Abstract
Efficient interbacterial transfer of streptomycete plasmid pIJ101 requires the pIJ101 tra gene, as well as a cis-acting plasmid function known as clt. Here we show that the minimal pIJ101 clt locus consists of a sequence no greater than 54 bp in size that includes essential inverted-repeat and direct-repeat sequences and is located in close proximity to the 3′ end of the korB regulatory gene. Evidence that sequences extending beyond the minimal locus and into the korB open reading frame influence clt transfer function and demonstration that clt-korB sequences are intrinsically curved raise the possibility that higher-order structuring of DNA and protein within this plasmid region may be an inherent feature of efficient pIJ101 transfer.
Plasmid-mediated conjugation is a widespread phenomenon within the gram-positive bacterial genus Streptomyces, and many conjugative plasmids have been isolated from a variety of Streptomyces species. There is evidence to suggest that the conjugation process in Streptomyces bacteria may be novel, including the fact that relatively few plasmid-borne genetic loci are required for conjugation to occur between streptomycete cells compared to other bacteria (11). For example, transfer of the 8,830-bp circular double-stranded Streptomyces lividans plasmid pIJ101 requires only one plasmid-encoded protein, the 70-kDa membrane-associated product (19) of the pIJ101 tra gene (14, 15). Tra is capable of mediating plasmid transfer by an undetermined mechanism either when expressed from a plasmid or when encoded by a tra gene copy that has been inserted into the host chromosome (20). Although Tra does not resemble proteins typically required for conjugative transfer of plasmids from gram-negative organisms, it does show intriguing homology to bacterial proteins that promote the movement or partitioning of chromosomal DNA during cellular processes that include sporulation (in Bacillus subtilis) (36) and cell division (in Escherichia coli) (1). Tra is also the only pIJ101-encoded factor required for chromosomal gene transfer during mating of streptomycete cells (15).
Efficient transfer of pIJ101 also requires a cis-acting locus termed clt. In conjunction with the chromosomally inserted tra gene, addition of the pIJ101 clt locus to transfer defective derivatives of either pIJ101 or non-pIJ101 replicons increases their conjugative transfer by two to three orders of magnitude (20). The clt locus was found to be contained on a 145-bp segment of pIJ101 that spans the 3′ end of the pIJ101 korB gene (Fig. 1) and extends into the intergenic region between the convergently transcribed korB and korA genes (20). Both korB and korA encode transcriptional repressors that have multiple regulatory functions; the KorB repressor, for example, controls expression from promoters for both the transfer-related kilB gene of pIJ101 and the korB gene (24, 25, 29, 37) and may also play an undefined role in regulation of plasmid replication (6). The KorA protein similarly represses transcription from the korA gene promoter and is also required for control of expression from the tra promoter (24, 25).
FIG. 1.
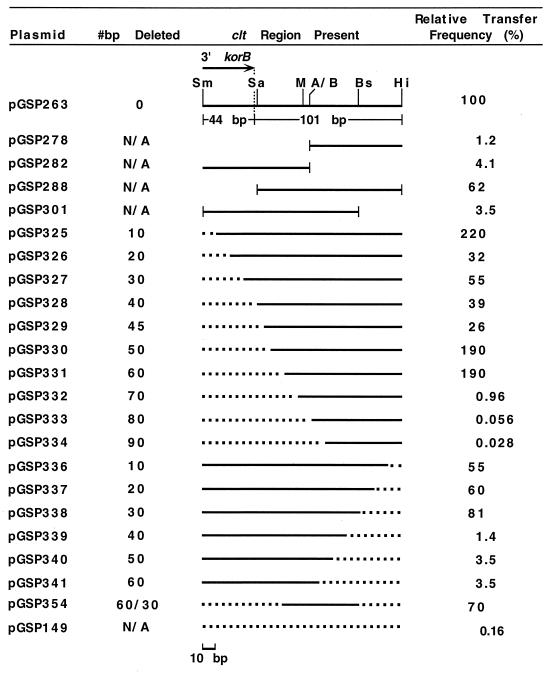
Relative transfer frequencies of clt region plasmid constructs. The physical map for the 145-bp clt+ sequence present on plasmid pGSP263 (20) is shown. Portions of this sequence that are within (44 bp) and downstream of (101 bp) the 3′ end of the korB ORF (small arrow) are indicated. For each additional construct, the solid horizontal line represents the portion of the 145-bp sequence present in that clone. Small vertical lines bracketing horizontal lines indicate cloning junctions in relation to the physical map. Nonbracketed ends for pGSP278 and pGSP282 indicate that pIJ101 sequences extending beyond the 145-bp region in the directions shown are present on these plasmids. For plasmids pGSP325 to pGSP354, the indicated incremental deletions (dotted-line portions) from either or both ends of the 145-bp clt+ insert of plasmid pGSP263 were constructed using a PCR-based method as described in the text. For constructs not derived by PCR, including the transfer-defective pIJ350 derivative pGSP149, which lacks the 145-bp clt+ region of pIJ101, the number of deleted base pairs is not applicable (N/A). Transfer of plasmids from S. lividans strain TK23.42 to strain TK23(pHYG1) was performed and quantified as described previously (19, 20), and relative transfer frequencies were determined by dividing the average ratio of transconjugants to donors from four independent matings involving a given plasmid by that same average ratio from four independent matings involving pGSP263 and then multiplying by 100%. Restriction sites for ApaI-Bsp120I (A/B), BspMI (Bs), HincII (Hi), MluI (M), Sau3AI (Sa), and SmaI (Sm) are indicated.
The only known mechanism for processing of circular, double-stranded plasmid DNA prior to its conjugative transfer involves site-specific endonucleolytic cleavage or nicking of a single strand of the plasmid within its origin of transfer (oriT) (16). This function is the only cis-acting locus known to be required for plasmid transfer and has been identified on numerous plasmids isolated mostly from gram-negative sources. While oriT regions vary from smaller than 50 bp to larger than 500 bp, they characteristically show a higher AT content than their surrounding DNA, have extensive direct and inverted nucleotide sequence repeats for protein binding, and are typically positioned in nontranscribed intergenic regions (e.g., overlapping divergent promoters that control the expression of transfer genes) (16). Although extensive homology of oriT sequences is found only in closely related plasmids, most oriT loci so far identified, including those from plasmids of gram-positive organisms (16) and one from a conjugative transposon of gram-positive origin (12), have been divided into three groups based on limited identity around their nick sites (16).
Cleavage at oriT, which is mediated by a plasmid-encoded enzyme (i.e., the relaxase) and normally involves other plasmid proteins, initially results in a membrane-associated DNA-multiprotein complex that includes the relaxase protein covalently attached to oriT at the site of nicking (16). Upon mating, interactions between such complexes and additional plasmid-encoded membrane proteins direct the translocation of the nicked strand by an undetermined mechanism through the membrane and into the recipient cell (7, 16), whereupon strand circularization and second-strand synthesis occur (32).
Since clt promotes efficient transfer of plasmid DNA in cis and does not appear to encode a protein (20), it may function analogously to oriT regions found on conjugative plasmids from other bacteria. Potential interactions occurring at clt would then be predicted to involve either the pIJ101 Tra protein or, given the paucity of plasmid genes required for streptomycete plasmid transfer (11), possibly one or more host-encoded factors. Interestingly, the bacterial proteins to which Tra is homologous, such as the B. subtilis SpoIIIE and E. coli FtsK proteins, participate in processes involving the intracellular movement of double-stranded DNA (17, 26, 35, 36). Thus, an intriguing possibility regarding the potential interaction between the Tra protein and clt locus of pIJ101 is the occurrence of a novel DNA-processing event (e.g., double-stranded DNA cleavage) that would somehow allow transfer of the plasmid in a unique double-stranded form. Another, perhaps conceptually less appealing possibility for clt function involves the interbacterial transfer of unprocessed, covalently closed circular pIJ101 molecules; in this case, clt would still serve as a site for productive interaction with transfer proteins but strand cleavage at clt would not result.
As a first step toward elucidating the role of clt in pIJ101 transfer, we have identified its minimal sequence determinants. The clt locus is composed of a region of pIJ101 no greater than 54 bp that maps just downstream of the korB gene. The locus shares some but not all of the characteristics typically associated with oriT sites, including the presence of inverted-repeat and direct-repeat regions, both of which are essential for full clt function (see below). Other data showing that sequences extending into the 3′ end of the korB open reading frame (ORF) can influence clt activity and that the clt plasmid region is intrinsically curved are consistent with the notion that three-dimensional structuring of protein and clt-korB DNA may occur during pIJ101 transfer. The strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| BL21(DE3) | Host for T7 polymerase/promoter-induced expression of pIJ101 Tra protein; F−hsdS (rB− mK−) gal with T7 RNA polymerase gene in chromosome | 27 |
| BRL2288 | Host for cloning; recA56 derivative of MC1061 [F−araD139 Δ(ara-leu)7679 ΔlacX74 galU galK hsdR2 (rK− mK−) mcrB1 rpsL] | Gibco BRL |
| DH10B | Host for cloning; F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 deoR recA1 endA1 araD139Δ(ara-leu)7697 galU galK λ−rpsL nupG | Gibco BRL |
| K38 | Host for T7 polymerase/promoter-induced expression of pIJ101 KorB protein | 22 |
| Streptomyces lividans | ||
| TK23 | spc-1 | 10 |
| TK23.42 | TK23 with the tra and korA genes of pIJ101 inserted into the chromosome | 20 |
| Plasmids | ||
| pGP1-2 | pACYC177 derivative with the T7 RNA polymerase gene under control of the inducible λ pL promoter and the heat-sensitive λ repressor gene cI857 | 28 |
| pGSP149 | pIJ350 with pSP72 inserted at the PstI site (bla gene transcribed in same direction as rep gene of pIJ101) | 20 |
| pGSP208 | pSP72 with the 2.0-kb NruI-FspI region of pIJ101 cloned as a blunt-end fragment at the BglII-SmaI sites (tra gene is in orientation to be expressed by T7 promoter) | 19 |
| pGSP226 | pSP72 with the 4.0-kb KpnI-BglII region of pHYG3 cloned at the same sites and with an XbaI linker at HincII (nt 6867) of pIJ101 | This study |
| pGSP250 | pGSP226 deleted from the SmaI site in the polylinker to SmaI (nt 7009) in korB | This study |
| pGSP260 | pSP72 with the 145-bp clt+ region of pGSP250 (i.e., nt 6868–7012 of pIJ101) cloned as a BamHI fragment at the BamHI site (korB sequences are toward the T7 promoter) | This study |
| pGSP260-Δ10L | pGSP260 with 10-bp insert deletion from HincII end | This study |
| pGSP260-Δ20L | pGSP260 with 20-bp insert deletion from HincII end | This study |
| pGSP260-Δ30L | pGSP260 with 30-bp insert deletion from HincII end | This study |
| pGSP260-Δ40L | pGSP260 with 40-bp insert deletion from HincII end | This study |
| pGSP260-Δ50L | pGSP260 with 50-bp insert deletion from HincII end | This study |
| pGSP260-Δ60L | pGSP260 with 60-bp insert deletion from HincII end | This study |
| pGSP260-Δ10R | pGSP260 with 10-bp insert deletion from SmaI end | This study |
| pGSP260-Δ20R | pGSP260 with 20-bp insert deletion from SmaI end | This study |
| pGSP260-Δ30R | pGSP260 with 30-bp insert deletion from SmaI end | This study |
| pGSP260-Δ40R | pGSP260 with 40-bp insert deletion from SmaI end | This study |
| pGSP260-Δ45R | pGSP260 with 45-bp insert deletion from SmaI end | This study |
| pGSP260-Δ50R | pGSP260 with 50-bp insert deletion from SmaI end | This study |
| pGSP260-Δ60R | pGSP260 with 60-bp insert deletion from SmaI end | This study |
| pGSP260-Δ70R | pGSP260 with 70-bp insert deletion from SmaI end | This study |
| pGSP260-Δ80R | pGSP260 with 80-bp insert deletion from SmaI end | This study |
| pGSP260-Δ90R | pGSP260 with 90-bp insert deletion from SmaI end | This study |
| pGSP260-Δ60RΔ30L | pGSP260 with 60- and 30-bp insert deletions from SmaI and HincII ends, respectively | This study |
| pGSP260-T35 | pGSP260 with Transprimer-1 inserted after nt 6924 of pIJ101 | This study |
| pGSP260-T38 | pGSP260 with Transprimer-1 inserted after nt 6923 of pIJ101 | This study |
| pGSP260-T42 | pGSP260 with Transprimer-1 inserted after nt 6926 of pIJ101 | This study |
| pGSP260-T95 | pGSP260 with Transprimer-1 inserted after nt 6935 of pIJ101 | This study |
| pGSP263 | pIJ350 with pGSP260 inserted at the PstI site (clt region in natural orientation relative to the pIJ101 rep gene) | 20 |
| pGSP272 | pGSP208 with the 145-bp clt+ region of pGSP260 cloned as a BamHI fragment in the BamHI site | This study |
| pGSP276 | pSP72 with the 0.8-kb ApaI-BglII region of pIJ101 cloned as a blunt-end fragment at the EcoRV site | This study |
| pGSP278 | pIJ350 with pGSP276 inserted at the PstI site | This study |
| pGSP280 | pSP72 with the 0.3-kb Bsp120I-SpeI region of pIJ101 cloned as a blunt-end fragment at the EcoRV site | This study |
| pGSP282 | pIJ350 with pGSP280 inserted at the PstI site | This study |
| pGSP287 | pSP72 derivative with the 99-bp Sau3AI-HincII clt+ region of pIJ101 cloned as an XbaI fragment at the XbaI site | This study |
| pGSP288 | pIJ350 with pGSP287 inserted at the PstI site | This study |
| pGSP290 | pSP72 containing the 1.0-kb PstI-BalI region of pIJ101 cloned as a blunt-end fragment at EcoRV (transcription of kilB is toward the T7 promoter) | 21 |
| pGSP299 | pSP72 with the ∼220-bp BspMI region of pGSP260 cloned as a blunt-end fragment at the EcoRV site | This study |
| pGSP300 | pSP72 with the 0.3-kb FspI-AvaI region of pGSP290 cloned as a blunt-end fragment at the EcoRV site (transcription from kilB promoter is toward the SP6 promoter) | This study |
| pGSP301 | pGSP149 with the ∼150-bp EcoRI fragment of pGSP299 (including nt 6899–7012 of pIJ101) cloned at the EcoRI site (a small deletion of duplicated polylinker sequences occurred) | This study |
| pGSP311 | pSP72 with the 121-bp PstI-BstEII fragment of pGSP300 containing the kilB promoter cloned at the EcoRV site | This study |
| pGSP325 | pGSP260-Δ10R with pIJ350 at the PstI site | This study |
| pGSP326 | pGSP260-Δ20R with pIJ350 at the PstI site | This study |
| pGSP327 | pGSP260-Δ30R with pIJ350 at the PstI site | This study |
| pGSP328 | pGSP260-Δ40R with pIJ350 at the PstI site | This study |
| pGSP329 | pGSP260-Δ45R with pIJ350 at the PstI site | This study |
| pGSP330 | pGSP260-Δ50R with pIJ350 at the PstI site | This study |
| pGSP331 | pGSP260-Δ60R with pIJ350 at the PstI site | This study |
| pGSP332 | pGSP260-Δ70R with pIJ350 at the PstI site | This study |
| pGSP333 | pGSP260-Δ80R with pIJ350 at the PstI site | This study |
| pGSP334 | pGSP260-Δ90R with pIJ350 at the PstI site | This study |
| pGSP336 | pGSP260-Δ10L with pIJ350 at the PstI site | This study |
| pGSP337 | pGSP260-Δ20L with pIJ350 at the PstI site | This study |
| pGSP338 | pGSP260-Δ30L with pIJ350 at the PstI site | This study |
| pGSP339 | pGSP260-Δ40L with pIJ350 at the PstI site | This study |
| pGSP340 | pGSP260-Δ50L with pIJ350 at the PstI site | This study |
| pGSP341 | pGSP260-Δ60L with pIJ350 at the PstI site | This study |
| pGSP344 | pGSP260-T35 with pIJ350 at the PstI site | This study |
| pGSP346 | pGSP260-T38 with pIJ350 at the PstI site | This study |
| pGSP347 | pGSP260-T42 with pIJ350 at the PstI site | This study |
| pGSP351 | pGSP260-T95 with pIJ350 at the PstI site | This study |
| pGSP354 | pGSP260-Δ60RΔ30L with pIJ350 at PstI site | This study |
| pHYG1 | Conjugally deficient Hygr deletion derivative of pIJ101 lacking all known transfer functions | 14 |
| pHYG3 | Conjugative Hygr derivative of pIJ101 | 20 |
| pIJ303 | Conjugative Tsrr derivative of pIJ101 | 15 |
| pIJ350 | Conjugally deficient Tsrr deletion derivative of pIJ101 lacking all known transfer functions | 15 |
| pSP72 | AprE. coli cloning vector; contains T7 promoter | Promega |
| pT7-6 | ColE1-based plasmid containing the T7 promoter | 28 |
| pT7-6-korB | pT7-6 with pIJ101 korB gene downstream of the T7 promoter | 29 |
| R1162 | Mobilizable E. coli plasmid | Richard Meyer |
| pUT1579 | R1162 with mutation that causes a tyrosine-to-phenylanine substitution at position 24 of the MobA protein | Richard Meyer |
Hygr, Tsrr, and Apr designate hygromycin, thiostrepton, and ampicillin resistance, respectively. nt, nucleotide. clt region sequences for the plasmids listed between (and including) pGSP325 and pGSP354 are in the natural orientation relative to the pIJ101 rep gene. Nucleotide positions are those of Kendall and Cohen (13).
Localization of the pIJ101 clt locus.
Previous DNA insertions (14, 20) in the region immediately downstream of korB on pIJ101 (Fig. 1) suggested that the clt locus spans the MluI site and possibly the adjacent ApaI/Bsp120I recognition sequence but does not appear to extend to the Sau3AI site located 2 bp beyond the korB ORF (20). To test these possibilities, portions of the region downstream of korB were inserted into pIJ350, a thiostrepton-resistant, transfer-defective pIJ101 derivative in which all transfer-related sequences are deleted (15). The resulting clones were tested in mating assays, as previously described (19, 20), for their ability to be transferred from S. lividans TK23.42, a strain containing chromosomal copies of the pIJ101 tra and korA genes (20), to S. lividans strain TK23 containing the hygromycin-resistant, transfer-defective pIJ101 derivative pHYG1 (14). Transfer frequencies for these plasmids were then determined as described previously (20), except that they were expressed relative to the frequency seen for plasmid pGSP263 (Fig. 1), a pIJ350 derivative which contains the 145-bp region of pIJ101 spanning the 3′ end of korB that was previously shown to contain clt (20).
Plasmids pGSP278 and pGSP282, whose inserts include clt region sequences downstream of korB either to the right or to the left, respectively, of the ApaI-Bsp120I restriction site as shown in Fig. 1, both showed significant reductions (i.e., by nearly 100- and 25-fold, respectively) in transfer frequency compared to pGSP263, although both still transferred at higher frequencies (i.e., by approximately 8- and 26-fold, respectively) than pGSP149, a pIJ350 derivative that lacks the 145-bp clt+ region present on pGSP263. Plasmid pGSP288, a derivative of pGSP263 which contains sequences only to the right of the Sau3AI site as shown in Fig. 1 and therefore lacks the entire korB portion of the parental plasmid, was moderately reduced by about 40% in its transfer frequency. These results, taken together with the previous data (20), thus implicate sequences spanning both the MluI and ApaI-Bsp120I restriction sites downstream of korB as comprising at least a portion of the clt locus while also suggesting that sequences extending into korB, although not part of the locus, may also contribute to clt activity, albeit less significantly.
To determine the clt locus endpoints downstream of korB, a series of incremental deletions from either or both ends of the 145-bp clt+ pGSP263 insert were constructed using a PCR-based protocol, and the resulting deletion derivatives were then tested as described above for plasmid transfer. While nested deletions of 44 bp or less from the SmaI end within korB would not remove sequences beyond the end of the korB ORF (Fig. 1) and therefore were not expected to yield information regarding the position of the downstream clt locus, such clones were nevertheless included in the deletion series since they were useful in evaluating the potential involvement of 3′ korB sequences in clt function.
Deletions from a single end of the pGSP263 insert were created using as a template plasmid pGSP260, the precursor of pGSP263 which contains the 145-bp clt+ region of pIJ101 cloned as a BamHI fragment in the E. coli vector pSP72 (Promega, Madison, Wis.), while deletions from both insert ends were created sequentially using pGSP260 as the initial template and a pGSP260 derivative deleted from a single end as the template for the second round of PCR. For each PCR, one primer was complementary to relevant insert-vector junction sequences minus the desired number of terminal insert bases (for deletions from the SmaI end, the first position of the insert was always preserved for cloning purposes) and the opposing primer was complementary to vector sequences near the other end of the insert. Following amplification for 30 cycles (1 cycle consisted of 94°C for 30 s, 37°C for 1 min, and 72°C for 2 min) using Pfu polymerase (Stratagene, La Jolla, Calif.) in the presence of 10% dimethyl sulfoxide, DNA was purified and digested to completion with BamHI and deleted derivatives of the 145-bp clt+ sequence were cloned as BamHI fragments into similarly digested pSP72 using standard molecular biology protocols (23). As judged by restriction mapping and automated sequencing, plasmids with deleted inserts in the same orientation as the 145-bp clt+ sequence of pGSP260 were then inserted in the proper orientation into the PstI site of Streptomyces plasmid pIJ350 to create specifically deleted versions of pGSP263 that were otherwise identical.
Beginning with insert deletions originating from the SmaI end (Fig. 1), removal of up to 60 bp (i.e., pGSP331), or to a position 17 bp downstream from the 3′ end of the korB ORF, occurred without dramatic effects on plasmid transfer frequency. However, deletion of an additional 10 bp (i.e., plasmid pGSP332) reduced transfer relative to pGSP263 by about 100-fold, to a level that was only approximately 6-fold higher than that of the transfer-defective plasmid pGSP149, which completely lacks the 145-bp clt+ region. Deletions of 80 bp (pGSP333) and 90 bp (pGSP334) (Fig. 1) from the SmaI end were judged to completely eliminate clt function, since the resulting plasmids transferred at relative frequencies (0.056 and 0.028%, respectively) no greater than the frequency seen for pGSP149 (0.16%).
Deletions from the opposing HincII end of the pGSP263 insert showed a similar pattern of clt activity: while removal of up to 30 bp (pGSP338) had only minor effects on plasmid transfer, deletion of 40, 50, or 60 bp significantly reduced the transfer frequencies of the resulting plasmids (pGSP339 to pGSP341, respectively) by approximately 30- to 70-fold relative to that of pGSP263 (Fig. 1). Taken together, our deletion data suggest that sequences critical for clt function lie within a region of pIJ101 no greater than 54 bp (i.e., the sequence between the opposing deletion endpoints present in plasmids pGSP331 and pGSP338) which begins 17 bp downstream from the korB ORF. Consistent with this assignment, plasmid pGSP354, which by deletion of 60 bp from the SmaI end of the pGSP263 insert and 30 bp from the HincII end retains only this 54-bp region, transferred at a relative frequency (70%) that was still comparable to that of pGSP263 (Fig. 1). We will refer to this 54-bp sequence as the minimal clt locus, which, in conjunction with the pIJ101 tra gene, results in an increase of approximately two orders of magnitude in transfer efficiency relative to transfer-defective plasmids (e.g., pGSP149). The term “minimal” has been included in the locus name since, as we show below, sequences extending beyond this sequence and into korB can influence clt function to lesser but nevertheless significant extents.
Characterization of the minimal clt locus.
As shown in Fig. 2, the minimal clt locus contains a single imperfect inverted-repeat sequence (designated IR) along with three copies of the direct-repeat sequence 5′-G(A/C)AAC (designated DR-1, DR-2, and DR-3). Given the possibility that clt may function analogously to oriT sites of other conjugative plasmids, some or all of these repeats may represent sites of functional interaction with proteins involved in the processing of pIJ101 for its subsequent transfer (16). Despite this similarity, however, it should also be noted that in contrast to known oriT regions (16), clt has an AT content that is not higher than that of its flanking sequences (data not shown), and it is located in a region of the plasmid known to be transcribed (5); further, none of the strongly conserved, albeit limited, sequence motifs that are characteristic of the three known groups of oriT sites (16) are present in the minimal clt locus (data not shown).
FIG. 2.
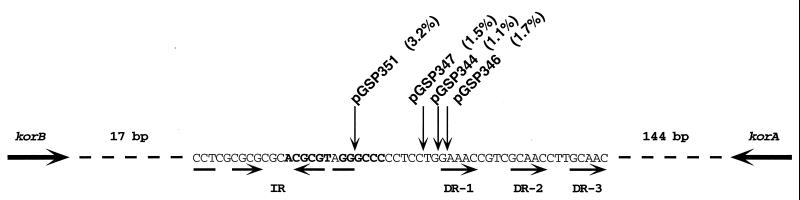
Localization and relative transfer frequencies of Transprimer transposon insertions within the minimal clt locus of plasmid pIJ101. As defined by the functional analysis described in the text, the 54-bp sequence comprising the minimal clt locus is shown. Small arrows below the sequence indicate the location and direction of one set of imperfect inverted repeats (IR), and three direct repeats (DR). Transprimer transposon insertions were isolated as described in the text at the positions indicated on plasmid pGSP260, which contains the the 145-bp clt+ region of pIJ101 (Fig. 1). Matings involving pGSP263 derivatives containing these insertions (i.e., pGSP344, pGSP346, pGSP347, and pGSP351) were performed and quantified as described in the legend to Fig. 1. Relative transfer frequencies (shown in parentheses for each plasmid) were also calculated as described in the legend to Fig. 1, using the results of four independent matings (except for pGSP347, where the results of two independent matings were used). The MluI and ApaI-Bsp120I recognition sequences within the minimal clt locus are in bold. The distances in base pairs from clt to the korB and korA ORFs (whose directions are indicated by the large arrows) are indicated.
That the IR sequence represents the single most important functional region within the minimal clt locus is implied by the results for plasmid pGSP333 (Fig. 1), in which nearly all of the IR sequence is deleted (i.e., to a position 20 bases within the left boundary of clt as shown in Fig. 2) and which, as noted above, is reduced in its transfer to frequencies not higher than those for a plasmid completely lacking clt region sequences (Fig. 1). This notion is also supported by a previous DNA insertion at the MluI site of pIJ101 (i.e., within the right arm of the inverted repeat of the clt locus as shown in Fig. 2), which resulted in a complete loss of plasmid transfer (20).
In contrast, deletions from the opposite (i.e., korA) direction that removed portions of the DR region (i.e., plasmids pGSP339 and pGSP340) reduced plasmid transfer significantly but not to the same extent as did the opposing deletions that removed the IR sequence (Fig. 1), a result that suggests that the DR sequences may play a relatively less significant role than the IR motif in overall clt function. Evidence supporting the notion that the DR-3 sequence, specifically, is involved in clt transfer function was provided by results obtained with plasmid pGSP301 (Fig. 1), an additional derivative of pGSP263 in which cleavage with the enzyme BspMI resulted in removal of 31 bp from the HincII end of the 145-bp clt+ sequence; such a deletion thus included removal of the 3′ C position of the 5′-GCAAC DR-3 sequence, as shown in Fig. 2, and thereby resulted in a nearly 30-fold reduction in transfer frequency for pGSP301 compared to pGSP263 (Fig. 1).
Previous DNA insertions (14) at the HaeIII site located downstream of korB on pIJ101 (this HaeIII site is within the ApaI-Bsp120I recognition sequence of the minimal clt locus) (Fig. 2) reduced pIJ101 transfer significantly, although not to the same extent as did the insertion at the nearby MluI site (20) within the IR region. Although the insertions at HaeIII occur within the minimal clt locus, they disrupt neither IR sequences nor DR sequences but, rather, add DNA to the portion located between them. To assess better the relative contribution of this intervening sequence to clt function, we isolated additional DNA insertions within this region of the minimal clt locus and tested their effects on clt transfer activity.
E. coli plasmid pGSP260, the precursor of pGSP263 which contains the 145-bp clt+ sequence (Fig. 1) cloned as a BamHI fragment in vector pSP72, was mutagenized using the GSP-1 genome-priming system (New England Biolabs, Beverly, Mass.) as specified by the manufacturer, and plasmids containing stable copies of kanamycin-resistant Transprimer-1 transposons were used to transform E. coli to both kanamycin and ampicillin resistance. To identify insertions within the clt-containing insert, plasmid DNA extracted from pooled transformant colonies was digested to completion with EcoRI and PstI and the fragment corresponding in size to clt insert sequences plus single randomly integrated transprimer insertions was purified, cloned into similarly digested pSP72 by standard procedures (23), and used to transform E. coli to dual ampicillin and kanamycin resistance. The position of the transposon on each of the individual plasmid clones was determined by restriction mapping and automated sequencing, and upon insertion of Streptomyces plasmid pIJ350 as described above for the deletion analysis, transposon-containing versions of pGSP263 were tested for plasmid transfer.
Using this method, we obtained four DNA insertions (Fig. 2) at unique positions between the IR and DR regions of the minimal clt locus (because of a 5-bp target site duplication upon integration of Transprimer transposons, the pGSP346 insertion leaves the DR-1 repeat intact relative to the rest of the DR region). Since these insertions caused 30- to 90-fold reductions in transfer frequency relative to pGSP263 (Fig. 2), these results demonstrate that either the exact sequence or the spacing between the two repeat regions is also important for clt function. Moreover, given that the transfer reductions for these insertions, including that of plasmid pGSP351, whose insertion is located just outside of the IR region, are reminiscent of the less severe reductions seen for deletions that removed the DR region (Fig. 1), these results provide additional complementary evidence that the IR motif is the most critical functional sequence within the minimal clt locus.
Nicking within oriT regions of plasmids of gram-positive bacteria has been demonstrated (3, 31) through expression of their relaxase proteins in E. coli cells that also contain cloned cognate oriT sequences on an E. coli plasmid replicon followed by isolation of transfer-related plasmid DNA-protein complexes (i.e., complexes that include plasmid molecules that are site-specifically nicked and covalently attached to their relaxase protein) (33) from these cells using purification procedures developed for E. coli. Plasmid pGSP272, a clt+ derivative of a plasmid (pGSP208) that was previously shown to allow inducible production of the pIJ101 Tra protein in E. coli cells expressing T7 RNA polymerase (19), was used to test whether Tra could site-specifically nick either strand of the clt locus in the heterologous host. Following induction of Tra protein expression as described previously (19), potentially processed pGSP272 plasmid DNA was isolated from induced cells and analyzed for nicking by a sensitive primer extension-based method previously described for E. coli plasmid R1162 (18). Under plasmid isolation and assay conditions that readily showed site-specific nicking of plasmid R1162 by its relaxase protein, no obvious nicking of either strand of the clt locus was seen in E. coli cells expressing the pIJ101 Tra protein (data not shown).
If clt indeed functions as an oriT where transfer-related nicking occurs, such processing, for reasons unknown, may only occur in vivo in the natural host, S. lividans, where Tra or a potential host-encoded factor may mediate the event. If such nicking at clt can be demonstrated, it will be interesting to determine whether this reaction, like pIJ101 transfer itself, is a temporally regulated event during the complex S. lividans life cycle (19) and whether it takes place only during surface growth of S. lividans, where conjugation readily occurs, or also in submerged culture, where plasmid transfer has not been detected (10).
To examine further the potential for interaction of the Tra protein with clt sequences, we performed gel shift assays involving 32P-labeled, linear double-stranded DNA containing the 145-bp clt+ region of pIJ101 (Fig. 1) along with Tra-containing crude extracts prepared (30) from either relevant E. coli cells containing plasmid pGSP208 or S. lividans cells containing the conjugative pIJ101 plasmid pIJ303 (15). As a control, we tested the previously described ability of the pIJ101 KorB protein present in identically prepared extracts of relevant E. coli or S. lividans cells to bind to radiolabeled kilB promoter sequences (29). Using protein binding and gel electrophoresis conditions that allowed the detection of KorB binding to kilB promoter determinants, no specific binding of clt DNA by Tra-containing E. coli or S. lividans cell extracts was observed (data not shown). Since no interactions were detected between linear clt DNA and proteins present within S. lividans cell extracts, we also therefore saw no evidence for binding of clt by any putative host factors that might be involved in pIJ101 transfer (20).
Sequences extending into korB influence clt function.
The moderate although consistent reduction in transfer frequency for plasmid pGSP288, the pGSP263 derivative with the korB portion of the original 145-bp clt+ sequence deleted (Fig. 1), prompted us to examine further the potential for involvement of 3′ korB sequences in clt function. While deletion of only 10 bp of korB sequence from the SmaI end of the pGSP263 insert (i.e., plasmid pGSP325) did not cause a reduction in transfer (Fig. 1), incremental deletions of 20, 30, and 40 bp of korB sequence, as well as deletion of 45 bp to a position just beyond the end of the korB ORF, all reduced transfer of the resulting plasmids (i.e., pGSP326 to pGSP329) to frequencies that ranged approximately two- to fourfold less than (Fig. 1), and greater than one standard deviation apart from (data not shown), the frequency seen for pGSP263. Thus, sequences extending as much as 33 bp into the 3′ end of the korB ORF (i.e., the deletion endpoint for pGSP325) appear to be capable of influencing clt transfer activity. An ancillary role in clt function for sequences extending beyond the minimal clt locus toward korB is also supported by the results for a previous DNA insertion (14) on pIJ101 at the Sau3AI site located just downstream from the korB ORF (Fig. 1), which is now known to be located between korB and clt and which was previously shown to cause an approximate threefold reduction in plasmid transfer (20).
Interestingly, deletions of 50 bp (pGSP330) and 60 bp (pGSP331) from the korB end of the pGSP263 insert that extend to positions further into the region between korB and clt had no obvious consequence on clt transfer function compared to the results seen for plasmids pGSP326 to pGSP329 (Fig. 1); thus, such additional deletion of DNA seemed to in effect restore transfer to frequencies comparable to those for pGSP263. While the basis for this apparent restoration effect remains undetermined, the results nevertheless indicate that clt sequences present on the deletion derivatives pGSP330 and pGSP331 can function as efficiently as the clt locus present on pGSP263 despite missing all or nearly all of their natural flanking sequences in the korB direction.
The clt-korB region of pIJ101 is intrinsically curved.
The observed contribution to the clt function of sequences outside of the minimal clt locus led us to consider the possibility, as has been analogously proposed for certain oriT loci (32), that higher-order structuring of DNA and protein within the clt-korB region may be an intrinsic feature of efficient pIJ101 transfer. If such a scenario is accurate, sequences within the korB ORF may, for example, play a role in ensuring proper structuring of clt DNA or perhaps serve as additional points of contact for transfer proteins interacting at clt. Since such three-dimensional structuring is believed to rely on specific sequence components, including intrinsically bent or curved DNA (32), we assessed the potential for curvature within the 145-bp clt+ region of pIJ101 using the algorithm of Goodsell and Dickerson (9) along with DNase I-based parameters (2) and the consensus bendability scale for DNA (8) as supplied by the bend-it server. This analysis predicted that the clt-korB region of pIJ101 contained significant curvature (i.e., several regions within the 145-bp clt+ sequence approached 6°/helical turn and thus exceeded the 5°/helical turn threshold level for experimentally determined curved sequences) (data not shown).
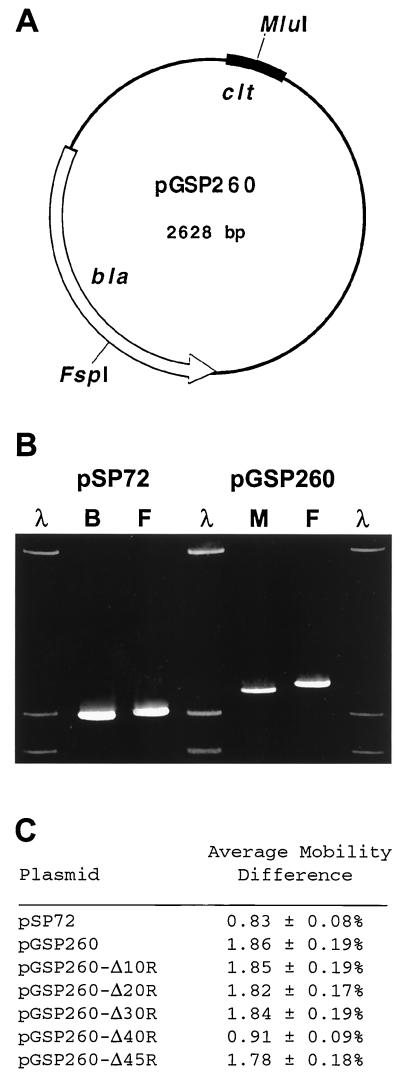
Using a protocol devised by Wu and Crothers (34) in which the gel mobility of linearized plasmid DNA containing a bent sequence motif is retarded when the bend is positioned close to the middle of the DNA molecule with respect to when it is present near the end, we tested directly for intrinsic curvature of clt-korB sequences. Plasmid pGSP260 (Fig. 3A), the 2.6-kb pSP72 derivative that contains the 145-bp clt+ region of pIJ101 at the BamHI site in the polylinker, was linearized with MluI within the insert (i.e., within potentially curved clt-korB sequences) or with FspI far away from the insert (and thus far from this same potentially bent region), and following electrophoresis through 5% nondenaturing polyacrylamide gels for 18 h at 80 V, migration distances of the linear molecules were determined. For a given gel, a mobility difference value (i.e., the ratio of the migration distance of MluI-digested pGSP260 minus the migration distance of FspI-digested pGSP260 to the migration distance of MluI-digested pGSP260) was calculated and, using the values from three separate experiments, an average mobility difference expressed as a percentage was determined. As a control, pSP72 vector DNA linearized either at BamHI, the site of cloning of clt-korB sequences in pGSP260, or at FspI was subjected to identical electrophoresis and then analyzed in a comparable manner.
FIG. 3.
Gel mobility assay to detect intrinsic curvature within the clt-korB region of pIJ101. (A) Physical and genetic map of plasmid pGSP260 as it pertains to gel mobility assays. Plasmid pGSP260 contains the 145-bp clt+ region of pIJ101 (Fig. 1) cloned as a BamHI fragment into the BamHI polylinker site of the E. coli vector pSP72. The relative positions of the MluI site (in clt) and FspI site (in the β-lactamase [bla] gene) are indicated. (B) Gel mobility assay. Plasmid pSP72 digested with either BamHI (B) or FspI (F) and pGSP260 digested with either MluI (M) or FspI (F) were electrophoresed on polyacrylamide gels, as described in the text, alongside HindIII-digested λ DNA size markers, and following electrophoresis, the gel was stained with ethidium bromide and saved as a digital image using an Eagle Eye II still video system (Stratagene). (C) Average mobility differences for pSP72 derivatives containing pIJ101 clt-korB sequences. For each gel mobility assay, the results for pGSP260 (or those for each of its deletion derivatives) were used to determine a mobility difference value, which, for pGSP260, was defined as the ratio of the migration distance of pGSP260 linearized with MluI minus the migration distance of FspI-digested pGSP260 to the migration distance of pGSP260 linearized with MluI. Similarly, a mobility difference value for pSP72 was calculated from each gel by determining the ratio of the migration distance of BamHI-digested pSP72 minus the migration distance of FspI-linearized pSP72 to the migration distance of BamHI-digested pSP72. The mobility difference values from three independent experiments were used to calculate an average mobility difference, expressed as a percentage ± standard deviation for each plasmid.
As shown in Fig. 3B, the somewhat impeded migration of FspI-digested pSP72 relative to pSP72 linearized with BamHI, which was determined to be an average mobility difference of 0.83 ± 0.08% (Fig. 3C), indicates that the vector itself shows some detectable curvature, perhaps involving polylinker cloning sequences; however, addition of the clt-korB region of pIJ101 caused a more obvious analogous disparity in migration between pGSP260 molecules linearized within clt and those linearized far away from the insert (Fig. 3B) and resulted in an average mobility difference for pGSP260 (Fig. 3C, 1.86 ± 0.19%) that was greater than twofold and was separated by at least four standard deviations from the value seen for pSP72. Using a standard curve calculated from accompanying λ HindIII-digested size markers (data not shown), linear pGSP260 molecules with curved clt-korB sequences in the middle appeared to be over 80 bp larger than identical molecules with clt-korB at the ends, and this apparent disparity in size was an increase of at least 50 bp over the moderate size disparity observed for pSP72 vector alone.
As indicated by the earlier deletion analysis, sequences within the 3′ end of the pIJ101 korB gene influence clt transfer function (Fig. 1). To determine whether these sequences also contribute to the observed bending of clt-korB DNA, we performed identical gel mobility experiments on pGSP260 derivatives with 10 through 45 bp deleted from the korB end of the 145-bp clt+ insert region (Fig. 1). The results (Fig. 3C) indicated that deletion of up to 30 bp appeared to have no effect on curvature since the average mobility differences for plasmids pGSP260-Δ10R, pGSP260-Δ20R, and pGSP260-Δ30R were unchanged relative to pGSP260. Deletions of 40 bp (pGSP260-Δ40R) and 45 bp (pGSP260-Δ45R) resulted in a significant decrease and increase, respectively, in average mobility difference, which may reflect changes in either the magnitude or direction of DNA bending (4). Deletions of 50 and 60 bp into the region between korB and the minimal clt locus (Fig. 1) on pGSP260 again caused significant reductions in average mobility differences for these clones relative to that of pGSP260 and so indicated that such deletions had additional effects on the curvature of this region (data not shown). These data therefore show that deletions extending to positions within and beyond korB gene sequences can affect the observed curvature of the clt-korB region. The apparent lack of correlation between the effects of deletions within korB on clt activity (Fig. 1) and their effects on DNA bending (Fig. 3C) may indicate that sequences beyond the minimal clt locus affect the function of clt by some mechanism other than simply through their contribution to the intrinsic curvature of this plasmid region.
Understanding the precise role of DNA curvature in clt transfer function awaits characterization of the relevant sequence elements that are responsible for this structural effect. Since our computer analysis predicted that several distinct portions of the clt-korB plasmid region may show intrinsic curvature (data not shown), there may be multiple determinants involved. Whether such DNA bending in turn contributes to the formation of higher-order protein-DNA complexes that are important for efficient pIJ101 transfer remains an interesting possibility to be explored.
Acknowledgments
Plasmids R1162 and pUT1579 were the kind gift of Richard Meyer. We thank Andromeda Daniel, Trent Lovette, and Shivani Prabhakar for construction of certain plasmids, and we thank Fred A. Rainey for assistance with automated sequencing. We are very grateful to Eric Achberger for helpful discussions about DNA curvature, advice regarding gel mobility assays, and critical reading of the manuscript.
This work was supported by National Science Foundation grant MCB-9604879 (to G.S.P.). M.D. is the recipient of a Louisiana Board of Regents graduate fellowship.
REFERENCES
- 1.Begg K J, Dewar S J, Donachie W D. A new Escherichia coli cell division gene, ftsK. J Bacteriol. 1995;177:6211–6222. doi: 10.1128/jb.177.21.6211-6222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brukner I, Sanchez R, Suck D, Pongor S. Sequence-dependent bending propensity of DNA as revealed by DNaseI: parameters for trinucleotides. EMBO J. 1995;14:1812–1818. doi: 10.1002/j.1460-2075.1995.tb07169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Climo M W, Sharma V K, Archer G L. Identification and characterization of the origin of conjugative transfer (oriT) and a gene (nes) encoding a single-stranded endonuclease on the staphylococcal plasmid pGO1. J Bacteriol. 1996;178:4975–4983. doi: 10.1128/jb.178.16.4975-4983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crothers D M, Haran T E, Nadeau J G. Intrinsically bent DNA. J Biol Chem. 1990;265:7093–7096. [PubMed] [Google Scholar]
- 5.Deng Z, Kieser T, Hopwood D A. Activity of a Streptomyces transcriptional terminator in Escherichia coli. Nucleic Acids Res. 1987;15:2665–2675. doi: 10.1093/nar/15.6.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng Z, Kieser T, Hopwood D A. “Strong incompatibility” between derivatives of the Streptomyces multi-copy plasmid pIJ101. Mol Gen Genet. 1988;214:286–294. doi: 10.1007/BF00337723. [DOI] [PubMed] [Google Scholar]
- 7.Dreiseikelmann B. Translocation of DNA across bacterial membranes. Microbiol Rev. 1994;58:293–316. doi: 10.1128/mr.58.3.293-316.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gabrielian A, Pongor S. Correlation of intrinsic DNA curvature with DNA property periodicity. FEBS Lett. 1996;393:65–68. doi: 10.1016/0014-5793(96)00855-1. [DOI] [PubMed] [Google Scholar]
- 9.Goodsell D S, Dickerson R E. Bending and curvature calculations in B-DNA. Nucleic Acids Res. 1994;22:5497–5503. doi: 10.1093/nar/22.24.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, England: John Innes Foundation; 1985. [Google Scholar]
- 11.Hopwood D A, Kieser T. Conjugative plasmids of Streptomyces. In: Clewell D B, editor. Bacterial conjugation. New York, N.Y: Plenum Press; 1993. pp. 293–311. [Google Scholar]
- 12.Jaworski D D, Clewell D B. A functional origin of transfer (oriT) on the conjugative transposon Tn916. J Bacteriol. 1995;177:6644–6651. doi: 10.1128/jb.177.22.6644-6651.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kendall K J, Cohen S N. Complete nucleotide sequence of the Streptomyces lividans plasmid pIJ101 and correlation of the sequence with genetic properties. J Bacteriol. 1988;170:4634–4651. doi: 10.1128/jb.170.10.4634-4651.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kendall K J, Cohen S N. Plasmid transfer in Streptomyces lividans: identification of a kil-kor system associated with the transfer region of pIJ101. J Bacteriol. 1987;169:4177–4183. doi: 10.1128/jb.169.9.4177-4183.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kieser T, Hopwood D A, Wright H M, Thompson C J. pIJ101, a multi-copy broad host-range Streptomyces plasmid: functional analysis and development of DNA cloning vectors. Mol Gen Genet. 1982;185:223–238. doi: 10.1007/BF00330791. [DOI] [PubMed] [Google Scholar]
- 16.Lanka E, Wilkins B M. DNA processing reactions in bacterial conjugation. Annu Rev Biochem. 1995;64:141–169. doi: 10.1146/annurev.bi.64.070195.001041. [DOI] [PubMed] [Google Scholar]
- 17.Liu G, Draper G C, Donachie W D. FtsK is a bifunctional protein involved in cell division and chromosome localization in Escherichia coli. Mol Microbiol. 1998;29:893–903. doi: 10.1046/j.1365-2958.1998.00986.x. [DOI] [PubMed] [Google Scholar]
- 18.Perwez T, Meyer R. MobB protein stimulates nicking at the R1162 origin of transfer by increasing the proportion of complexed plasmid DNA. J Bacteriol. 1996;178:5762–5767. doi: 10.1128/jb.178.19.5762-5767.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pettis G S, Cohen S N. Plasmid transfer and expression of the transfer (tra) gene product of plasmid pIJ101 are temporally regulated during the Streptomyces lividans life cycle. Mol Microbiol. 1996;19:1127–1135. doi: 10.1046/j.1365-2958.1996.493986.x. [DOI] [PubMed] [Google Scholar]
- 20.Pettis G S, Cohen S N. Transfer of the pIJ101 plasmid in Streptomyces lividans requires a cis-acting function dispensable for chromosomal gene transfer. Mol Microbiol. 1994;13:955–964. doi: 10.1111/j.1365-2958.1994.tb00487.x. . (Corrigendum, 16:170, 1995.) [DOI] [PubMed] [Google Scholar]
- 21.Pettis G S, Prakash S. Complementation of conjugation functions of Streptomyces lividans plasmid pIJ101 by the related Streptomyces plasmid pSB24.2. J Bacteriol. 1999;181:4680–4685. doi: 10.1128/jb.181.15.4680-4685.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Russel M, Model P. Replacement of the fip gene of Escherichia coli by an inactive gene cloned on a plasmid. J Bacteriol. 1984;159:1034–1039. doi: 10.1128/jb.159.3.1034-1039.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Stein D S, Cohen S N. Mutational and functional analysis of the korA and korB gene products of Streptomyces plasmid pIJ101. Mol Gen Genet. 1990;222:337–344. doi: 10.1007/BF00633838. [DOI] [PubMed] [Google Scholar]
- 25.Stein D S, Kendall K J, Cohen S N. Identification and analysis of transcriptional regulatory signals for the kil and kor loci of Streptomyces plasmid pIJ101. J Bacteriol. 1989;171:5768–5775. doi: 10.1128/jb.171.11.5768-5775.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steiner W, Liu G, Donachie W D, Kuempel P. The cytoplasmic domain of FtsK protein is required for resolution of chromosome dimers. Mol Microbiol. 1999;31:579–583. doi: 10.1046/j.1365-2958.1999.01198.x. [DOI] [PubMed] [Google Scholar]
- 27.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 28.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tai J T-N, Cohen S N. The active form of the KorB protein encoded by the Streptomyces plasmid pIJ101 is a processed product that binds differentially to the two promoters it regulates. J Bacteriol. 1993;175:6996–7005. doi: 10.1128/jb.175.21.6996-7005.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson J, Rae S, Cundliffe E. Coupled transcription-translation in extracts of Streptomyces lividans. Mol Gen Genet. 1984;195:39–43. doi: 10.1007/BF00332721. [DOI] [PubMed] [Google Scholar]
- 31.Wang A, Macrina F L. Streptococcal plasmid pIP501 has a functional oriT site. J Bacteriol. 1995;177:4199–4206. doi: 10.1128/jb.177.15.4199-4206.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilkins B, Lanka E. DNA processing and replication during plasmid transfer between Gram-negative bacteria. In: Clewell D B, editor. Bacterial conjugation. New York, N.Y: Plenum Press; 1993. pp. 105–136. [Google Scholar]
- 33.Willetts N, Wilkins B. Processing of plasmid DNA during bacterial conjugation. Microbiol Rev. 1984;48:24–41. doi: 10.1128/mr.48.1.24-41.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu H-M, Crothers D M. The locus of sequence-directed and protein-induced bending. Nature. 1984;308:509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- 35.Wu L J, Errington J. Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science. 1994;264:572–575. doi: 10.1126/science.8160014. [DOI] [PubMed] [Google Scholar]
- 36.Wu L J, Lewis P J, Allmansberger R, Hauser P M, Errington J. A conjugation-like mechanism for prespore chromosome partitioning during sporulation in Bacillus subtilis. Genes Dev. 1995;9:1316–1326. doi: 10.1101/gad.9.11.1316. [DOI] [PubMed] [Google Scholar]
- 37.Zaman S, Richards H, Ward J. Expression and characterisation of the korB gene product from the Streptomyces lividans plasmid pIJ101 in Escherichia coli and determination of its binding site on the korB and kilB promoters. Nucleic Acids Res. 1992;20:3693–3700. doi: 10.1093/nar/20.14.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]