Abstract
By multiplex reverse transcription-PCR, we demonstrate that the SoxRS response, which protects cells against superoxide toxicity, is triggered also by hydrogen peroxide. SoxR-dependent inductions of 7.3-, 7.6-, 4.6-, 2.2-, and 2.6-fold were quantified for soxS, micF, sodA, inaA, and fpr transcripts, respectively. This finding suggests an extensive and tight connectivity between different regulatory pathways in the Escherichia coli response to oxidative stress.
Key regulators of adaptive responses to hydrogen peroxide (H2O2) and superoxide anion (O2·−) are OxyR and SoxR together with SoxS, respectively (recently reviewed in reference 11). H2O2 oxidizes the transcription factor OxyR, leading to the formation of an intramolecular disulfide bond (17). Oxidized OxyR then induces the transcription of a set of genes, including katG (hydroperoxidase I), dps (a nonspecific DNA binding protein), ahpCF (alkyl hydroperoxide reductase) and gorA (glutathione reductase). Superoxide-generating compounds, such as paraquat, activate the transcription factor SoxR by the univalent oxidation of the 2Fe-2S clusters of the protein through an unknown mechanism (11). Oxidized SoxR then induces the expression of a second transcription factor, SoxS, which in turn activates the transcription of a set of genes. Among the SoxRS-regulated genes are micF (a regulatory RNA), sodA (manganese superoxide dismutase), inaA (unknown function), and fpr (NADPH- ferredoxin reductase) (10, 11). Redox-cycling agents also activate the syntheses of proteins that are induced by H2O2 and controlled by OxyR, due to the spontaneous and superoxide dismutase-mediated conversion of O2·− to H2O2 (4, 15). In contrast, it is commonly believed that H2O2 is unable to switch on the SoxRS regulon expression (4, 8, 14, 15).
We have devised a multiplex reverse transcription-PCR for the simultaneous quantitation of the in vivo transcription of more than 10 different target genes (2) (M. Manchado, C. Michán, M. Cousinou, G. Dorado and C. Pueyo, unpublished data). In this protocol, all target genes, a housekeeping gene, and one or two external standards are amplified in the same reaction tube. Specific fluorescent primers are used, and amplification products are analyzed with a DNA sequencer. Putative variations in the expression of the housekeeping gene are controlled by the external standards. Expressions of the targets relative to the reference (or to one of the external standards) are measured. Recently, we have experimentally demonstrated that our methodology fulfills all theoretical requirements for precise quantification of both induction and repression of gene transcription (M. Manchado, C. Michán, M. Cousinou, G. Dorado and C. Pueyo, unpublished data). Because of the PCR amplification step, our method displays a much higher sensitivity than those of current techniques for mRNA quantitation, such as Northern blotting or primer extension analyses. Here, we used this sensitive experimental approach to investigate if H2O2 is able to trigger the expression of the SoxRS regulon, in order to better define the coordination between the OxyR and SoxRS regulatory networks of Escherichia coli.
H2O2 induces sodA transcription.
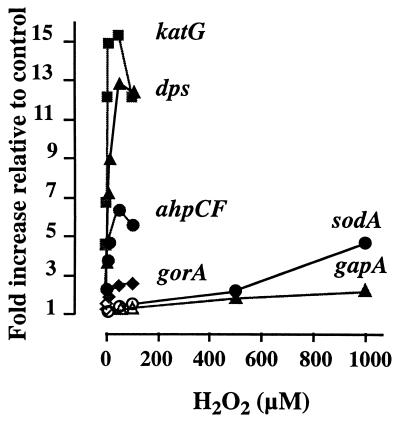
The putative activation of the SoxRS regulon by H2O2 was first investigated by examining the expression of the sodA gene in wild-type cells exposed to a wide range of H2O2 concentrations (varying from 0.25 to 1,000 μM) (Fig. 1). While the lower-dose treatments (from 1 to 100 μM H2O2) induced specifically the transcription of selected OxyR-regulated genes (katG, dps, ahpCF, gorA), the higher-dose treatments (≥500 μM H2O2) resulted in the activation of sodA. In contrast with previous data (14), inductions of 2.1- and 4.7-fold were readily seen for sodA mRNA in response to 500 and 1,000 μM H2O2, respectively, immediately after the addition of the oxidant (Fig. 1).
FIG. 1.
H2O2 induces sodA transcription. Wild-type cells grown in M9 minimal medium (optical density at 600 nm of 0.2) were treated with the H2O2 concentration (μM) indicated in the abscissa. Samples were collected immediately (<1 min) after the addition of the oxidant. RNA purification, cDNA synthesis, and multiplex PCRs were carried out as described previously (7). An exogenous fragment of the gene (CYP1A) coding for cytochrome P4501A from Liza aurata (9) was coamplified with the target genes and the reference gapA gene. The fluorescence signal of each PCR product was compared to that of CYP1A (noncompetitor heterologous standard). Data were from an average of eight multiplexed PCR amplifications. Values from treated samples were divided by those from the corresponding control (unexposed bacteria). Statistical comparisons were done by an analysis of variance test. Significant increments are indicated by filled-in symbols.
In previous studies (2, 7, 9), the amounts of the mRNAs of interest were determined with reference to the mRNA level of the gapA gene (used as an internal standard), which codes for a key enzyme of the glycolytic and gluconeogenesis pathways (d-glyceraldehyde-3-phosphate dehydrogenase). As discussed elsewhere (M. Manchado, C. Michán, M. Cousinou, G. Dorado and C. Pueyo, unpublished data), it cannot be taken for granted that the so-called housekeeping gene will maintain a steady level of expression under all circumstances. Indeed, we observed a small (≤twofold), though statistically significant, increase in gapA transcription in response to ≥500 μM H2O2. Thus, to circumvent possible underestimations of transcriptional inductions, all data in this work were compared to an external standard. A noncompetitor heterologous standard was used for Fig. 1, and a competitor homologous standard was used for Fig. 2 and 3. As described elsewhere (M. Manchado, C. Michán, M. Cousinou, G. Dorado and C. Pueyo, unpublished data), both types of external standards are equally useful for monitoring changes in the expression levels of the genes, but the competitor has the additional advantage of using the same pair of primers that amplifies the reference gene.
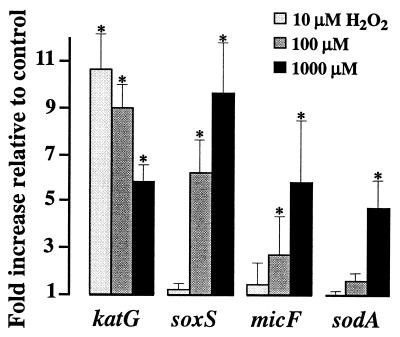
FIG. 2.
H2O2 induces SoxRS regulon transcription. Treatments and analyses were as described in the legend to Fig. 1. A laboratory-engineered fragment (gapA*) of the housekeeping gene was used as an external standard (M. Manchado, C. Michán, M. Cousinou, G. Dorado, and C. Pueyo, unpublished data). The fluorescence signal of each PCR product was compared to that of gapA* (competitor homologous standard). Error bars were estimated from the corresponding standard error of the mean values. Significant increments are marked with an asterisk.
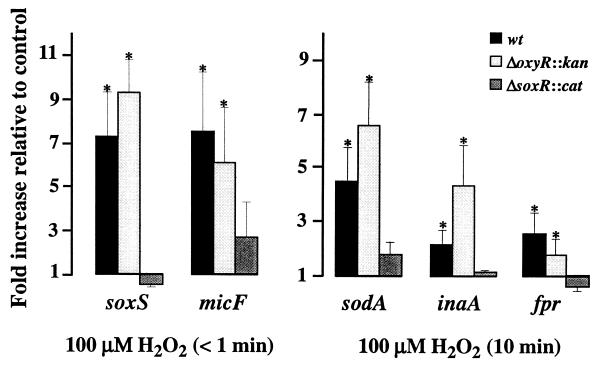
FIG. 3.
SoxR regulates the induction of SoxRS regulon transcription by H2O2. Wild-type, ΔoxyR::kan, and ΔsoxR::cat bacteria were treated with 100 μM H2O2. Samples were collected immediately (<1 min) or at 10 min after the addition of H2O2. The fluorescence signal of each PCR product was compared to that of a competitor homologous standard (gapA*). See the legends to Fig. 1 and 2 for more details.
SoxRS regulon activation by H2O2 depends on SoxR.
The results above and the particularly complex multiregulated transcription of sodA (13) prompted us to investigate the effect of H2O2 treatment on the expression of other SoxRS-regulated genes, such as soxS, micF, inaA, and fpr (Fig. 2 and 3). H2O2 at a 100 μM concentration increased instantaneously the transcript levels of soxS and micF (Fig. 2), whereas 5- to 10-fold-higher concentrations were required to induce significantly the sodA expression (Fig. 1 and 2). Nonetheless, significant increments in sodA transcription and also in inaA and fpr transcription were observed when bacteria were exposed to 100 μM H2O2 for 10 min (Fig. 3). These results clearly demonstrate that H2O2 stress conditions that are regularly used to induce the OxyR response (e.g., references 1, 12 and 17) strongly activate the in vivo transcription of the SoxRS regulon. Therefore, induction levels of 7.3- and 7.6-fold (<1 min of treatment) and of 4.6-, 2.2-, and 2.6-fold (10 min of treatment) were quantified for soxS, micF, sodA, inaA, and fpr, respectively, in response to 100 μM H2O2 (Fig. 3). Interestingly, the activation of the SoxRS-regulated genes by H2O2 was seen concomitantly with a decline in the amounts of the OxyR-regulated transcripts (e.g., the katG transcript levels shown in Fig. 2).
Figure 3 provides further evidence that the induction of soxS, micF, sodA, inaA, and fpr expression by H2O2 was abolished by the introduction of the ΔsoxR9::cat mutation (16), indicating a strict dependence on a functional SoxR regulator. In contrast, this transcriptional up-regulation was preserved in the strain with the ΔoxyR::kan mutation (6), indicating that OxyR is not involved in the SoxR-mediated response to H2O2. In fact, generally, the induction ratios of the SoxRS-regulated genes by H2O2 were somewhat higher in the ΔoxyR::kan mutant than in the wild-type strain, which might be attributed to the inability of OxyR-defective bacteria to induce katG transcription (7), i.e., the H2O2 breakdown catalyzed by hydroperoxidase I.
The activation of the SoxRS regulon by H2O2 stress in a SoxR-dependent manner might be an indirect result, depending on the formation of an H2O2-generated signal that is sensed by SoxR rather than on H2O2 itself. In fact, a crucial unanswered question concerns the nature of the signal(s) sensed by SoxR. Another possibility is that H2O2 somehow could interfere with the unknown system that keeps SoxR in its reduced, inactive form in vivo, e.g., by depleting the electron source for the recently discovered SoxR reductase (5). Indeed, we have demonstrated (9) that both glutaredoxin and thioredoxin pathways, which consume NADPH, are triggered in the OxyR-mediated response to H2O2. Whatever the mechanism, the H2O2-mediated induction of the SoxRS response may reflect the existence of multiple pathways for SoxR activation and deactivation, as discussed by others (3). This versatility would let the SoxR system respond to a wide range of environmental and intracellular changes indicative of possible oxidative stress. The multiplex reverse transcription-PCR methodology used in this work for quantification of transcription will be of relevance in further experiments. Nevertheless, quantifications at the protein level will also be necessary in order to unravel the relationships between mRNA production and protein synthesis.
Acknowledgments
M.M. and C.M. contributed equally to this paper, and both should be considered first authors.
This work was supported by grant PB98-1627 (DGES) and by Junta de Andalucía (group CVI 0187). M.M. was the recipient of a predoctoral fellowship from the Spanish Ministry of Education and Culture (MEC).
REFERENCES
- 1.Altuvia S, Almiron M, Huisman G, Kolter R, Storz G. The dps promoter is activated by OxyR during growth and by IHF and ςS in stationary phase. Mol Microbiol. 1994;13:265–272. doi: 10.1111/j.1365-2958.1994.tb00421.x. [DOI] [PubMed] [Google Scholar]
- 2.Gallardo-Madueño R, Leal J F, Dorado G, Holmgren A, Lopez-Barea J, Pueyo C. In vivo transcription of nrdAB operon and of grxA and fpg genes is triggered in Escherichia coli lacking both thioredoxin and glutaredoxin 1 or thioredoxin and glutathione, respectively. J Biol Chem. 1998;273:18382–18388. doi: 10.1074/jbc.273.29.18382. [DOI] [PubMed] [Google Scholar]
- 3.Gaudu P, Dubrac S, Touati D. Activation of SoxR by overproduction of desulfoferrodoxin: multiple ways to induce the soxRS regulon. J Bacteriol. 2000;182:1761–1763. doi: 10.1128/jb.182.6.1761-1763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greenberg J T, Demple B. A global response induced in Escherichia coli by redox-cycling agents overlaps with that induced by peroxide stress. J Bacteriol. 1989;171:3933–3939. doi: 10.1128/jb.171.7.3933-3939.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kobayashi K, Tagawa S. Isolation of reductase for SoxR that governs an oxidative response regulon from Escherichia coli. FEBS Lett. 1999;451:227–230. doi: 10.1016/s0014-5793(99)00565-7. [DOI] [PubMed] [Google Scholar]
- 6.Kullik I, Stevens J, Toledano M B, Storz G. Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for DNA binding and multimerization. J Bacteriol. 1995;177:1285–1291. doi: 10.1128/jb.177.5.1285-1291.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michán C, Manchado M, Dorado G, Pueyo C. In vivo transcription of the Escherichia coli oxyR regulon as a function of growth phase and in response to oxidative stress. J Bacteriol. 1999;181:2759–2764. doi: 10.1128/jb.181.9.2759-2764.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nunoshiba T, Hidalgo E, Amabile Cuevas C F, Demple B. Two-stage control of an oxidative stress regulon: the Escherichia coli SoxR protein triggers redox-inducible expression of the soxS regulatory gene. J Bacteriol. 1992;174:6054–6060. doi: 10.1128/jb.174.19.6054-6060.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prieto-Álamo M J, Jurado J, Gallardo-Madueño R, Monje-Casas F, Holmgren A, Pueyo C. Transcriptional regulation of glutaredoxin and thioredoxin pathways and related enzymes in response to oxidative stress. J Biol Chem. 2000;275:13398–13405. doi: 10.1074/jbc.275.18.13398. [DOI] [PubMed] [Google Scholar]
- 10.Rosner J L, Slonczewski J L. Dual regulation of inaA by the multiple antibiotic resistance (Mar) and superoxide (SoxRS) stress response systems of Escherichia coli. J Bacteriol. 1994;176:6262–6269. doi: 10.1128/jb.176.20.6262-6269.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Storz G, Imlay J A. Oxidative stress. Curr Opin Microbiol. 1999;2:188–194. doi: 10.1016/s1369-5274(99)80033-2. [DOI] [PubMed] [Google Scholar]
- 12.Tao K, Makino K, Yonei S, Nakata A, Shinagawa H. Molecular cloning and nucleotide sequencing of oxyR, the positive regulatory gene of a regulon for an adaptive response to oxidative stress in Escherichia coli: homologies between OxyR protein and a family of bacterial activator proteins. Mol Gen Genet. 1989;218:371–376. doi: 10.1007/BF00332397. [DOI] [PubMed] [Google Scholar]
- 13.Touati D. Superoxide dismutases in bacteria and pathogen protists. In: Scandalios J G, editor. Oxidative stress and the molecular biology of antioxidant defenses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1997. pp. 447–493. [Google Scholar]
- 14.Touati D. Transcriptional and posttranscriptional regulation of manganese superoxide dismutase biosynthesis in Escherichia coli, studied with operon and protein fusions. J Bacteriol. 1988;170:2511–2520. doi: 10.1128/jb.170.6.2511-2520.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walkup L K, Kogoma T. Escherichia coli proteins inducible by oxidative stress mediated by the superoxide radical. J Bacteriol. 1989;171:1476–1484. doi: 10.1128/jb.171.3.1476-1484.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu J, Weiss B. Two-stage induction of the soxRS (superoxide response) regulon of Escherichia coli. J Bacteriol. 1992;174:3915–3920. doi: 10.1128/jb.174.12.3915-3920.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng M, Aslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]