Abstract
A temperature-sensitive mutant of Mycobacterium smegmatis was characterized that contains a mutation in ddlA, the gene encoding d-alanine:d-alanine ligase. Enzymatic assays using recombinant proteins and d-cycloserine susceptibility indicate that the A365V mutation in the SMEG35 DdlA protein causes a reduction in enzymatic activity in vitro and in vivo.
A nearly universal component of bacterial cell walls is peptidoglycan, a macromolecule that is composed of polysaccharide chains that are cross-linked by short peptide bridges. Peptidoglycan gives the bacterial cell its characteristic shape and prevents the cell from lysing due to high internal osmotic pressure. Our understanding of how peptidoglycan is synthesized in bacteria is derived mostly from work done with Escherichia coli in which a number of temperature-sensitive mutants have been isolated that are defective in the biosynthesis of peptidoglycan at 42°C (10, 11, 13, 14, 20). Two hallmarks of these mutants are cell lysis at the nonpermissive temperature and suppression of the temperature-sensitive phenotype on media containing osmotic stabilizers (10, 11, 13, 14, 20).
We previously described the generation of a bank of temperature-sensitive mutants of Mycobacterium smegmatis mc2155 (2, 4). One of the mutants, SMEG35, exhibits the two phenotypic characteristics associated with E. coli mutants defective in peptidoglycan biosynthesis. First, SMEG35 cells grown at 30°C to an optical density at 600 nm of 0.5 and then shifted to 42°C lyse after one doubling time, as evidenced by a visual clearing of the culture and the appearance of flocculent material (data not shown). Second, the temperature-sensitive phenotype of SMEG35 can be suppressed on growth medium containing either 0.5 M sucrose or 0.2 M NaCl (data not shown).
The bacterial strains and plasmids used in this study are listed in Table 1. To identify the mutated gene, SMEG35 was complemented with an M. smegmatis genomic cosmid library as previously described (2). A sublibrary was constructed from the complementing cosmid pAEB222 and the E. coli-mycobacterial shuttle vector pMD30 as described previously (2). The nucleotide sequence was determined for the insert of the smallest complementing subclone pAEB224 with an ABI310 automated sequencer (PE Biosystems, Foster City, Calif.) and gene-specific primers. The 1,346-bp sequence of pAEB224 contains a single open reading frame that encodes a protein of 373 amino acids. Database searches using BLAST (1) indicate that the predicted gene product has similarity to a number of d-alanine:d-alanine ligases (Ddls) from gram-negative and gram-positive bacteria. Among the database matches, the M. smegmatis gene product is most similar to the Mycobacterium tuberculosis H37Rv DdlA homolog encoded by the Rv2981c gene (3), with 83% similarity. While some bacteria, such as E. coli, have two Ddls, DdlA and DdlB (21), M. tuberculosis (and by inference M. smegmatis) has only one, DdlA (3). Amino acid alignments with Clustal W (19) indicate that a number of residues are conserved between M. smegmatis DdlA and the Ddl proteins of representative gram-negative and gram-positive bacteria, including those amino acid residues that are mechanistically important (8, 17; data not shown).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant features | Source or reference |
|---|---|---|
| Strains | ||
| M. smegmatis | ||
| mc2155 | ept1 ddlA+ | 16 |
| SMEG35 | ept1 ddlA1(Ts) | This study |
| E. coli XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 SupE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene, La Jolla, Calif. |
| Plasmids | ||
| pBluescript SK− | E. coli cloning vector | Stratagene |
| pMD30 | E. coli-mycobacterial shuttle vector | 7 |
| pQE31 | Protein expression vector | Qiagen, Chatsworth, Calif. |
| pAEB222 | M. smegmatis-derived cosmid containing ddlA | This study |
| pAEB224 | 1.4-kb Sau3AI fragment containing ddlA cloned into pMD30 | This study |
| pAEB242 | Wild-type ddlA gene cloned into pQE31 | This study |
| pAEB243 | SMEG35 ddlA1 gene cloned into pQE31 | This study |
The presence of a mutation in the ddlA gene of SMEG35 was confirmed by sequencing of the mutant allele. The gene was amplified from the SMEG35 genome with Pfu DNA polymerase (Stratagene) and the primers 5′-TTGTGACTGCCCCGAACC-3′ (forward) and 5′-CGAAAAACCCGTCGAGCC-3′ (reverse) in a PCR mixture supplemented with 5% formamide. Sequence analysis revealed a single mutation at bp 1095 of ddlA that changes a C to a T on the top strand. This mutation results in an alanine-to-valine substitution at amino acid 365 of Ddl, close to the C terminus of the protein. The alanine that is mutated in the SMEG35 Ddl is a nonconserved amino acid residue that does not correspond to any of the amino acids that were previously shown to be important for function in the Ddls of other bacteria.
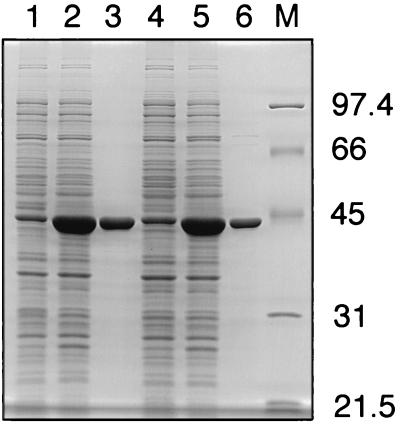
In E. coli, DdlA and DdlB synthesize d-alanyl-d-alanine, a dipeptide used in the biosynthesis of the peptidoglycan pre- cursor UDP-N-acetylmuramoyl-l-alanyl-d-isoglutamyl-meso- diaminopimelyl-d-alanyl-d-alanine (20). Since the specific reaction catalyzed by the two Ddls is 2 d-alanine + ATP → d-alanine:d-alanine + ADP + Pi, enzymatic activity can be assayed by quantitating the d-alanine-dependent liberation of free phosphate from ATP (5, 21). We found that this method lacked the sensitivity required to measure Ddl activity in crude mycobacterial extracts. Therefore, to study the consequences of the A365V mutation in SMEG35 DdlA, the wild-type and mutant DdlA proteins were overexpressed in E. coli. To make the protein expression constructs, wild-type and SMEG35 ddlA genes were first amplified as described above, except that two nucleotides (underlined) were reversed in the sequence of the forward primer (5′-TTGTGCATGCCCCGAACC-3′) to introduce an SphI site. The PCR products were cloned into the EcoRV site of pBluescript SK− (Stratagene), and then the resulting constructs were digested with SphI and HindIII to liberate the 1.3-kb insert. The DNA fragments from the wild type and SMEG35 were cloned into the corresponding sites of pQE31 to create pAEB242 and pAEB243, respectively. Plasmid pQE31 is an E. coli vector that allows for the overexpression of N-terminal His6-tagged proteins from a phage T5 promoter containing two lac operator sequences (Qiagen, Chatsworth, Calif.). Pilot inductions were performed with E. coli cells at 30°C with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 2 h. Analysis of cellular protein contents indicated that both the wild-type and mutant DdlA proteins were overexpressed to high levels in E. coli under these conditions (Fig. 1).
FIG. 1.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of protein overexpression and purification. Lanes: 1 and 2, protein profiles of E. coli containing DNA constructs encoding the wild-type DdlA protein uninduced (lane 1) and induced with IPTG (lane 2); 3, purified wild-type DdlA protein; 4 and 5, protein profiles of E. coli containing DNA constructs encoding the A365V protein from SMEG35 uninduced (lane 4) and induced with IPTG (lane 5); 6, purified A365V protein; M, molecular mass markers (sizes given to the right in kilodaltons).
The temperature sensitivity of SMEG35 could result from thermolability of the mutant protein, a general folding defect (as described previously for EcoRI endonuclease and phage P22 tail spike endorhamnosidase [9, 15]), or a general reduction in specific activity resulting in a mutant protein that cannot keep up with the increased metabolic demands of growth at higher temperatures. To address this, the wild-type and mutant DdlA proteins were purified and tested for enzymatic activity at different temperatures. For these experiments, the proteins were overexpressed at 30°C, since under these conditions, the two proteins are synthesized at similar levels and have comparable (and high) levels of solubility (>90%); no inclusion bodies were observed. When expressed at higher temperatures, less expression of the mutant protein was observed, and a greater fraction of it was insoluble relative to the wild-type protein (data not shown). While these observations are consistent with the mutant possessing altered folding proteins, they also complicate any comparison of enzyme activities when isolated at higher temperatures.
The His6-tagged wild-type and mutant DdlA proteins were isolated from E. coli cells induced with 1 mM IPTG for 2 h and purified by nickel affinity chromatography. Proteins were isolated under native conditions by using 10 mM HEPES (pH 8.0)-buffered solutions supplemented with 5% glycerol and 5 mM β-mercaptoethanol according to the manufacturer's recommendations (Qiagen), and the specific activities were measured with a phosphate release assay. We observed that the wild-type DdlA protein has a high level of specific activity at all temperatures tested, although it is about twofold higher at 30°C than at 42°C. The specific activity of the wild-type protein at 37°C is somewhat higher (8- to 19-fold) than that reported previously for the DdlA and DdlB proteins of E. coli and Salmonella enterica serovar Typhimurium (5, 21), suggesting that it is unlikely that the inclusion of the His tag has introduced any significantly deleterious property to the protein or that a substantial portion of the purified protein is inactive.
The specific activities of the mutant protein show a response to temperature similar to that seen with wild-type DdlA, with a temperature optimum at 30°C and approximately twofold less activity at 42°C than at 30°C (Table 2). These observations indicate that the mutant protein is no more thermolabile than wild-type DdlA. Thus, thermolability of DdlA is unlikely to account for the temperature sensitivity of SMEG35. However, we also observed that at all three temperatures, A365V DdlA has almost 30-fold less activity than the wild-type protein, and the activity of both proteins was linear with time, indicating that the assay conditions were not limiting (data not shown). It therefore seems more likely that the mutant substitution in A365V DdlA interferes with the general catalytic properties of the protein, although we cannot rule out additional defects resulting from altered protein folding. We note, however, that the alanine at position 365 in DdlA is not well conserved among Ddl proteins and has not been previously described as a catalytically important residue.
TABLE 2.
d-Alanine:d-alanine ligase activities
| Temp (°C) | Sp act (μmol h−1 mg of protein−1)a
|
|
|---|---|---|
| Wild type | A365V | |
| Ambient | 294 | 10.4 |
| 30 | 410 | 14 |
| 37 | 236.8 | 10.4 |
| 42 | 207.2 | 7.6 |
Each value shown is the average of three independent assays. The values obtained varied by no more than 7%, except for those obtained for the A365V protein at 42°C, which varied by 20%.
The specific activities of the DdlA proteins described above indicate that there is a significant reduction in DdlA activity in the mutant protein even at the permissive temperature. To determine if the Ddl activity is low in SMEG35 relative to that in the wild type at 30°C in vivo, we examined the susceptibility of the two strains to d-cycloserine, a drug that specifically inhibits the activity of the Ddl enzymes in bacteria, including mycobacteria (6, 16). When the MIC of d-cycloserine for both strains was determined, it was found that the growth of SMEG35 was inhibited >90% on medium containing 50 μg/ml of the drug, while 200 μg/ml of the drug was required for the same response by the wild type. The MIC of other antimycobacterial drugs, such as rifampin, ethambutol, and isoniazid, for SMEG35 was similar to that for the wild type. These observations are consistent with the A365V mutant protein having reduced enzymatic activity relative to wild-type DdlA at 30°C.
Taken together, these data suggest that the temperature sensitivity of SMEG35 is due to a defect in peptidoglycan biosynthesis that is mediated by a mutation in ddlA. The A365V substitution in the SMEG35 DdlA protein apparently causes either a catalytic or a folding defect that results in a reduction in enzymatic activity at all temperatures, such that the temperature sensitivity of SMEG35 does not result from a thermolabile DdlA protein. Since M. smegmatis mc2155 grows about three times faster at 42°C than it does at 30°C (2), we favor the alternative hypothesis that the reduced activity of the mutant DdlA protein is insufficient to meet the metabolic demands of faster growth at the higher temperature, where the overall level of peptidoglycan synthesis is significantly higher.
Nucleotide sequence accession number.
The DNA sequence of the M. smegmatis ddlA gene has been deposited in GenBank under accession no. AFO77728.
Acknowledgments
We thank Jeffery Brodsky and Karla Fullner for helpful suggestions.
This work was supported by grant AI37848 from the National Institutes of Health to G.F.H. and an American Lung Association Fellowship to A.E.B.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belanger A E, Hatfull G F. Exponential-phase glycogen recycling is essential for growth in Mycobacterium smegmatis. J Bacteriol. 1999;181:6670–6678. doi: 10.1128/jb.181.21.6670-6678.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Englmeir K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail M A, Rajandream M-A, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 4.Connell N D, Belanger A E. Mutants and mutagenesis. In: Hatfull G F, Jacobs W R Jr, editors. Molecular genetics of mycobacteria. Washington, D.C.: American Society for Microbiology; 2000. pp. 257–263. [Google Scholar]
- 5.Daub E L, Zawadzke L E, Botstein D, Walsh C T. Isolation, cloning and sequencing of the Salmonella typhimurium ddlA gene with purification and characterization of its product, d-alanine:d-alanine ligase (ADP forming) Biochemistry. 1988;27:3701–3708. doi: 10.1021/bi00410a027. [DOI] [PubMed] [Google Scholar]
- 6.David H L, Takyama K, Goldman D S. Susceptibility of mycobacterial d-alanyl-d-alanine synthetase to d-cycloserine. Am Rev Respir Dis. 1969;100:579–581. doi: 10.1164/arrd.1969.100.4.579. [DOI] [PubMed] [Google Scholar]
- 7.Donnelly-Wu M, Jacobs W R, Jr, Hatfull G F. Superinfection immunity of mycobacteriophage L5: applications for genetic transformation of mycobacteria. Mol Microbiol. 1993;7:407–417. doi: 10.1111/j.1365-2958.1993.tb01132.x. [DOI] [PubMed] [Google Scholar]
- 8.Fan C P, Moews P C, Walsh C T, Knox J R. Vancomycin resistance: structure of the d-alanine:d-alanine ligase at 2.3 Å resolution. Science. 1994;266:439–443. doi: 10.1126/science.7939684. [DOI] [PubMed] [Google Scholar]
- 9.Haase-Pettingell C, King J. Prevalence of temperature-sensitive folding mutations in the parallel beta coil domain of the phage P22 tailspike endorhamnosidase. J Mol Biol. 1997;267:88–102. doi: 10.1006/jmbi.1996.0841. [DOI] [PubMed] [Google Scholar]
- 10.Lugtenberg E J J, de Haas-Menger L, Ruyters W H M. Murein synthesis and identification of cell wall precursors of temperature-sensitive lysis mutants of Escherichia coli. J Bacteriol. 1972;109:326–335. doi: 10.1128/jb.109.1.326-335.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lugtenberg E J J, van Schijndel-van Dam A. Temperature-sensitive mutants of Escherichia coli K-12 with low activities of the l-alanine adding enzyme and the d-alanyl-d-alanine adding enzyme. J Bacteriol. 1972;110:35–40. doi: 10.1128/jb.110.1.35-40.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin F, Sharples G J, Lloyd R G, Eiler S, Moras D, Gangloff J, Eriani G. Characterization of a thermosensitive Escherichia coli aspartyl-tRNA synthetase mutant. J Bacteriol. 1997;179:3691–3696. doi: 10.1128/jb.179.11.3691-3696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuzawa H, Matsuhashi M, Oka A, Sugino Y. Genetic and biochemical studies on cell wall peptidoglycan synthesis in Escherichia coli K-12. Biochem Biophys Res Commun. 1969;36:682–689. doi: 10.1016/0006-291x(69)90360-x. [DOI] [PubMed] [Google Scholar]
- 14.Miyakawa T, Matsuzawa H, Matsuhashi M, Sugino Y. Cell wall peptidoglycan mutants of Escherichia coli K-12: existence of two clusters of genes, mra and mrb, for cell wall peptidoglycan biosynthesis. J Bacteriol. 1972;112:950–958. doi: 10.1128/jb.112.2.950-958.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muir R S, Flores H, Zinder N D, Model P, Soberon X, Heitman J. Temperature-sensitive mutants of the EcoRI endonuclease. J Mol Biol. 1997;274:722–737. doi: 10.1006/jmbi.1997.1419. [DOI] [PubMed] [Google Scholar]
- 16.Neuhaus F C, Lynch J L. The enzymatic synthesis of d-alanyl-d-alanine. III. On the inhibition of d-alanyl-d-alanine synthetase by the antibiotic d-cycloserine. Biochemistry. 1964;3:471–480. doi: 10.1021/bi00892a001. [DOI] [PubMed] [Google Scholar]
- 17.Shi Y, Walsh C T. Active site mapping of Escherichia colid-Ala-d-Ala ligase by structure-based mutagenesis. Biochemistry. 1995;34:2768–2776. doi: 10.1021/bi00009a005. [DOI] [PubMed] [Google Scholar]
- 18.Snapper S, Melton R, Keiser T, Jacobs W R., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 19.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Heijenoort J. Murein synthesis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Shaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 1025–1034. [Google Scholar]
- 21.Zawadzke L E, Bugg T D H, Walsh C T. Existence of two d-alanine:d-alanine ligases in Escherichia coli: cloning and sequencing of the ddlA gene and purification and characterization of the DdlA and DdlB enzymes. Biochemistry. 1991;30:1673–1682. doi: 10.1021/bi00220a033. [DOI] [PubMed] [Google Scholar]