Abstract
In recent years, microRNAs (miRNAs) have garnered increasing attention for their potential implications in cancer pathogenesis, functioning either as oncogenes or tumor suppressors. Notably, angiosarcoma, along with various other cardiovascular tumors such as lipomas, rhabdomyomas, hemangiomas, and myxomas, has shown variations in the expression of specific miRNA subtypes. A substantial body of evidence underscores the pivotal involvement of miRNAs in the genesis of angiosarcoma and certain cardiovascular tumors. This review aims to delve into the current literature on miRNAs and their prospective applications in cardiovascular malignancies, with a specific focus on angiosarcoma. It comprehensively covers diagnostic methods, prognostic evaluations, and potential treatments while providing a recapitulation of angiosarcoma’s risk factors and molecular pathogenesis, with an emphasis on the role of miRNAs. These insights can serve as the groundwork for designing randomized control trials, ultimately facilitating the translation of these findings into clinical applications. Moving forward, it is imperative for studies to thoroughly scrutinize the advantages and disadvantages of miRNAs compared to current diagnostic and prognostic approaches in angiosarcoma and other cardiovascular tumors. Closing these knowledge gaps will be crucial for harnessing the full potential of miRNAs in the realm of angiosarcoma and cardiovascular tumor research.
Keywords: microRNA, angiosarcoma, cancer, cardiovascular tumor, tumor suppressor, oncogenesis
1. Introduction
MicroRNAs (miRNAs) are a class of small, non-coding, single-stranded RNAs that regulate gene expression at the post-transcriptional level, impacting crucial cellular processes like proliferation, differentiation, and apoptosis (1–4). This regulatory influence extends to an estimated 60% of human genes, highlighting their widespread impact (5). Consequently, miRNAs are attracting significant interest as potential targets for the prevention, diagnosis, and treatment of complex human diseases (6–8). Emerging evidence underscores the critical role of miRNAs in maintaining cellular homeostasis by tightly regulating diverse biological functions (9, 10). Notably, miRNAs can function as either oncogenes or tumor suppressors, playing a pivotal role in cancer biology (11–13). Dysregulation of miRNA expression is frequently observed in various types of cancer (12, 14). These alterations contribute to cancer hallmarks by enhancing proliferation signals, evading growth inhibitors, resisting cell death, promoting invasiveness, and stimulating angiogenesis (15). The profound influence of miRNAs on these crucial cancer-related processes suggests their immense potential for future research and therapeutic development.
Angiosarcoma is a rare and aggressive soft-tissue sarcoma characterized by a high propensity for local recurrence, distant metastasis, and poor prognosis if not diagnosed early (16). Endothelial cells lining blood and lymphatic vessels give rise to angiosarcoma, contributing to its frequent lymph node and systemic metastases (17, 18). While primarily affecting the skin and soft tissues, particularly the head and neck region, angiosarcoma also represents the most common form of differentiated cardiac malignancy, accounting for approximately 10–15% of primary cardiac tumors (16, 17, 19). Despite advancements in cancer therapy, angiosarcoma management remains challenging, highlighting the need for a deeper understanding of its molecular underpinnings and the development of novel therapeutic strategies (18). The rarity of this disease translates to a lack of established treatment protocols. While localized surgery offers the best chance of cure, technical challenges can limit its feasibility in some cases (20). Emerging evidence suggests that various microRNA subtypes might influence angiosarcoma development and progression (21). For instance, tumor cells often exhibit upregulation of miR-210, leading to the downregulation of its putative targets, ephrin A3 and E2F3 (22). Additionally, miRNAs hold promise as potential diagnostic biomarkers for angiosarcoma. Exosomal miR-5684 and miR-125b-5p are significantly downregulated in angiosarcoma patients compared to healthy controls (23). Furthermore, dysregulation of miRNAs like miR-222, miR-199a, miR-195, and miR-125b has been observed in angiosarcoma, potentially influencing cell cycle proteins and contributing to cell cycle deregulation (24). Thus, targeting these aberrantly expressed miRNAs represents a promising therapeutic approach for improved treatment outcomes and survival rates in angiosarcoma patients.
Given the emerging recognition that microRNAs (miRNAs) exhibit differential expression patterns across various cancer types, grades, and clinical courses, these molecules hold significant promise as diagnostic, prognostic, and even therapeutic tools (15). Therefore, this review delves into angiosarcoma, a rare and aggressive soft-tissue sarcoma. We will first provide a concise overview of angiosarcoma risk factors and molecular pathogenesis. Following this foundation, we will critically evaluate the current evidence regarding the role of miRNAs in angiosarcoma. Our focus will be on the potential of miRNAs as future targets for angiosarcoma diagnosis, prognosis, and therapy. Finally, we will broaden the scope to discuss the potential significance of miRNAs in other cardiovascular tumors.
2. Risk factors and molecular pathogenesis of angiosarcoma
Angiosarcoma is an uncommon and aggressive soft tissue sarcoma, accounting for 1-2% of all diagnosed sarcomas (25). This section presents a comprehensive summary of the occurrence, development, diagnosis, prediction of outcomes, survival rates, and treatment choices for angiosarcoma. Angiosarcomas can manifest at any age but are predominantly detected in older individuals, with a median age of onset ranging from 60 to 71 years (25). The annual prevalence is approximately 1-2 cases per million individuals (26), with an incidence rate adjusted for age ranging from 1-4% (27).
The development of angiosarcoma is intricate and needs to be completely comprehended. Nevertheless, certain variables that increase the risk of angiosarcoma have been identified, including long-term lymphedema, previous radiation therapy, and exposure to environmental substances that might cause cancer (28). Evidence suggests that genetic predispositions, such as POT1 gene mutations, play a role in familial angiosarcoma cases (29). Diagnosis of angiosarcoma might be challenging due to its infrequency and lack of identifiable clinical symptoms. Diagnostic procedures such as ultrasound, CT, and MRI visualize internal structures. However, a conclusive diagnosis involves examining tissue samples under a microscope and confirming with immunohistochemistry tests. Markers such as CD31, CD34, and Factor-VIII indicate the origin of vascular endothelial cells (25). Prognostication is influenced by several parameters, including the size, location, resectability, and existence of tumor metastases at the time of diagnosis (28). However, the survival rates for angiosarcoma exhibit significant variability. The reported total 5-year survival rate ranges from 27% to 40%, with median survival durations varying from 6 to 16 months (27). Patients diagnosed with high-grade tumors or those who already have metastasized illness upon presentation have a meager chance of a good prognosis (25). Surgical excision, frequently accompanied by adjuvant radiation, is the cornerstone of treatment for localized angiosarcoma (25). Chemotherapy, using anthracycline or taxane-based regimens, is commonly employed to treat instances that cannot be operated on or cases that have spread to other body parts. Current advancements in targeted treatments and immunotherapy are potential therapeutic alternatives, while the specific order and combination with traditional medications are still actively studied (30).
2.1. Risk factors
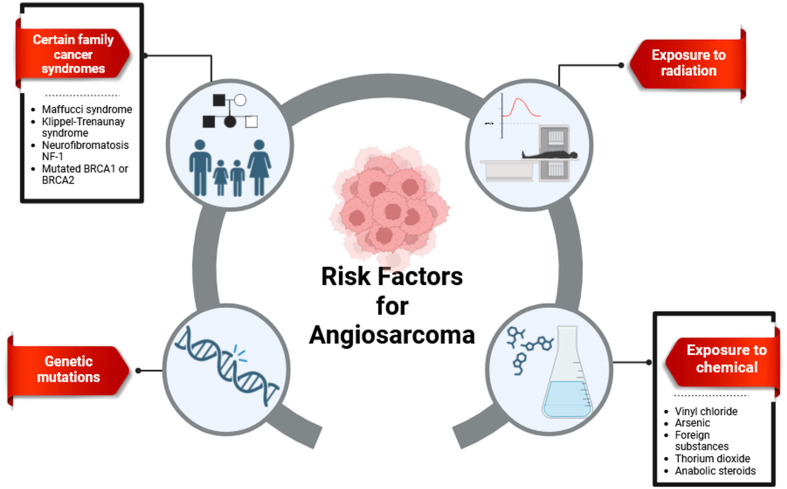
The etiology of angiosarcoma remains unclear; however, several well-established risk factors have been identified. These include exposure to radiation, chronic lymphedema, and carcinogens such as thorium dioxide, vinyl chloride, and arsenic. Genetic disorders also contribute to the constellation of risk factors for angiosarcoma (31, 32), as illustrated in Figure 1 .
Figure 1.
Several risk factors for angiosarcoma (adapted from Ni et al. (33) (33).
2.1.1. Radiation
The link between radiation exposure and increased cancer risk is well-established. Chronic lymphedema and radiation-induced genetic changes are potential contributing factors to radiation-associated malignancies (34, 35). Notably, radiation therapy for early-stage sarcomas can paradoxically lead to radiation-induced sarcomas, a significant subtype of secondary sarcomas (36, 37). While the exact link between radiation and angiosarcoma development needs further investigation, studies suggest a potential association between higher radiation doses, longer lifespans after treatment, and the risk of radiation-induced angiosarcoma (38, 39). Previous radiation exposure might also be a risk factor (40). However, the overall risk of radiation-induced angiosarcoma remains low when balanced against the benefits of radiotherapy (25).
2.1.2. Chronic lymphedema
A well-documented association exists between angiosarcoma and long-term chronic lymphedema, a condition known as Stewart-Treves syndrome (STS) (41). STS predominantly affects women, particularly following adjuvant radiation and breast surgery (41, 42). The use of adjuvant radiation during treatment for the primary malignancy is believed to contribute to STS progression (43). Other potential causes of lymphedema that might elevate angiosarcoma risk include idiopathic, congenital, traumatic, or filarial etiologies (44). Approximately 5% of angiosarcomas are attributed to STS, typically presenting within 5-15 years after surgery and radiation (43). Unfortunately, STS is associated with a poor prognosis, with a median expected survival of around ten months (45). The mechanisms underlying the progression from different forms of persistent lymphedema to secondary angiosarcomas remain poorly understood. Hypotheses suggest that mutations in tumor suppressor genes like MYC or P53 might play a co-factor role (45, 46).
2.1.3. Environmental carcinogens
Despite the unclear etiology of approximately 75% of hepatic angiosarcomas (47), exposure to specific environmental carcinogens has been implicated as a potential risk factor. These carcinogens include vinyl chloride monomer (VCM), a component in polyvinyl chloride production, iatrogenic exposure to the radioactive contrast agent Thorotrast used in radiological examinations, and chronic arsenic consumption (48, 49).
2.1.4. Genetic disorders
Approximately 3% of primary angiosarcomas are classified as subtypes associated with familial and genetic disorders. Klippel-Trenaunay syndrome (KTS) is a notable example, characterized by congenital malformations involving blood vessels, soft tissues, and bones. Other genetic disorders linked to angiosarcoma risk include Ollier disease and Recklinghausen neurofibromatosis. Interestingly, these familial predispositions are not specific to angiosarcoma and have also been associated with Xeroderma pigmentosa, Maffucci disease, and bilateral retinoblastoma (50).
2.2. Molecular pathogenesis
Angiosarcoma pathogenesis is a complex process driven by a multitude of genetic and epigenetic alterations. Expanding upon the groundwork established in Section 2.1, this section provides a more comprehensive analysis of the precise molecular mechanisms that govern the development of angiosarcoma. Recent studies have brought attention to the intricate relationship that exists among epigenetic dysregulation, mutations in genes, and signaling pathways in the development of angiosarcoma.
2.2.1. Dysregulation of angiogenic pathways and tumor suppressors
Angiosarcoma exhibits significant molecular heterogeneity (31, 51). Notably, studies have reported high-level amplification at the 8q24.21 chromosomal locus and significant MYC overexpression in secondary cutaneous angiosarcomas (52). These findings highlight the importance of reliable methods for MYC detection, such as fluorescent in situ hybridization (FISH) or immunohistochemistry, to differentiate cutaneous angiosarcomas from atypical vascular lesions, which are typically MYC-negative (53).
Recent research sheds light on the role of upstream signaling pathways in MYC overexpression. MYC-positive cutaneous angiosarcomas demonstrate overexpression of atypical Protein Kinase C lambda/iota (aPKCλ). Beyond its established functions in cell polarity and cancer progression, aPKCλ appears to influence endothelial cell proliferation in angiosarcoma. Mechanistically, aPKCλ phosphorylates the endothelial transcription factor FoxO1 at Ser218, leading to its inactivation and preventing its binding to the miR-34c promoter. miR-34c normally acts as a negative regulator of MYC expression; thus, its downregulation by aPKCλ-mediated FoxO1 inactivation contributes to MYC overexpression (54).
Interestingly, aPKCλ can upregulate PD-L1 expression in tumor cells, potentially influencing the trajectory of cancerous cells (55). Selective targeting of aPKCλ with specific inhibitors alongside miR-34c mimics warrants further investigation for their therapeutic potential (18).
2.2.2. Mutation landscape and signaling pathway aberrations
Angiosarcoma development is also characterized by distinct mutation patterns in primary and secondary cutaneous subtypes. Secondary cutaneous angiosarcomas frequently harbor mutations in genes like KIT, ICOS, FLT4, and RASGRP3, while primary cutaneous angiosarcomas exhibit mutations in TP53, KRAS, and BRAF among others. Interestingly, primary cutaneous angiosarcomas are also associated with mutations in fusion genes like NUP160-SCL43A3 and PTPRB/VE-PTP (56, 57).
Primary cutaneous angiosarcoma is frequently associated with TP53 loss-of-function mutations. The p53 signaling pathway, activated by various cellular stresses, plays a critical role in tumor suppression (58). Given its function in cell cycle regulation, p53 mutations are prevalent across diverse cancers, including angiosarcoma (59). Notably, p53 inhibits angiogenesis by downregulating vascular endothelial growth factor (VEGF). However, Mouse Double Minute 2 Homolog (MDM2) can negatively regulate p53 activity, leading to increased VEGF expression and promoting angiosarcoma formation (60, 61). Consequently, the presence of anti-p53 antibodies in serum might serve as a potential biomarker for angiosarcoma diagnosis or prognosis (62).
The MAPK signaling pathway is implicated in the pathogenesis of cutaneous angiosarcoma. Dai et al. reported that BRAF inhibitor therapy, while effective in some cancers, can paradoxically activate the MAPK pathway and lead to secondary malignancies like RET-mutant cutaneous angiosarcomas (63). Furthermore, Murali et al. (56) identified frequent mutations in CDKN2A (26%) and TP53 (35%), both affecting the MAPK pathway, suggesting its potential as a therapeutic target. Given the high prevalence of these mutations, specific inhibitors targeting the MAPK pathway might offer a promising treatment approach for aggressive angiosarcoma subtypes (56).
2.2.3. Epigenetic dysregulation
Epigenetic changes, such as adjustments in DNA methylation patterns and histone modifications, are being increasingly acknowledged as significant determinants in the genesis of cancer (64). DNA methylation is an epigenetic alteration that has a significant correlation with the regulation of gene expression. The significance of changes in DNA methylation in the development of tumors motivates our efforts to unravel the human epigenome (65). Previous research on DNA methylation in angiosarcoma has focused solely on the tumor suppressor gene cyclin-dependent kinase inhibitor 2A (CDKN2A, or INK4A/ARF). This has shown that most sporadic liver angiosarcoma cases have hypermethylation of the CDKN2A (p16INK4A) promoter (66). The majority of radiation-associated angiosarcomas, according to Mentzel et al., have lost H3K27me3 expression; however, endothelial cells in benign and atypical vascular lesions that have formed after prior radiation therapy stained positively for H3K27me3. When tested for H3K27me3, the sporadic angiosarcomas showed inconsistent staining. One other diagnostic tool that may be used to identify radiation-associated angiosarcomas is the loss of H3K27me3 (67). While research on epigenetic modifications in angiosarcoma is ongoing, some studies suggest their potential role. For instance, Sirtuins (SIRT1-7), a class of NAD+-dependent deacetylases, have emerged as contributors to various age-related pathologies like cancer, neurodegeneration, and metabolic disorders (68, 69). Interestingly, angiosarcoma cells exhibit decreased SIRT7 expression, which correlates with suppressed cell proliferation and invasion. Furthermore, SIRT7 can counteract the tumor suppressive effects of miR-340, highlighting its complex role in angiosarcoma (21).
2.2.4. Other molecular players in angiosarcoma pathogenesis
Beyond the established roles of angiogenic pathways and tumor suppressor genes, several other molecular players contribute to angiosarcoma development. The E2F family of transcription factors involved in cell cycle progression, survival, and differentiation, are also implicated in angiosarcoma pathogenesis. This highlights the intricate network of molecular pathways involved in this malignancy (70).
Protein tyrosine phosphatase receptor type B (PTPRB) encodes VE-PTP, a phosphatase that inhibits vascular endothelial growth factor receptors (VEGFR) 1-3 in endothelial cells. Given the established overexpression of VEGFRs in angiosarcomas (71, 72), targeting VE-PTP represents a promising therapeutic strategy. Furthermore, amplification of the FLT4 gene, encoding VEGFR3 (73), has been observed in approximately 25% of secondary angiosarcomas alongside MYC amplification, leading to increased FLT4 mRNA expression (74). These findings suggest that targeting FLT4 might be another potential avenue for angiosarcoma treatment.
ATRX loss and reduced expression of Notch1 and Notch2 have also been documented in angiosarcomas. Interestingly, reduced Notch1 expression is associated with advanced disease and cutaneous origin, while lower Notch2 levels correlate with poorer disease-specific outcomes. Moreover, studies suggest a suppressive role for Notch1 in liver endothelial cell function, potentially contributing to hepatic angiosarcoma development (75). Furthermore, the suppression of Notch1 in liver endothelial cells contributed to the development of hepatic angiosarcomas (76).
3. MicroRNAs and angiosarcoma
MicroRNAs, also known as miRNAs, are a type of tiny RNA molecules that play a crucial role in regulating gene expression by specifically targeting messenger RNAs (mRNAs) (77). MiRNAs control physiological functions, including cell necrosis, apoptosis, and active secretion through exosome microvesicles (78). The deregulation of miRNA expression is a common observation in tumorigenesis, as miRNA loci are often linked to chromosomal regions containing DNA amplifications, deletions, or translocations (79). Emerging evidence suggests a complex interplay between miRNAs and angiosarcoma. KCa3.1, a calcium-activated potassium channel, regulates endothelial proliferation and angiogenesis. Its expression is significantly upregulated upon repression of miR-497-3p. In vivo studies using xenotransplant models demonstrated that TRAM-34, KCa3.1 inhibitor, or miR-497-5p mimetics could impair the growth of angiosarcoma cell lines [59]. Furthermore, biallelic deletion of Dicer1, a crucial enzyme for miRNA biogenesis, has been associated with the development of highly aggressive angiosarcomas [60]. This observation highlights the potential of utilizing antagonistic miRNAs or miRNA mimetics for angiosarcoma therapy [8]. Garcia-Heredia et al. examined miRNA expression in 17 sarcomas and 6 nontumor mesenchymal tissues and compared them to the IMR90 cell line to find differentially expressed miRNAs. They discovered 81 miRNAs (23 down, 49 up) (19).
Accordingly, antagonistic miRNAs or miRNA mimetics, with a specific application toward angiosarcoma, could prove valuable in angiosarcoma therapy in the future (18). The intricate interplay between miRNAs and cancer cells in angiosarcoma, with the potential for therapeutic interventions and diagnostic or prognostic markers, will be discussed in the next section in detail. The characteristics of miRNAs exhibited in cancers may vary depending on the tumor tissue, as these molecules possess both oncogenic and tumor-suppressive properties (80) as follows:
3.1. Tumor suppressor
While miRNA deregulation is a common feature in cancers, some miRNAs exhibit tumor-suppressive properties in angiosarcoma (81, 82). miR-214, for instance, acts as a tumor suppressor by targeting the COP1-p53 pathway, thereby restoring the activity of the p53 tumor suppressor gene and promoting programmed cell death in angiosarcoma cells, restoring the activity of p53, a crucial gene that inhibits tumor growth. These findings indicate that targeting the miR-214-COP1-p53 axis is a promising and innovative approach for treating canine hemangiosarcoma and human angiosarcoma (83). Similarly, miR-340 has been shown to inhibit the proliferation and infiltration of angiosarcoma cells by targeting SIRT7, further highlighting its potential as a tumor suppressor miRNA in this aggressive malignancy (21). Reduced cell migration and tumor development were additional outcomes of miR-497 overexpression. An angiosarcoma cell line’s RNA-sequencing data, expression data from AS patients, and target prediction algorithms were utilized to identify therapeutically significant target genes, allowing us to investigate the tumor suppression mechanism of miR-497 (82).
3.2. Oncogenic
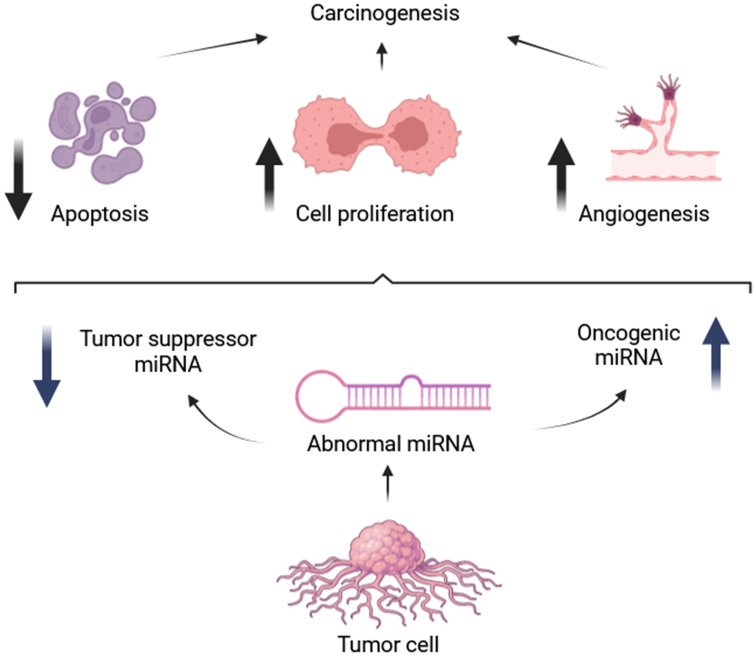
Angiosarcomas, like many cancers, exhibit dysregulation of miRNAs, which can function as both oncogenes and tumor suppressors (84). This characteristic opens avenues for exploring miRNAs as therapeutic targets. Strategies include mimicking tumor-suppressive miRNAs or inhibiting oncogenic miRNAs to manipulate their influence on cancer development (85–87). For example, miR-210 has been shown to promote angiosarcoma cell proliferation by targeting ephrin A3 and E2F3, proteins involved in cell growth and division. Studies revealed upregulation of these target proteins alongside miR-210 overexpression in tumor cells, while knockdown of these targets significantly reduced angiosarcoma cell numbers, suggesting an oncogenic role for miR-210 in this context (22). Interestingly, miR-340 appears to possess a dual role in tumorigenesis, acting as both a tumor suppressor and an oncogene depending on the cellular context (21). Angiosarcoma is frequently associated with mutations in Dicer1, a ribonuclease enzyme essential for processing precursor miRNAs (pre-miRNAs) into mature functional miRNAs, and also has been implicated in angiosarcoma pathogenesis (88, 89). Studies demonstrated that biallelic Dicer1 deletion can lead to aggressive and metastatic angiosarcoma, even in the absence of additional oncogenic mutations (90). Studies in mice have demonstrated that biallelic loss of Dicer1 predisposes endothelial cells to angiosarcoma formation, highlighting the critical role of DICER1 in maintaining proper miRNA biogenesis (90, 91). This finding strengthens the link between miRNA alterations and angiosarcoma development (82). Furthermore, Hanna et al. reported that mice with Dicer1-deleted angiosarcomas exhibit overexpression of target genes for miR-23, a miRNA known to regulate cell cycle progression. The increased expression of miR-23 promotes S phase entry by enhancing the activity of cell cycle regulators CCND1 and CDK4/6 (90). This finding highlights the crucial role of miRNAs in preventing angiosarcoma and suggests that disruption of this regulatory mechanism can promote tumorigenesis (90). In summary, miRNAs act primarily as negative regulators of gene expression. However, excessive levels of oncogenic miRNAs can promote tumor development by stimulating cell proliferation and inhibiting apoptosis, as illustrated in Figure 2 (93).
Figure 2.
Roles of oncogenic and tumor suppressive microRNAs [adapted from Hata et al. (2015)] (92).
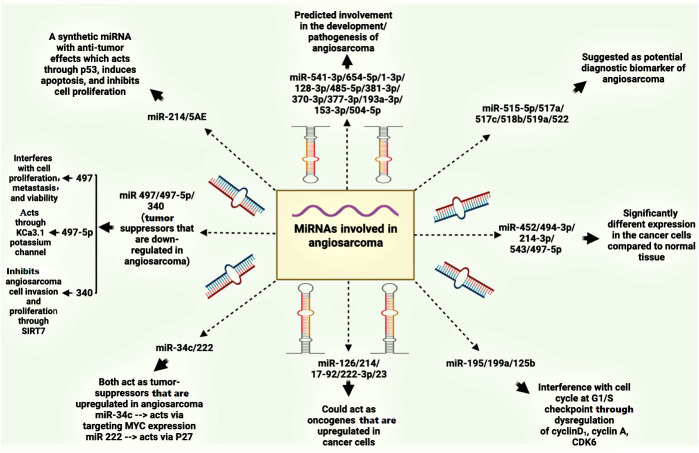
The expanding role of miRNAs in cancer diagnosis, prognosis, and therapy has spurred research into their potential applications in angiosarcoma (94). Several miRNAs have emerged as candidates for angiosarcoma biomarkers. Upregulation of miR-541-3p/miR-654-5p, miR-1-3p, miR-128-3p, miR-485-5p, miR-381-3p, miR-370-3p, miR-377-3p, miR-193a-3p, miR-153-3p, and miR-504-5p (95). Additionally, miRNAs such as miR-515-5p, miR-515-3p, miR-517a, miR-517c, miR-518b, miR-519a, and miR-522 have been suggested as potential diagnostic biomarkers for angiosarcoma due to their overexpression in angiosarcomas (96). These findings are depicted in Figure 3 .
Figure 3.
MiRNAs involved in angiosarcoma.
Heishima et al. reported downregulation of miR-214 and overexpression of COP1 in canine hemangiosarcoma, a model for human angiosarcoma [71]. In this context, miR-214 functions as a tumor suppressor by restoring p53 activity and promoting apoptosis, thereby inhibiting invasion and migration (83, 97). However, studies have revealed unexpectedly high plasma levels of miR-214 and miR-126 in human angiosarcoma patients [83]. Intriguingly, miR-214 is hypothesized to possess anti-angiogenic properties within cells but exert angiogenic effects when released extracellularly, suggesting the involvement of target cells and co-factors like microvesicle proteins in its activity (98). This complex interplay highlights the potential of circulating miR-214 as a precise diagnostic biomarker for tumors, including angiosarcoma (99).
Several reveals altered expression of several miRNAs in the serum of dogs with splenic masses compared to healthy controls. Interestingly, miR-214 levels are specifically elevated in angiosarcoma tissues compared to other splenic masses (100). Yoshikawa et al. applied miR-214/5AE, a synthetic miR-214, to mice with canine angiosarcoma. They found that synthetic miR-214, through the overexpression of cleaved caspase-3 and p53, can induce apoptosis and inhibit cell proliferation (101). On the other hand, Except for glioblastoma, bladder cancer, and angiosarcoma, miR-210 is up-regulated in nearly all of the cancer types that have been studied (102).
miR-497-5p has emerged as a tumor suppressor in angiosarcoma, inhibiting cell proliferation, metastasis, and viability (82). Furthermore, Benton et al. discovered that miR-497 overexpression drastically decreased cell migration and tumor development (82).While KCa3.1 was previously identified as a target of miR-497-5p, with studies demonstrating its ability to suppress tumor invasion by over 70% (81), recent research suggests the presence of additional targets. Specifically, Cdk6, Ccnd2, and Vat1 have been identified as potential downstream effectors of miR-497-5p in angiosarcoma, highlighting the complex regulatory network it governs (82).
Studies show that vinyl chloride exposure alters miRNA expression in rat hepatocytes. This exposure decreases the expression of tumor-suppressive miRNAs like miR-195, miR-199a, and miR-125b, while increasing miR-222, an oncogenic miRNA. These changes in miRNA levels lead to altered activity of target proteins involved in cell cycle regulation, such as cyclin D1, cyclin A, CDK6, and p27. Interestingly, unlike in the liver, serum miRNA expression remains unaffected by vinyl chloride exposures. This highlights the potential of miRNAs in liver tissue, but not serum, as early diagnostic markers for vinyl chloride-induced liver cancers (24).
Several miRNAs contribute to angiosarcoma development by regulating key cellular processes. Nakashima et al. demonstrated that miR-210 downregulation in angiosarcoma, compared to cherry angiomas, leads to increased protein levels of its targets, ephrinA3 and E2F3, both involved in cell proliferation. miR-210 primarily regulates translation, rather than transcription, of these factors, and its decrease results in reduced cell proliferation in angiosarcoma cells (22). Wang et al. also showed in their study that miR-340 inhibits angiosarcoma cell invasion and proliferation by targeting the 3’-UTR region of SIRT7 in angiosarcoma cells (21). MYC amplification, observed in both primary and secondary angiosarcomas, can also indirectly influence angiosarcoma development [88]. MYC amplification can lead to decreased expression of THBS1 through the upregulation of the miR-17-92 cluster, thereby promoting the angiogenic phenotype of the disease (103). Interestingly, FoxO1, a transcription factor, appears to play a complex role in angiosarcoma. While FoxO1 suppression leads to reduced c-MYC expression and cell proliferation, it also results in decreased miR-34 expression. FoxO1 binding to DNA is crucial for transcribing genes involved in c-MYC signaling and miR-34c expression. Mutations in the FoxO1 protein (FoxO1Ser218) can impair its ability to activate miR-34, leading to MYC overexpression and increased cell proliferation in primary cutaneous angiosarcomas (54).
4. MicroRNAs and other cardiovascular system-related tumors
While this paper has predominantly discussed the role of miRNAs in angiosarcomas, emerging evidence indicates their crucial regulatory role in several prevalent cardiovascular malignancies. The following are six instances of miRNAs and the functions they play in cancers of the cardiovascular system:
miR-485-5p (7q22.1): miR-485-5p is an important microRNA that has shown roles in tumors and the cardiovascular system. It affects how the cardiovascular system looks and works. Heart fibroblast activation can be enhanced by miR-485-5p deficiency (104).
Maternally expressed gene 3 (MEG3): MEG3 is an lncRNA that plays a role in various events, including apoptosis, inflammation, oxidative stress, and endoplasmic reticulum stress, directly or through competitive binding with miRNA. Cardiovascular disorders and cancers are among the many conditions it has a role (105).
Circular RNAs: Single-stranded, covalently closed non-coding RNA molecules called circRNAs are being studied as gene expression epigenetic controllers (13). They are class of RNA molecules formed by the back-splicing of precursor messenger RNAs. Recently, circRNAs have garnered interest for their potential to regulate gene expression via sponging microRNAs, interacting with RNA-binding proteins, controlling transcription and splicing, and protein translation (106). In addition to being involved in a wide range of biological processes, they have strong associations with several illnesses, including cancer, diabetes, disorders of the neurological system, and cardiovascular disease. They have several potential uses, such as biomarkers, RNA-binding protein interactions, and miRNA sponges (107). For instance, Nakashima et al. found that circ_0024169/CUL5 ratio may be a diagnostic biomarker for vascular malignancies, whereas circ_0024169 levels may be a prognosis marker for angiosarcoma (108).
Ferroptosis-related genes: Ferroptosis is a kind of cell death not caused by apoptosis and is linked to several cardiovascular disorders. The regulatory network of mRNA-miRNA interactions including genes associated with ferroptosis, plays a crucial role in the occurrence of myocardial ischemia-reperfusion damage (109).
Nonvalvular Endovascular Infections: They typically occur in conjunction with intravascular devices and prosthetic material. They may include primary cardiac tumors, such as myxomas. Common risk factors encompass cardiovascular disease, diabetes mellitus, and cancer (110).
KIAA1429 (m6A Methyltransferase): m6A methyltransferase is essential for RNA metabolism and is implicated in several biological processes, such as cell proliferation, differentiation, and death. The dysregulation of KIAA1429 has been linked to a range of disorders, including cardiovascular diseases and cancer (111).
The following sections will delve into the engagement of miRNAs in these diverse cardiovascular tumors.
4.1. Rhabdomyosarcoma
Numerous miRNAs intricately regulate rhabdomyosarcoma, with notable tumor suppressors including miR-206, miR-1, miR-29, and miR-26a (112–126). A conspicuous reduction in miR-206 levels is observed within rhabdomyosarcoma tissues and cells. The elevated presence of miR-206 induces myogenic differentiation and impedes tumor growth by targeting essential genes like c-Met, PAX3, and G6PD (114–116, 118–122). By suppressing the expression of c-Met and PAX3, miR-1 also acts as a tumor suppressor, resulting in cell cycle arrest and autophagic cell death (115, 117, 118, 124). Inhibiting tumor growth, miR-29 targets explicitly cyclin D2 and E2F7 (112, 118, 126), and miR-26a, downregulated in this context, exerts tumor-suppressive effects by reducing EZH2 expression (113, 125). These miRNAs impede proliferation, induce apoptosis and differentiation, and target pivotal oncogenes, suppressing rhabdomyosarcoma progression.
Emerging evidence suggests a multifaceted tumor-suppressive role for miR-206 in rhabdomyosarcoma, making it a promising candidate for future therapeutic development (114–116, 118–122). Restoration of miR-206 expression holds therapeutic potential, warranting further investigation into targeted delivery methods for rhabdomyosarcoma treatment (114). Hence, miRNAs assume a crucial role in rhabdomyosarcoma through their diverse mechanisms by functioning as tumor suppressors.
4.2. Hemangioma
miRNAs play a diverse regulatory role in hemangioma progression by targeting key signaling pathways. Notably, miR-130a exhibits a consistent upregulation in hemangioma tissues and cell lines compared to normal controls. Functional studies revealed that inhibiting miR-130a significantly reduces proliferation and tumor growth by targeting TFPI2 and modulating the FAK/PI3K/Rac1/Mdm2 pathway, highlighting its potential as a therapeutic target (127). Propranolol, a beta-blocker medication, has been shown to decrease miR-382 levels in hemangiomas, leading to reduced tumor progression through activation of the PTEN/AKT/mTOR pathway (128). DNA methylation alterations are also implicated in hemangioma development. The combination of DNMT3A inhibition and miR-206 overexpression has been shown to promote extracellular matrix deposition and impede malignant transformation of hemangioma endothelial cells, suggesting a potential therapeutic approach (129). Beyond protein-coding genes, miRNAs can interact with long non-coding RNAs (lncRNAs) to regulate hemangioma development. An inverse correlation exists between the expression of lncRNA NEAT1 and miR-361-5p in hemangioma tissues. Knockdown of NEAT1 restrains hemangioma endothelial cell proliferation and migration by sequestering miR-361-5p and modulating VEGFA expression (130). Similarly, miR-125a-3p acts as a negative regulator of hemangioma endothelial cells by interacting with the oncogenic lncRNA CASC9, controlling proliferation, migration, and invasion (131). Signaling pathways crucial for development and differentiation are also miRNA targets in hemangioma. Suppression of miR-203, a regulator of Notch signaling, promotes proliferation and inhibits apoptosis in hemangioma endothelial cells (132). Furthermore, reduced levels of miR-497-5p enhance proliferation and diminish ferroptosis (iron-dependent cell death) through modulation of the Notch2 pathway (133). Therefore, various microRNAs intricately regulate hemangioma progression by interacting with different genes and pathways. MiR-130a, in particular, stands out due to its consistent upregulation across hemangiomas, making it a promising candidate for therapeutic targeting.
4.3. Cardiac myxoma
In cardiac myxoma, miR-217 acts as a tumor suppressor by negatively regulating the oncogenic IL-6 gene. Therapeutic modulation of miR-217 levels may provide a potential avenue for managing this primary heart tumor (134). Table 1 summarizes the mechanisms by which various miRNAs affect distinct types of cardiovascular tumors.
Table 1.
Action mechanism of various miRNAs on distinct types of cardiovascular tumors.
| Cardiovascular system-related tumor | MicroRNA expression status | Influence/mechanism(s) | Model | Ref. |
|---|---|---|---|---|
| Angiosarcoma | miR-541-3p/654-5p/1-3p/128-3p/485-5p/381-3p/370-3p/377-3p/193a-3p/153-3p/504-5p (Up-regulated) | • Integration of expression profiles with miRNA expression data indicated probable important miRNA-gene regulator connections for each sarcoma subtype. • The anticipated targets HMCN1, NKX2-2, SCNN1G, and SOX2 were increased in Ewing’s sarcoma and fibromatosis samples, where miR-182-5p is downregulated. |
ex vivo | (95) |
| miR-515-5p/517a/517c/518b/519a/522 (Overexpressed) | Overall, S-MED serves as a valuable resource for understanding the miRNA expression patterns and potential mechanisms involved in angiosarcoma. | ex vivo | (135) | |
| miR-126 (oncogene), (Up-regulated) | • TP53 loss is selective in pCAS, while myc amplification/overexpression is indicative of sCAS. Both pCAS and sCAS exhibit considerable molecular heterogeneity. • Affected genes and their molecular regulators, including miRNAs, may serve as future drug targets. |
in situ | (18) | |
| miR-452/494-3p/214-3p/543/497-5p (Up-regulated) |
• Serum microRNAs differed between dogs with splenic tumors and healthy dogs with histologically normal spleens. | in vivo | (100) | |
| miR-497 (Tumor suppressor), (Down-regulated) |
• miR-497-5p (miR-497) significantly suppresses cell viability in AS. • miR-497 overexpression leads to reduced cell migration and tumor formation in AS and directly leads to reduced cell migration and tumor formation in AS, and can regulate VAT1, suggesting that VAT1 may promote cell migration in AS • miR-497 mimic transfection increases apoptosis in AS and hemangioendothelioma cell lines. |
in vitro and in vivo | (82) | |
| miR-195/199a/125b (Tumor suppressor), (Down-regulated) miR-222 (Tumor suppressor), (Down-regulated initially, then Up-regulated) |
• Under Vinyl chloride (VC) exposure, hepatocellular carcinoma and angiosarcoma miRNA-222, miRNA-199a, miRNA-195, and miRNA-125b levels varied dynamically. MiRNA-222 first reduced and then grew, while miRNA-199a, 195, and 125b increased and then declined. Cell cycle distribution and expression of cell cycle-related proteins (p27, cyclinA, cyclinD1, and CDK6) changed. • These miRNAs’ serum levels remained unchanged. • Cell cycle-related proteins can be deregulated by VC-induced dynamic changes in miR-222, miR-199a, miR-195, and miR-125b. These miRNAs may be early indicators for VC-induced cancer. |
in vivo | (24) | |
| miR-340 (Tumor suppressor), (Down-regulated) |
• miR-340 expression was considerably lower in angiosarcoma than controls. • Overexpression of miR-340 decreased angiosarcoma cell proliferation and invasion. • Sirtuin 7 (SIRT7) may be a miR-340 target gene. • Overexpression of SIRT7 increased angiosarcoma cell proliferation and invasion and partially reversed miR-340’s anticancer impact. |
in vitro and ex vivo | (21) | |
| miR-210 (Tumor suppressor), (Down-regulated) |
• miR-210 was downregulated in vivo and in vitro in angiosarcoma cells. • E2F3 and ephrin A3 were upregulated in angiosarcoma tumor cells. • Angiosarcoma cell numbers decreased significantly after E2F3 or ephrin A3 knockdown. • Angiosarcoma patients not significantly differed from healthy controls in serum miR-210 levels. |
in vitro and ex vivo | (22) | |
| miR-17-92 (oncogene), (Up-regulated) miR-18a and miR-19a (Down-regulated) |
• The miR-17-92 cluster was significantly elevated in MYC-amplified AS compared to AS without amplification and the control group. • In AS with MYC amplification, miR-18a and miR-19a from the miR-17-92 cluster down-regulated THBS1 and CTGF gene mRNA expression. • By inhibiting clusterin production via the TGF-b signaling pathway, the miR-17-92 cluster promotes angiogenesis. • CCN family member CTGF was downregulated in MYC-amplified AS, suggesting its role in angiogenesis suppression in this sarcoma subtype. |
In vitro and in situ | (103) | |
| miR-34c (Tumor suppressor), (Up-regulated) |
• aPKCλ phosphorylates FoxO1’s DNA binding domain, promoting endothelial cell proliferation and c-Myc expression. • Angiosarcoma cell growth and c-Myc expression decrease with aPKC inhibition. |
In vitro, and in vivo | (54) | |
| miR-23 (Up-regulated) | • In rat and human angiosarcoma, miR-23 target genes (Ccnd1, Adam19, Plau, and Wsb1) that increase invasiveness and metastasis were enriched. | In vitro, in vivo and in situ | (90) | |
| Canine angiosarcoma | miR-214/5AE, (synthetic) (Anti-tumor effects) |
• p53, induces apoptosis, and inhibits cell proliferation | in vivo | (101) |
| Canine hemangiosarcoma | miR-214, (Tumor suppressor) (Downregulated) |
• Through p53-regulated gene transcription, miR-214 restoration decreased cell proliferation and triggered apoptosis in canine hemangiosarcoma cell lines. • HSA cell lines overexpressed COP1, and knocking it down caused apoptosis and upregulated p53-regulated genes. • COP1 knockdown induced apoptosis in HSA cells by up-regulating p53-regulated gene expression, similar to the results obtained using miR-214-transfection. |
in vitro | (83) |
| Cardiac myxoma (CM) | miR-217, (Tumor suppressor) (Down-regulated) |
• miR-217 expression is downregulated in CM tissues and is inversely correlated with IL-6 mRNA expression. • Overexpression of miR-217 reduces primary CM cell growth and induces apoptosis. • miR-217 directly targets IL-6, as shown by dual luciferase experiments. • IL-6 downregulation by siRNA mimics miR-217’s tumor-suppressive effects in CM. • The anti-proliferative and pro-apoptotic impact of miR-217 overexpression in CM cells is reversed by IL-6 restoration. |
in situ | (134) |
| Hemangioma | miR-203, (Tumor suppressor) (Down-regulated) |
• Knockdown of MEG8 upregulates miR-203 expression. • HemECs’ MEG8 knockdown is reversed by MiR-203 silencing. • The MEG8 knockdown reduces JAG1 and Notch1 expression in HemECs. |
in vitro | (132) |
| miR-452 (oncogene) (Up-regulated) |
• Expression higher in serum of dogs with splenic hemangiosarcoma • Increasing EGFR and Akt signaling • EGFR supports anchorage-independent growth and metastasis in canine hemangiosarcoma. |
in vivo | (100) | |
| miR-497-5p, (Tumor suppressor) (Down-regulated) |
• Silencing MEG8 inhibited the proliferation of HemECs, increased the level of MDA in HemECs, decreased the expression of SLC7A11 and GPX4 in HemECs, but no effect on the level of AIFM2. • Blocking miR-497-5p reversed the effects of MEG8 loss on cell viability, MDA level, and expression levels of NOTCH2, SLC7A11, and GPX4 in HemECs. |
in vitro | (133) | |
| miR-361-5p, (Tumor suppressor) (Down-regulated) |
• NEAT1 expression was higher in HA tissues, notably proliferative phase HAs, than in skin. MiR-361-5p expression reduced in HA tissues, especially proliferating HAs. NEAT1 functioned as a competing endogenous RNA to regulate VEGFA expression by sponging miR-361-5p. • Downregulation of NEAT1 dramatically reduced HemEC viability, PCNA expression, and migration while increased apoptosis and caspase-3 activity. • Knockdown of NEAT1 suppressed the proliferation and migration, but facilitated the apoptosis of HemECs. • NEAT1 enhanced the proliferation and migration, and decreased the apoptosis of HemECs via negatively regulating miR-361-5p. |
In vitro | (130) | |
| miR-125-3p, (Tumor suppressor) (Down-regulated) |
• CASC9 knockdown significantly inhibited proliferation, migration, and invasion of HDECs. CASC9 knockdown reduced cyclinD1, N-cadherin, Twist, and MMP2 translation. Also, CASC9 knockdown decreased Twist, MMP2, cyclinD1, and N-cadherin translation. • CASC9 interacts with miR-125a-3p/Nrg1 to regulate cellular functions. miR-125a-3p can reverse the effect of CASC9 on proliferation, migration, and invasion of HDECs. • CASC9 over-expression exerted the opposite effect, promoting proliferation, migration, and invasion of HDECs. • CASC9 over-expression exerted the opposite effect, promoting proliferation, migration, and invasion of HDECs. |
In vitro and in vivo | (131) | |
| Infantile Hemangioma (IH) | miR-382, (Tumor suppressor) (Up-regulated) |
• miR-382 is overexpressed in IH • miR-382 downregulates PTEN • miR-382 activates AKT/mTOR signaling • Promoting IH progression |
In vitro | (128) |
| miR-206, (Tumor suppressor) (Down-regulated) |
• MiR-206 downregulated in proliferative hemangioma suppressed HemEC malignancy via modulating ECM-related genes. • In vitro and in vivo research showed that miR-206 increases ECM accumulation by targeting DNMT3A, preventing HemEC malignancy and alleviating IH. • Overexpression of DNMT3A prevented miR-206 mimic from inhibiting HemECs and regulating ECM. |
In vitro and in vitro | (129) | |
| Rhabdomyosarcoma | miR-206, (Tumor suppressor) (Down-regulated) |
• miR-206 modulates over 700 genes to restore differentiation in RMS cells. • In ERMS cells, silencing G6PD, a miR-206 target, inhibits proliferation and soft agar growth. G6PD overexpression does not affect miR-206’s pro-differentiation impact. • DCA improves miR-206-induced RMS cell growth inhibition and maintains it after de-induction. This shows combining differentiation-inducing and metabolism-directed techniques may be advantageous. |
in vitro | (114, 120) |
| miR-1/-206/-29 (Tumor suppressor) (Down-regulated) |
• Cell cycle gene CCND2 is regulated by miR-1, -206, and -29. • Similar to CCND2, miR-29 targets E2F7, a cell cycle regulator. • Cell function study demonstrates that miR-29 overexpression downregulates several cell cycle genes, promotes partial G1 arrest, and decreases cell proliferation. |
in vitro and ex vivo | (118) | |
| miR-26a (Tumor suppressor) (Down-regulated) |
• This study reveals deregulation of miR-26a and Ezh2 in rhabdomyosarcoma samples and cell lines, suggesting their potential involvement in rhabdomyosarcoma pathogenesis. | in vitro | (113) | |
| miR-1/206 | • miR-1/206 decreased c-Met expression in rhabdomyosarcoma and may be a strong tumor suppressor. • Inhibition of miR-1/206 may cause abnormal cell growth and migration, causing rhabdomyosarcoma. |
in vitro and ex vivo | (136) | |
| miR-29 (Tumor suppressor) (Down-regulated) |
• miR-29a suppressed subcutaneous xenograft carcinogenesis in nude mice and GEFT mRNA and protein expression in transplanted tumors. | in vitro and ex vivo | (126) | |
| miR-26a (Tumor suppressor) (Down-regulated) |
• Plasma levels of muscle-specific miR-206 were substantially higher in RMS patients than healthy donors. • In an independent larger cohort of patients (validation set), digital droplet PCR detected reduced miR-26a and miR-30b/c levels. |
ex vivo | (125) |
AS, Angiosarcoma; CAS, Cutaneous angiosarcoma; CM, Cardiac myxoma; DNMT3A, DNA Methyltransferase 3A; IH, Infantile Hemangioma; MDA, malondialdehyde; pCAS, spontaneous cutaneous angiosarcoma.
5. Future directions and conclusion
The current review highlights the diverse roles of miRNAs in angiosarcoma and other cardiovascular tumors. Despite the promising findings, it is evident that further research is imperative before miRNAs can be seamlessly integrated into clinical practice as diagnostic or prognostic markers or therapeutic targets. Future investigations should extend beyond angiosarcomas to explore miRNA expression and interactions in other cardiovascular tumors that have received less attention. A crucial aspect is understanding how miRNAs contribute to the molecular biology of these tumors and comparing their potential benefits or drawbacks against current diagnostic and prognostic approaches.
As previously discussed, examples such as miR-217 suppressing the carcinogenic IL-6 gene in cardiac myxoma and miR-1 inhibiting c-Met and PAX3 to suppress tumor growth in rhabdomyosarcoma showcase the diverse roles of miRNAs in different tumors. Additionally, miR-340 targeting SIRT7’s 3’-UTR to decrease angiosarcoma cell invasion and proliferation demonstrates the specificity of miRNA actions. Nonetheless, the levels of miR-214 were significantly higher in angiosarcoma tissues compared to non-angiosarcoma tissues. Elucidating the precise mechanisms, interactions, and potential side effects of therapeutic tools targeting miRNAs is essential for translating these findings into clinical applications. This knowledge will provide the foundation for designing randomized control trials and facilitating the clinical application of miRNA-based interventions. Despite the progress made, there remains a significant gap in our understanding before miRNAs can be considered reliable diagnostic or prognostic markers or plausible therapeutic targets in clinical settings.
Overall, this review emphasizes the potential of miRNAs as diagnostic, prognostic, and therapeutic tools in cardiovascular tumors, including angiosarcoma, a challenging soft-tissue sarcoma. Furthermore, it explores the potential contributions of miRNAs to other cardiovascular tumors, proposing their utility as diagnostic, prognostic markers, and therapeutic targets. Although the area is progressing, our comprehension of the complex function of miRNAs in cardiovascular malignancies is still in its nascent phase, requiring further research to reveal their therapeutic usefulness and underlying processes.
Author contributions
AM: Writing – original draft, Validation. AA: Supervision,Writing – original draft. NE: Writing – original draft. SR: Writing – original draft. NR: Writing – original draft. SD: Writing – original draft, Visualization. FF: Supervision, Writing – original draft. ZN: Writing – original draft. RA: Writing – review & editing. HM: Supervision, Writing – review & editing.
Acknowledgments
Figures was created with “BioRender.com”
Funding Statement
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Abbreviations
miR, Micro Ribonucleic acid; STS, Stewart-Treves syndrome; VCM, vinyl chloride monomer; FISH, fluorescent in situ hybridization; aPKCλ, atypical Protein Kinase C lambda/iota; FoxO1, Forkhead box protein O1; PD-L1, Programmed death-ligand 1; VEGF, vascular endothelial growth factor; MDM2, Mouse double minute 2 homolog; MAPK, Mitogen-activated protein kinase; CDKN2A, Cyclin-dependent kinase inhibitor 2A; THBS1, thrombospondin 1; VE-PTP, vascular endothelial protein tyrosine phosphatase; FLT4, fms-related receptor tyrosine kinase 4; NEAT1, Nuclear Enriched Abundant Transcript 1; lncRNA, Long non-coding RNA.
Conflict of interest
Author AM was employed by Kimia Andisheh Teb Medical and Molecular Research Laboratory Co.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1. Motedayyen H, Ghotloo S, Saffari M, Sattari M, Amid R. Evaluation of microRNA-146a and its targets in gingival tissues of patients with chronic periodontitis. J periodontol. (2015) 86:1380–5. doi: 10.1902/jop.2015.150319 [DOI] [PubMed] [Google Scholar]
- 2. Yang F, Ning Z, Ma L, Liu W, Shao C, Shu Y, et al. Exosomal miRNAs and miRNA dysregulation in cancer-associated fibroblasts. Mol Cancer. (2017) 16:148. doi: 10.1186/s12943-017-0718-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Turk A, Calin GA, Kunej T. MicroRNAs in leukemias: A clinically annotated compendium. Int J Mol Sci. (2022) 23:1–16. doi: 10.3390/ijms23073469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goleij P, Babamohamadi M, Rezaee A, Sanaye PM, Tabari MAK, Sadreddini S, et al. Types of RNA therapeutics. Prog Mol Biol Trans Sci. (2024) 203:41–63. doi: 10.1016/bs.pmbts.2023.12.022 [DOI] [PubMed] [Google Scholar]
- 5. Smolarz B, Durczynski A, Romanowicz H, Szyllo K, Hogendorf P. miRNAs in cancer (Review of literature). Int J Mol Sci. (2022) 23:1–18. doi: 10.3390/ijms23052805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ghotloo S, Motedayyen H, Amani D, Saffari M, Sattari M. Assessment of micro RNA-146a in generalized aggressive periodontitis and its association with disease severity. J periodontal Res. (2019) 54:27–32. doi: 10.1111/jre.12538 [DOI] [PubMed] [Google Scholar]
- 7. Han GS, Gao Q, Peng LZ, Tang J. Hessian Regularized [Formula: see text]-Nonnegative Matrix Factorization and Deep Learning for miRNA-Disease Associations Prediction. Interdiscip Sci. (2024) 16(1):176–91. doi: 10.1007/s12539-023-00594-8 [DOI] [PubMed] [Google Scholar]
- 8. Goleij P, Sanaye PM, Rezaee A, Tabari MAK, Arefnezhad R, Motedayyen H. RNA therapeutics for kidney injury. (2024) 204:69–95. doi: 10.1016/bs.pmbts.2023.12.007 [DOI] [PubMed] [Google Scholar]
- 9. Liu B, Li J, Cairns MJ. Identifying miRNAs, targets and functions. Brief Bioinform. (2014) 15:1–19. doi: 10.1093/bib/bbs075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sattari M, Taheri RA, Arefnezhad R, Motedayyen H. The expression levels of MicroRNA-146a, RANKL and OPG after non-surgical periodontal treatment. BMC Oral Health. (2021) 21:1–7. doi: 10.1186/s12903-021-01883-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Varrone F, Caputo E. The miRNAs role in melanoma and in its resistance to therapy. Int J Mol Sci. (2020) 21:1–28. doi: 10.3390/ijms21030878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Arefnezhad R, Roghani-Shahraki H, Motedayyen H, Rezaei-Tazangi F. The function of microRNAs in the normal and abnormal ovarian activities: a focus on microRNA-21. Int J Fertility Sterility. (2024) 18(2). doi: 10.22074/IJFS.2023.1985792.1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mazziotta C, Badiale G, Cervellera CF, Tognon M, Martini F, Rotondo JC. Regulatory mechanisms of circular RNAs during human mesenchymal stem cell osteogenic differentiation. Theranostics. (2024) 14:143–58. doi: 10.7150/thno.89066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Palanichamy JK, Rao DS. miRNA dysregulation in cancer: towards a mechanistic understanding. Front Genet. (2014) 5:54. doi: 10.3389/fgene.2014.00054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. (2016) 1:15004. doi: 10.1038/sigtrans.2015.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gaballah AH, Jensen CT, Palmquist S, Pickhardt PJ, Duran A, Broering G, et al. Angiosarcoma: clinical and imaging features from head to toe. Br J Radiol. (2017) 90:20170039. doi: 10.1259/bjr.20170039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tambe SA, Nayak CS. Metastatic angiosarcoma of lower extremity. Indian Dermatol Online J. (2018) 9:177–81. doi: 10.4103/idoj.IDOJ_92_17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goerdt LV, Schneider SW, Booken N. Cutaneous angiosarcomas: molecular pathogenesis guides novel therapeutic approaches. J Dtsch Dermatol Ges. (2022) 20:429–43. doi: 10.1111/ddg.14694 [DOI] [PubMed] [Google Scholar]
- 19. García-Heredia JM, Pérez M, Verdugo-Sivianes EM, Martínez-Ballesteros MM, Ortega-Campos SM, Carnero A. A new treatment for sarcoma extracted from combination of miRNA deregulation and gene association rules. Signal Transduction Targeted Ther. (2023) 8:231. doi: 10.1038/s41392-023-01470-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Krishnan T, Pettersson G, Mukherjee R, Singhal N. Cardiac angiosarcoma: A diagnostic and therapeutic challenge. J Cardiol cases. (2020) 22:90–3. doi: 10.1016/j.jccase.2020.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang X, Song Y. MicroRNA-340 inhibits the growth and invasion of angiosarcoma cells by targeting SIRT7. Biomed Pharmacother. (2018) 103:1061–8. doi: 10.1016/j.biopha.2018.04.148 [DOI] [PubMed] [Google Scholar]
- 22. Nakashima S, Jinnin M, Kanemaru H, Kajihara I, Igata T, Okamoto S, et al. The role of miR-210, E2F3 and ephrin A3 in angiosarcoma cell proliferation. Eur J Dermatol. (2017) 27:464–71. doi: 10.1684/ejd.2017.3084 [DOI] [PubMed] [Google Scholar]
- 23. Zhang Z, Tang Y, Song X, Xie L, Zhao S, Song X. Tumor-derived exosomal miRNAs as diagnostic biomarkers in non-small cell lung cancer. Front Oncol. (2020) 10:560025. doi: 10.3389/fonc.2020.560025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pan W, Yu S, Jia J, Hu J, Jie L, Zhang P, et al. Deregulation of the cell cycle and related microRNA expression induced by vinyl chloride monomer in the hepatocytes of rats. Toxicol Ind Health. (2021) 37:365–76. doi: 10.1177/07482337211015591 [DOI] [PubMed] [Google Scholar]
- 25. Cao J, Wang J, He C, Fang M. Angiosarcoma: a review of diagnosis and current treatment. Am J Cancer Res. (2019) 9:2303–13. [PMC free article] [PubMed] [Google Scholar]
- 26. Batouli A, Fairbrother SW, Silverman JF, Muniz Mde L, Taylor KB, Welnick MA, et al. Primary splenic angiosarcoma: clinical and imaging manifestations of this rare aggressive neoplasm. Curr Probl Diagn Radiol. (2016) 45:284–7. doi: 10.1067/j.cpradiol.2015.07.004 [DOI] [PubMed] [Google Scholar]
- 27. Yan Q, Fernandez RA, Elmi M, Gelfond J, Davies MG. Outcomes of interventions for angiosarcoma. Front Surg. (2022) 9. doi: 10.3389/fsurg.2022.819099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Buehler D, Rice SR, Moody JS, Rush P, Hafez GR, Attia S, et al. Angiosarcoma outcomes and prognostic factors: a 25-year single institution experience. Am J Clin Oncol. (2014) 37:473–9. doi: 10.1097/COC.0b013e31827e4e7b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Simonin-Wilmer I, Ossio R, Leddin EM, Harland M, Pooley KA, Martil de la Garza MG, et al. Population-based analysis of POT1 variants in a cutaneous melanoma case-control cohort. J Med Genet. (2023) 60:692–6. doi: 10.1136/jmg-2022-108776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Florou V, Wilky BA. Current management of angiosarcoma: recent advances and lessons from the past. Curr Treat Options Oncol. (2021) 22:61. doi: 10.1007/s11864-021-00858-9 [DOI] [PubMed] [Google Scholar]
- 31. Shon W, Billings SD. Cutaneous Malignant vascular neoplasms. Clin Lab Med. (2017) 37:633–46. doi: 10.1016/j.cll.2017.06.004 [DOI] [PubMed] [Google Scholar]
- 32. Florou V, Wilky BA. Current and future directions for angiosarcoma therapy. Curr Treat Options Oncol. (2018) 19:14. doi: 10.1007/s11864-018-0531-3 [DOI] [PubMed] [Google Scholar]
- 33. Ni Q, Shang D, Peng H, Roy M, Liang G, Bi W, et al. Primary angiosarcoma of the small intestine with metastasis to the liver: a case report and review of the literature. World J Surg Oncol. (2013) 11:242. doi: 10.1186/1477-7819-11-242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yin M, Wang W, Drabick JJ, Harold HA. Prognosis and treatment of non-metastatic primary and secondary breast angiosarcoma: a comparative study. BMC Cancer. (2017) 17:295. doi: 10.1186/s12885-017-3292-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Verdura V, Di Pace B, Concilio M, Guastafierro A, Fiorillo G, Alfano L, et al. A new case of radiation-induced breast angiosarcoma. Int J Surg Case Rep. (2019) 60:152–5. doi: 10.1016/j.ijscr.2019.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Virtanen A, Pukkala E, Auvinen A. Angiosarcoma after radiotherapy: a cohort study of 332,163 Finnish cancer patients. Br J Cancer. (2007) 97:115–7. doi: 10.1038/sj.bjc.6603805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Disharoon M, Kozlowski KF, Kaniowski JM. Case 242: radiation-induced angiosarcoma. Radiology. (2017) 283:909–16. doi: 10.1148/radiol.2017150456 [DOI] [PubMed] [Google Scholar]
- 38. Plichta JK, Hughes K. Radiation-induced angiosarcoma after breast-cancer treatment. N Engl J Med. (2017) 376:367. doi: 10.1056/NEJMicm1516482 [DOI] [PubMed] [Google Scholar]
- 39. Alves I, Marques JC. Radiation-induced angiosarcoma of the breast: a retrospective analysis of 15 years’ experience at an oncology center. Radiologia Bras. (2018) 51:281–6. doi: 10.1590/0100-3984.2017.0129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Seo CJ, Lek SM, Tan GHC, Teo M. Radiation-associated peritoneal angiosarcoma. BMJ Case Rep. (2017) 2017:bcr–2016-217887. doi: 10.1136/bcr-2016-217887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pereira ES, Moraes ET, Siqueira DM, Santos MA. Stewart treves syndrome. Bras Dermatol. (2015) 90:229–31. doi: 10.1590/abd1806-4841.20153685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Peterson CB, Beauregard S. Radiation-induced breast angiosarcoma: case report and clinical approach. J Cutan Med Surg. (2016) 20:304–7. doi: 10.1177/1203475416631525 [DOI] [PubMed] [Google Scholar]
- 43. Cui L, Zhang J, Zhang X, Chang H, Qu C, Zhang J, et al. Angiosarcoma (Stewart-Treves syndrome) in postmastectomy patients: report of 10 cases and review of literature. Int J Clin Exp Pathol. (2015) 8:11108–15. [PMC free article] [PubMed] [Google Scholar]
- 44. Mesli SN, Ghouali AK, Benamara F, Taleb FA, Tahraoui H, Abi-Ayad C. Stewart-treves syndrome involving chronic lymphedema after mastectomy of breast cancer. Case Rep Surg. (2017) 2017:4056459. doi: 10.1155/2017/4056459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Berebichez-Fridman R, Deutsch YE, Joyal TM, Olvera PM, Benedetto PW, Rosenberg AE, et al. Stewart-treves syndrome: A case report and review of the literature. Case Rep Oncol. (2016) 9:205–11. doi: 10.1159/000445427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Benmansour A, Laanaz S, Bougtab A. Stewart-Treves syndrome: a case report. Pan Afr Med J. (2014) 19:2. doi: 10.11604/pamj.2014.19.2.4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tripke V, Heinrich S, Huber T, Mittler J, Hoppe-Lotichius M, Straub BK, et al. Surgical therapy of primary hepatic angiosarcoma. BMC Surg. (2019) 19:5. doi: 10.1186/s12893-018-0465-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chaudhary P, Bhadana U, Singh RA, Ahuja A. Primary hepatic angiosarcoma. Eur J Surg Oncol. (2015) 41:1137–43. doi: 10.1016/j.ejso.2015.04.022 [DOI] [PubMed] [Google Scholar]
- 49. Choi JH, Ahn KC, Chang H, Minn KW, Jin US, Kim BJ. Surgical treatment and prognosis of angiosarcoma of the scalp: A retrospective analysis of 14 patients in a single institution. BioMed Res Int. (2015) 2015:321896. doi: 10.1155/2015/321896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Berry SA, Peterson C, Mize W, Bloom K, Zachary C, Blasco P, et al. Klippel-trenaunay syndrome. Am J Med Genet. (1998) 79:319–26. doi: 10.1002/(ISSN)1096-8628 [DOI] [PubMed] [Google Scholar]
- 51. Mullin C, Clifford CA. Histiocytic sarcoma and hemangiosarcoma update. Vet Clin North Am Small Anim Pract. (2019) 49:855–79. doi: 10.1016/j.cvsm.2019.04.009 [DOI] [PubMed] [Google Scholar]
- 52. Manner J, Radlwimmer B, Hohenberger P, Mössinger K, Küffer S, Sauer C, et al. MYC high level gene amplification is a distinctive feature of angiosarcomas after irradiation or chronic lymphedema. Am J Pathol. (2010) 176:34–9. doi: 10.2353/ajpath.2010.090637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Motaparthi K, Lauer SR, Patel RM, Vidal CI, Linos K. MYC gene amplification by fluorescence in situ hybridization and MYC protein expression by immunohistochemistry in the diagnosis of cutaneous angiosarcoma: Systematic review and appropriate use criteria. J Cutan Pathol. (2021) 48:578–86. doi: 10.1111/cup.13912 [DOI] [PubMed] [Google Scholar]
- 54. Riddell M, Nakayama A, Hikita T, Mirzapourshafiyi F, Kawamura T, Pasha A, et al. aPKC controls endothelial growth by modulating c-Myc via FoxO1 DNA-binding ability. Nat Commun. (2018) 9:5357. doi: 10.1038/s41467-018-07739-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kawamura A, Kawamura T, Riddell M, Hikita T, Yanagi T, Umemura H, et al. Regulation of programmed cell death ligand 1 expression by atypical protein kinase C lambda/iota in cutaneous angiosarcoma. Cancer Sci. (2019) 110:1780–9. doi: 10.1111/cas.13981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Murali R, Chandramohan R, Moller I, Scholz SL, Berger M, Huberman K, et al. Targeted massively parallel sequencing of angiosarcomas reveals frequent activation of the mitogen activated protein kinase pathway. Oncotarget. (2015) 6:36041–52. doi: 10.18632/oncotarget.v6i34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ronchi A, Cozzolino I, Zito Marino F, De Chiara A, Argenziano G, Moscarella E, et al. Primary and secondary cutaneous angiosarcoma: Distinctive clinical, pathological and molecular features. Ann Diagn Pathol. (2020) 48:151597. doi: 10.1016/j.anndiagpath.2020.151597 [DOI] [PubMed] [Google Scholar]
- 58. Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. (2009) 137:413–31. doi: 10.1016/j.cell.2009.04.037 [DOI] [PubMed] [Google Scholar]
- 59. Naka N, Tomita Y, Nakanishi H, Araki N, Hongyo T, Ochi T, et al. Mutations of p53 tumor-suppressor gene in angiosarcoma. Int J Cancer. (1997) 71:952–5. doi: [DOI] [PubMed] [Google Scholar]
- 60. Zietz C, Rössle M, Haas C, Sendelhofert A, Hirschmann A, Stürzl M, et al. MDM-2 oncoprotein overexpression, p53 gene mutation, and VEGF up-regulation in angiosarcomas. Am J Pathol. (1998) 153:1425–33. doi: 10.1016/S0002-9440(10)65729-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhou S, Gu L, He J, Zhang H, Zhou M. MDM2 regulates vascular endothelial growth factor mRNA stabilization in hypoxia. Mol Cell Biol. (2011) 31:4928–37. doi: 10.1128/MCB.06085-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kiyohara M, Aoi J, Kajihara I, Otuka S, Kadomatsu T, Fukushima S, et al. Serum anti-p53 autoantibodies in angiosarcoma. J Dermatol. (2020) 47:849–54. doi: 10.1111/1346-8138.15416 [DOI] [PubMed] [Google Scholar]
- 63. Dai J, Kunder CA, Chu EY, Chan EF, Egan CL, Novoa RA. Development of RET mutant cutaneous angiosarcoma during BRAF inhibitor therapy. J Cutan Pathol. (2017) 44:1053–6. doi: 10.1111/cup.13024 [DOI] [PubMed] [Google Scholar]
- 64. Weidema ME, Van De Geer E, Koelsche C, Desar IME, Kemmeren P, Hillebrandt-Roeffen MHS, et al. DNA methylation profiling identifies distinct clusters in angiosarcomas. Clin Cancer Res. (2020) 26:93–100. doi: 10.1158/1078-0432.CCR-19-2180 [DOI] [PubMed] [Google Scholar]
- 65. Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. (2010) 70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2 [DOI] [PubMed] [Google Scholar]
- 66. Weihrauch M, Markwarth A, Lehnert G, Wittekind C, Wrbitzky R, Tannapfel A. Abnormalities of the ARF-p53 pathway in primary angiosarcomas of the liver. Hum Pathol. (2002) 33:884–92. doi: 10.1053/hupa.2002.126880 [DOI] [PubMed] [Google Scholar]
- 67. Mentzel T, Kiss K. Reduced H3K27me3 expression in radiation-associated angiosarcoma of the breast. Virchows Arch. (2018) 472:361–8. doi: 10.1007/s00428-017-2242-8 [DOI] [PubMed] [Google Scholar]
- 68. Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol: Mech Dis. (2010) 5:253–95. doi: 10.1146/annurev.pathol.4.110807.092250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Toiber D, Sebastian C, Mostoslavsky R. Characterization of nuclear sirtuins: molecular mechanisms and physiological relevance. Histone Deacetylases: Biol Clin Implication. (2011) 206:189–224. [DOI] [PubMed] [Google Scholar]
- 70. Chan YC, Banerjee J, Choi SY, Sen CK. miR-210: The master hypoxamir. Microcirculation. (2012) 19:215–23. doi: 10.1111/j.1549-8719.2011.00154.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Itakura E, Yamamoto H, Oda Y, Tsuneyoshi M. Detection and characterization of vascular endothelial growth factors and their receptors in a series of angiosarcomas. J Surg Oncol. (2008) 97:74–81. doi: 10.1002/jso.20766 [DOI] [PubMed] [Google Scholar]
- 72. Hasenstein JR, Kasmerchak K, Buehler D, Hafez GR, Cleary K, Moody JS, et al. Efficacy of Tie2 receptor antagonism in angiosarcoma. Neoplasia. (2012) 14:131–40. doi: 10.1593/neo.111770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Antonescu C. Malignant vascular tumors–an update. Mod Pathol. (2014) 27 Suppl 1:S30–38. doi: 10.1038/modpathol.2013.176 [DOI] [PubMed] [Google Scholar]
- 74. Cornejo KM, Deng A, Wu H, Cosar EF, Khan A, St Cyr M, et al. The utility of MYC and FLT4 in the diagnosis and treatment of postradiation atypical vascular lesion and angiosarcoma of the breast. Hum Pathol. (2015) 46:868–75. doi: 10.1016/j.humpath.2015.02.014 [DOI] [PubMed] [Google Scholar]
- 75. Panse G, Chrisinger JS, Leung CH, Ingram DR, Khan S, Wani K, et al. Clinicopathological analysis of ATRX, DAXX and NOTCH receptor expression in angiosarcomas. Histopathology. (2018) 72:239–47. doi: 10.1111/his.13337 [DOI] [PubMed] [Google Scholar]
- 76. Dill MT, Rothweiler S, Djonov V, Hlushchuk R, Tornillo L, Terracciano L, et al. Disruption of Notch1 induces vascular remodeling, intussusceptive angiogenesis, and angiosarcomas in livers of mice. Gastroenterology. (2012) 142:967–977 e962. doi: 10.1053/j.gastro.2011.12.052 [DOI] [PubMed] [Google Scholar]
- 77. Liu J, Yang T, Huang Z, Chen H, Bai Y. Transcriptional regulation of nuclear miRNAs in tumorigenesis (Review). Int J Mol Med. (2022) 50:235–45. doi: 10.3892/ijmm [DOI] [PubMed] [Google Scholar]
- 78. Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol. (2014) 11:145–56. doi: 10.1038/nrclinonc.2014.5 [DOI] [PubMed] [Google Scholar]
- 79. Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci. (2004) 101:2999–3004. doi: 10.1073/pnas.0307323101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. (2004) 116:281–97. doi: 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 81. Chen Y, Kuang D, Zhao X, Chen D, Wang X, Yang Q, et al. miR-497-5p inhibits cell proliferation and invasion by targeting KCa3.1 in angiosarcoma. Oncotarget. (2016) 7:58148–61. doi: 10.18632/oncotarget.v7i36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Benton A, Terwilliger E, Moriarty NM, Liu B, Murphy A, Maluvac H, et al. Target gene regulatory network of miR-497 in angiosarcoma. bioRxiv. (2023) 4:1–33. doi: 10.1101/2023.09.24.559218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Heishima K, Mori T, Sakai H, Sugito N, Murakami M, Yamada N, et al. MicroRNA-214 promotes apoptosis in canine hemangiosarcoma by targeting the COP1-p53 axis. PloS One. (2015) 10:e0137361. doi: 10.1371/journal.pone.0137361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Teo AYT, Lim VY, Yang VS. MicroRNAs in the pathogenesis, prognostication and prediction of treatment resistance in soft tissue sarcomas. Cancers. (2023) 15:577. doi: 10.3390/cancers15030577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discovery. (2014) 13:622–38. doi: 10.1038/nrd4359 [DOI] [PubMed] [Google Scholar]
- 86. Rupaimoole R, Calin GA, Lopez-Berestein G, Sood AK. miRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discovery. (2016) 6:235–46. doi: 10.1158/2159-8290.CD-15-0893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discovery. (2017) 16:203–22. doi: 10.1038/nrd.2016.246 [DOI] [PubMed] [Google Scholar]
- 88. Foulkes WD, Priest JR, Duchaine TF. DICER1: mutations, microRNAs and mechanisms. Nat Rev Cancer. (2014) 14:662–72. doi: 10.1038/nrc3802 [DOI] [PubMed] [Google Scholar]
- 89. Loh JW, Lee JY, Lim AH, Guan P, Lim BY, Kannan B, et al. Spatial transcriptomics reveal topological immune landscapes of Asian head and neck angiosarcoma. Commun Biol. (2023) 6:461. doi: 10.1038/s42003-023-04856-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hanna JA, Drummond CJ, Garcia MR, Go JC, Finkelstein D, Rehg JE, et al. Biallelic Dicer1 loss mediated by aP2-Cre drives angiosarcoma. Cancer Res. (2017) 77:6109–18. doi: 10.1158/0008-5472.CAN-17-1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hanna JA, Langdon CG, Garcia MR, Benton A, Lanman NA, Finkelstein D, et al. Genetic context of oncogenic drivers dictates vascular sarcoma development in aP2-Cre mice. J Pathol. (2022) 257:109–24. doi: 10.1002/path.5873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Hata A, Lieberman J. Dysregulation of microRNA biogenesis and gene silencing in cancer. Sci Signal. (2015) 8:re3. doi: 10.1126/scisignal.2005825 [DOI] [PubMed] [Google Scholar]
- 93. Lim HJ, Yang JL. Regulatory roles and therapeutic potential of microRNA in sarcoma. Crit Rev Oncol Hematol. (2016) 97:118–30. doi: 10.1016/j.critrevonc.2015.08.001 [DOI] [PubMed] [Google Scholar]
- 94. Pichler M, Calin GA. MicroRNAs in cancer: from developmental genes in worms to their clinical application in patients. Br J Cancer. (2015) 113:569–73. doi: 10.1038/bjc.2015.253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Sarver AE, Sarver AL, Thayanithy V, Subramanian S. Identification, by systematic RNA sequencing, of novel candidate biomarkers and therapeutic targets in human soft tissue tumors. Lab Invest. (2015) 95:1077–88. doi: 10.1038/labinvest.2015.80 [DOI] [PubMed] [Google Scholar]
- 96. Subramanian S, Kartha RV. MicroRNA-mediated gene regulations in human sarcomas. Cell Mol Life Sci. (2012) 69:3571–85. doi: 10.1007/s00018-012-1127-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Liao GB, Li XZ, Zeng S, Liu C, Yang SM, Yang L, et al. Regulation of the master regulator FOXM1 in cancer. Cell Commun Signal. (2018) 16:57. doi: 10.1186/s12964-018-0266-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Heishima K, Mori T, Ichikawa Y, Sakai H, Kuranaga Y, Nakagawa T, et al. MicroRNA-214 and microRNA-126 are potential biomarkers for Malignant endothelial proliferative diseases. Int J Mol Sci. (2015) 16:25377–91. doi: 10.3390/ijms161025377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Heishima K, Ichikawa Y, Yoshida K, Iwasaki R, Sakai H, Nakagawa T, et al. Circulating microRNA-214 and -126 as potential biomarkers for canine neoplastic disease. Sci Rep. (2017) 7:2301. doi: 10.1038/s41598-017-02607-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Grimes JA, Robinson KR, Bullington A-CM, Schmiedt JM. Identification of serum microRNAs with differential expression between dogs with splenic masses and healthy dogs with histologically normal spleens. Am J Veterinary Res. (2021) 82:659–66. doi: 10.2460/ajvr.82.8.659 [DOI] [PubMed] [Google Scholar]
- 101. Yoshikawa R, Maeda A, Ueno Y, Sakai H, Kimura S, Sawadaishi T, et al. Intraperitoneal administration of synthetic microRNA-214 elicits tumor suppression in an intraperitoneal dissemination mouse model of canine hemangiosarcoma. Veterinary Res Commun. (2022) 46:447–57. doi: 10.1007/s11259-021-09869-1 [DOI] [PubMed] [Google Scholar]
- 102. Khalilian S, Bijanvand A, Abedinlou H, Ghafouri-Fard S. A review on the role of miR-210 in human disorders. Pathol Res Pract. (2023) 241:154244. doi: 10.1016/j.prp.2022.154244 [DOI] [PubMed] [Google Scholar]
- 103. Italiano A, Thomas R, Breen M, Zhang L, Crago AM, Singer S, et al. The miR-17-92 cluster and its target THBS1 are differentially expressed in angiosarcomas dependent on MYC amplification. Genes Chromosomes Cancer. (2012) 51:569–78. doi: 10.1002/gcc.21943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Zhang D, Nie Q, Liu Y, Li Z, Xie W, Jiang L. Mir-485-5p on the morphology and function of cardiovascular system. Cell Mol Biol (Noisy-le-grand). (2022) 68:117–23. doi: 10.14715/cmb/2022.68.1.15 [DOI] [PubMed] [Google Scholar]
- 105. Li Z, Gao J, Sun D, Jiao Q, Ma J, Cui W, et al. LncRNA MEG3: Potential stock for precision treatment of cardiovascular diseases. Front Pharmacol. (2022) 13:1045501. doi: 10.3389/fphar.2022.1045501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Su M, Xiao Y, Ma J, Tang Y, Tian B, Zhang Y, et al. Circular RNAs in Cancer: emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol Cancer. (2019) 18:90. doi: 10.1186/s12943-019-1002-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Wang L, Meng X, Li G, Zhou Q, Xiao J. Circular RNAs in cardiovascular diseases. Adv Exp Med Biol. (2018) 1087:191–204. [DOI] [PubMed] [Google Scholar]
- 108. Nakashima S, Jinnin M, Ide M, Kajihara I, Igata T, Harada M, et al. A potential significance of circ_0024169 down regulation in angiosarcoma tissue. Intractable Rare Dis Res. (2019) 8:129–33. doi: 10.5582/irdr.2019.01034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Wang Z, He Z, Xuan Q, Zhang Y, Xu J, Lin J, et al. Analysis of the potential ferroptosis mechanism and multitemporal expression change of central ferroptosis-related genes in cardiac ischemia-reperfusion injury. Front Physiol. (2022) 13:934901. doi: 10.3389/fphys.2022.934901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Kearney RA, Eisen HJ, Wolf JE. Nonvalvular infections of the cardiovascular system. Ann Intern Med. (1994) 121:219–30. doi: 10.7326/0003-4819-121-3-199408010-00010 [DOI] [PubMed] [Google Scholar]
- 111. Zhang X, Li MJ, Xia L, Zhang H. The biological function of m6A methyltransferase KIAA1429 and its role in human disease. PeerJ. (2022) 10:e14334. doi: 10.7717/peerj.14334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Wang H, Garzon R, Sun H, Ladner KJ, Singh R, Dahlman J, et al. NF-κB–YY1–miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. (2008) 14:369–81. doi: 10.1016/j.ccr.2008.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Ciarapica R, Russo G, Verginelli F, Raimondi L, DonFrancesco A, Rota R, et al. Deregulated expression of miR-26a and Ezh2 in rhabdomyosarcoma. Cell Cycle. (2009) 8:172–5. doi: 10.4161/cc.8.1.7292 [DOI] [PubMed] [Google Scholar]
- 114. Taulli R, Bersani F, Foglizzo V, Linari A, Vigna E, Ladanyi M, et al. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J Clin Invest. (2009) 119:2366–78. doi: 10.1172/JCI38075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Yan D, Dong XD, Chen X, Wang L, Lu C, Wang J, et al. MicroRNA-1/206 targets c-met and inhibits rhabdomyosarcoma development*. J Biol Chem. (2009) 284:29596–604. doi: 10.1074/jbc.M109.020511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Missiaglia E, Shepherd C, Patel S, Thway K, Pierron G, Pritchard-Jones K, et al. MicroRNA-206 expression levels correlate with clinical behaviour of rhabdomyosarcomas. Br J Cancer. (2010) 102:1769–77. doi: 10.1038/sj.bjc.6605684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Rao PK, Missiaglia E, Shields L, Hyde G, Yuan B, Shepherd CJ, et al. Distinct roles for miR-1 and miR-133a in the proliferation and differentiation of rhabdomyosarcoma cells. FASEB J. (2010) 24:3427. doi: 10.1096/fj.09-150698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Li L, Sarver AL, Alamgir S, Subramanian S. Downregulation of microRNAs miR-1,-206 and-29 stabilizes PAX3 and CCND2 expression in rhabdomyosarcoma. Lab Invest. (2012) 92:571–83. doi: 10.1038/labinvest.2012.10 [DOI] [PubMed] [Google Scholar]
- 119. MacQuarrie KL, Yao Z, Young JM, Cao Y, Tapscott SJ. miR-206 integrates multiple components of differentiation pathways to control the transition from growth to differentiation in rhabdomyosarcoma cells. Skeletal Muscle. (2012) 2:1–14. doi: 10.1186/2044-5040-2-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Coda DM, Lingua MF, Morena D, Foglizzo V, Bersani F, Ala U, et al. SMYD1 and G6PD modulation are critical events for miR-206-mediated differentiation of rhabdomyosarcoma. Cell Cycle. (2015) 14:1389–402. doi: 10.1080/15384101.2015.1005993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Ciesla M, Marona P, Kozakowska M, Jez M, Seczynska M, Loboda A, et al. Heme oxygenase-1 controls an HDAC4-miR-206 pathway of oxidative stress in rhabdomyosarcoma. Cancer Res. (2016) 76:5707–18. doi: 10.1158/0008-5472.CAN-15-1883 [DOI] [PubMed] [Google Scholar]
- 122. Hanna J, Garcia M, Go J, Finkelstein D, Kodali K, Pagala V, et al. PAX7 is a required target for microRNA-206-induced differentiation of fusion-negative rhabdomyosarcoma. Cell Death Dis. (2016) 7:e2256–6. doi: 10.1038/cddis.2016.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Chen J, Zhang K, Xu Y, Gao Y, Li C, Wang R, et al. The role of microRNA-26a in human cancer progression and clinical application. Tumor Biol. (2016) 37:7095–108. doi: 10.1007/s13277-016-5017-y [DOI] [PubMed] [Google Scholar]
- 124. Sugito N, Taniguchi K, Kuranaga Y, Ohishi M, Soga T, Ito Y, et al. Cancer-specific energy metabolism in rhabdomyosarcoma cells is regulated by microRNA. Nucleic Acid Ther. (2017) 27:365–77. doi: 10.1089/nat.2017.0673 [DOI] [PubMed] [Google Scholar]
- 125. Tombolan L, Millino C, Pacchioni B, Cattelan M, Zin A, Bonvini P, et al. Circulating miR-26a as potential prognostic biomarkers in pediatric rhabdomyosarcoma. Front Genet. (2020) 11:606274. doi: 10.3389/fgene.2020.606274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Wang Y, Zhang L, Pang Y, Song L, Shang H, Li Z, et al. MicroRNA-29 family inhibits rhabdomyosarcoma formation and progression by regulating GEFT function. Am J Transl Res. (2020) 12:1136–54. [PMC free article] [PubMed] [Google Scholar]
- 127. Gao F, Wang F-G, Liu R-R, Xue F, Zhang J, Xu G-Q, et al. Epigenetic silencing of miR-130a ameliorates hemangioma by targeting tissue factor pathway inhibitor 2 through FAK/PI3K/Rac1/mdm2 signaling. Int J Oncol. (2017) 50:1821–31. doi: 10.3892/ijo.2017.3943 [DOI] [PubMed] [Google Scholar]
- 128. de Falco B, Incerti G, Bochicchio R, Phillips TD, Amato M, Lanzotti V. Metabolomic analysis of Salvia hispanica seeds using NMR spectroscopy and multivariate data analysis. Ind Crops Products. (2017) 99:86–96. doi: 10.1016/j.indcrop.2017.01.019 [DOI] [Google Scholar]
- 129. Wu M, Chen Y, Feng L, Dai H, Fang S, Xu J. MiR-206 promotes extracellular matrix accumulation and relieves infantile hemangioma through targeted inhibition of DNMT3A. Cell Cycle. (2021) 20:978–92. doi: 10.1080/15384101.2021.1919820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Yu X, Liu X, Wang R, Wang L. Long non-coding RNA NEAT1 promotes the progression of hemangioma via the miR-361-5p/VEGFA pathway. Biochem Biophys Res Commun. (2019) 512:825–31. doi: 10.1016/j.bbrc.2019.03.084 [DOI] [PubMed] [Google Scholar]
- 131. Li X, Chen B, Chi D, Zhang Y, Jiang W. lncRNA CASC9 regulates cell migration and invasion in hemangioma endothelial cells by targeting miR-125a-3p/Nrg1. Onco Targets Ther. (2019) 12:423–32. doi: 10.2147/OTT [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Hu Z, Liu X, Guo J, Zhuo L, Chen Y, Yuan H. Knockdown of lncRNA MEG8 inhibits cell proliferation and invasion, but promotes cell apoptosis in hemangioma, via miR−203−induced mediation of the Notch signaling pathway. Mol Med Rep. (2021) 24:1–8. doi: 10.3892/mmr [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Ma Q, Dai X, Lu W, Qu X, Liu N, Zhu C. Silencing long non-coding RNA MEG8 inhibits the proliferation and induces the ferroptosis of hemangioma endothelial cells by regulating miR-497-5p/NOTCH2 axis. Biochem Biophys Res Commun. (2021) 556:72–8. doi: 10.1016/j.bbrc.2021.03.132 [DOI] [PubMed] [Google Scholar]
- 134. Zhang J, Wang C, Xu H. miR-217 suppresses proliferation and promotes apoptosis in cardiac myxoma by targeting Interleukin-6. Biochem Biophys Res Commun. (2017) 490:713–8. doi: 10.1016/j.bbrc.2017.06.106 [DOI] [PubMed] [Google Scholar]
- 135. Sarver AL, Phalak R, Thayanithy V, Subramanian S. S-MED: sarcoma microRNA expression database. Lab Invest. (2010) 90:753–61. doi: 10.1038/labinvest.2010.53 [DOI] [PubMed] [Google Scholar]
- 136. Yan D, Dong Xda E, Chen X, Wang L, Lu C, Wang J, et al. MicroRNA-1/206 targets c-Met and inhibits rhabdomyosarcoma development. J Biol Chem. (2009) 284:29596–604. doi: 10.1074/jbc.M109.020511 [DOI] [PMC free article] [PubMed] [Google Scholar]