Abstract
Oncolytic viruses (OVs), a group of replication-competent viruses that can selectively infect and kill cancer cells while leaving healthy cells intact, are emerging as promising living anticancer agents. Unlike traditional drugs composed of non-replicating compounds or biomolecules, the replicative nature of viruses confer unique pharmacokinetic properties that require further studies. Despite some pharmacokinetics studies of OVs, mechanistic insights into the connection between OV pharmacokinetics and antitumor efficacy remain vague. Here, we characterized the pharmacokinetic profile of oncolytic virus M1 (OVM) in immunocompetent mouse tumor models and identified the JAK‒STAT pathway as a key modulator of OVM pharmacokinetics. By suppressing the JAK‒STAT pathway, early OVM pharmacokinetics are ameliorated, leading to enhanced tumor-specific viral accumulation, increased AUC and Cmax, and improved antitumor efficacy. Rather than compromising antitumor immunity after JAK‒STAT inhibition, the improved pharmacokinetics of OVM promotes T cell recruitment and activation in the tumor microenvironment, providing an optimal opportunity for the therapeutic outcome of immune checkpoint blockade, such as anti-PD-L1. Taken together, this study advances our understanding of the pharmacokinetic-pharmacodynamic relationship in OV therapy.
Key words: Pharmacokinetics, Oncolytic virus, JAK‒STAT, Anticancer
Graphical abstract
This study reveals the pharmacokinetics of OVM, as a replicable living drug, in various tumor models. Amelioration of early OVM pharmacokinetics results in stronger oncolytic efficacy and antitumor immunity via targeting JAK‒STAT pathway (figure was created with Biorender.com).
1. Introduction
Oncolytic viruses (OVs) are replicative viruses that preferentially kill cancer cells in vitro and in vivo while leaving non-neoplastic cells intact. OVs can convert the “cold” tumor into “hot” tumor, and are emerging as a class of prominent living drugs for cancer treatment1,2. More than 50 OVs from at least ten virus families, including DNA virus and RNA virus, are approved or being tested against various solid tumors in clinical trials3. Talimogene laherparepvec is the only OV granted approval both in the USA and Europe for treatment of melanoma patients with injectable but unresectable lesions in the skin and lymph nodes4,5.
Given the critical correlation of pharmacokinetics with the clinical outcome, in-depth understandings of OVs’ pharmacokinetic profiles are essential. These understandings may aid the development of safer and more effective OVs, help the identification of patients who are most likely to respond to the treatment, and guide the rational design of combination therapies6. Nonetheless, the classical pharmacokinetic considerations (absorption, distribution, metabolism, and elimination, ADME) are only in parts applicable to the OV products, simply due to their self-replicative property, and only a few descriptive OV pharmacokinetic studies have been published. Modulators of OV pharmacokinetics remain to be identified7.
Oncolytic virus M1 (OVM) is a clinically relevant OV whose safety profile and anticancer potential have been demonstrated in a variety of preclinical in vitro cell systems and animal models, including cynomolgus monkeys8, 9, 10. OVM is currently undergoing Phase I clinical trials in China and Japan, and has been granted Orphan Drug Designation by the US Food and Drug Administration for the treatment of liver cancer and malignant glioma. OVM is a small, enveloped, positive-sense single-stranded RNA virus with an 11.7 kb genome encoding four non-structural proteins and five structural proteins. We previously demonstrated that OVM can selectively replicate in and kill tumor cells via intravenous injection11,12. We also investigated the biodistribution of OVM in normal rats, cynomolgus monkeys, and tumor-bearing immunocompromised mice13. However, little is known regarding the viral replication and distribution of OVM in tumor-bearing immunocompetent mice, and key modulators of the pharmacokinetics of OVM are still not clear.
In this study, we delineated the pharmacokinetics profile of OVM as a kind of living drug in tumor-bearing immunocompetent mice, identified the determinants of OVM pharmacokinetics, and further enhanced its antitumor effect and immune activation by improving the pharmacokinetics.
2. Material and methods
2.1. Cell culture and virus
Cell lines were purchased from American Type Culture Collection and Shanghai Institute of Cell Biology. Cells were cultured in DMEM and RPMI-1640 supplemented with 10% (v/v) FBS and 1% penicillin/streptomycin (Life Technologies). OVM was grown in Vero cells. Virus titer was determined by CCID50 assay using BHK-21 cells and converted to PFU. All cell lines were tested negative for mycoplasma by MycoGuard mycoplasma PCR detection kit (MPD-T-050, GeneCopoeia).
2.2. Antibodies and reagents
Antibodies used in this study are listed as follows: JAK1 (3344, Cell Signaling Technology, USA, 1:1000), phosphorylated JAK1 (66,245, Cell Signaling Technology, 1:1000), JAK2 (3230, Cell Signaling Technology, 1:1000), phosphorylated JAK2 (3771, Cell Signaling Technology, 1:1000), STAT1 (14,994, Cell Signaling Technology, 1:1000), phosphorylated STAT1 (9167, Cell Signaling Technology, 1:1000), STAT3 (9139, Cell Signaling Technology, 1:1000), phosphorylated STAT3 (9145, Cell Signaling Technology, 1:1000), Ms CD45 BV510 (568,891, BD Bioscience, USA, 1:100), Ms CD3e PerCP-Cy5.5 (551,163, BD Bioscience, 1:100), FITC anti-mouse CD4 (100,406, Biolegend, USA, 1:100), Ms CD8a Alexa 700 (557,759, BD Bioscience, 1:100), Fixable Viability Stain 780 (565,388, BD Bioscience, 1:500), GAPDH (AP0063, Bioworld, USA, 1:10,000), α-tubulin (ARG65693, Arigo Biolaboratories, China, 1:5000), OVM E1 (produced by Beijing Protein Innovation, China, 1:2000). Compounds used in this study (MCC950, Amlexanox, Fludarabine, Ruxolitinib) were purchased from Selleckchem. Mice were i.p. injected with 1 mg anti-IFNAR1 (clone MAR1-5A3; BioXcell), 2 mg anti-CSF1R (clone AFS98; BioXcell), 200 μg anti-NK1.1 (PK-136; BioXcell), or 200 μg anti-Ly6G (clone 1A8; BioXcell) Abs 1 day before OVM injection, and further, the antibody treatment continued on Days 1, 3 after OVM injection.
2.3. In vivo assay in animal models
This study was approved by the Animal Ethical and Welfare Committee of Sun Yat-sen University. C57BL/6 N mice were bought from Charles River, China. For the subcutaneous xenograft model, B16-F10 (106 cells/mouse) or Pan02 (3 × 106 cells/mouse) were inoculated subcutaneously into the hind flanks of 4-week-old female C56BL/6 N mice. After 7 days, when palpable tumors developed (∼50 mm3), mice were randomly grouped to receive different treatments. For pharmacokinetic studies, single or five daily intravenous injections of OVM (3 × 106 PFU/dose/day) with or without three intraperitoneal injections of ruxolitinib (100 mg/kg) were administered. Efficacy measurements were not included in pharmacokinetic studies. For therapeutic studies, five daily intravenous injections of OVM (6 × 105 PFU/dose/day) with or without three intraperitoneal injections of ruxolitinib (100 mg/kg) were received. Tumor length and width were measured every 3–4 days, and the volume was calculated according to the formula (length × width2)/2. To optimize the detection of the host transcriptional response, we reduced the viral dose from 3 × 106 PFU to 1 × 105 PFU for RNA-seq analysis. The higher dose yielded high viral RNA and low host RNA, obscuring robust detection of differentially expressed host genes. Importantly, the Tmax was equivalent between both viral doses, indicating preserved viral replication kinetics and host response induction despite the 30-fold lower dose. Thus, the 1 × 105 PFU dose balances host and viral RNA while maintaining host response kinetics equivalent to the higher dose.
2.4. qRT-PCR
Total RNA was extracted using TRIzol (Life Technologies), and reverse transcription was performed from 3 μg total RNA using oligo(dt) and RevertAid Reverse Transcriptase (Thermo Scientific) according to the manufacturer's recommendation. Quantitative PCR was performed with SuperReal PreMix SYBR Green (TIANGEN) using an Applied Biosystems 7500 Fast Real-Time PCR System (Life Technologies). All genes were normalized to β-actin. Amplification primers (Thermo Fisher) were:
OVM Q3S1 sense: GGGATTCACTACACCTGCTTAGAC
OVM Q3S1 antisense: GCTGACTCTGTCTGCGTAACC
OVM Q3S1 probe: CTCTCATCAGCAGCGAGCCTCCT
OVM NS1 sense: GTTCCAACAGGCGTCACCATC
OVM NS1 antisense: ACACATTCTTGTCTAGCACAGTCC
IFN-α sense: TCTGATGCAGCAGGTGGG
IFN-α antisense (AGGGCTCTCCAGACTTCTGCTCTG)
IFN-β sense (CAGCTCCAAGAAAGGACGAAC)
IFN-β antisense (GGCAGTGTAACTCTTCTGCAT)
IFN-γ sense (ATGAACGCTACACACTGCATC)
IFN-γ antisense (CCATCCTTTTGCCAGTTCCTC)
Granzyme B sense (TCTCGACCCTACATGGCCTTA)
Granzyme B antisense (TCCTGTTCTTTGATGTTGTGGG)
2.5. Western blot analyses
Cells were lysed using the T-PER Mammalian Protein Extraction Reagent (Thermo Scientific), and sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed. Membranes were visualized on a ChemiDoc XRS + System (Bio-Rad) using Immobilon Western Chemiluminescent HRP Substrate (Millipore).
2.6. RNA sequencing and data processing
Total RNA was extracted from the tumors using Trizol (Invitrogen, Carlsbad, CA, USA) according to the manual and sent to BGI (The Beijing Genomics Institute) for further processing and RNA-seq analysis. In brief, total RNA was qualified and quantified using a Nano Drop and Agilent 2100 bioanalyzer (Thermo Fisher Scientific, MA, USA). RNA-seq libraries were prepared using the Illumina TruSeq RNA Sample Preparation v2 Guide (Illumina Part # 15026495), then the pair end 100 bases reads were generated on BGIseq500 platform (BGI-Shenzhen, China).
The raw reads were aligned to the hg38 reference genome via TopHat (version 2.1.0) with the default parameters14. The gene count mapped reads with the parameter “-s no -a 20” using the HTSeq program15. Gene count normalization and differential expression analysis were performed using the DESeq package16.
GSEA was performed to identify significantly altered gene sets in comparison to the tumors treated with OVM for 0 or 96 h. Analysis was carried out with the GSEA package (v.3.0) (Broad Institute), following the protocol described by Reimand et al.17,18 Briefly, genes were ranked to generate gene lists according to their expression changes, which was in the comparison of two groups as the figure described. All these pre-ranked gene lists were then used as an input, while gene datasets (KEGG) were used as a reference. To calculate the P values for each pathway, 1000 random permutations were performed. Only gene sets with P < 0.05 and FDR <0.25 were considered as significantly enriched. Single sample GSEA score was conducted to evaluate the comprehensive expression level of a single gene set using the R package GSVA19.
2.7. Flow cytometry
Tumor tissues were harvested, and single-cell homogenates were prepared by using a mouse tumor dissociation kit (Miltenyi Biotec) and passed through a 40-μm strainer before the red blood cells were lysed. Surface markers were stained with the following antibodies at 4 °C for 30 min: CD45-BV510 (563,891, BD), CD3-PerCP-Cy5.5 (551,163, BD), CD4-FITC (100,406, BioLegend), CD8-Alexa Fluor 700 (557,959, BD).
2.8. ELISA
ELISA Mouse IL-6 (LIANKE BIOTECH, EK206/3), IL-10 (LIANKE BIOTECH, EK210/4), IFN-γ (LIANKE BIOTECH, EK280/3) and TNF-α (LIANKE BIOTECH, EK282/4) in serum were analyzed according to the manufacturer's instructions.
2.9. Statistical analysis
All statistical analyses were performed using GraphPad Prism software 8.0 (RRID: SCR_002798) (GraphPad Software Inc., San Diego, CA, USA), R 3.4.0 (https://www.r-project.org), for GSEA analysis and SPSS 18.0 software (RRID: SCR_002865) (IBM SPSS Statistics Inc., Chicago, IL, China). All sample sizes and statistical methods were indicated in the corresponding figure legend. No statistical methods were used to predetermine sample size. No data we generated were excluded. If the data were normally distributed (by Shapiro–Wilk test) and homoscedastic (by Bartlett's test), Student's t test (for two groups) and one-way analysis of variance (ANOVA) (more than two groups) were used to test the mean difference. The statistical significance of GSEA analysis was determined by unpaired Student's t test. All statistical tests were two-sided, and the animal experiments were randomized by a table of random digits. Bars show the mean ± SD or SEM of three independent repeated experiments. Significant differences were accepted if the P value was <0.05.
3. Results
3.1. Preclinical pharmacokinetic profile of intravenously delivered OVM
To investigate the preclinical pharmacokinetics of the OVM, we subcutaneously grafted B16-F10 or Pan02 cancer cells in immunocompetent mice and intravenously injected one dose of OVM (3 × 106 PFU per mouse). The genomic viral RNA of OVM in tumor and normal organs were quantified by RT-qPCR every day. In both tumor models, significantly higher levels of OVM were observed in tumor than in normal tissues, validating the tumor selectivity of OVM (Fig. 1A and C). In tumor sites, OVM levels peaked within 96 or 48 h (Tmax) after intravenous infusion and then declined within 24 or 48 h. The peak OVM level (Cmax) in B16-F10 or Pan02 tumor was 32,765,250 ± 17,609,452 or 128,998 ± 47,457 copies/μg total RNA. Given the fact that the Tmax of OVM is much longer than canonical drugs (usually within minutes to hours), we consider it to be replication-dependent rather than distribution (Fig. 1B and D). After that, the OVM levels declined rapidly from peak to bottom (lower than detection threshold) in both models, and the half-life (t1/2) of OVM in B16-F10 or Pan02 model was 12 or 19.353 h respectively, suggesting a quick elimination of OVM in tumor sites. The area under the curve (AUC), representing the extent of exposure to OVM, was also calculated. It is noteworthy that both Cmax and AUC were much higher in B16-F10 tumor model than Pan02, which is consistent with the in vivo antitumor efficacy of OVM in these models (Fig. 1B and D).
Figure 1.
Pharmacokinetic profiles of OVM in syngeneic tumor mouse models (the OVM injection dose was 3 × 106 PFU). (A, C, E, G, I) The change of the OVM RNA copies in tumor and normal organs over time, n = 3. (B, D, F, H, J) Main parameters of pharmacokinetics in tumor is shown, n = 3.
We further analyzed OVM biodistribution in tumor-bearing mice after 5 daily intravenous injections, in comparison to a single injection. Similar to the single injection group, OVM exhibited tumor-selective replication after multiple injections (Fig. 1E–H). However, we observed an earlier time to reach maximum OVM titer (Tmax) of 72 h post first injection, compared to 96 h for the single injection group in B16-F10 tumor model (Fig. 1E–F). This suggests that repeated dosing leads to accelerated intratumoral viral accumulation. While in Pan02 tumor model, key pharmacokinetic parameters like Tmax, Cmax, and AUC were comparable between single and multiple OVM injections (Fig. 1G–H). Notably, there were no significant differences in viral biodistribution in normal tissues, including heart, liver, spleen, lung, kidney, intestine, and brain, between single and multiple injection groups at matched timepoints post-injection. This indicates that repeated OVM injections do not increase off-target infection.
To enhance clinical relevance, we assessed the pharmacokinetic profile of OVM utilizing a B16-F10 lung metastasis model. The results illustrate that OVM sustains tumor-selective accumulation within a metastasis model. Moreover, the exposure levels were similar between the metastasis and subcutaneous models (Fig. 1I and J). This indicates that the selective tumor delivery and exposure of OVM is preserved in metastatic disease, further highlighting its translational promise.
In general, the pharmacokinetic profile of OVM shows that it has high tumor tropism in vivo, and the pharmacokinetic characteristics may affect therapeutic efficacy.
3.2. Peak intratumoral OVM level accompanies with activation of inflammatory response
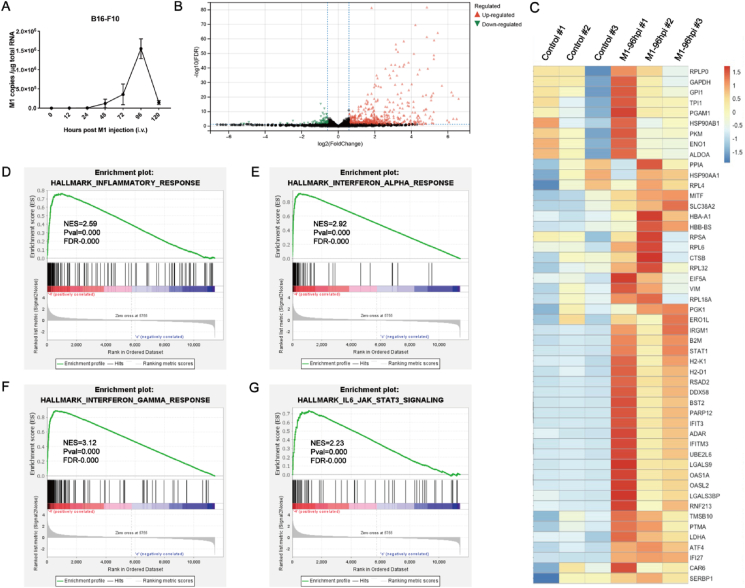
The rapid decline of OVM level in tumor sites suggests that the elimination of OVM occurs in tumor rather than canonical metabolic organs such as liver and kidney. To reveal the regulators of pharmacokinetics of OVM, especially the elimination process, we performed RNA sequencing at Tmax of OVM by using B16-F10 tumor tissues (Fig. 2A). The RNA sequencing results revealed that 1017 genes were significantly upregulated while 247 genes were downregulated, and a considerable number of upregulated genes were immune-related genes (Fig. 2B). Among the most upregulated genes, more than 60% are associated with inflammatory response (Fig. 2C). GSEA analysis showed that OVM treatment significantly activated inflammatory response, interferon-α (IFN-α), interferon-γ (IFN-γ) and JAK‒STAT signaling pathways, suggesting these pathways are involved in OVM elimination (Fig. 2D–G).
Figure 2.
Peak intratumoral OVM level accompanies with activation of inflammatory response. (A) C57BL/6 mice were implanted subcutaneously in the right flank with B16-F10 cells and treated intravenously with OVM, the OVM injection dose was 1 × 105 PFU (n = 5). The tumors were harvested at 0 and 96 h after OVM administration, the total RNA was extracted, and RNA sequencing was then performed. (B) Volcano plot in tumor tissue is shown. (C) Gene enrichment heatmap in tumor tissue. (D–G) GSEA results for the inflammatory response (D), IFN-α response (E), IFN-γ response (F), and JAK‒STAT signaling (G) are shown.
All the results described above demonstrate that OVM induces an inflammatory response, which may in turn suppress viral replication and promote viral elimination. Unlike traditional drugs which are usually metabolized by liver enzymes, the pharmacokinetics of OVM may be regulated by inflammatory pathways.
3.3. Inhibiting JAK‒STAT pathway increases intratumoral Cmax and AUC of OVM
We used four small molecule inhibitors of inflammatory response, including MCC950 (a selective inhibitor of NLRP3), amlexanox (TBK1 and IKK-ε inhibitor), fludarabine (STAT1 inhibitor), ruxolitinib (JAK1 and JAK2 inhibitor), to identify the key regulatory pathway of OVM pharmacokinetics.
While targeting the inflammasome pathway with 25 mg/kg MCC950 significantly increased the intratumoral viral RNA at 96 h post OVM administration, it had no effect at 120 h post infection (Fig. 3A). Similarly, inhibition of NFκB signaling with 100 mg/kg amlexanox resulted in 6 fold upregulation of viral RNA in tumor only at 120 h post OVM inoculation but not 96 h post infection (Fig. 3B). Fludarabine, however, did not affect the replication of OVM in tumor site (Fig. 3C). Unlike the above three inhibitors, suppression of JAK1/2 pathways with 100 mg/kg ruxolitinib remarkably increased the intratumoral viral RNA levels up to 150 folds at both 96 and 120 h post OVM injection (Fig. 3D). Therefore, we postulated that JAK1/2 pathways may play critical roles in regulating the pharmacokinetics of OVM.
Figure 3.
Inhibiting JAK‒STAT pathway increases intratumoral Cmax and AUC of OVM (3 × 106 PFU, single dose). (A–D) Effects of two concentrations of MCC950 (A), amlexanox (B), fludarabine (C), and ruxolitinib (D) on the replication of OVM in tumor, n = 3. (E and F) Western blot image (left) of JAK‒STAT pathway and IRF-1, IRF-9 expression and quantitative statistics (right), n = 3. (G–H) IFN-α (G) and IFN-β (H) mRNA level of B16-F10 tumor with OVM alone or combined with ruxolitinib administration, n = 5. (I) Western blot image (left) of OVM protein expression and quantitative statistics (right), n = 4. (J–M) Pharmacokinetic profiles of OVM combined neutralizing or depleting antibodies in B16-F10 models, n = 3. (N–O) OVM replication curve and AUC in B16-F10 tumor (N) or Pan02 tumor (O) when OVM alone or combined with ruxolitinib administration, n = 5. (P) Copy number of OVM in different tissues when OVM alone or combined with ruxolitinib administration at 96 hpi, n = 3. (Q) OVM RNA copy number in normal and tumor tissues at 21 days post-treatment initiation, n = 3. hpi, h post infection. AUC, area under curve. Data are reported as the mean ± SD. ns, no statistical difference. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001.
Ruxolitinib is a small molecule inhibitor of both JAK1 and JAK2, targeting which leads to the inhibition of phosphorylation of downstream signal molecule STATs. We found that ruxolitinib mainly blocks JAK/STAT pathway by inhibiting the phosphorylation of JAK1 and JAK2 at 48 hpi (Fig. 3E), resulting in significant decrease in phosphorylation of STAT1 and STAT3 (Fig. 3F). Subsequently, the expression of IRF-1, IRF-9 and type I IFNs (IFN-Is) were downregulated (Fig. 3F–H), which in turn increase of the expression of viral E1 protein in the tumor at 96 h (Fig. 3I). We further elucidated the roles of JAK‒STAT-IFN signaling and specific immune cells in modulating intratumoral OVM replication. Specifically, we found that IFNAR1-neutralizing antibodies significantly increased the Cmax and AUC of OVM, confirming the importance of IFN signaling in intratumoral virus clearance. Moreover, the JAK inhibitor ruxolitinib did not further increase intratumoral virus amount when combined with IFNAR1 blockade, indicating its effect is based on inhibiting IFN signaling (Fig. 3J–L). We also identified the involvement of specific innate immune cells by using depleting antibodies targeting macrophage (CSF1R), neutrophil (Ly6G), and NK cell (NK1.1). Depletion of macrophage and neutrophil accelerated virus replication and shifted Tmax earlier to 72 h post infection, whereas NK cell depletion increased Cmax. Taken together, these results demonstrate that all three major innate immune cell types participate in the rapid intratumoral clearance of OVM (Fig. 3K–M).
Next, we examined whether ruxolitinib affects the pharmacokinetics of OVM in B16-F10 and Pan02 tumors. The results showed that ruxolitinib could significantly enhance the intratumoral Cmax and AUC of OVM (Fig. 3N and O). These results suggest that targeting JAK1/2 with ruxolitinib can increase the pharmacokinetics of OVM in tumor.
We further investigated if ruxolitinib has an impact on viral replication in normal tissues given that it increases OVM replication in tumor tissues. The results showed that ruxolitinib did not increase the replication of OVM in normal heart, liver, spleen, lung, kidney, brain, and intestine (Fig. 3P), suggesting a highly selective improvement of OVM replication. In addition, we measured viral RNA copy number in normal and tumor tissues at 21 days post-treatment initiation. All copy numbers were low (<100 copies/μg RNA) (Fig. 3Q), indicating effective viral clearance.
3.4. Inhibiting JAK‒STAT pathway improves the therapeutic efficacy of OVM
We have previously evaluated the therapeutic effect of M1 alone using a higher dose in syngeneic mouse tumor models, which demonstrated potent therapeutic effects. In the following study, we examined whether the improved pharmacokinetics of OVM were associated with enhanced antitumor activity. To investigate whether ruxolitinib can enhance the therapeutic effect of M1, we used a low dose of M1 (6 × 105 PFU/day) in combination with ruxolitinib, which may have resulted in a relatively modest response with M1 alone. Mice with subcutaneous B16-F10 or Pan02 tumors were treated with OVM, ruxolitinib, or combination of the two (Fig. 4A and E). Consistent with the increase of intratumoral virus level, the combination of OVM and ruxolitinib resulted in a significantly stronger inhibition of tumor growth in both B16-F10 and Pan02 tumors compared with either monotherapy (Fig. 4B and F). Moreover, the combination therapy significantly prolonged overall survival in both tumor models (Fig. 4C and G). Despite using a 50-fold higher OVM dose (3 × 107 PFU/dose), we observed no additional benefit when combined with ruxolitinib (Supporting Information Fig. S1), suggesting that OVM replication augmentation by ruxolitinib may outweigh the incremental impacts of higher OVM doses.
Figure 4.
Inhibiting JAK‒STAT pathway improves the therapeutic efficacy of OVM in immunocompetent mice (6 × 105 PFU/dose/day, five daily injections). (A, E) C57BL/6 mice were implanted subcutaneously in the right flank with B16-F10 (A) or Pan02 (E) cells on day 0 and treated intravenously with control or OVM once per day on Days 6–10 (A) or Days 8–12 (E). Ruxolitinib was treated 3 times. (B, F) Tumor growth curves in B16-F10 (C) or Pan02 (F) tumor-bearing mice are shown, n = 6. (C, G) Survival curves in B16-F10 (C) or Pan02 (G) tumor-bearing mice are shown, n = 10. (D, H) Body weight changes in different treatment groups of B16-F10 (D) and Pan02 (H) tumor-bearing mouse models, n = 6. (I) H&E staining of heart, liver, spleen, lung, kidney, and intestine of B16-F10 tumor-bearing mice 21 days post treatment with OVM alone or OVM combined with ruxolitinib. Scale bar, 100 μm ns, no statistical difference; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001.
Considering the potential toxicities caused by ruxolitinib or the elevated virus replication, we next tested the safety profile of ruxolitinib combined with OVM. First, we monitored the body weight of mice during the whole process. No significant difference in body weight was observed between the combination group and the other three groups (Fig. 4D and H). Subsequently, we performed H&E staining on heart, liver, spleen, lung, kidney, and intestine of B16-F10 tumor-bearing mice at 21 days post first dose of OVM to assess whether the combination therapy caused pathological damage in vital organs. The results showed that there was no obvious pathological damage in all tested tissues in the OVM alone group and combined with ruxolitinib group (Fig. 4I). In addition, we detected the hematologic toxicity and cytokine release related syndrome. The results showed that there was no significant difference in blood routine indicators and expression of inflammatory factors between the two groups (Supporting Information Fig. S2, Table S1). The aforementioned findings demonstrate that ruxolitinib enhances OVM's pharmacokinetics and antitumor efficacy while preserving OVM's safety profile.
In addition, we inoculated human melanoma A375 cells in nude mice to evaluate the antitumor effectiveness in immune-defect model. Intraperitoneal injection of the ruxolitinib resulted in amplified viral RNA of OVM (Fig. 5A). Accordingly, ruxolitinib evidently potentiated the antitumor activity of OVM in that both the tumor volume and tumor weight were significantly inhibited (Fig. 5B–D). More importantly, even in immunocompromised nude mice, weight loss was not observed after treatment with combination of ruxolitinib and OVM, implying the high safety of this combination therapy (Fig. 5E).
Figure 5.
Inhibiting JAK‒STAT pathway improves the therapeutic efficacy of OVM in mouse model implanted with human melanoma cells (6 × 105 PFU/dose/day, five daily injections). (A) OVM copies in A375 human melanoma model tumor, n = 4. (B and C) Tumor growth curve and T/C (%) in A375 tumor-bearing mice are shown, n = 6. (E) A375 tumor-bearing mouse weight curve is shown, n = 6. (D) A375 tumors and quantitative statistics are shown. Group Ruxolitinib + M1 had 1/6 tumor regression, n = 6. T/C (%) = TRTV/CRTV × 100; TRTV, RTV in treatment group; CRTV, RTV in isotype control group; RTV: relative tumor volume; RTV = Vt/V0; Vt, tumor volume after treatment; V0, tumor volume before treatment. ns, no statistical difference; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001.
3.5. Inhibiting JAK‒STAT pathway boosts the OVM-induced intratumoral T cell infiltration
IFN-Is are emerging as critical drivers of antitumor immunity20, which is essential for OVM's therapeutic activity21. To determine whether the inhibition of IFN-Is by ruxolitinib may undermine the induction of antitumor immune response by OVM, we analyzed the tumor microenvironment (TME) by flow cytometry (Gating strategy is shown in Supporting Information Fig. S3). On Day 7 after virotherapy, the infiltration of total T cells (CD3+), CD4+ T cells, and CD8+ T cells were slightly increased by OVM monotherapy and were remarkably elevated by OVM plus ruxolitinib combination therapy (Fig. 6A–C), indicating that ruxolitinib did not weaken the antitumor immune response induced by OVM. Instead, the increase of intratumoral replication of OVM by ruxolitinib further recruited even more T cells to the TME. We also examined the systemic immune response in spleen. Results showed that OVM alone can significantly increase the number of total T cells, and the addition of ruxolitinib did not cause further changes (Fig. 6D–F).
Figure 6.
Inhibiting JAK‒STAT pathway boosts the OVM-induced intratumoral T cell infiltration and activation. (A–C) The infiltration of CD45+ CD3+ T cells(A), CD8+ T cell (B), and CD4+ T cells (C) in B16-F10 model tumor was detected by flow cytometry, n = 4. (D–F) The infiltration of CD45+ CD3+ T cells (D), CD8+ T cell (E), and CD4+ T cells (F) in spleen of B16-F10 model, n = 4. (G and H) The mRNA level of IFN-I in B16-F10 model tumor after 36 h (G) or 120 h (H) post OVM alone or combined ruxolitinib administration, n = 3. (I) The mRNA level of Granzyme B and IFNγ in B16-F10 model tumor after 36 h (I) or 120 h (J) post OVM alone or combined ruxolitinib administration, n = 3. ns, no statistical difference; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001.
Mechanistic studies showed that although the expressions of IFN-α and IFN-β were significantly downregulated by ruxolitinib at 36 h post the first dose of OVM, they were substantially increased at 120 h post first infection of OVM, that is 48 h after the last dose of ruxolitinib (Fig. 6G–H). These findings prompted us to speculate that the inhibition of IFN-Is by ruxolitinib at the early infection stage promotes virus replication, which in turn stimulates higher levels of IFN-Is once ruxolitinib is withdrawn. Next, we detected the expression of antitumor factors regulated by JAK‒STAT pathway and found that the expression of Granzyme B was significantly down-regulated in 36 h (Fig. 6I–J).
3.6. PD-L1 antibody enhances the antitumor efficacy of OVM and ruxolitinib combination therapy
In our previous studies, we found that OVM can upregulate the expression of PD-L1 in tumors and anti-PD-L1 antibody can increase the therapeutic effect of OVM. Considering that ruxolitinib can increase the AUC of OVM, which may further promote the expression of PD-L1 in tumor. We first detected the expression of PD-L1 in tumor after the combination of ruxolitinib and OVM. As expected, the OVM monotherapy upregulated the expression of PD-L1, and the combination of ruxolitinib and OVM resulted in even higher expression (Fig. 7A). Therefore, we investigated the combination of OVM, ruxolitinib and PD-L1 antibody in B16-F10 tumor models (Fig. 7B). The triple therapy further enhanced antitumor activity and overall survival compared with the dual-agent treatments (Fig. 7C and D). It is worth noting that no significant difference in body weight was observed between control group and treatment groups, indicating the safety profile of the triple therapy (Fig. 7E). Taken together, our data reveal that PD-L1 antibody can synergize with OVM and ruxolitinib to suppress tumor growth in vivo.
Figure 7.
PD-L1 antibody enhances the anti-tumor efficacy of OVM and ruxolitinib combination therapy (6 × 105 PFU/dose/day, five daily injections). (A) The mRNA level of PD-L1 in B16-F10 tumor-bearing model after OVM alone or combined ruxolitinib administration, n = 3. (B) C57BL/6 mice were implanted subcutaneously in the right flank with B16-F10 cells on day 0 and treated intravenously with control (n = 10) or OVM (n = 10) once per day on Days 6–10. Ruxolitinib and PD-L1 antibody were respectively treated 3 times. (C–E) Tumor growth curve (C, n = 6), weight of mice (D, n = 6), survival curve of tumor-bearing mice (E, n = 10) in each group are shown. ns, no statistical difference; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001.
4. Discussion
To uncover host factors modulating OVM pharmacokinetic profile in immunocompetent mice, we performed a genome-wide transcriptomic analysis. Among the significantly enriched pathways, we identified JAK‒STAT pathway as a key determinant of OVM pharmacokinetics. Inhibition of JAK‒STAT signaling improves OVM pharmacokinetics, resulting in potentiated oncolytic activity and stronger antitumor immunity, while maintaining a tolerable safety profile.
Unlike the non-replicative canonical drugs, our pharmacokinetic study confirmed that the living drug OVM preferentially replicates in tumor sites in immunocompetent mouse models. Although there was no difference between neoplastic and non-neoplastic tissues in the distribution phase just after drug administration, OVM was strictly enriched in the tumor site in the following days until virus was eliminated, ensuring its safety profile22. This tumor selectivity also makes OVM eligible for intravenous administration in future clinical applications which is needed to achieve control of disseminated cancers. Our results further suggest that OVM pharmacokinetics have significant tumor type selectivity, which may impact dosing, pharmacokinetic characteristics, and therapeutic outcomes. This preferential tumor selection that alters OVM dosing is a valuable research area to guide efficient clinical OVM application.
The pharmacokinetic profile of canonical drugs and their regulatory mechanisms have long been studied in depth, however, little is known about the pharmacokinetics of living drugs. Investigating these key determinants can help to improve the pharmacokinetic characteristics of living drugs, prolong their half-lives, increase the drug AUC, and thus may enhance the clinical outcome. Here, we found that the intratumoral half-life of OVM was regulated by the JAK‒STAT pathway which was significantly induced by intravenous delivery of OVM. Inhibition of JAK‒STAT pathway with ruxolitinib, a prescription medicine used to treat polycythemia vera, myelofibrosis, and acute/chronic graft-versus-host disease, can increase the Cmax and AUC of OVM, enhancing its anticancer efficacy23. The JAK‒STAT‒IFN pathway plays a crucial role in the immune response to viral infections. The pathway is activated by interferons (IFNs), which are cytokines produced by host cells in response to viral infections. The activation of the JAK‒STAT‒IFN pathway leads to the expression of a variety of genes that are involved in the antiviral response, including genes that encode proteins involved in the regulation of the immune response, such as cytokines and chemokines. Innate immune cells, including macrophages, neutrophils, and NK cells, are important sources and responders of IFNs. Although important for OVM pharmacokinetics, the only inhibition of JAK‒STAT pathway may not be sufficient to optimize the outcome of OVM treatment. In our opinion, targeting multiple OVM pharmacokinetic modulators simultaneously can help unleashing the full potential of OVM-based therapies. Many of these modulators remain to be identified. For example, while ruxolitinib did not alter tumor sizes in our models, tumor size itself may impact OVM pharmacokinetics through physical factors and immune cell composition. Future studies are needed to elucidate how size-dependent changes in the tumor microenvironment influence OVM dissemination and efficacy. Though numerous clinical and nonclinical pharmacokinetic studies have been conducted on OV, the pharmacokinetic mechanisms that affect OV are still not well understood. Further research is required to elucidate the underlying mechanisms that govern the pharmacokinetics of OV.
Although promising, the changes in viral pharmacokinetics may also increase the potential risk of causing damage to normal tissues. Therefore, pharmacokinetics needs to be well-tuned to balance efficacy and toxicology24,25. In our study, we observed a tolerable safety profile of OVM and ruxolitinib combination therapy during the whole treatment process by monitoring body weight and performing histopathological tests of vital organs. While our study provides evidence that viral replication is suppressed in normal tissues, including lung tissue, an insightful limitation was raised regarding our inability to directly analyze viral replication in paraneoplastic regions adjacent to lung metastases. This identifies an interesting question for future work with optimized sampling and detection methods.
It is also well-reported that repeated administration of virus would induce neutralizing antibodies to reduce the level of infectious virus and hamper the antitumor effect. More studies are still needed to illustrate the influence of neutralizing antibodies on the pharmacokinetics of OVM26, 27, 28. Additionally, OVM replication over time by titrating infectious viruses in blood and tissues should be evaluated.
Growing evidence supports that IFN-Is are indispensable for the activation of antitumor immune response, so we concerned the inhibition of JAK‒STAT-IFN pathway may dampen the antitumor immunity which is one of the mechanisms of action of OVM. Fortunately, instead of suppressing immunity, the increased viral replication boosted an even stronger immune response after ruxolitinib withdrawal, possibly owing to the short half-life of ruxolitinib.
5. Conclusions
In conclusion, our study reveals JAK‒STAT pathway, rather than the classical hepatic drug-metabolizing enzyme system, as a key determinant of the OVM pharmacokinetic profile. The discovery of inflammatory pathways as host factors involved in OVM pharmacokinetics may facilitate the identification of other modulators of OVM pharmacokinetics. This will not only deepen our understanding of OVM pharmacokinetics but also pave the way for the identification of therapeutic targets to improve clinical response and patient survival with OVM-based therapies.
Declaration of Generative AI and AI-assisted technologies in the writing process
During the preparation of this work, the authors used Claude in order to improve language and readability. After using this tool/service, the authors reviewed and edited the content as needed and took full responsibility for the content of the publication.
Author contributions
Jun Hu, Wenbo Zhu, Jiankai Liang, Jing Cai, Guangmei Yan, and Yuan Lin designed the research study; Jingyi Tan, Jiayu Zhang, Cheng Hu, Gongwei Wang, Qianyao Ren, Chaoqun Wang, and Zexin Zeng performed the experiments; Jingyi Tan, Jiayu Zhang and Cheng Hu analyzed the data, and Jingyi Tan, Jiayu Zhang and Yuan Lin wrote the paper.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgments
This study was supported by National Key R&D Program of China (No. 2021YFA0909800, China), Guangdong Basic and Applied Basic Research Foundation (Nos. 2022B1515020056, 2021A1515011881, 2023A1515010737, China), Leading team for entrepreneurship in Guangzhou, Guangdong Province (No. 201809020004, China), Fundamental Research Funds for the Central Universities (No. 22ykqb12, China), Pioneering talents project of Guangzhou Development Zone, Guangdong Province (2020-L036, China), Natural Science Foundation of Guangdong Province (No. 2022A1515011056, China).
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2024.03.007.
Appendix A. Supporting information
The following is the Supporting information to this article.
References
- 1.Macedo N., Miller D.M., Haq R., Kaufman H.L. Clinical landscape of oncolytic virus research in 2020. J Immunother Cancer. 2020;8 doi: 10.1136/jitc-2020-001486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang W., Liu S., Dai P., Yang N., Wang Y., Giese R.A., et al. Elucidating mechanisms of antitumor immunity mediated by live oncolytic vaccinia and heat-inactivated vaccinia. J Immunother Cancer. 2021;9 doi: 10.1136/jitc-2021-002569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hemminki O., Dos Santos J.M., Hemminki A. Oncolytic viruses for cancer immunotherapy. J Hematol Oncol. 2020;13:84. doi: 10.1186/s13045-020-00922-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pol J., Kroemer G., Galluzzi L. First oncolytic virus approved for melanoma immunotherapy. OncoImmunology. 2016;5 doi: 10.1080/2162402X.2015.1115641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corrigan P.A., Beaulieu C., Patel R.B., Lowe D.K. Talimogene Laherparepvec: an oncolytic virus therapy for melanoma. Ann Pharmacother. 2017;51:675–681. doi: 10.1177/1060028017702654. [DOI] [PubMed] [Google Scholar]
- 6.Andtbacka R.H.I., Amatruda T., Nemunaitis J., Zager J.S., Walker J., Chesney J.A., et al. Biodistribution, shedding, and transmissibility of the oncolytic virus talimogene laherparepvec in patients with melanoma. EBioMedicine. 2019;47:89–97. doi: 10.1016/j.ebiom.2019.07.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parra-Guillen Z.P., Freshwater T., Cao Y., Mayawala K., Zalba S., Garrido M.J., et al. Mechanistic modeling of a novel oncolytic virus, V937, to describe viral kinetic and dynamic processes following intratumoral and intravenous administration. Front Pharmacol. 2021;12 doi: 10.3389/fphar.2021.705443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu J., Cai X.F., Yan G. Alphavirus M1 induces apoptosis of malignant glioma cells via downregulation and nucleolar translocation of p21WAF1/CIP1 protein. Cell Cycle. 2009;8:3328–3339. doi: 10.4161/cc.8.20.9832. [DOI] [PubMed] [Google Scholar]
- 9.Li K., Zhang H., Qiu J., Lin Y., Liang J., Xiao X., et al. Activation of cyclic adenosine monophosphate pathway increases the sensitivity of cancer cells to the oncolytic virus M1. Mol Ther. 2016;24:156–165. doi: 10.1038/mt.2015.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H., Lin Y., Li K., Liang J., Xiao X., Cai J., et al. Naturally existing oncolytic virus M1 is nonpathogenic for the nonhuman primates after multiple rounds of repeated intravenous injections. Human Gene Ther. 2016;27:700–711. doi: 10.1089/hum.2016.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu W., Liang J., Tan J., Guo L., Cai J., Hu J., et al. Real-time visualization and quantification of oncolytic M1 virus in vitro and in vivo. Human Gene Ther. 2021;32:158–165. doi: 10.1089/hum.2020.273. [DOI] [PubMed] [Google Scholar]
- 12.Lin Y., Zhang H., Liang J., Li K., Zhu W., Fu L., et al. Identification and characterization of alphavirus M1 as a selective oncolytic virus targeting ZAP-defective human cancers. Proc Natl Acad Sci U S A. 2014;111:E4504–E4512. doi: 10.1073/pnas.1408759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai J., Zhu W., Lin Y., Zhang S., Chen X., Gong S., et al. Systematic characterization of the biodistribution of the oncolytic virus M1. Human Gene Ther. 2020;31:1203–1213. doi: 10.1089/hum.2020.114. [DOI] [PubMed] [Google Scholar]
- 14.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anders S., Pyl P.T., Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reimand J., Isserlin R., Voisin V., Kucera M., Tannus-Lopes C., Rostamianfar A., et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat Protoc. 2019;14:482–517. doi: 10.1038/s41596-018-0103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boukhaled G.M., Harding S., Brooks D.G. Opposing roles of type I interferons in cancer immunity. Annu Rev Pathol. 2021;16:167–198. doi: 10.1146/annurev-pathol-031920-093932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y., Cai J., Liu W., Lin Y., Guo L., Liu X., et al. Intravenous injection of the oncolytic virus M1 awakens antitumor T cells and overcomes resistance to checkpoint blockade. Cell Death Dis. 2020;11:1062. doi: 10.1038/s41419-020-03285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krens S.D., Lassche G., Jansman F.G.A., Desar I.M.E., Lankheet N.A.G., Burger D.M., et al. Dose recommendations for anticancer drugs in patients with renal or hepatic impairment. Lancet Oncol. 2019;20:e200–e207. doi: 10.1016/S1470-2045(19)30145-7. [DOI] [PubMed] [Google Scholar]
- 23.Verstovsek S., Mesa R.A., Gotlib J., Levy R.S., Gupta V., DiPersio J.F., et al. A double-blind, placebo-controlled trial of ruxolitinib for myelofibrosis. N Engl J Med. 2012;366:799–807. doi: 10.1056/NEJMoa1110557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le T.M.D., Yoon A.R., Thambi T., Yun C.O. Polymeric systems for cancer immunotherapy: a review. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.826876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carter M.E., Koch A., Lauer U.M., Hartkopf A.D. Clinical trials of oncolytic viruses in breast cancer. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.803050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melero I., Castanon E., Alvarez M., Champiat S., Marabelle A. Intratumoural administration and tumour tissue targeting of cancer immunotherapies. Nat Rev Clin Oncol. 2021;18:558–576. doi: 10.1038/s41571-021-00507-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hemminki O., Dos Santos J.M., Hemminki A. Oncolytic viruses for cancer immunotherapy. J Hematol Oncol. 2020;13:84. doi: 10.1186/s13045-020-00922-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rahman M.M., McFadden G. Oncolytic viruses: newest frontier for cancer immunotherapy. Cancers (Basel) 2021;13:5452. doi: 10.3390/cancers13215452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.