Abstract
The cbsA gene encoding the collagen-binding S-layer protein of Lactobacillus crispatus JCM5810 was expressed in L. casei ATCC 393T. The S-protein was not retained on the surface of the recombinant bacteria but was secreted into the medium. By translational fusion of CbsA to the cell wall sorting signal of the proteinase, PrtP, of L. casei, CbsA was presented at the surface, rendering the transformants able to bind to immobilized collagens.
Lactobacillus is one of the genera belonging to the broad group of lactic acid bacteria (LAB). LAB are widely used in the manufacture of several fermented foods and beverages and are considered to be safe or are generally recognized as safe microorganisms. Several lactobacilli are natural inhabitants of the gastrointestinal and urogenital tracts of humans and animals, and considerable evidence has implicated them in a variety of potentially beneficial roles within the host. Lactobacilli have been reported to protect against infection, e.g., by modulating the immune system (7, 23). This property and the capacity to colonize mucosal surfaces have prompted efforts aimed at their use as vaccine delivery vehicles for oral immunization (14, 21). Although the molecular basis of their so-called probiotic properties are not very well understood, adhesion to the mucosa is considered a prerequisite for their survival and establishment in the intestinal tract (10, 24). Surface-located molecules such as lipoteichoic acid (26), lectin-like molecules (17), and secreted proteins (1, 5) have been identified as adhesins which specifically interact with different receptor moieties in the intestinal tissue.
S-layers are crystalline monolayers formed from single protein monomers (S-protein) that self-assemble into multimeric units to form an array covering the entire cell as the outermost envelope (2, 16). The role of S-protein in adherence to host tissues has been confirmed for the S-layer of the fish pathogen Aeromonas salmonicida (6). However, adhesive properties of S-layers in probiotic lactobacilli remain poorly characterized. The S-layer of Lactobacillus acidophilus has been shown to be involved in the interaction with avian epithelial cells (25), whereas other authors reported that the S-layer proteins of L. crispatus BG2FO4 (formerly classified as L. acidophilus) and L. acidophilus NCFM/N2 did not participate in adherence of these strains to human Caco-2 cells (9). The S-layer protein of L. crispatus JCM5810 (CbsA) was shown to bind to collagens and human subintestinal extracellular matrix (30). As described for pathogenic bacteria, these binding abilities may promote bacterial colonization (31). In this report, we describe the expression of the cbsA gene in L. casei ATCC393T, which lacks an S-layer, and an attempt to transfer the collagen-binding phenotype displayed by this protein to a bacterium that is not able either to bind or to colonize the gastrointestinal tract.
Expression cassettes.
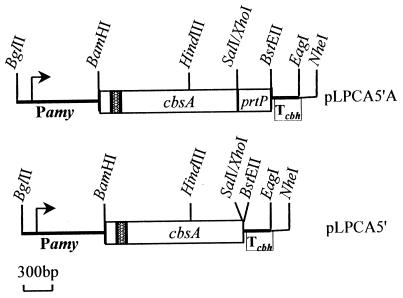
The expression cassettes in plasmids pLPCA5′ and pLPCA5′A are the same except for the presence of an anchor sequence in pLPCA5′A (Fig. 1). In both plasmids, the promoterless cbsA gene was cloned in Lactobacillus-Escherichia coli shuttle vector pLPM11 under the control of the inducible α-amylase promoter of L. amylovorus. pLPM11 is a derivative of pLP402t which lacks the β-glucuronidase gene (20). In both vectors, the insert starts 128 bp upstream of the ATG codon and includes the signal sequence and the entire CbsA-encoding region. A computer analysis predicted that the 128-bp 5′-untranslated region would form a hairpin structure (ΔG, −26.7 kcal/mol). This might render mRNA more stable and therefore might result in higher translation efficiency, as the untranslated leader sequence of S-protein mRNA in L. acidophilus ATCC 4356 was shown to be involved in efficient S-protein production (3). To obtain a vector with CbsA fused to the anchor sequence (pLPCA5′A), the cbsA gene was amplified by PCR from L. crispatus JCM5810 chromosomal DNA with the primers A1F (5′-GCGGATCCTCTAGACTACTACCTCATGAGAG-3′; starts 128 nucleotides upstream of ATG) and A3SalR (5′ GCGAATTCGTCGACAAAGTTTGAAGCCTTTACGTAAG-3′; ends before stop codon). BamHI-XbaI and EcoRI-SalI restriction sites (underlined) were introduced to facilitate further cloning. Since these oligonucleotides are complementary to the highly conserved regions in cbsA and in cbsB, the silent S-layer protein-encoding gene of this strain (27), a mixture of both genes was obtained. cbsB was eliminated by digestion with NheI, which specifically cuts this gene, and a SalI site was introduced just before the stop codon of cbsA in pTUAT to fuse cbsA in frame to the coding sequence of the cell wall anchor of the prtP gene of L. casei (14). Fusion of the anchor to CbsA was expected to cause its covalent linkage to the cell wall and surface exposure. To generate a vector without the anchor sequence (pLPCA5′), a BamHI-SalI DNA fragment encoding mature CbsA was cloned in pTUT (14). A BamHI-HindIII fragment of the 5′ end of cbsA was replaced with the corresponding fragment of pTUAT-cbsA, which comprises the untranslated region and signal sequence. The structure of the vectors was verified by DNA sequencing. Finally, BamHI-NheI cassettes from pTUAT-cbsA and pTUT-cbsA were transferred to expression vector pLPM11. To circumvent instability in E. coli caused by the presence of actively expressed cbsA, L. casei ATCC 393T was transformed with ligation mixtures (19). In this host, the plasmids could be stably maintained.
FIG. 1.
Schematic drawing of the cassettes designed for cbsA expression in L. casei ATCC 393T. Relevant restriction sites are shown. Lactobacillus-E. coli shuttle vector pLPM11 (20) was used to clone cbsA under the control of the inducible α-amylase promoter (Pamy). The insert contained the ribosome binding site and signal secretion sequence of cbsA (shaded area). prtP, cell wall anchoring sequence; Tcbh, transcription terminator sequence.
Production of CbsA by L. casei ATCC 393T.
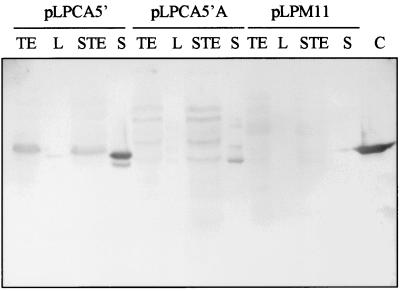
Putative L. casei transformants harboring either pLPCA5′ or pLPCA5′A were first assayed for CbsA production. Colonies were streaked on nitrocellulose filters placed on API (API 50 CHL; Biomerieux, Marcy l'Etoile, France) agar plates. The promoter activity was induced by the presence in the medium of galactose (1%, wt/vol). After overnight incubation, filters were extensively washed and incubated with polyclonal rabbit serum against CbsA. A strong positive reaction was detected in all transformants carrying cbsA, and no cross-reaction was observed with L. casei transformed with the vector, pLPM11 (data not shown). Positive transformants were subjected to further analysis to locate CbsA in different culture fractions. Cells from exponentially growing cultures in API medium were collected by centrifugation, washed with phosphate-buffered saline solution (PBS), and adjusted to an optical density at 590 nm of 1.0 (cell suspension). The supernatant was precipitated with trichloroacetic acid, dissolved in urea buffer (6 M urea in 50 mM Tris HCl, pH 8), and extensively dialyzed against water at 4°C. The precipitate formed (1/100 of the initial volume [trichloroacetic acid fraction]) was dissolved in urea buffer. Surface-associated proteins were extracted by 5 M LiCl (LiCl fraction) (13). Total cell extracts of either LiCl-treated or nontreated cells were obtained by disruption with glass beads (0.4 mm) in the presence of urea and boiled for 15 min. The soluble proteins were collected after centrifugation. The different fractions, corresponding to 1 ml of the initial cell suspension, were subjected to standard sodium dodecyl sulfate (SDS)–10% polyacrylamide gel electrophoresis (PAGE) (12) and electroblotted to polyvinylidene difluoride membranes (Millipore). Standard Western analysis was performed to specifically detect CbsA with polyclonal rabbit anti-CbsA serum (Fig. 2).
FIG. 2.
Production of CbsA by induced cultures of L. casei 393T transformed with expression plasmids pLPCA5′ and pLPCA5′A. Different culture fractions were subjected to standard SDS-PAGE and immunoblotted with rabbit polyclonal CbsA antiserum. L. casei/pLPM11 was used as a negative control. Lanes: TE, total protein extract; L, protein extracted by 5 M LiCl; STE, total protein extract from stripped cells; S, TCA-precipitated culture supernatant; C, CbsA control sample extracted from 0.12 ml of L. crispatus JCM 5810 cells.
In L. casei 393/pLPCA5′, most of the CbsA was recovered from the culture medium whereas only minor quantities were extracted from the bacterial cell surface by LiCl (Fig. 2). Electron microscopic analysis of the CbsA recovered from the supernatant of 393/pLPCA5′ showed the presence of small crystalline sheets, like those observed for CbsA from L. crispatus (data not shown) and for CbsA produced by E. coli (27), indicating that the assembly properties of CbsA produced by L. casei were unaltered.
To determine the amount of CbsA secreted, several dilutions of the culture medium and of purified CbsA were used to coat a microtiter plate for an enzyme-linked immunosorbent assay at 4°C overnight. Plates were blocked for 1 h at 37°C in PBS containing 1% bovine serum albumin (BSA) and washed twice with PBS–0.05% Tween 20. CbsA was detected with rabbit CbsA antiserum and horseradish peroxidase-conjugated anti-rabbit immunoglobulin G. The amount of CbsA present in the medium, determined from a standard curve, was 10 to 15 mg/liter. As judged by its migration on SDS-PAGE, CbsA secreted by L. casei seemed to be properly processed without further modification, although some breakdown products could be observed (Fig. 2, lane S). The protein found in cell extracts may represent unprocessed CbsA still containing the signal sequence. In pLPCA5′A-containing bacteria, CbsA was found mainly in the cell extract (Fig. 2, lanes TE and STE). The occurrence of peptides with a molecular weight higher than that of CbsA was attributed to the presence of cell wall fragments associated with CbsA, since they were not detected after treatment with mutanolysin (data not shown). Furthermore, small amounts of CbsA were also detected in the culture medium of 393/pLPCA5′A, most likely due to some cell lysis. The total amount of CbsA produced by 393/pLPCA5′A was smaller than that produced by 393/pLPCA5′, presumably due to saturation of the target sites for anchoring of the protein and/or competition with the host proteinase P. As expected, no CbsA was detected in any fraction of the negative control, L. casei 393/pLPM11.
Surface presentation of CbsA in L. casei/pLPCA5′A.
Antiserum raised against CbsA was used to confirm the surface location of CbsA in L. casei 393/pLPCA5′A by immunofluorescence microscopy. A strong signal was detected on cells of L. crispatus and of L. casei expressing cbsA::prtP. No reaction was obtained with L. casei 393/pLPCA5′ or 393/pLPM11 (data not shown).
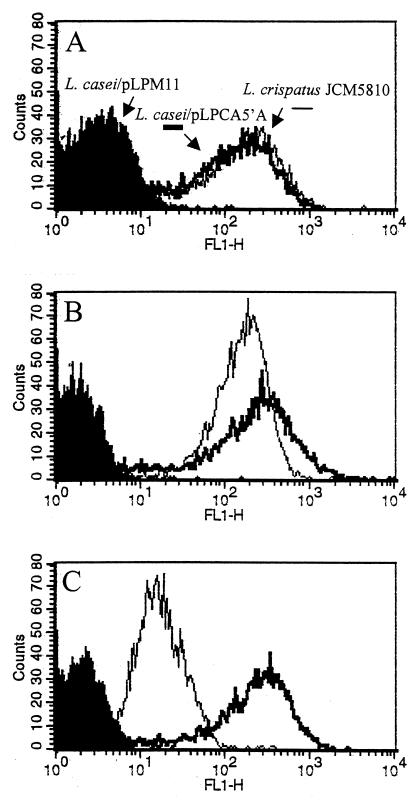
Flow cytometric analysis further confirmed the presence of CbsA on the L. casei cell surface (Fig. 3A). The intensity of the signal measured on individual cells of L. casei/pLPCA5′A was, surprisingly, as high as that on L. crispatus JCM5810 cells. This might suggest that equal numbers of CbsA molecules are exposed on both bacteria. It has been estimated that approximately 5 × 105 S-layer monomers are needed to cover an entire cell (28). In experiments aimed at presenting antigens at the bacterial surface, the numbers of molecules found invariably were much lower. Maassen et al. (14) have estimated that 1.4 × 103 tetanus toxin fragment C molecules were present on the L. casei cell surface when the same expression plasmid, host, and anchor sequence were used. Other authors have reported up to 104 molecules when using other proteins and different anchor partners (18, 29). In accordance with these reports, Western blots of total cell extracts of equal numbers of cells of L. casei/pLPCA5′A and the S-layered L. crispatus strain showed that the quantity of CbsA produced by L. casei/pLPCA5′A was considerably less than that found in L. crispatus (data not shown). An explanation for these conflicting observations would be that the antiserum used for immunodetection, which was raised against His-tagged CbsA produced by E. coli, probably contained antibodies that do not react with S-protein when it is present at the L. crispatus cell surface. Therefore, the strong fluorescence-activated cell sorter (FACS) signal observed for L. casei/pLPCA5′A cells might be due to the reaction with epitopes that are not accessible on L. crispatus. To ascertain whether the same CbsA epitopes are exposed on L. casei/pLPCA5′A as on L. crispatus, we repeated the FACS analysis with antibodies that specifically recognize S-protein epitopes displayed at the L. crispatus cell surface (anti-S-CbsA) and with antibodies reacting with regions of S-protein that are not exposed when S-protein is packed as an S-layer (anti-NS-CbsA). Under denaturing conditions, both antibody fractions reacted equally with CbsA in Western blots (data not shown). With anti-S-CbsA, the FACS profile for L. casei/pLPCA5′A cells was similar to that observed with anti-CbsA while a more homogeneous distribution was observed for L. crispatus JCM5810 cells compared with anti-CbsA serum (compare Fig. 3A and B). As expected, a strongly reduced signal was detected when anti-NS-CbsA antibodies were used to detect CbsA in L. crispatus whereas L. casei/pLPCA5′A cells displayed an equally strong signal (Fig. 3C). From these results, it is apparent that more S-protein epitopes are accessible on recombinant L. casei cells than on L. crispatus cells. This may be caused by the less dense packing of CbsA molecules on the L. casei cell surface compared with that on the L. crispatus cell surface, where CbsA is arranged as a closely packed layer. Furthermore, anchoring of CbsA to the cell wall may have prevented proper folding and/or exposure of the protein at the bacterial surface, rendering monomers unable to form a true S-layer.
FIG. 3.
Flow cytometry analysis of L. casei transformants. CbsA was detected on the cell surface by incubation of the cells with different rabbit polyclonal antibody fractions for CbsA and stained with fluorescein isothiocyanate-labeled goat anti-rabbit serum. These fractions were obtained by adsorbing the CbsA antiserum to approximately 1010 exponentially growing L. crispatus JCM5810 cells for 1 h at 4°C. Unbound antibodies (anti-NS-CbsA) were collected in the supernatant after centrifugation and were filtered through a 0.22-μm-pore-size membrane. Antibody-coated bacteria were washed twice with PBS–0.05% Tween 20, and the surface-specific CbsA antibodies (anti-S-CbsA) were eluted by incubation in 0.1 M glycine–0.5% BSA (pH 2.5) for 1 h at 4°C. After centrifugation, the eluted antibodies were neutralized with 1 M Tris HCl (pH 9) and filtered. Panels: A, anti-CbsA; B, anti-S-CbsA; C, anti-NS-CbsA. L. casei/pLPM11 was used as a negative control.
Collagen-binding ability of cbsA-expressing L. casei.
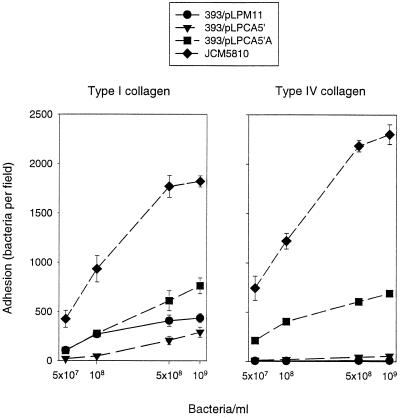
The capacity of L. crispatus JCM5810 and L. casei transformants to bind in vitro to collagen types I and IV was evaluated as described before (29). Briefly, 5 × 107, 1 × 108, 5 × 108, and 1 × 109 cells were applied on diagnostic slides previously coated with 2.5 pmol of collagen type I or IV or BSA, respectively. After 2 h of incubation, loosely attached bacteria were removed by extensive rinsing with PBS. Samples were stained with methylene blue and analyzed by light microscopy. As shown in Fig. 4, only wild-type L. crispatus and L. casei/pLPCA5′A, where CbsA was surface located, attached significantly to collagen-coated surfaces. The efficiency of binding to both collagens was higher for L. crispatus than for L. casei/pLPCA5′A. This difference may reflect the smaller amount of S-protein and/or an altered CbsA configuration on the L. casei cell surface. Besides, it should be kept in mind that lactobacilli recognize collagens via multiple, specific and nonspecific, interactions (15). To confirm that the binding phenotype displayed by L. crispatus JCM5810 relies solely on CbsA, an analysis of isogenic mutants lacking an S-layer would be needed. However, inactivation of S-layer protein-encoding genes in L. crispatus and related organisms by reverse genetics or growth in the presence of antibodies could not be achieved (our unpublished observations).
FIG. 4.
Adherence of L. crispatus JCM5810 and L. casei transformants to immobilized type I or IV collagen on glass. Means of bacterial numbers in 20 randomly chosen microscopic fields of 1.4 × 104 μm2 are shown. Bars indicate standard deviations. None of the bacteria adhered to BSA-coated slides (data not shown).
Heterologous expression of S-layer protein-encoding genes and assembly of their products as an S-layer onto the bacterial surface have not yet been achieved. As shown before for Lactococcus (4) and confirmed in this study for L. casei, non-S-layer bacteria can efficiently produce and secrete S-proteins but the proteins do not interact with the cell surface. Recent findings have outlined the role of secondary cell wall polymers as anchors for several S-layer proteins (8, 11, 22). Preliminary experiments have shown that CbsA produced in L. acidophilus is firmly retained at the bacterial surface, suggesting that CbsA may be anchored to heterologous S-layered hosts through the same anchoring structure as the host S-layer protein.
Our results show, for the first time, that the collagen-binding capacity of an S-protein can be transferred to another LAB host. This may be considered a first step in the analysis of the role of this adhesin in adherence to and colonization of host tissues by lactobacilli. Furthermore, expression of cbsA mutants and hybrid S-protein molecules with different collagen-binding properties with the system described in this study might help to delimit the collagen-binding domain(s) within CbsA and to define key amino acids in S-layer formation.
Acknowledgments
We thank F. Tielen for technical assistance with flow cytometric analyses and R. Leer and N. van Luijk for helpful discussions.
This work was supported by EC grant 961758.
REFERENCES
- 1.Adlerberth I, Ahrne S, Johansson M L, Molin G, Hanson L A, Wold A E. A mannose-specific adherence mechanism in Lactobacillus plantarum conferring binding to the human colonic cell line HT-29. Appl Environ Microbiol. 1996;62:2244–2251. doi: 10.1128/aem.62.7.2244-2251.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beveridge T J. Bacterial S-layers. Curr Opin Struct Biol. 1994;4:204–212. [Google Scholar]
- 3.Boot H J, Kolen C P, Andreadaki F J, Leer R J, Pouwels P H. The Lactobacillus acidophilus S-layer protein gene expression site comprises two consensus promoter sequences, one of which directs transcription of stable mRNA. J Bacteriol. 1996;178:5388–5394. doi: 10.1128/jb.178.18.5388-5394.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Callegari M L, Riboli B, Sanders J W, Cocconcelli P S, Kok J, Venema G, Morelli L. The S-layer gene of Lactobacillus helveticus CNRZ 892: cloning, sequence and heterologous expression. Microbiology. 1998;144:719–726. doi: 10.1099/00221287-144-3-719. [DOI] [PubMed] [Google Scholar]
- 5.Chauvière G, Coconnier M-H, Kerneis S, Fourniat J, Servin A L. Adhesion of human Lactobacillus acidophilus strain LB to human enterocyte-like Caco-2 cells. J Gen Microbiol. 1992;138:1689–1696. doi: 10.1099/00221287-138-8-1689. [DOI] [PubMed] [Google Scholar]
- 6.Doig P, Emödy L, Trust T J. Binding of laminin and fibronectin by the trypsin-resistant major structural domain of the crystalline virulence surface array protein of Aeromonas salmonicida. J Biol Chem. 1992;267:43–49. [PubMed] [Google Scholar]
- 7.Dunne C, Murphy L, Flynn S, O'Mahony L, O'Halloran S, Feeney M, Morrissey D, Thornton G, Fitzgerald G, Daly C, Kiely B, Quigley E M, O'Sullivan G C, Shanahan F, Collins J K. Probiotics: from myth to reality. Demonstration of functionality in animal models of disease and in human clinical trials. Antonie Van Leeuwenhoek. 1999;76:279–292. [PubMed] [Google Scholar]
- 8.Egelseer E M, Leitner K, Jarosch M, Hotzy C, Zayni S, Sleytr U B, Sára M. The S-layer proteins of two Bacillus stearothermophilus wild-type strains are bound via their N-terminal region to a secondary cell wall polymer of identical chemical composition. J Bacteriol. 1998;180:1488–1495. doi: 10.1128/jb.180.6.1488-1495.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greene J D, Klaenhammer T R. Factors involved in adherence of lactobacilli to human Caco-2 cells. Appl Environ Microbiol. 1994;60:4487–4494. doi: 10.1128/aem.60.12.4487-4494.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Havenaar R, Huis in 't Veld J H J. Probiotics: a general view. In: Wood B J B, editor. The lactic acid bacteria. I. The lactic acid bacteria in health and disease. Amsterdam, The Netherlands: Elsevier Applied Science; 1992. pp. 151–170. [Google Scholar]
- 11.Ilk N, Kosma P, Puchberger M, Egelseer E M, Mayer H F, Sleytr U B, Sára M. Structural and functional analyses of the secondary cell wall polymer of Bacillus sphaericus CCM 2177 serving as an S-layer-specific anchor. J Bacteriol. 1999;181:7643–7646. doi: 10.1128/jb.181.24.7643-7646.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 13.Lortal S, van Heijenoort J, Gruber K, Sleytr U B. S-layer of Lactobacillus helveticus ATCC 12046: isolation, chemical characterization and reformation after extraction with lithium chloride. J Gen Microbiol. 1992;138:611–618. [Google Scholar]
- 14.Maassen C B, Laman J D, den Bak-Glashouwer M J, Tielen F J, van Holten-Neelen J C, Hoogteijling L, Antonissen C, Leer R J, Pouwels P H, Boersma W J, Shaw D M. Instruments for oral disease-intervention strategies: recombinant Lactobacillus casei expressing tetanus toxin fragment C for vaccination or myelin proteins for oral tolerance induction in multiple sclerosis. Vaccine. 1999;17:2117–2128. doi: 10.1016/s0264-410x(99)00010-9. [DOI] [PubMed] [Google Scholar]
- 15.McGrady J A, Butcher W G, Beighton D, Switalski L M. Specific and charge interactions mediate collagen recognition by oral lactobacilli. J Dent Res. 1995;74:649–657. doi: 10.1177/00220345950740020501. [DOI] [PubMed] [Google Scholar]
- 16.Messner P, Sleytr U B. Crystalline bacterial cell-surface layers. Adv Microbiol Physiol. 1992;33:213–275. doi: 10.1016/s0065-2911(08)60218-0. [DOI] [PubMed] [Google Scholar]
- 17.Mukai T, Arihara K, Itoh H. Lectin-like activity of Lactobacillus acidophilus strain JCM 1026. FEMS Microbiol Lett. 1992;77:71–74. doi: 10.1016/0378-1097(92)90134-a. [DOI] [PubMed] [Google Scholar]
- 18.Piard J C, Hautefort I, Fischetti V A, Ehrlich S D, Fons M, Gruss A. Cell wall anchoring of the Streptococcus pyogenes M6 protein in various lactic acid bacteria. J Bacteriol. 1997;179:3068–3072. doi: 10.1128/jb.179.9.3068-3072.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Posno M, Leer R J, van Lujk N, van Giezen M J F, Heuvelmans P T H M, Lokman B C, Pouwels P H. Incompatibility of lactobacillus vectors with replicons derived from small cryptic lactobacillus plasmids and segregational instability of the introduced vectors. Appl Environ Microbiol. 1991;57:1822–1828. doi: 10.1128/aem.57.6.1822-1828.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pouwels P H, Leer R J, Boersma W J A. The potential of Lactobacillus as a carrier for oral immunization: development and preliminary characterization of vector systems for targeted delivery of antigens. J Biotechnol. 1996;44:183–192. doi: 10.1016/0168-1656(95)00140-9. [DOI] [PubMed] [Google Scholar]
- 21.Pouwels P H, Leer R J, Shaw M, Heijne den Bak-Glashouwer M J, Tielen F D, Smit E, Martinez B, Jore J, Conway P L. Lactic acid bacteria as antigen delivery vehicles for oral immunization purposes. Int J Food Microbiol. 1998;41:155–167. doi: 10.1016/s0168-1605(98)00048-8. [DOI] [PubMed] [Google Scholar]
- 22.Ries W, Hotzy C, Schocher I, Sleytr U B, Sára M. Evidence that a secondary cell wall polymer recognizes the N-terminal part of the S-layer protein from Bacillus stearothermophilus PV72/p2. J Bacteriol. 1997;179:3892–3898. doi: 10.1128/jb.179.12.3892-3898.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salminen S, von Wright A, Morelli L, Marteau P, Brassart D, de Vos W M, Fonden R, Saxelin M, Collins K, Mogensen G, Birkeland S E, Mattila-Sandholm T. Demonstration of safety of probiotics—a review. Int J Food Microbiol. 1998;44:93–106. doi: 10.1016/s0168-1605(98)00128-7. [DOI] [PubMed] [Google Scholar]
- 24.Savage D C. Associations of indigenous microorganisms with gastrointestinal epithelial surfaces. In: Hentges D J, editor. Human intestinal microflora in health and disease. New York, N.Y: Academic Press, Inc.; 1983. pp. 55–78. [Google Scholar]
- 25.Schneitz C, Nuotio L, Lounatmad K. Adhesion of Lactobacillus acidophilus to avian intestinal epithelial cells mediated by the crystalline bacterial cell surface layer (S-layer) J Appl Bacteriol. 1993;74:290–294. doi: 10.1111/j.1365-2672.1993.tb03028.x. [DOI] [PubMed] [Google Scholar]
- 26.Sherman L A, Savage D C. Lipoteichoic acids in lactobacillus strains that colonize the mouse gastric epithelium. Appl Environ Microbiol. 1986;52:302–304. doi: 10.1128/aem.52.2.302-304.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sillanpää J, Martínez B, Antikainen J, Toba T, Kalkkinen N, Tankka S, Lounatmaa K, Keränen J, Höök M, Westerlund-Wikström B, Pouwels P H, Korhonen T K. Characterization of the collagen-binding S-layer protein CbsA of Lactobacillus crispatus. J Bacteriol. 2000;182:6440–6450. doi: 10.1128/jb.182.22.6440-6450.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sleytr U B, Messner P. Crystalline surface layers in procaryotes. J Bacteriol. 1988;170:2891–2897. doi: 10.1128/jb.170.7.2891-2897.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strauss A, Gotz F. In vivo immobilization of enzymatically active polypeptides on the cell surface of Staphylococcus carnosus. Mol Microbiol. 1996;21:491–500. doi: 10.1111/j.1365-2958.1996.tb02558.x. [DOI] [PubMed] [Google Scholar]
- 30.Toba T, Virkola R, Westerlund B, Björkman Y, Sillanpää J, Vartio T, Kalkkinen N, Korhonen T K. A collagen-binding S-layer protein in Lactobacillus crispatus. Appl Environ Microbiol. 1995;61:2467–2471. doi: 10.1128/aem.61.7.2467-2471.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Westerlund B, Korhonen T K. Bacterial proteins binding to the mammalian extracellular matrix. Mol Microbiol. 1993;9:687–694. doi: 10.1111/j.1365-2958.1993.tb01729.x. [DOI] [PubMed] [Google Scholar]