the NRC (Registration number: 19033).
Table 1.
Demographic and clinical data of autistic subjects
| Data | Number (%) |
|---|---|
| Sex | |
| Males | 55 (79.7) |
| Females | 14 (20.3) |
| Age class | |
| Children | 56 (81.1) |
| Adolescents | 13 (18.9) |
| Severity | |
| Mild | 34 (49.3) |
| Moderate | 22 (31.8) |
| Severe | 13 (18.9) |
| Consanguinity | |
| Positive | 19 (27.5) |
| Negative | 50 (72.5) |
| EEG abnormalities | |
| Positive | 28 (40.6) |
| Negative | 41 (59.4) |
| GIT abnormalities | |
| Positive | 44 (63.8) |
| Negative | 25 (36.2) |
| Sleep disorder | |
| Positive | 11 (16) |
| Negative | 58 (84) |
GIT Gastrointestinal
Telomere Length Measurement
DNA was extracted from peripheral blood leukocytes using standard salting out protocol. 36B4 was used as reference single-copy gene. 80 n of extracted DNA was subjected to qPCR using HERA SYBR green master mix (Willowfort, UK) and primers for telomere (Ujvari et al., 2012) and 36B4 (Willeit et al., 2014) (Primers sequences are listed in Table 2). Reaction was incubated for 2 min at 95 °C and then amplified over 40 cycles of 10 s at 95 °C and 30 s at 60 °C (for telomeres) or 56 °C (for 36B4). The Reactions were run in light cycler 480 II (Roch). For quality control, A no-template control and duplicate calibrator samples were used in all runs to allow for a comparison of the results across all runs (Svikle et al., 2022). RTL was reflected by T/S values (telomere/36B4 ratio): T/S = 2−∆Ct (∆Ct = Ct telomere − Ct36B4).
Table 2.
Sequence of primers used in qPCR and MSP
| Name | Sequence 5 → 3 | Refs. |
|---|---|---|
| Telomere F | GGTTTTTGAGGGTGAGGGTGAGGGTGAGGGTGAGGGT | Ujvari et al. (2012) |
| Telomer R | TCCCGACTATCCCTATCCCTATCCC TATCCCTATCCCTA | |
| 36B4 F | CAGCAAGTGGGAAGGTGTAATCC | Willeit et al. (2014) |
| 36B4 R | CCCATTCTATCATCAACGGGTACAA | |
| LINE-1 M-F | GAGGTATTGTTTTATTTGGGAAGC | |
| LINE-1 M-R | TACTAACAATCAACGAAATTCCGTA | |
| LINE-1 U-F | AGGTATTGTTTTATTTGGGAAGTGT | |
| LINE-1 U-R | TACTAACAATCAACAAAATTCCATA |
F Forward, R Reverse, M Methylated, U Un-methylated
LINE-1 Methylation Analysis
The extracted DNA was subjected to bisulfite modification using the EpiTect Bisulfite kit (Qiagen). The Initial starter amount of DNA was 1.3 µg and the final elution volume was 40 µl. Bisulfite-treated DNA was subjected to methylation specific PCR (MSP), in which two distinct sets of primers (methylated and unmethylated) were employed and amplified. Each MSP reaction was conducted in 15 µl reaction using 7.5 µl of maxima SYBR green master mix (Thermo Scientific), 2 µl of treated DNA, and 150 nM of each primer. The percentage of methylation was calculated using the CT method follows: % meth = 100/[1 + 2ΔCt (meth. − un−meth.)]%. The LINE-1 sequence was downloaded from NCBI (https://www.ncbi.nlm.nih.gov/) (L19088.1). Primers were designed using the free online methprimer tool (http://www.urogene.org/methprimer) which identified two CpG islands with 371 and 101 bp in length, and the primers were designed to span 209 and 210 bp of the first island. The number of CpG sites covered by each primer are zero for methylated forward, two for methylated reverse, one for unmethylated forward and two for unmethylated reverse (sequences are listed in Table 2).
Statistical Analysis
The statistical analysis was carried out using IBM SPSS statistics (Statistical Package for Social Sciences) software version 18.0, IBM Corp., Chicago, USA, 2009. Descriptive statistics were done for quantitative data as minimum and maximum of the range as well as mean ± SD. A Modified Z-score test was used to detect outliers of age. Difference of RTL and LINE-1 methylation percentage between different groups (controls and autistic subjects, males and females, children and adolescents and positive and negative consanguinity) was determined by independent T test. We applied univariate linear model with gender and age as covariates to adjust sex and age effect. In this model, age is reclassified into four groups with corresponding scores (score1, 2–5 years; score 2, 5.1–10 years; score 3, 10.1–15 years; and score 4, 15.1–20 years). Also, sex was scored as a categorical variable as 1 (males) or 2 (females). ANOVA was utilized to compare RTL and LINE-1 methylation levels between different severity groups (mild, moderate, and severe). To determine the effectiveness of RTL and LINE-1 methylation as a discriminatory biomarker for autistic patients, the receiver operating characteristic (ROC) curve was analyzed. Next, Binary logistic regression was used to analyze the combined ROC curve of both biomarkers. The correlation between the two biomarkers was calculated using Pearson’s correlation coefficient. Spearman’s correlation coefficient was employed to determine the degree of correlation between RTL or LINE-1 methylation level and other characteristics, such as age, sex, consanguinity, severity, EEG, sleep disorders, and gastrointestinal disturbance. The level of significance was set at P value < 0.05.
Results
RTL as a Biomarker for Autism
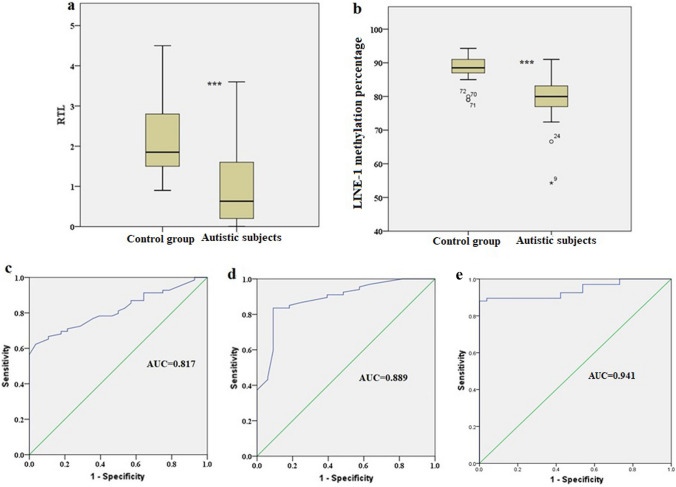
RTL is significantly shorter in autistic patients (1, ± 0.97) compared to the group (2.1; ± 0.93), P < 0.001. We have applied ROC curve analysis to test the predictive ability of RTL to distinguish autistic individuals from normal subjects based on the value of area under the curve (AUC), which is considered more discriminative at more higher value (maximum value is 1 and minimum value is 0.5) (Hajian-Tilaki, 2013). Based on our data, RTL is considered a predictive biomarker for autism; AUC = 0.817 and P < 0.001 (Table 3 and Fig. 1). By comparing RTL between children and adolescents, no statistically significant difference was found (P = 0.880). However, the mean of RTL increased in cases with positive consanguinity (1.3; ± 1) than in cases with negative consanguinity (0.78; ± 0.7) (P = 0.058), and the Spearman’s correlation coefficient between RTL and consanguinity was not statistically significant: r = 0.249 and P = 0.053. Similarly, there was no significant association between RTL and sex: r = 0.228 and P = 0.074 (Table 4).
Table 3.
ROC curve analysis and distribution of RTL and LINE-1methylation percentage between autistic and control subjects
| Biomarker | ROC curve | Independent T test | Adjusted P value* | ||||
|---|---|---|---|---|---|---|---|
| AUC | P value | Mean (± SD) | P value | 95% CI | |||
| Lower | Upper | ||||||
| RTL | |||||||
| Controls (N = 28) | 0.817 | P < .001 | 2.1 (0.93) | P < .001 | − 1.58869 | − .73010 | 0.005 |
| Cases (N = 69) | 1 (0.97) | ||||||
| LINE-1 methylation percentage | |||||||
| Controls (N = 33) | 0.889 | P < .001 | 88.2 (3.8) | P < .001 | − 10.61465 | − 6.12981 | 0.005 |
| Cases (N = 67) | 79.8 (5.8) | ||||||
| Combined | 0.941 | P < .001 | |||||
Significance at P < 0.05
AUC Area under curve
*P value after adjustment of age and sex using univariate linear model
Fig. 1.
Altered RTL and LINE-1 methylation level in autistic patients. a and b represent boxplot for distribution of RTL (a) and LINE-1 methylation percentage (b) in autistic and control groups. c, d and e showing ROC curve analysis of RTL (c), LINE-1 methylation percentage (d) and the combined biomarkers (e) for autism prediction
Table 4.
Absence of association between RTL or LINE-1 methylation percentage and age class, sex and consanguinity
| Independent T test | Spearman correlation | |||
|---|---|---|---|---|
| Mean (± SD) | P | r | P | |
| RTL | ||||
| Age class | ||||
| Children | 0.96 (± 0.9) | 0.88 | − 0.029 | 0.824 |
| Adolescents | 0.91 (± 0.99) | |||
| Sex | ||||
| Males | 0.87 (± 0.9) | 0.2 | 0.228 | 0.074 |
| Females | 1.2 (± 0.86) | |||
| Consanguinity | ||||
| + ve | 1.4 (± 1.1) | 0.058 | 0.249 | 0.053 |
| − ve | 0.79 (± 0.76) | |||
| LINE-1 methylation percentage | ||||
| Age class | ||||
| Children | 79.7 (± 6) | 0.69 | − 0.006 | 0.965 |
| Adolescent | 78.9 (± 6) | |||
| Sex | ||||
| Males | 79.8 (± 6) | 0.45 | ||
| Females | 78.3 (± 5.3) | − 0.076 | 0.558 | |
| Consanguinity | ||||
| + ve | 80.7 (± 5.7) | 0.36 | 0.051 | 0.697 |
| − ve | 79.1 (± 6) | |||
Significance at P < 0.05
r correlation coefficient, + ve positive consanguinity, − ve negative consanguinity
LINE- Methylation Level 1 as a Biomarker for Autism
Our data revealed that autistic subjects have a decreased LINE-1 methylation percentage (79.8%; ± 5.8) than the control group (88.2%; ± 3.8), P < 0.001 (Table 3). By applying ROC curve, LINE-1 methylation percentage is identified as a biomarker for autism: AUC = 0.889 and P < 0.001 (Fig. 1). By comparing LINE-1 methylation percentage between children and adolescents, as well as between males and females, no significant difference was found; P = 0.698 and 0.455, respectively. According to Pearson’s correlation analysis, no significant association was found between LINE-1 methylation percentage and age: r = − 0.06 and P = 0.96. Furthermore, the Spearman’s correlation coefficient showed that there is no significant association between LINE-1 methylation percentage and sex (r = − 0.076 and P = 0.558) or consanguinity (r = 0.051 and P = 0.69, Table 4).
RTL and LINE-Methylation Percentage not Associated with Autism Severity
Regarding severity, there was no difference in the distribution of RTL and LINE-1 methylation percentage among mild, moderate, and severe subjects (P = 0.488 and 0.067, respectively). Also, no correlation was found between RTL or LINE-1 methylation percentage and severity, EEG abnormalities, GIT disturbance or sleep disorder (Table 5).
Table 5.
Absence of association between RTL or LINE-1 methylation percentage and patient’s clinical characteristics
| Comparative means* | Spearman correlation | |||
|---|---|---|---|---|
| Mean (± SD) | P | r | P | |
| RTL | ||||
| Severity | ||||
| Mild | 1(± 0.9) | |||
| Moderate | 0.9 (± 0.6) | 0.9 | 0.067 | 0.609 |
| Severe | 0.8 (± 1.2) | |||
| EEG abnormalities | ||||
| + VE | 0.8 (± 0.9) | 0.156 | 0.235 | 0.082 |
| − VE | 1.1 (± 0.9) | |||
| GIT disorders | ||||
| + VE | 0.9 (± 0.9) | 0.68 | 0.046 | 0.738 |
| − VE | 1 (± 1) | |||
| Sleep disorder | ||||
| + VE | 1.1 (± 1) | 0.59 | − 0.080 | 0.556 |
| − VE | 0.9 (± 0.9) | |||
| LINE-1 methylation percentage | ||||
| Severity | ||||
| Mild | 79.6 (± 5) | |||
| Moderate | 78.1 (± 8) | 0.26 | − 0.173 | 0.190 |
| Severe | 81.9 (± 3.5) | |||
| EEG abnormalities | ||||
| + VE | 80.6 (± 5.3) | 0.17 | − 0.122 | 0.376 |
| − VE | 78.4 (± 6.5) | |||
| GIT disorders | ||||
| + VE | 80.1 (± 5.5) | 0.23 | − 0.092 | 0.503 |
| − VE | 78.1 (± 6.8) | |||
| Sleep disorder | ||||
| + VE | 79.7 (± 5.7) | 0.82 | − 0.084 | 0.540 |
| − VE | 79.3 (± 6) | |||
*Means are compared by ANOVA in case of severity, and by independent T test in case of EEG abnormalities, GIT disorders and sleep disorder. Significance at P < 0.05
Correlation Between RTL and LINE-1 Methylation in Autistic Subjects
We applied Pearson correlation analysis to the data of RTL and LINE-1 methylation to determine the existence of association between their decrease in autistic subjects and found that the correlation is significant (r = 0.439 and P < 0.001). Additionally, logistic regression analysis for the combined ROC curve of RTL and LINE-1 methylation revealed an AUC = 0.941 and a significance level of P < 0.001.
Discussion
As indicated by twin studies, autism represent one of the most heritable developmental disorders and genetic factors are key players in its pathogenesis (Tick et al., 2016), however, fundamental environmental factors have been reported to have a considered implication (Hallmayer et al., 2011; Li et al., 2014). This study focused on two factors related to genetic and environmental burden. The first is altered TL which was found to increase genetic variation, involved in genomic instability, and reflect genetic predisposition (Hackett et al., 2001; Li et al., 2014), and its alteration may be attributed to genetic factors (Asghar et al., 2015; Nersisyan et al., 2019; Shubin & Greider, 2020) and environmental signals (Romano et al., 2013). Secondly, methylation percentage of the index of global methylation, LINE-1, which was reported to play a role in the neurogenesis and survival of neuronal progenitors due to its activation via the canonical WNT pathway (Kuwabara et al., 2009; Tangsuwansri et al., 2018). The involvement of both TL and LINE-1 in genomic stability, led us to explore the presence of a correlation between them in mediating autistic behaviour.
Shortening of TL induces cells to undergo senescence or apoptosis. Many causes are responsible for this shortening of telomere, including reduced telomerase expression, telomere trimming by removal of telomere loop (T-loop), and inflammation which enhance cell proliferation and cause oxidative damage to telomeric DNA and its associated proteins (Eitan et al., 2014). TL is also related to smoking, obesity and air pollution and its shortening has been linked to several neuropsychiatric disorders, such as schizophrenia, Alzheimer’s disease (Hackenhaar et al., 2021) and attention-deficit/hyperactivity disorder (Costa Dde et al., 2015). By comparing RTL between autistic patients and control subjects in our study, a significant decrease was revealed suggesting the potential role of telomere length in mediating autistic behaviour. The ability of RTL to act as biomarker for autism is suggested by the roc curve analysis with high significance P value and AUC. However, no correlation was found between RTL and severity of autism. The relationship between shortened TL and severe sensory symptoms was demonstrated in a previous study, which also found that cognitive functions, rather than autistic traits, are associated with TL in the parents of ASD children, however, in ASD patients, cognitive function and TL were not related (Lewis et al., 2020).
The promoter region of LINE-1 has CpG residues, these residues are regulated by Methyl-CpG binding Protein 2 (MeCP2), which has been implicated in a severe developmental disorder with autistic phenotypes and Rett syndrome (Tangsuwansri et al., 2018). Our data revealed a significant correlation between autism and a significantly decreased LINE-1 methylation percentage compared to control subjects. In addition, the high AUC determined by ROC curve analysis indicates high predictive ability of LINE-1 methylation percentage for autism. Our results differ from those of previous study which used pyrosequencing to analyze LINE-1 in DNA extracted from blood of autistic patients and found slight non-significant decrease in LINE-1 methylation (Garcia-Ortiz et al., 2021). The variance in outcome may be attributable to the precise positions of CpG sites analyzed or the method of analysis. Previous research utilized lymphoblastoid cell lines demonstrated that INE-1-inserted differentially expressed genes are associated with ASD-related mechanisms, such as sex hormone receptor signalling and axon guidance signalling. LINE-1 methylation was also found to be reduced in cell lines derived from patients with severe language impairment (Tangsuwansri et al., 2018). In our study, no association was observed between of LINE-1 methylation and severity, EEG abnormalities, or GI disorders.
LINE-1 hypomethylation is suggested to decrease the methylation of sub-telomeric regions, which are associated with short TL (Wong et al., 2014). In 2017, a study on healthy adolescent showed an association between lower levels of global DNA methylation and shorter TL which may decrease genome stability and raise the susceptibility to diseases (Dong et al., 2017). Considering that both RTL and LINE-1 methylation contribute to vulnerability of autism, we applied Pearson correlation to examine the association between RTL and LINE-1 methylation in autistic patients and found that this association is confirmed and statistically significant. Moreover, logistic regression analysis revealed an increase in the predictive efficacy of the combined biomarkers for autism as indicated by the increase in AUC value. These findings suggest the involvement of LINE-1 hypomethylation and shorter TL in the decreased genomic stability in autistic individuals (Williams et al., 2013) and the ability of using them as predictive biomarkers for autism. Additionally, these results support the previous findings which suggest that LINE-1 hypomethylation is linked to telomere shortening, whereas DNA methylation is one of the mechanisms that control TL and maintenance, and telomere dysfunction can cause LINE-1 retrotransposition (Chang et al., 2015).
Altogether, the study has concluded that RTL and LINE-1 methylation percentage are reduced in autism without affecting its severity. However, the study has been carried out on a small number of subjects, thus, in the future research we plan to increase the number of subjects and analyze the expression of the two open reading frames (ORF1 and ORF2) of LINE-1 and genes involved in regulating the methylation machinery in autism.
Acknowledgments
The authors are grateful to Science and Technology Development Fund (STDF) for financial support of this study. Special thanks to Human Genetics and Genome Research Institute, National Research Centre for providing all facilities to accomplish this research. Also, we thank the parents and guardians of the all patients who participated in this study.
Author Contributions
SS involved in study design, experimental analysis, data collection and manuscript writing. EA involved in diagnosis of patients and severity scoring.
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB). This work is funded by Science and Technology Development Fund (STDF), Egypt (Grant No. 33432).
Declarations
Conflict of interest
Authors declare that there is no conflict of interest.
Ethical Approval
The study was approved by the ethical research committee of the NRC (Registration number: 19033).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Asghar M, Bensch S, Tarka M, Hansson B, Hasselquist D. Maternal and genetic factors determine early life telomere length. Proceedings of the Biological Sciences. 2015;282:20142263. doi: 10.1098/rspb.2014.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belancio VP. LINE-1 activity as molecular basis for genomic instability associated with light exposure at night. Mobile Genetic Elements. 2015;5:1–5. doi: 10.1080/2159256X.2015.1037416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HB, Zou JZ, He C, Zeng R, Li YY, Ma FF, Liu Z, Ye H, Wu JX. Association between long interspersed nuclear element-1 methylation and relative telomere length in Wilms tumor. Chinese Medical Journal (engl) 2015;128:3055–3061. doi: 10.4103/0366-6999.169071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa Dde S, Rosa DV, Barros AG, Romano-Silva MA, Malloy-Diniz LF, Mattos P, de Miranda DM. Telomere length is highly inherited and associated with hyperactivity-impulsivity in children with attention deficit/hyperactivity disorder. Frontiers in Molecular Neuroscience. 2015;8:28. doi: 10.3389/fnmol.2015.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Huang Y, Gutin B, Raed A, Dong Y, Zhu H. Associations between global DNA methylation and telomere length in healthy adolescents. Science and Reports. 2017;7:4210. doi: 10.1038/s41598-017-04493-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitan E, Hutchison ER, Mattson MP. Telomere shortening in neurological disorders: An abundance of unanswered questions. Trends in Neurosciences. 2014;37:256–263. doi: 10.1016/j.tins.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyke W, Velinov M. FMR1 and autism, an intriguing connection revisited. Genes (basel) 2021 doi: 10.3390/genes12081218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ortiz MV, de la Torre-Aguilar MJ, Morales-Ruiz T, Gomez-Fernandez A, Flores-Rojas K, Gil-Campos M, Martin-Borreguero P, Ariza RR, Roldan-Arjona T, Perez-Navero JL. Analysis of global and local DNA methylation patterns in blood samples of patients with autism spectrum disorder. Frontiers in Pediatrics. 2021;9:685310. doi: 10.3389/fped.2021.685310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarrera S, Fiorito G, Onland-Moret NC, Russo A, Agnoli C, Allione A, Di Gaetano C, Mattiello A, Ricceri F, Chiodini P, Polidoro S, Frasca G, Verschuren MWM, Boer JMA, Iacoviello L, van der Schouw YT, Tumino R, Vineis P, Krogh V, Panico S, Sacerdote C, Matullo G. Gene-specific DNA methylation profiles and LINE-1 hypomethylation are associated with myocardial infarction risk. Clinical Epigenetics. 2015;7:133. doi: 10.1186/s13148-015-0164-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenhaar FS, Josefsson M, Adolfsson AN, Landfors M, Kauppi K, Hultdin M, Adolfsson R, Degerman S, Pudas S. Short leukocyte telomeres predict 25-year Alzheimer’s disease incidence in non-APOE epsilon4-carriers. Alzheimers Research & Therapy. 2021;13:130. doi: 10.1186/s13195-021-00871-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JA, Feldser DM, Greider CW. Telomere dysfunction increases mutation rate and genomic instability. Cell. 2001;106:275–286. doi: 10.1016/S0092-8674(01)00457-3. [DOI] [PubMed] [Google Scholar]
- Hajian-Tilaki K. Receiver Operating Characteristic (ROC) curve analysis for medical diagnostic test evaluation. Caspian Journal of Internal Medicine. 2013;4:627–635. [PMC free article] [PubMed] [Google Scholar]
- Hallmayer J, Cleveland S, Torres A, Phillips J, Cohen B, Torigoe T, Miller J, Fedele A, Collins J, Smith K, Lotspeich L, Croen LA, Ozonoff S, Lajonchere C, Grether JK, Risch N. Genetic heritability and shared environmental factors among twin pairs with autism. Archives of General Psychiatry. 2011;68:1095–1102. doi: 10.1001/archgenpsychiatry.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homs A, Codina-Sola M, Rodriguez-Santiago B, Villanueva CM, Monk D, Cusco I, Perez-Jurado LA. Genetic and epigenetic methylation defects and implication of the ERMN gene in autism spectrum disorders. Translational Psychiatry. 2016;6:e855. doi: 10.1038/tp.2016.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura R, Nakata M, Funabiki Y, Suzuki S, Awaya T, Murai T, Hagiwara M. An epigenetic biomarker for adult high-functioning autism spectrum disorder. Science and Reports. 2019;9:13662. doi: 10.1038/s41598-019-50250-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara T, Hsieh J, Muotri A, Yeo G, Warashina M, Lie DC, Moore L, Nakashima K, Asashima M, Gage FH. Wnt-mediated activation of NeuroD1 and retro-elements during adult neurogenesis. Nature Neuroscience. 2009;12:1097–1105. doi: 10.1038/nn.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CR, Taguinod F, Jepsen WM, Cohen J, Agrawal K, Huentelman MJ, Smith CJ, Ringenbach SDR, Braden BB. Telomere length and autism spectrum disorder within the family: Relationships with cognition and sensory symptoms. Autism Research. 2020;13:1094–1101. doi: 10.1002/aur.2307. [DOI] [PubMed] [Google Scholar]
- Li Z, Tang J, Li H, Chen S, He Y, Liao Y, Wei Z, Wan G, Xiang X, Xia K, Chen X. Shorter telomere length in peripheral blood leukocytes is associated with childhood autism. Science and Reports. 2014;4:7073. doi: 10.1038/srep07073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maenner MJ, Shaw KA, Baio J, EdS WA, Patrick M, DiRienzo M, Christensen DL, Wiggins LD, Pettygrove S, Andrews JG, Lopez M, Hudson A, Baroud T, Schwenk Y, White T, Rosenberg CR, Lee LC, Harrington RA, Huston M, Hewitt A, PhD EA, Hall-Lande J, Poynter JN, Hallas-Muchow L, Constantino JN, Fitzgerald RT, Zahorodny W, Shenouda J, Daniels JL, Warren Z, Vehorn A, Salinas A, Durkin MS, Dietz PM. Prevalence of autism spectrum disorder among children aged 8 years—Autism and developmental disabilities monitoring network, 11 sites, United States, 2016. MMWR Surveillance Summaries. 2020;69:1–12. doi: 10.15585/mmwr.ss6904a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nersisyan L, Hopp L, Loeffler-Wirth H, Galle J, Loeffler M, Arakelyan A, Binder H. Telomere length maintenance and its transcriptional regulation in lynch syndrome and sporadic colorectal carcinoma. Frontiers in Oncology. 2019;9:1172. doi: 10.3389/fonc.2019.01172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A, Rauch TA, Pfeifer GP, Hu VW. Global methylation profiling of lymphoblastoid cell lines reveals epigenetic contributions to autism spectrum disorders and a novel autism candidate gene, RORA, whose protein product is reduced in autistic brain. The FASEB Journal. 2010;24:3036–3051. doi: 10.1096/fj.10-154484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano GH, Harari Y, Yehuda T, Podhorzer A, Rubinstein L, Shamir R, Gottlieb A, Silberberg Y, Pe'er D, Ruppin E, Sharan R, Kupiec M. Environmental stresses disrupt telomere length homeostasis. PLoS Genetics. 2013;9:e1003721. doi: 10.1371/journal.pgen.1003721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpyleva S, Melnyk S, Pavliv O, Pogribny I, Jill James S. Overexpression of LINE-1 retrotransposons in autism brain. Molecular Neurobiology. 2018;55:1740–1749. doi: 10.1007/s12035-017-0421-x. [DOI] [PubMed] [Google Scholar]
- Shubin CB, Greider CW. The role of Rif1 in telomere length regulation is separable from its role in origin firing. eLife. 2020 doi: 10.7554/eLife.58066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JA, Chaiken MF, Wang F, Price CM. Maintaining the end: Roles of telomere proteins in end-protection, telomere replication and length regulation. Mutation Research. 2012;730:12–19. doi: 10.1016/j.mrfmmm.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Poschmann J, Cruz-Herrera Del Rosario R, Parikshak NN, Hajan HS, Kumar V, Ramasamy R, Belgard TG, Elanggovan B, Wong CCY, Mill J, Geschwind DH, Prabhakar S. Histone acetylome-wide association study of autism spectrum disorder. Cell. 2016;167(5):1385–1397. doi: 10.1016/j.cell.2016.10.031. [DOI] [PubMed] [Google Scholar]
- Svikle Z, Pahirko L, Zarina L, Baumane K, Kardonaite D, Radzeviciene L, Daugintyte-Petrusiene L, Balciuniene VJ, Verkauskiene R, Tiscuka A, Rovite V, Sjakste N, Sokolovska J. Telomere lengths and serum proteasome concentrations in patients with type 1 diabetes and different severities of diabetic retinopathy in Latvia and Lithuania. Journal of Clinical Medicine. 2022 doi: 10.3390/jcm11102768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swets M, Zaalberg A, Boot A, van Wezel T, Frouws MA, Bastiaannet E, Gelderblom H, van de Velde CJ, Kuppen PJ. Tumor LINE-1 methylation level in association with survival of patients with stage II colon cancer. International Journal of Molecular Sciences. 2016 doi: 10.3390/ijms18010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangsuwansri C, Saeliw T, Thongkorn S, Chonchaiya W, Suphapeetiporn K, Mutirangura A, Tencomnao T, Hu VW, Sarachana T. Investigation of epigenetic regulatory networks associated with autism spectrum disorder (ASD) by integrated global LINE-1 methylation and gene expression profiling analyses. PLoS ONE. 2018;13:e0201071. doi: 10.1371/journal.pone.0201071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tick B, Bolton P, Happe F, Rutter M, Rijsdijk F. Heritability of autism spectrum disorders: A meta-analysis of twin studies. Journal of Child Psychology and Psychiatry. 2016;57:585–595. doi: 10.1111/jcpp.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonacci A, Bagnato G, Pandolfo G, Billeci L, Sansone F, Conte R, Gangemi S. MicroRNA cross-involvement in autism spectrum disorders and atopic dermatitis: A literature review. Journal of Clinical Medicine. 2019 doi: 10.3390/jcm8010088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujvari B, Pearse AM, Taylor R, Pyecroft S, Flanagan C, Gombert S, Papenfuss AT, Madsen T, Belov K. Telomere dynamics and homeostasis in a transmissible cancer. PLoS ONE. 2012;7:e44085. doi: 10.1371/journal.pone.0044085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willeit P, Raschenberger J, Heydon EE, Tsimikas S, Haun M, Mayr A, Weger S, Witztum JL, Butterworth AS, Willeit J, Kronenberg F, Kiechl S. Leucocyte telomere length and risk of type 2 diabetes mellitus: New prospective cohort study and literature-based meta-analysis. PLoS ONE. 2014;9:e112483. doi: 10.1371/journal.pone.0112483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams EL, Casanova MF, Switala AE, Li H, Qiu M. Transposable elements occur more frequently in autism-risk genes: Implications for the role of genomic instability in autism. Translational Neuroscience. 2013;4:172–202. doi: 10.2478/s13380-013-0113-6. [DOI] [Google Scholar]
- Wong JY, De Vivo I, Lin X, Grashow R, Cavallari J, Christiani DC. The association between global DNA methylation and telomere length in a longitudinal study of boilermakers. Genetic Epidemiology. 2014;38:254–264. doi: 10.1002/gepi.21796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousef AM, Roshdy EH, Abdel Fattah NR, Said RM, Atia MM, Hafez EM, Mohamed AE. Prevalence and risk factors of autism spectrum disorders in preschool children in Sharkia, Egypt: A community-based study. Middle East Current Psychiatry. 2021;28:36. doi: 10.1186/s43045-021-00114-8. [DOI] [Google Scholar]
- Zhao Y, Sfeir AJ, Zou Y, Buseman CM, Chow TT, Shay JW, Wright WE. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell. 2009;138:463–475. doi: 10.1016/j.cell.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]