Abstract
In a human immunodeficiency virus type 1 (HIV-1)-infected individual, immune-pressure-mediated positive selection operates to maintain the antigenic polymorphism on the gp120 third variable (V3) loop. Recently, we suggested on the basis of sequencing C2/V3 segments from an HIV-1 subtype E-infected family that a V3 sequence lineage group of the non-syncytium-inducing (NSI) variants (group 1) was relatively resistant to positive selection pressure (35). To better understand the relationship between the intensity of positive selection pressure and cell tropism of the virus, we determined the linkage between each V3 genotype and its function of directing coreceptor preference and MT2 cell tropism. The biological characterization of a panel of V3 recombinant viruses showed that all of the group 1 V3 sequences could confer an NSI/CCR5-using (NSI/R5) phenotype on HIV-1LAI, whereas the group 2 V3 sequence, which was more positively charged than the group 1 sequence, dictated mainly a syncytium-inducing, CXCR4-using (SI/X4) phenotype. Phylogenetic analysis of C2/V3 sequences encoding group 1 or 2 V3 suggested that the variants carrying group 1 V3 are the ancestors of the intrafamilial infection and persisted in the family, while the variants carrying group 2 V3 evolved convergently from the group 1 V3 variants during disease progression in the individuals. Finally, a statistical test showed that the V3 sequence that could dictate an NSI/R5 phenotype had a synonymous substitution rate significantly higher than the nonsynonymous substitution rate. These data suggest that V3 sequences of the subtype E NSI/R5 variants are more resistant to positive selection pressure than those of the SI/X4 variants.
During the course of infection of an animal with a pathogen, amino acid sequence polymorphism is often generated in the surface antigenic sites. The mechanism to maintain this polymorphism is explained by positive Darwinian selection, which is thought to be caused by the immune-pressure-mediated host-parasite struggle referred to as antigenic drift (38–40, 43, 51). In this regard, it is generally accepted that a high degree of sequence polymorphism in the third variable (V3) loop within env gp120 of Human immunodeficiency virus type 1 (HIV-1) is maintained by positive selection, since the element consists of a major epitope for neutralizing antibodies (31, 47). Indeed, nonsynonymous substitution per nonsynonymous site (Ka) exceeded synonymous substitution per synonymous site (Ks) in the V3 loop (43, 58), and this tendency was enhanced with the duration of the immunocompetent period of infected individuals (23).
On the other hand, the V3 loop element consists of a critical determinant for the target cell type preference of the virus (6, 41, 48). The loop specifies the coreceptor usage of HIV-1 (4, 5), probably via interaction with the coreceptor molecules (1, 15, 52). Therefore, nonsynonymous mutation in the V3 loop can result in generation of either defective virus or virus which alters cell tropism. When such an alteration is disadvantageous to the virus, the V3 sequence would be selected as necessary to maintain the function of the V3 element and thereby decrease amino acid polymorphism of this region. In addition, the V3 loop of an non-syncytium-inducing (NSI) variant appears to be hidden from the neutralizing antibody (2), which may decrease the intensity of the positive selection pressure of NSI-dictating V3 sequences. In consequence, the amino acid substitution rate of a group of V3 loops, which use certain coreceptors or prefer certain cell types, may be lower than the rate for other types of V3 loop. Two distinct V3 genotypes associated with different cell tropisms and diversity are known to consistently evolve in HIV-1-infected individuals (3, 18, 21, 25, 53).
Recently, we reported an intrafamilial infection case in which a single source of HIV-1 subtype E of Thai origin infected the father (NH1), who transmitted the virus to the mother (NH2), who then transmitted the virus to her child (NH3) (35, 36). On the basis of genetic analyses of C2/V3 clonal sequences, we showed that two major V3 lineage groups had evolved in the family members. Group 1 was maintained with low variation in all three family members regardless of the clinical state or length of infection, whereas group 2 was present only in symptomatic individuals and was more positively charged and diverse than group 1. Only virus isolates carrying the group 2 V3 sequences infected and induced syncytia in MT2 cells. Interestingly, only the group 2 V3 region showed a significantly higher Ka/Ks ratio than 1 (35). These data suggest that the HIV-1 variant possessing the homogeneous V3 element and exhibiting the NSI phenotype persisted in infected individuals independent of clinical status under the weak positive selection pressure.
To better understand the relationship between the intensity of positive selection pressure and cell tropism of the virus, we ascertained the role of the group 1 and 2 V3 sequences in dictating coreceptor usage and the NSI or syncytium-inducing (SI) phenotype of the virus, using an HIV-1LAI subtype E V3 recombinant system (34). In addition, we estimated the evolutionary process of HIV-1 variants carrying each V3 sequence by constructing the phylogeny of C2/V3 clones from which the V3 sequences were derived. Furthermore, we examined the intensity of positive selection pressure on the V3 loop by comparing the synonymous and nonsynonymous substitutions occurring in each evolutionary process. Results from these analyses suggest that the subtype E NSI, CCR5-using (NSI/R5) variants, which were responsible for the establishment and the persistence of the HIV-1 infection, were more resistant to the positive selection pressure on the V3 loop than the SI/CXCR4 variant.
MATERIALS AND METHODS
Source of env C2/V3 sequences.
A total of 86 clonal nucleotide sequences encoding an open reading frame of the env C2/V3 region were used for these analyses. These sequences were derived from uncultured peripheral blood mononuclear cells (PBMCs) from the three members of a subtype E-infected Japanese family that consisted of a male index patient (NH1), the female spouse of NH1 (NH2), and their child (NH3) (36). NH1 had no history of blood transfusion, surgical operation, or homosexual activity but had sexual contacts with female prostitutes in Thailand in 1989 and 1990. NH2 had no documented risk factors for HIV-1 infection other than sexual contacts with NH1. NH3 was born to NH2 in June 1991. The PBMCs of NH1, NH2, and NH3 were collected in June 1993, when NH1 had developed AIDS but NH2 and NH3 remained asymptomatic. Follow-up collection was done for NH2 on March 1996 (NH2-II) and on January 1997 (NH2-III) after she had developed AIDS. Among the 86 clones, 82 clones had unique sequences along the C2/V3 region. Details of epidemiological and clinical information of the family members, as well as the cloning and sequencing methods, are provided in previous reports (35, 36).
Construction of the full-length HIV-1 recombinant DNA clones.
In previous studies, a total of 22 deduced amino acid sequences of the V3 loop were detected in this family (35, 36). For each V3 loop, HIV-1LAI subtype E V3 recombinant DNA clones were constructed as previously described (34). Briefly, 22 unique V3 sequences representing nine group 1 sequences (A1 to A9) and 13 group 2 sequences (B1 to B13) were selected. A StuI-to-NheI recombinant DNA fragment (456 bp) encoding the subtype E V3 and pLAI (28) flanking sequence was generated by the overlap extension method (17), digested with StuI and NheI, and cloned back into the full-length HIV-1 subtype B infectious molecular clone, pLAI (28), as shown in Fig. 1. The nucleotide sequences of the PCR-amplified regions and around all junctions for cloning were verified with an ABI PRISM 310 automated sequencer (Perkin-Elmer, Norwalk, Conn.). By this method, only the sequence between the two cysteine residues flanking the V3 loop was altered.
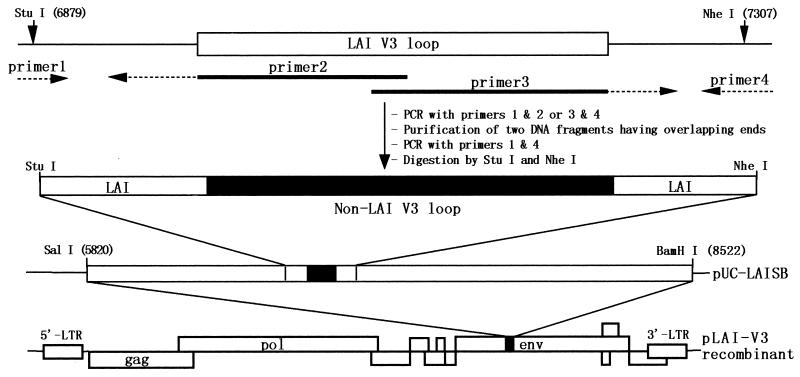
FIG. 1.
Scheme for construction of recombinant HIV-1LAI DNAs encoding V3 loop sequences from the NH family. Overlapping primers 2 and 3 and outer primers 1 and 4 were used to generate recombinant DNA segments having the non-LAI V3 and LAI flanking sequences by the overlap extension method (17). The products were digested with StuI and NheI and cloned into the gp120 subclone (pUC-LAISB) made from pLAI (28). Subsequently, the SalI-BamHI fragment of pUC-LAISB was cloned into pLAI to reconstitute a full-length HIV-1 molecular clone. Nucleotide sequences of outside primers 1 and 4 are described in a previous report (34). LTR, long terminal repeat.
Preparation of the cell-free recombinant virus stocks.
Recombinant and parental LAI viruses were prepared as previously described (34). Briefly, 5 × 105 HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM) with 10% (vol/vol) heat-immobilized fetal bovine serum (FBS) and transfected with 20 μg of pLAI-NHV3 DNA by the calcium phosphate coprecipitation method. The culture supernatants were collected at 48 or 72 h after transfection, filtered (0.45-μm-pore-size filter), and kept at −152°C until use. The reverse transcriptase (RT) activity of the transfection supernatant was measured in a standard assay as previously described (49).
HIV infections in HOS-CD4+ cell lines and MT2 cells.
HOS-CD4+ cell lines (7) were infected with the V3 recombinant viruses in triplicate in 96-well plates as previously described (34). Briefly, 5 × 103 HOS-CD4+ cells expressing human CCR5 (HOS-CD4-CCR5) or CXCR4 (HOS-CD4-CXCR4) were incubated in 0.1 ml of serially diluted cell-free transfection supernatant containing 2.5 × 106 (twofold dilution) to 2.0 × 104 (250-fold dilution) cpm of RT activity for 24 h at 37°C, washed, and grown in 0.2 ml of DMEM with 10% FBS and puromycin (1.0 μg/ml). Half of the volume of the culture medium was replaced by fresh medium every 2 days during the 12 days after infection, stored at −80°C, and analyzed for RT activity (49). The highest dilution required to produce an RT-positive culture was taken as the endpoint, and HIV-1 titers of HOS-CD4+ cells were indicated by tissue culture infective dose per 5 × 106 cpm of the RT counts.
Infections of MT2 cells with the V3 recombinant viruses were carried out in duplicate in 48-well plates as previously described (34). Briefly, 105 MT2 cells were incubated in 0.13 ml of the cell-free transfection supernatant containing 7.5 × 105 32P cpm of RT activity for 24 h at 37°C and then cultured in 2.1 ml of RPMI 1640 with 10% (vol/vol) FBS. Half of the volume of the culture medium was replaced by fresh medium every 2 days during the 12-day period after infection, stored at −80°C, and subjected to the RT assay. Syncytium formation was monitored daily under a light microscope.
Evolutionary analysis of the C2/V3 molecular clones from the family.
A total of 19 available C2/V3 sequences of HIV-1 subtype E (26) from Thailand (TN235, TN239, TN241, TN242, TN2432, TN244, TH0065, 92TH011, CM238, CM240, 92TH022.4, 93TH966.8, 93TH975.15, and 93TH976.17), the Central African Republic (CARMBA, CARELO, CAR4017, and CAR4071), and England (94-11643) were used for the outgroup of the clonal C2/V3 sequences from the family. Nucleotide sequences of 82 clones were aligned with the outgroup sequences by using CLUSTAL W, version 1.4 (46), and then corrected by hand to ensure that gaps did not alter the reading frame. The aligned nucleotide sequence data were then truncated to align reading frames, and maximum-likelihood trees were generated by using the DNAML program of PHYLIP, version 3.572 (11), as well as the baseml program of PAML, version 1.4 (54). We also constructed the neighbor-joining (NJ) tree (33) with 100 bootstrap replicates (10, 16) from the matrix of numbers of nucleotide substitutions per site based on Tajima and Nei's method (45). Distance matrices and the NJ trees were computed by MEGA (22), version 1.02.
The adaptive evolution of the virus lineage was analyzed by comparing synonymous and nonsynonymous substitutions for the putative ancestral sequences as described by Zhang et al. (57). Briefly, the ancestral nucleotide sequences at the interior nodes of the likelihood tree were inferred by the Bayesian method (55), using the baseml program of PAML, version 1.4 (54). The ancestral and the derived sequences were aligned by using CLUSTAL W, version 1.4 (46), and divided into two regions, the V3 sequence (105 bp) and the flanking region (217 bp). We then estimated the numbers of synonymous (s) and nonsynonymous (n) substitutions and synonymous (S) and nonsynonymous (N) sites between each sequence, using SNAP (14) by the method described by Nei and Gojobori (27) in addition to Jukes-Cantor corrections. Adaptive evolution of the V3 loop and its flanking region were analyzed by Fisher's exact test, given the null hypothesis of neutral evolution, i.e., n/s is identical to N/S.
Nucleotide sequence accession numbers.
The sequence data of the clones have been registered in the DDBJ database with accession no. AB014775 to AB014874.
RESULTS
Structural characteristics of the subtype E V3 loops from the family.
In a previous study, V3 sequences from the NH family were divided into two genotypic subgroups, groups 1 and 2, on the basis of the presence of basic amino acid substitutions, phylogeny, and the extent of sequence variation (35). Group 1 was characterized by the presence of a GPGQ motif at the tip of the V3 loop and by the lack of basic amino acid substitutions with respect to the consensus of the subtype E NSI V3 loop from 21 NSI virus isolates in the early 1990s in Thailand (Fig. 2). These sequences were similar to each other or to the NSI consensus and were found in all of the family members independent of the clinical stages of infection or in NSI virus isolates from the family members (35). Group 2 was characterized by the presence of a GPGR motif at the tip of the V3 loop and by the presence of the basic amino acid substitution(s) to the NSI consensus (Fig. 2). Group 2 was more diversified and was found only in individuals with AIDS or in SI virus isolates (35).
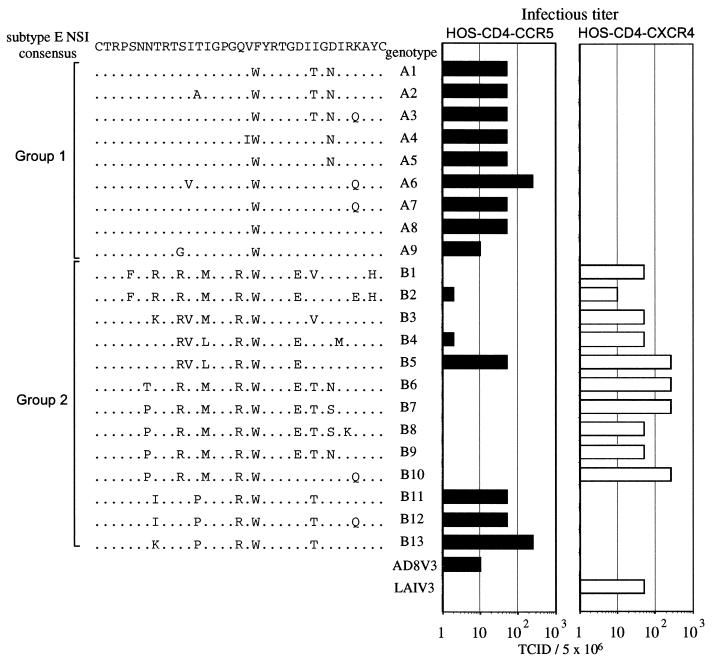
FIG. 2.
Infective doses of recombinant viruses in HOS-CD4 cell lines. HOS-CD4 cells expressing CCR5 or CXCR4 were infected with viruses containing decreasing numbers of RT counts (2.5 × 106, 5.0 × 105, 1.0 × 105, and 2.0 × 104 32P cpm) over 24 h. The highest dilution required to produce an RT-positive culture on day 7 after infection was taken as the endpoint, and HIV-1 titers were expressed as 2, 10, 50, and 250 tissue culture infective doses per 5 × 106 32P cpm RT activity. Infectious titers of the V3 recombinant viruses are shown as bar graphs at the right; deduced amino acid sequences and V3 genotypes of the recombinant viruses are shown at the left. Dots in the amino acid alignment represent residues identical to a subtype E NSI consensus sequence made from 21 subtype E NSI isolates (8, 19).
Coreceptor usage and MT2 cell tropism of the recombinant viruses encoding the V3 loops from the family.
A panel of HIV-1LAI V3 recombinant viruses was generated by the overlap extension method and was successfully used to characterize the biological function of the subtype E V3 sequence in virus entry (20, 34). RT activities per unit volume of the transfection supernatant was similar among the parental and recombinant viruses, peaking on days 2 and 3 (1.5 × 104 to 2.0 × 104 cpm/μl of culture supernatant), suggesting that the V3 replacement did not deleteriously affect the processing of the Gag/Pol precursor, virion assembly, and budding, as suggested previously (34).
A total of 22 recombinant viruses were tested for coreceptor usage. Transfection supernatants containing equal amounts of RT activity were serially diluted and used to infect HOS-CD4-CCR5 or HOS-CD4-CXCR4 cells. As shown in Fig. 2, all of the group 1 V3 recombinant viruses (A1 to A9) efficiently infected the HOS-CD4-CCR5 cells, having infectious titers comparable to that of the AD8 V3 control recombinant virus carrying the subtype B NSI V3. However, none of the viruses infected the HOS-CD4-CXCR4 cells. In contrast, most of the group 2 recombinant viruses (10 of 13) replicated efficiently in the HOS-CD4-CXCR4 cells, having infectivity comparable to that of the LAI V3 control virus (Fig. 2, 1 to B10). Among the group 2 V3 recombinant viruses, three (B2, B4, and B5) could infect both the HOS-CD4-CCR5 and HOS-CD4-CXCR4 cells, and three viruses (B11, B12, and B13) could infect only HOS-CD4-CCR5 cells.
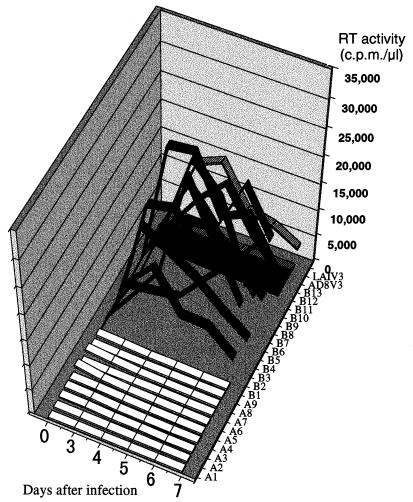
All of the recombinant viruses which used CXCR4 for infection of HOS-CD4 cells consistently and efficiently replicated in MT2 cells and induced syncytia within 3 to 7 days after infection (Fig. 3, 1 to B10). RT activities in the virus-producing culture supernatants reached a peak within 7 days of infection, ranging from 14,500 to 34,500 cpm per μl of culture supernatant. In contrast, all of the recombinant viruses which used CCR5 in preference to CXCR4 could not replicate in MT2 cells, showing an NSI phenotype (Fig. 3, A1 to A9 and B11 to B13).
FIG. 3.
Replication kinetics of recombinant viruses in the MT2 cell line. Virus stocks used to infect cells were generated by transfecting HeLa cells with each sample of recombinant plasmid DNA. MT2 cells were infected with supernatant containing equal amounts of RT activity, and progeny virus production was monitored by measuring the RT activity released into the culture medium at the indicated time points. The RT activity of each recombinant virus at each time point is represented in the three-dimensional graph by white (viruses encoding group 1 V3), black (viruses encoding group 2 V3), and hatched (controls) bands.
Evolutionary relationship of the quasispecies in the family.
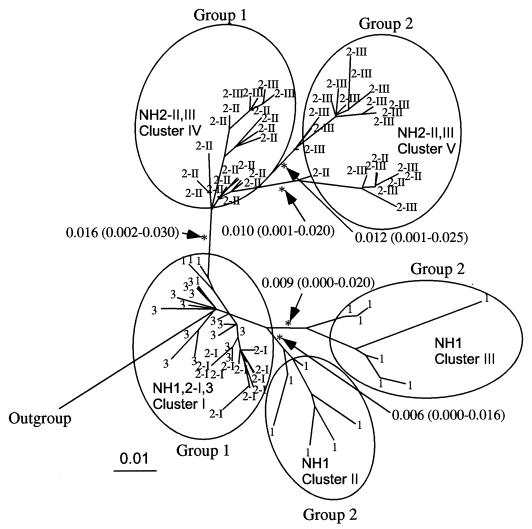
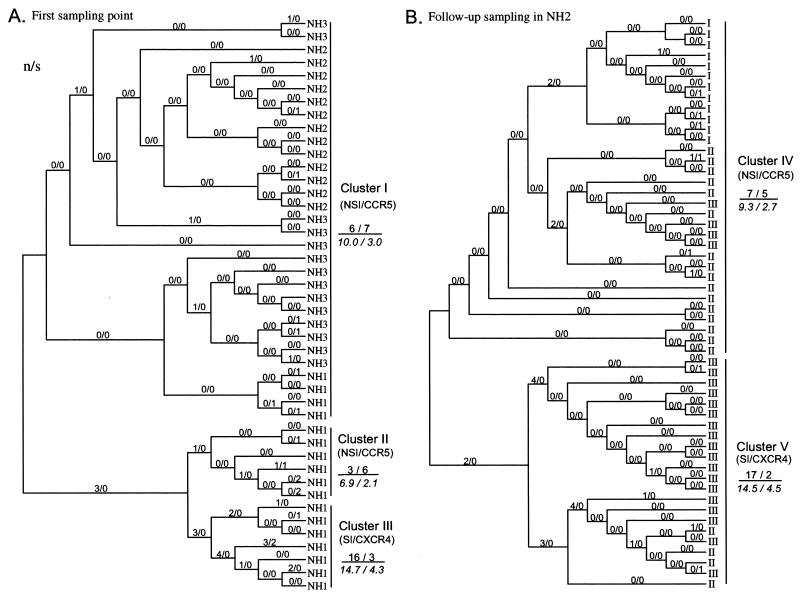
Phylogenetic analysis of the env C2/V3 and p17/p24 gag nucleotide sequences indicated that a single source of the HIV-1 subtype E of Thai origin had infected the family (35). To further estimate the evolutionary relationship of the HIV-1 variants carrying the group 1 and 2 V3 sequences, a phylogenetic tree of the env C2/V3 region which encoded each V3 sequence was constructed for 82 independent sense clones by the maximum-likelihood method with the PHYLIP package and rooted with the outgroup sequences of subtype E from Thailand, the Central African Republic, and England (Fig. 4). The C2/V3 sequences from the family members were divided into six subtrees. Statistics for the branch length in the maximum-likelihood tree showed that all of the branches dividing the subtrees were significant (Fig. 4, arrows). On the basis of the phylogeny, we categorized the 82 C2/V3 sequences into five evolutionarily closely related groups called clusters (Fig. 4, clusters I to V).
FIG. 4.
Maximum-likelihood tree of the nucleotide sequences of C2/V3 clones (324 bp) from uncultured PBMCs of the family members at different sampling points. The tree was estimated by using the DNAML program in PHYLIP, version 3.572. Nineteen available sequences of HIV-1 subtype E with various geographic origins (26) were added as the outgroup. Numbers with arrows represent lengths and confidence limits of branches in the likelihood analysis. Branches with asterisks are significantly positive (P < 0.01).
As shown in Fig. 4, cluster I consisted of all sequences from NH2 and NH3 at the asymptomatic stage, as well as four sequences from NH1 with AIDS. Cluster I contained a node from the outgroups, i.e., the putative origin of the virus-initiated intrafamilial infection. Clusters II and III contained six and seven sequences, respectively, both of which were from NH1 with AIDS. These clusters derived from the same ancestor as cluster I, with similar evolutionary distances. Cluster IV contained 19 sequences of NH2 with AIDS (NH2-II and NH2-III). Cluster IV had a distant ancestor from cluster II and III, although the ancestor also belonged to cluster I. Cluster V contained two closely related subtrees, which included 13 and 9 sequences from NH2 with AIDS (NH2-II and NH2-III). They were merged into one evolutionary related group (a cluster) because of their similarity in the origin and biological activities of V3 within the subtrees (see below). Thus, the phylogenetic relationship suggested that the C2/V3 sequences of the family originated from cluster I and evolved along with the two genealogical pathways, one including clusters II and III from NH1 and the other including clusters IV and V from NH2 with AIDS. The maximum-likelihood tree computed by the PAML program (54), as well as the NJ tree from the substitution matrix calculated by Tajima and Nei's method (45), presented the same topology as in Fig. 4 (data not shown).
Summary of relationship between V3 genotype, V3 phenotype, and C2/V3 lineages.
Table 1 summarizes the relationship between the V3 genotypes, the biological activities of V3 (ability to confer an NSI/R5 or an SI/X4 phenotype), and the C2/V3 lineages from which the V3 sequences were derived (clusters I to V). All of the group 1 V3 functioned to determine an NSI/CCR5 phenotype of the virus. Among the group 1 V3, A1 to A5 were linked to the cluster I clones found in all of the family members at the first sampling point, while A6 to A9 were linked to the cluster IV clones found in NH2 at the symptomatic stages, which were 3 to 4 years after the first sampling point. Although the group 1 V3 uniformly conferred an NSI/R5 phenotype on the virus regardless of the C2/V3 clusters, the biological activity of the group 2 V3 showed a variation. Although the group 2 V3 genotype from clusters III and V (B1 to B3 and B4 to B10, respectively) conferred a CXCR4 preference on the virus, three genotypes from cluster II (B11, B12, and B13) were found to dictate the CCR5 but not the CXCR4 phenotype. These genotypes lacked a basic substitution at position 11 (serine to arginine), whereas the other group 2 genotypes had this type of substitution (Fig. 2). In addition, three sequences from clusters III and V (B2, B4, and B5) could dictate both CXCR4 and CCR5 usage.
TABLE 1.
Relationship between V3 genotypes, phenotypes, and C2/V3 lineages
| V3 genotype | V3 group | V3 phenotype | C2/V3 lineage | Frequency in specimensa
|
||||
|---|---|---|---|---|---|---|---|---|
| Asymptomatic
|
AIDS
|
|||||||
| NH2-I | NH3 | NH1 | NH2-II | NH2-III | ||||
| A1 | 1 | NSI/R5 | I | 11 | 4 | 2 | ||
| A2 | 1 | NSI/R5 | I | 1 | ||||
| A3 | 1 | NSI/R5 | I | 2 | ||||
| A4 | 1 | NSI/R5 | I | 1 | ||||
| A5 | 1 | NSI/R5 | I | 10 | ||||
| A6 | 1 | NSI/R5 | IV | 1 | ||||
| A7 | 1 | NSI/R5 | IV | 10 | ||||
| A8 | 1 | NSI/R5 | IV | 1 | ||||
| A9 | 1 | NSI/R5 | IV | 3 | 4 | |||
| B1 | 2 | SI/X4 | III | 2 | ||||
| B2 | 2 | SI/Dual | III | 1 | ||||
| B3 | 2 | SI/X4 | III | 3 | ||||
| B4 | 2 | SI/dual | V | 3 | ||||
| B5 | 2 | SI/dual | V | 10 | ||||
| B6 | 2 | SI/X4 | V | 1 | ||||
| B7 | 2 | SI/X4 | V | 3 | 2 | |||
| B8 | 2 | SI/X4 | V | 1 | ||||
| B9 | 2 | SI/X4 | V | 2 | ||||
| B10 | 2 | SI/X4 | V | 1 | ||||
| B11 | 2 | NSI/R5 | II | 2 | ||||
| B12 | 2 | NSI/R5 | II | 1 | ||||
| B13 | 2 | NSI/R5 | II | 3 | ||||
Arranged by clinical status. NH2-I, NH2-II, and NH2-III represent follow-up samples from NH2 collected in June 1993, March 1996, and January 1997, respectively.
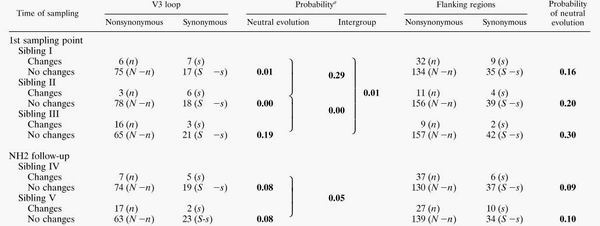
Tests of adaptive evolution on the V3 loop and its flanking region.
To infer the putative sequences at the interior nodes of the phylogenetic tree, we reconstituted the maximum-likelihood phylogeny of the C2/V3 region by using the PAML program (54). The topology of the tree was virtually identical to that in Fig. 4. We then computed the number of synonymous (s) and nonsynonymous (n) substitutions, as well as the numbers of the synonymous (S) and nonsynonymous (N) sites occurring in the evolutionary process in each branch in the phylogenetic tree; this was carried out separately for the V3 region (Fig. 5) and the flanking region (data not shown). The estimated number of S and N for the V3 region ranged between 23.5 and 26 and between 79 and 81.5, respectively, and the mean N/S was 3.27. Using the method described by Zhang et al. (57), we analyzed the adaptive evolution of V3 and the flanking regions by comparing s and n with their expected numbers under the hypothesis of neutral evolution. Figure 5 shows that the observed ratio of the total number of n/s in the V3 sequence was lower than the expected ratio in the lineage of clusters I, II, and IV, which encoded V3 sequences to dictate an NSI/R5 virus phenotype. In contrast, the observed n/s ratio was higher than the expected ratio at the lineage of clusters III and V, which encoded an SI/X4 or SI/X4R5 phenotype (Fig. 5). Thirty-six percent of the nonsynonymous substitutions plotted in Figure 5 occurred in positions 27 and 32 of V3 loop regardless of lineage (9 of 15 and 8 of 32 at NSI/R5 and SI/X4 lineages, respectively; P = 0.001), whereas no nonsynonymous substitution occurred in position 9, except for two substitutions which caused coreceptor alteration.
FIG. 5.
Numbers of synonymous (s) and nonsynonymous (n) nucleotide substitutions per sequence for each branch of the phylogenetic tree of C2/V3 region. The values of n and s are given as n/s for each branch above the line. The n and s values at the right of the tree represent the sum of n and s for all branches in a lineage, respectively. Expected numbers for the sums of n and s [(n + s)N/(N + S) and (n + s)S/(N + S), respectively] are given in italics below the line. (A) Sequences collected from NH1, NH2, and NH3 in June 1993. (B) Sequences collected in the follow-up study of NH2 at June 1993, March 1996, and January 1997.
Using Fisher's exact test, we analyzed the positive and negative selection forces within each C2/V3 lineage (Table 2). Negative selection was suggested for the V3 sequence in cluster I (P = 0.01) and II (P = 0.00), while neutral evolution could not be rejected for V3 in the other clusters. Tests for intercluster difference in the n/s ratio showed that cluster III had a significantly higher ratio than the other clusters (Table 2, P = 0.01 and P = 0.00 for cluster III to cluster I and for cluster III to cluster II, respectively), while the ratio between clusters I and II and between clusters IV and V were not significantly different. Contrary to the V3 loop element, the test for neutrality of the flanking region showed that none of the results for the clusters could reject the null hypothesis (Table 2, flanking region). These data indicated that a significant purifying selection had acted on only the V3 sequence of the evolutionary process in clusters I and II.
TABLE 2.
Numbers of synonymous and nonsynonymous substitutions per sequence for each C2/V3 lineage on a V3 loop and its flanking regions
 |
DISCUSSION
Although the neo-Darwinist concept of adaptive evolution was established nearly a half-century ago, it has been difficult to demonstrate molecular evidence for adaptive evolution. If a molecule of an organism has an element interacting with the environment of the organism, a change in the environment should increase sequence variation of the gene encoding this element. In a virus, the elements interacting with the host molecules during either virus replication or competition with the host's immune system are candidates for adaptive evolution. In this context, it is interesting to examine the evolution of the HIV-1 env gp120 V3 loop sequence and function, because the element is involved in determining cell tropism (1, 3, 15, 48, 50, 52) and because it simultaneously maps to an epitope for neutralizing antibodies (31, 47). In the present study, we have provided evidence that the mode of evolution of the V3 element is indeed affected by environmental factors such as coreceptor usage or host cell type. This is the first molecular evidence that adaptive evolution of a virus follows the environmental choice of the virus.
In a previous report, we suggested that the group 1 and group 2 V3 loops may dictate a virus's preference for usage of CCR5 and CXCR4, respectively (35). In the present study, we found a direct association between the V3 genotype and its function in a subtype E recombinant system (34). Infection of HOS-CD4 cells with the V3 recombinant viruses showed that all of the group 1 V3 loops were able to confer CCR5 and not CXCR4 usage on HIV-1LAI (Fig. 2). All of the recombinant viruses in group 1 consistently failed to replicate on MT2 cells (Fig. 3). These results indicate that the group 1 V3 loops dictate the NSI/R5 phenotype of a virus. This, in turn, suggests that the group 1 V3 sequences are derived from NSI/V3 viruses in the NH family. This conclusion is consistent with our previous observations that the group 1 V3 sequences were similar or identical to those of the NSI virus isolates from the NH family and that predominated in asymptomatic individuals (35).
In contrast to the group 1 V3 loops, the major type of group 2 V3 loops, which accompanied basic amino acid substitutions, dictated the SI phenotype and CXCR4 preference of the virus (Fig. 2 and 3). Among the 10 group 2 genotypes associated with CXCR4 usage, 3 (B2, B4, and B5) could also dictate CCR5 usage, suggesting that some group 2 variants were derived from dualtropic viruses which could use both CCR5 and CXCR4. The data are consistent with the observation that variants from the late stages of infection often expand the range of coreceptor usage (37, 44). Analysis of the V3 recombinant viruses also revealed that three group 2 genotypes (B11, B12, and B13) dictated only an NSI/CCR5 phenotype. These V3 elements were less positively charged than the other group 2 sequences (35) and lacked a basic substitution at position 11 in the V3 loop (Fig. 2), which plays a critical role in determining the SI/CXCR4 phenotype of viruses of subtypes B (6, 12, 13) and E (20). Thus, we concluded that 10 group 2 V3 genotypes that had a basic substitution at position 11 were all derived from SI/CXCR4 variants, whereas three group 2 V3 genotypes that lacked the particular substitution were derived from NSI/CCR5 variants in the family.
Previously, we suggested that the variants possessing group 1 V3 loops persisted in infected individuals independent of their clinical status, while the variants carrying group 2 V3 loops emerged during disease progression (35). In the present study, we have further extended this finding and attempted to ascertain the intrafamilial genealogy of variants carrying group 1 and 2 V3 sequences, using the env C2/V3 tree. The analysis showed that a cluster of C2/V3 sequences encoding the group 1 V3 (Fig. 4, cluster I) contained the ancestor node rooted by subtype E outgroups, suggesting that the cluster I variants had a close relationship to a variant establishing infections in this family. The analysis also revealed that a cluster of C2/V3 sequences encoding group 1 V3 loops from NH2 with AIDS (Fig. 4, cluster IV) directly originated from cluster I, which consists of C2/V3 sequences from NH1, NH2, and NH3. Because all of the group 1 V3 loops were exclusively associated with an NSI/R5 phenotype of the virus, the above evolutionary relationship suggests that the NSI/R5 variants were involved in person-to-person transmission and persistent infection in this family. Thus, the finding for HIV-1 subtype B infection that the NSI variant plays a critical role in sustaining infections in a human population (24, 32, 59) seems to be applicable also to this HIV-1 subtype E infection.
Furthermore, env C2/V3 phylogeny clearly showed that the group 2 V3 from the father (NH1) with AIDS and the group 2 V3 from the mother (NH2) with AIDS had evolved independently within each individual (Fig. 4, clusters III and V). This result suggests that the SI variants exhibiting similar coreceptor preference had evolved convergently in different individuals. Of note is that clusters III and V were shown to be derived from the NSI/R5 variants (Fig. 4, clusters I and IV, respectively), suggesting that the SI/X4 variants found in NH1 and NH2 at the late stages of infection (35) had indeed evolved from the NSI/R5 variants that had predominated in the early stages of infection rather than emerged via the outgrowth of SI/X4 variants that might have existed in the primary infection. Thus, these data support the notion that the convergent evolution of the SI/X4 phenotype from the NSI/R5 phenotype had occurred during progression of the disease in subtype E-infected individuals (35).
Taken together, the present analyses of the V3 function and C2/V3 lineages indicate the following evolutionary processes of the HIV-1 subtype E quasispecies in the family. First, the NSI variants carrying the group 1 V3 sequences (cluster I) had initiated infections in the three family members, NH1, NH2, and NH3. In NH1, the group 1 V3 NSI variants had evolved into the three subgroups: the NSI/R5 variants carrying V3 similar to the ancestral group 1 sequences (cluster I); the NSI/R5 variants divergent from cluster I and having V3 sequences with a single basic substitution from the ancestral group 1 sequence (cluster II); and the SI/X4 variants carrying the V3 sequences with two or three basic substitutions (cluster III). In NH2, the group 1 V3 NSI variants (cluster I) had evolved into two subgroups, the NSI/CCR5 variants carrying the group 1 sequences (cluster IV) and the SI/CXCR4 variants carrying the group 2 V3 with two or three basic substitutions (cluster V).
Detailed statistical analysis of synonymous and nonsynonymous substitution in this study revealed that the intensity of the positive selection force on the V3 sequence was related to the V3 phenotype lineage (Fig. 5; Table 2). All of the lineages in which V3 could dictate an SI/X4 phenotype showed an n/s ratio of V3 sequence higher than that obtained with the NSI/R5 lineages or to the ratio from the neutral hypothesis (Fig. 5, clusters III and V). These data suggest that the positive selection had occurred in the V3 sequences of SI/X4 variants. Although the neutral evolution of the V3 sequence cannot be rejected by the statistical test (Table 2), we interpret this statistical insignificance to be the result of the purifying selection which was occurring concurrently on the V3 region. Because the V3 loop is a critical determinant in specifying the coreceptor usage of the virus, many residues in this region would be subject to functional constraint. Unequal distribution of nonsynonymous substitutions in the V3 lineages may support this assumption. This antagonistic effect on the V3 sequence variation may be responsible for the statistically insignificant results in the present study. Similarly, nonsignificant results obtained with a similar analytical method were also observed in a study of primate lysozymes (57), in which these enzymes were thought to undergo positive selection on the basis of relatively high Ka/Ks ratio as seen in the V3 sequences of SI/X4 variants.
In contrast to the SI/X4 lineages, all of the lineages carrying the NSI/CCR5 phenotype-linked V3 sequences showed a lower n/s ratio independent of the clinical stages of infection compared to the ratio from the neutral hypothesis (Fig. 5, clusters I, II, and IV). Moreover, the statistical test showed that the Ka/Ks ratio is significantly lower than 1 in two of three lineages (Table 2, clusters I and II). These results indicate that a neutral evolution maintained a nucleotide variation of the V3 region in the NSI/R5 variants, and most of the nonsynonymous mutations had deleterious effects (i.e., purifying selection) in these variants. Unlike the case for the V3 sequence, the n/s ratios of regions flanking the various V3 sequences were similar to each other independent of the virus lineage or phenotype (Table 2). The ratio was lower than that for the V3 region in the SI/X4 lineages and higher than that for the V3 region in the NSI/R5 V3 lineages. This dissimilarity between the n/s ratio of V3 and the n/s ratio of its flanking region, together with the difference in the n/s ratio of V3 among the lineages, suggests that the V3 sequence of the SI/X4 variant had undergone a relatively strong positive selection compared with the flanking region, whereas that of the NSI/R5 variant had been maintained by strong purifying selection compared to the flanking region.
There are several reports that are relevant to the present work. The diversity of subtype E V3 sequences worldwide shows a bimodal pattern, reflecting the presence of two V3 subpopulations with distinct variation (9). The difference seems to correlate with the extent of positive charge in V3 and biological properties of the virus; a highly homogeneous set of V3 sequences were less positively charged and derived from asymptomatic carriers with an NSI variant, whereas more divergent sequences were more positively charged and derived from AIDS patients with SI variants (9, 56). Interestingly, this relationship between divergent pattern of V3 sequence, positive charge in V3, and biological property of virus seems to hold true for other subtypes. Subtype D has a highly diverse set of V3 compared to other HIV-1 group M subtypes, which correlates with increased positive charge in V3 (9). In contrast, subtype C has a less diverse and positively charged set of V3 than other subtypes and exhibits predominantly an NSI/CCR5 phenotype (9, 29, 30). Thus, our finding that V3 loop sequences are subjected to positive selection only when they dictate the SI/X4 phenotype of virus may be applicable to infections of HIV-1 group M subtypes.
Why do subtype E NSI/R5 variants identified in this study appear to be more resistant to positive selection pressure than the SI/X4 variants? First, it is possible that these NSI/R5 variants are more resistant to neutralization by antibodies against the V3 loop. It has been suggested that V3 loops of NSI variants are very poorly exposed to neutralizing antibodies (2). This may allow the NSI variants to survive under humoral immune pressure against their V3 sequences, while some SI variants may be eliminated by neutralizing antibodies recognizing their V3 loops. This difference in fitness between the NSI/R5 and SI/X4 variants may in turn lead to the difference in the number of substitutions driven by positive selection.
Second, even if the intensity of positive selection pressure is comparable among NSI/R5 and SI/X4 variants, the functional constraint to interact with the CXCR4 molecule may be much weaker than the constraint to interact with the CCR5 molecule. It was reported that the V1/V2 region of a dualtropic variant could confer on a macrophagetropic virus the ability to use CXCR4, even if the maximum efficiency of the CXCR4 usage was obtained by a combination of V1/V2 and V3 loop replacement (4). The V1/V2 ability to dictate the CXCR4 choice of virus may rescue a virus that has a V3 loop with a deleterious mutation of the CXCR4-dictating function, leading this region to be relatively changeable. Furthermore, the V3 loop sequences of laboratory NSI strains selected for growth in macrophages in vitro are generally highly conserved, whereas those of the laboratory SI strains selected for growth in the transformed T-cell lines are still highly variable (42). This implies that the NSI/R5 variants are more sensitive to mutations in the V3 loop than the SI/X4 variants in the absence of immune pressure. To determine why the V3 sequences of the NSI/R5 variants are more resistant to positive selection pressure than those of the SI/X4 variants, experiments to assess the neutralization sensitivity and intensity of the functional constraint of the HIV-1LAI-NHV3 recombinant viruses are in progress.
In summary, this study demonstrates that the V3 sequence of the HIV-1 subtype E NSI/CCR5 variants were more resistant to positive selection pressure than those of the SI/X4 variants and had evolved according to the neutral theory in the infection of the Japanese family. It is not clear whether it is applicable to another subtype or to a subtype E virus with very different V3 sequence. Further studies using more follow-up specimens from epidemiologically unrelated individuals and more extensive genetic elements are needed to confirm the generality of the present observations.
ACKNOWLEDGMENTS
We thank Keith Peden for providing pLAI, and we thank Naruya Saito for providing information about the statistical analysis of adaptive evolution. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: HOS-CD4 cells and HOS-CD4 cell lines expressing CXCR4 or CCR5, from Nathaniel Landau.
This work was supported by grants from the Ministry of Health and Welfare of Japan, the Ministry of Education, Science and Culture of Japan, and the Science and Technology Agency of the Japanese Government.
REFERENCES
- 1.Bieniasz P D, Fridell R A, Aramori I, Ferguson S S, Caron M G, Cullen B R. HIV-1-induced cell fusion is mediated by multiple regions within both the viral envelope and the CCR-5 co-receptor. EMBO J. 1997;16:2599–2609. doi: 10.1093/emboj/16.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bou-Habib D C, Roderiquez G, Oravecz T, Berman P W, Lusso P, Norcross M A. Cryptic nature of envelope V3 region epitopes protects primary monocytotropic human immunodeficiency virus type 1 from antibody neutralization. J Virol. 1994;68:6006–6013. doi: 10.1128/jvi.68.9.6006-6013.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chesebro B, Wehrly K, Nishio J, Perryman S. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J Virol. 1992;66:6547–6554. doi: 10.1128/jvi.66.11.6547-6554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho M W, Lee M K, Carney M C, Berson J F, Doms R W, Martin M A. Identification of determinants on a dualtropic human immunodeficiency virus type 1 envelope glycoprotein that confer usage of CXCR4. J Virol. 1998;72:2509–2515. doi: 10.1128/jvi.72.3.2509-2515.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 6.De Jong J J, De Ronde A, Keulen W, Tersmette M, Goudsmit J. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J Virol. 1992;66:6777–6780. doi: 10.1128/jvi.66.11.6777-6780.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 8.De Wolf, F., E. Hogervorst, J. Goudsmit, E. M. Fenyo, H. Rubsamen-Waigmann, H. Holmes, B. Galvao-Castro, E. Karita, C. Wasi, S. D. Sempala, E. Baan, F. Zorgdrager, V. Lukashov, S. Osmanov, C. Kuiken, M. Cornelissen, and WHO Network for HIV Isolation and Characterization. 1994. Syncytium-inducing and non-syncytium-inducing capacity of human immunodeficiency virus type 1 subtypes other than B: phenotypic and genotypic characteristics. AIDS Res. Hum. Retroviruses 10:1387–1400. [DOI] [PubMed]
- 9.Dighe P K, Korber B, Foley B T. Global variation in the HIV-1 V3 region. In: Korber B, Hahn B, Foley B T, Mellors J, Leitner T, Myers G, McCutchan F, Kuiken C, editors. Human retrovirus and AIDS. III. Los Alamos, N. Mex: Theoretical Biology and Biophysics, Los Alamos National Laboratory; 1997. pp. 74–206. [Google Scholar]
- 10.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 12.Fouchier R A, Brouwer M, Broersen S M, Schuitemaker H. Simple determination of human immunodeficiency virus type 1 syncytium-inducing V3 genotype by PCR. J Clin Microbiol. 1995;33:906–911. doi: 10.1128/jcm.33.4.906-911.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fouchier R A, Groenink M, Kootstra N A, Tersmette M, Huisman H G, Miedema F, Schuitemaker H. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66:3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ganeshan S, Dickover R E, Korber B T, Bryson Y J, Wolinsky S M. Human immunodeficiency virus type 1 genetic evolution in children with different rates of development of disease. J Virol. 1997;71:663–677. doi: 10.1128/jvi.71.1.663-677.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill C M, Deng H, Unutmaz D, Kewalramani V N, Bastiani L, Gorny M K, Zolla-Pazner S, Littman D R. Envelope glycoproteins from human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus can use human CCR5 as a coreceptor for viral entry and make direct CD4-dependent interactions with this chemokine receptor. J Virol. 1997;71:6296–6304. doi: 10.1128/jvi.71.9.6296-6304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hillis D M, Bull J J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol. 1993;42:182–192. [Google Scholar]
- 17.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 18.Holmes E C, Zhang L Q, Simmonds P, Ludlam C A, Brown A J. Convergent and divergent sequence evolution in the surface envelope glycoprotein of human immunodeficiency virus type 1 within a single infected patient. Proc Natl Acad Sci USA. 1992;89:4835–4839. doi: 10.1073/pnas.89.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ichimura H, Kliks S C, Visrutaratna S, Ou C Y, Kalish M L, Levy J A. Biological, serological, and genetic characterization of HIV-1 subtype E isolates from northern Thailand. AIDS Res Hum Retroviruses. 1994;10:263–269. doi: 10.1089/aid.1994.10.263. [DOI] [PubMed] [Google Scholar]
- 20.Kato K, Sato H, Takebe Y. Role of naturally occurring basic amino acid substitutions on viral coreceptor usage and cellular tropism in human immunodeficiency virus type 1 subtype E envelope V3 loop. J Virol. 1999;73:5520–5526. doi: 10.1128/jvi.73.7.5520-5526.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuiken C L, de Jong J J, Baan E, Keulen W, Tersmette M, Goudsmit J. Evolution of the V3 envelope domain in proviral sequences and isolates of human immunodeficiency virus type 1 during transition of the viral biological phenotype. J Virol. 1992;66:4622–4627. doi: 10.1128/jvi.66.7.4622-4627.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar S, Tamura K, Nei M. MEGA: molecular evolutionary genetics analysis, version 1.01. University Park, Pa: The Pennsylvania State University; 1993. [Google Scholar]
- 23.Lukashov V V, Kuiken C L, Goudsmit J. Intrahost human immunodeficiency virus type 1 evolution is related to length of the immunocompetent period. J Virol. 1995;69:6911–6916. doi: 10.1128/jvi.69.11.6911-6916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McNearney T, Hornickova Z, Markham R, Birdwell A, Arens M, Saah A, Ratner L. Relationship of human immunodeficiency virus type 1 sequence heterogeneity to stage of disease. Proc Natl Acad Sci USA. 1992;89:10247–10251. doi: 10.1073/pnas.89.21.10247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milich L, Margolin B, Swanstrom R. V3 loop of the human immunodeficiency virus type 1 Env protein: interpreting sequence variability. J Virol. 1993;67:5623–5634. doi: 10.1128/jvi.67.9.5623-5634.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myers G, Korber B, Hahn B, Jeang K-T, Mellors J, McCutchan F, Henderson L, Pavlakis G. Human retroviruses and AIDS 1995: a compilation and analysis of nucleic acid and amino acid sequences. Group T-10. Los Alamos, N.Mex: Los Alamos National Laboratory; 1995. [Google Scholar]
- 27.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 28.Peden K, Emerman M, Montagnier L. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology. 1991;185:661–672. doi: 10.1016/0042-6822(91)90537-l. [DOI] [PubMed] [Google Scholar]
- 29.Peeters M, Vincent R, Perret J L, Lasky M, Patrel D, Liegeois F, Courgnaud V, Seng R, Matton T, Molinier S, Delaporte E. Evidence for differences in MT2 cell tropism according to genetic subtypes of HIV-1: syncytium-inducing variants seem rare among subtype C HIV-1 viruses. J Acquired Immune Defic Syndr Hum Retrovirol. 1999;20:115–121. doi: 10.1097/00042560-199902010-00002. [DOI] [PubMed] [Google Scholar]
- 30.Ping L H, Nelson J A, Hoffman I F, Schock J, Lamers S L, Goodman M, Vernazza P, Kazembe P, Maida M, Zimba D, Goodenow M M, Eron J J, Jr, Fiscus S A, Cohen M S, Swanstrom R. Characterization of V3 sequence heterogeneity in subtype C human immunodeficiency virus type 1 isolates from Malawi: underrepresentation of X4 variants. J Virol. 1999;73:6271–6281. doi: 10.1128/jvi.73.8.6271-6281.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Profy A T, Salinas P A, Eckler L I, Dunlop N M, Nara P L, Putney S D. Epitopes recognized by the neutralizing antibodies of an HIV-1-infected individual. J Immunol. 1990;144:4641–4647. [PubMed] [Google Scholar]
- 32.Roos M T, Lange J M, de Goede R E, Coutinho R A, Schellekens P T, Miedema F, Tersmette M. Viral phenotype and immune response in primary human immunodeficiency virus type 1 infection. J Infect Dis. 1992;165:427–432. doi: 10.1093/infdis/165.3.427. [DOI] [PubMed] [Google Scholar]
- 33.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 34.Sato H, Kato K, Takebe Y. Functional complementation of the envelope hypervariable V3 loop of human immunodeficiency virus type 1 subtype B by the subtype E V3 loop. Virology. 1999;257:491–501. doi: 10.1006/viro.1999.9670. [DOI] [PubMed] [Google Scholar]
- 35.Sato H, Shiino T, Kodaka N, Taniguchi K, Tomita Y, Kato K, Miyakuni T, Takebe Y. Evolution and biological characterization of human immunodeficiency virus type 1 subtype E gp120 V3 sequences following horizontal and vertical virus transmission in a single family. J Virol. 1999;73:3551–3559. doi: 10.1128/jvi.73.5.3551-3559.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sato H, Taniguchi K, Tomita Y, Shiino T, Miyakuni T, Takebe Y. Evidence for the selective pressure to reduce heterogeneity of HIV-1 subtype E envelope V3-loop sequences in an intrafamilial infection case. AIDS. 1997;11:396–397. [PubMed] [Google Scholar]
- 37.Scarlatti G, Tresoldi E, Bjorndal A, Fredriksson R, Colognesi C, Deng H K, Malnati M S, Plebani A, Siccardi A G, Littman D R, Fenyo E M, Lusso P. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 38.Scholtissek C. The genes coding for the surface glycoproteins of influenza A viruses contain a small conserved and a large variable region. Virology. 1979;93:594–597. doi: 10.1016/0042-6822(79)90264-2. [DOI] [PubMed] [Google Scholar]
- 39.Scholtissek C. Molecular aspects of the epidemiology of virus disease. Experientia. 1987;43:1197–1201. doi: 10.1007/BF01945523. [DOI] [PubMed] [Google Scholar]
- 40.Seibert S A, Howell C Y, Hughes M K, Hughes A L. Natural selection on the gag, pol, and env genes of human immunodeficiency virus 1 (HIV-1) Mol Biol Evol. 1995;12:803–813. doi: 10.1093/oxfordjournals.molbev.a040257. [DOI] [PubMed] [Google Scholar]
- 41.Shioda T, Levy J A, Cheng-Mayer C. Small amino acid changes in the V3 hypervariable region of gp120 can affect the T-cell-line and macrophage tropism of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1992;89:9434–9438. doi: 10.1073/pnas.89.20.9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shpaer E G, Delwart E L, Kuiken C L, Goudsmit J, Bachmann M H, Mullins J I. Conserved V3 loop sequences and transmission of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 1994;10:1679–1684. doi: 10.1089/aid.1994.10.1679. [DOI] [PubMed] [Google Scholar]
- 43.Simmonds P, Balfe P, Ludlam C A, Bishop J O, Brown A J. Analysis of sequence diversity in hypervariable regions of the external glycoprotein of human immunodeficiency virus type 1. J Virol. 1990;64:5840–5850. doi: 10.1128/jvi.64.12.5840-5850.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simmons G, Wilkinson D, Reeves J D, Dittmar M T, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, Clapham P R. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tajima F, Nei M. Estimation of evolutionary distance between nucleotide sequences. Mol Biol Evol. 1984;1:269–285. doi: 10.1093/oxfordjournals.molbev.a040317. [DOI] [PubMed] [Google Scholar]
- 46.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vogel T, Kurth R, Norley S. The majority of neutralizing Abs in HIV-1-infected patients recognize linear V3 loop sequences. Studies using HIV-1MN multiple antigenic peptides. J Immunol. 1994;153:1895–1904. [PubMed] [Google Scholar]
- 48.Westervelt P, Trowbridge D B, Epstein L G, Blumberg B M, Li Y, Hahn B H, Shaw G M, Price R W, Ratner L. Macrophage tropism determinants of human immunodeficiency virus type 1 in vivo. J Virol. 1992;66:2577–2582. doi: 10.1128/jvi.66.4.2577-2582.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Willey R L, Smith D H, Lasky L A, Theodore T S, Earl P L, Moss B, Capon D J, Martin M A. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J Virol. 1988;62:139–147. doi: 10.1128/jvi.62.1.139-147.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Willey R L, Theodore T S, Martin M A. Amino acid substitutions in the human immunodeficiency virus type 1 gp120 V3 loop that change viral tropism also alter physical and functional properties of the virion envelope. J Virol. 1994;68:4409–4419. doi: 10.1128/jvi.68.7.4409-4419.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolinsky S M, Korber B T, Neumann A U, Daniels M, Kunstman K J, Whetsell A J, Furtado M R, Cao Y, Ho D D, Safrit J T. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science. 1996;272:537–542. doi: 10.1126/science.272.5261.537. [DOI] [PubMed] [Google Scholar]
- 52.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 53.Yamashita A, Yamamoto N, Matsuda J, Koyanagi Y. Cell type-specific heterogeneity of the HIV-1 V3 loop in infected individuals: selection of virus in macrophages and plasma. Virology. 1994;204:170–179. doi: 10.1006/viro.1994.1521. [DOI] [PubMed] [Google Scholar]
- 54.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. CABIOS. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 55.Yang Z, Kumar S, Nei M. A new method of inference of ancestral nucleotide and amino acid sequences. Genetics. 1995;141:1641–1650. doi: 10.1093/genetics/141.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu X F, Wang Z, Beyrer C, Celentano D D, Khamboonruang C, Allen E, Nelson K. Phenotypic and genotypic characteristics of human immunodeficiency virus type 1 from patients with AIDS in northern Thailand. J Virol. 1995;69:4649–4655. doi: 10.1128/jvi.69.8.4649-4655.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang J, Kumar S, Nei M. Small-sample tests of episodic adaptive evolution: a case study of primate lysozymes. Mol Biol Evol. 1997;14:1335–1338. doi: 10.1093/oxfordjournals.molbev.a025743. [DOI] [PubMed] [Google Scholar]
- 58.Zhang L, Diaz R S, Ho D D, Mosley J W, Busch M P, Mayer A. Host-specific driving force in human immunodeficiency virus type 1 evolution in vivo. J Virol. 1997;71:2555–2561. doi: 10.1128/jvi.71.3.2555-2561.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu T, Mo H, Wang N, Nam D S, Cao Y, Koup R A, Ho D D. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]