Abstract
The explosion and abundance of digital data could facilitate large-scale research for psychiatry and mental health. Research using so-called “real world data”—such as electronic medical/health records—can be resource-efficient, facilitate rapid hypothesis generation and testing, complement existing evidence (e.g. from trials and evidence-synthesis) and may enable a route to translate evidence into clinically effective, outcomes-driven care for patient populations that may be under-represented. However, the interpretation and processing of real-world data sources is complex because the clinically important ‘signal’ is often contained in both structured and unstructured (narrative or “free-text”) data. Techniques for extracting meaningful information (signal) from unstructured text exist and have advanced the re-use of routinely collected clinical data, but these techniques require cautious evaluation. In this paper, we survey the opportunities, risks and progress made in the use of electronic medical record (real-world) data for psychiatric research.
Subject terms: Psychiatric disorders, Depression, Bipolar disorder
Introduction
Psychiatry covers a vast heterogeneous group of mental disorders, manifesting as unusual mental or behavioural patterns that can impact an individual. Psychiatric research has increased rapidly to help in understanding the mechanisms of disease and treatments of multiple mental health and neurological disorders. With the growth of large-scale data, such as electronic medical records (EMR), research into psychiatric disorders can benefit from this and can provide multiple opportunities in psychiatric research that will produce evidence that could be incorporated into standards and guidelines. This, in turn, will directly impact clinical decision-making and, ultimately, the patient benefit.
Electronic medical (health) records (EMR) contain data describing clinical interactions, administrative, medico-legal, diagnostic, intervention, prescribing and investigations collected for the purposes of providing routine clinical care. In psychiatry (unlike other medical specialities), detailed clinical data is most often in unstructured, narrative “free text” and depending on the healthcare system, other clinical data (e.g. structured data recording the results of investigations and prescribing) will be available to varying degrees. Rather than representing unadulterated “real world” data, the potential for EMRs to provide relevant, reliable and rich data varies depending on the application; for example, reusing EMR data for predicting child and adolescent mental health problems after first contact with services [1] demonstrated limited utility. An often unrecognised problem with EMR data—particularly as a source of observational, retrospective cohort data—is that the content reflects treatment as usual (i.e. extracted prescribing data will likely display indication biases), the culture of the institution and its practitioners (e.g. unstructured narrative data might reflect the mixing of administrative, medico-legal and clinical data) and the institution’s implementation of an EMR platform [2, 3] (for example, whether the pathology EMR system in use at the same hospital are linked meaningfully to the central EMR being used for research data extraction) [4].
There is still a common consensus that randomised control trials (RCTs) are the gold standard to provide causal evidence for the efficacy, effectiveness and benefits of interventions, and for inferential modelling of risk factors for mental illness. However, RCTs can be expensive, time-consuming, unethical to conduct and generally have short follow-up times compared to observational studies. Some argue that this delivers evidence lacking generalisability to patients and their presentations in routine clinical care and excludes those patients whose risk formulation excludes them from clinical trials. Therefore, evidence derived from EMR-based research has the potential to complement evidence from controlled trials, especially when considering health equity and reproducibility [5–7]. Furthermore, causal inference methods are being introduced to address some of the biases in observational research using EMR data.
This narrative review concentrates on how research on neuropsychiatric disorders (such as depression, bipolar, schizophrenia, anxiety, eating disorders and dementia) can utilise big data such as EMRs to generate evidence to inform clinical decision-making and, importantly, improve patient outcomes. Examples of types of data that can be utilised, examples of use, and the benefits and limitations of EMR will be discussed. Finally, a summary of how EMR can be further advanced, such as the use of genetics and data triangulation will be discussed to further help optimise EMR for psychiatry research.
Data sources for large-scale psychiatric research
There is a vast variety of data sources that can be used for large-scale research in psychiatry. Before designing a study, it is important to understand different data sources and their strengths and limitations to ensure a research question can firstly be answered and then without significant biases. Broadly speaking, large-scale data resources can be roughly grouped into three types (Fig. 1).
Fig. 1. Examples of potential data sources for psychiatric research.
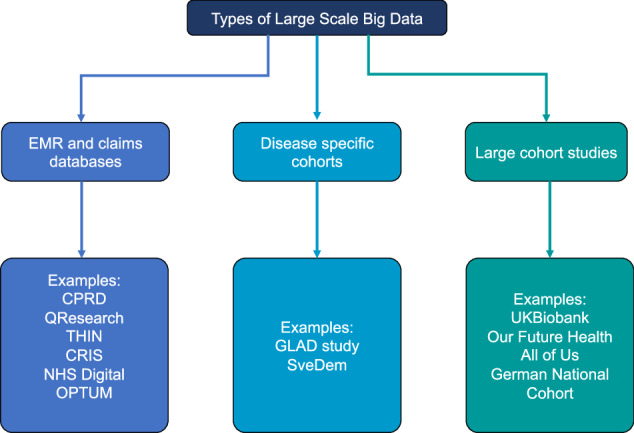
CPRD: Clinical Practice Research Datalink [174], QResearch (https://www.qresearch.org/), THIN: The health improvement network (https://www.the-health-improvement-network.com/), CRIS: Clinical Record Interactive Search, OPTUM (https://www.optum.com/), NHS Digital (https://digital.nhs.uk/), GLAD study: Genetic Links to Anxiety and Depression Study [175], SveDem: The Swedish Dementia Registry [176], UK Biobank [177], Our Future Health (https://ourfuturehealth.org.uk/), All of Us (https://allofus.nih.gov/), German National Cohort [178]. EMR and claims databases contain a variety of data formats which can be classified as structured or unstructured [69]. Structured data includes information such as age and gender, measurements such as blood pressure readings, height and also diagnosis codes, laboratory tests and medication prescribing. Whereas unstructured text includes narrative data such as clinical notes (e.g. biopsychosocial formulations, differential diagnoses, mental state examinations and risk formulations). Compared to narrative, unstructured data, structured data is easier to process with little pre-processing because it is stored in a standardised format. EHR and claims databases have vast patient numbers covering all diseases and disorders, giving the opportunity to look at psychiatric conditions and their comorbid diseases.
Disease registries contain patients with a specific condition and collect patient information longitudinally [8]. As the early and accurate diagnosis of psychiatric conditions is essential for better disease monitoring and management, registries represent a valuable tool for studying the known risk factors, as well as identifying new risk factors and markers that may help improve the accuracy of diagnostic procedures in psychiatry [8]. Disease registries also allow insights into medication use and their effectiveness and adverse effects in managing mental health conditions. Therefore, disease registries play an important part in improving health outcomes for patients and reducing healthcare costs [9].
Large population cohort studies contain large sample sizes and extensive phenotypic, imaging and biological measurements, including genetics [10]. Due to the large number of participants, this allows researchers to investigate psychiatric conditions with sufficient statistical power. With genotyping carried out for these large cohort studies this allows for the complex relationship of multiple small-effect genetic and environmental influences of psychiatric conditions to be studied [11]. One of the caveats of some large population studies is the potential lack of representativeness [12] and diversity, particularly for those with mental health conditions [13]. Other data sources that are potentially important for psychiatry research include data collected from wearables, mobile phones and social media platforms [14–18].
The current uses of EMR to enhance psychiatry research
EMR is used to generate a wide variety of evidence to inform and improve patient care ranging from using curated EMR data for epidemiology to identifying novel risk factors, opportunities for innovation in treatments and predictive analytics for those at risk and/or treatment response. The main uses related to the psychiatry field are discussed below.
Comparative effectiveness studies
Comparative effectiveness research using EMR can provide evidence to improve patient care and reduce healthcare costs. This is done by comparing the benefits and harms of alternative treatments or methods to prevent, diagnose, and treat a variety of health conditions [19, 20]. There are a variety of study designs that can be implemented to understand the effects of different mental health disorders, such as anxiety and depression, on quality of life before and after diagnosis [21], as well as the effectiveness of different medications [22–24] and different treatment regimes [25] for a variety of mental illnesses [26–28]. For neurodegenerative conditions such as dementia, there is also a growing body of evidence using EMR to investigate the potential benefits and harms of licensed medications [29–34]. As there is evidence that common diseases such as diabetes and hypertension are probable risk factors [35], this suggests that treatments for these conditions may influence cognitive decline and potentially modify dementia risk. On the other hand other anticholinergic medications [36, 37] and benzodiazepines [38, 39], may accelerate decline or increase the risk of dementia.
Descriptive studies
Descriptive studies quantify features of the health of a population of interest. This leads to knowledge that could generate hypotheses for aetiologic research and inform action in the population it concerns [40, 41]. The use of descriptive studies can be used to estimate the burden of disease in a population at a certain point in time or over time (e.g. incidence and prevalence). For psychiatry, descriptive studies can be used to ascertain if there have been changes in trends of mental health disorders such as depression [42] and anxiety [43] as they present to healthcare services or within certain populations of patients with chronic diseases, mental health conditions [44] and life-limiting diseases such as cancer [45]. This can help develop strategies that could mitigate and treat those with mental health conditions and descriptive epidemiology has been vital to understanding the impact of the COVID-19 pandemic on mental health [46–48]. Other types of descriptive studies entail describing drug utilisation and adverse drug reactions to medications [49, 50]. These studies can provide information regarding potential over-, under- or mis-prescribing of medications leading to poorer patient outcomes, particularly in high-risk populations such as those with mental health or neurological conditions [49, 51, 52].
Prediction modelling
Predictive modelling attempts to complement evidence-based medical practice by providing methods for using clinical data to estimate an individual’s probability of, e.g. experiencing benefit or harm from a treatment, experiencing an outcome (prognosis) or having a diagnosis [53]. A critical stage in developing predictive models is external validation and calibration of a tentative model, ideally in a prospective evaluation. EMRs are often conceived as ideal data sources for predictive model development and, sometimes, validation; but currently, there is limited evidence for the robustness of predictive models in psychiatric applications more generally, for example, in a systematic review of risk prediction models [54], of 89 studies, only 29 had been subjected to external validation and 1 study was considered for implementation.
Common clinical domains for predictive modelling include suicide risk [55], diagnostic trajectories [56], treatment outcomes in depression [57] and identification of dementia cases [58]. Notably, many well-designed and implemented models (e.g. those with robust validation) have tended to use national registry data (rather than EMR-derived data). Whilst individual studies using EMR data have shown promise [59–61], there is little synthesised evidence demonstrating the value of EMRs for predictive modelling. Registry data is (importantly) different from EMR data (even if one federates a number of organisation’s individual EMRs) because registries are samples of the whole population, whereas EMRs are selection-biassed (i.e. only people who are unwell and require input from services will be visible in EMRs).
Challenges and opportunities with using electronic medical records for large-scale psychiatry research
One of the most important considerations when utilising EMR for research is that it is collected for healthcare and not for research purposes. It is important to understand this when using EMRs for research because they contain a vast amount of data that reflects medico-legal and administrative concerns, rather than being clinically relevant.
The Big Data Paradox
Big data can be characterised by its variety, volume, velocity, and veracity [62, 63]. In context, EMR can be considered “big data” (due to its variety, volume and veracity) containing information in the order of thousands to millions of patients. The large number of patients and coverage of clinical conditions allow opportunities to study rare events or disorders (i.e. exploiting volume, variety and veracity) encountered in “real-world” clinical practice [64]. However, EMR is collected to support healthcare delivery and services, which gives rise to heterogeneity in the data collected. The volume of EMR datasets promises large sample sizes but this often leads to an assumption that derived error and uncertainty estimates will be necessarily more precise. However, this commonly received wisdom does not always hold; the “big data paradox” [65, 66] describes how increasing the sample size alone does not guarantee a more precise estimate of e.g. sample averages. In studies of survey data, vaccine uptake and the prediction and tracking of flu [67, 68], large sample sizes yielded misleadingly narrow uncertainty estimates leading to biased population inferences. We should be mindful of the quality, heterogeneity, and problem difficulty that are all functions of the data used, how it is collected, and the specific application or re-use of that data [65].
The dominance of unstructured text in electronic medical records in psychiatry
Unstructured data, such as free text, requires considerable pre-processing and, usually, domain expertise and human annotation. A major problem with clinical free text is the language used by clinicians is often idiosyncratic, with frequent abbreviations (sometimes, with parochial meaning such as the names of clinical services), and varied medical vocabularies [69]. Drug names, for example, often have different brand names in different national territories or “class” nomenclature (i.e. “antidepressant”) depending on the institution, requiring ontologies to be developed for mapping between synonymous terms (e.g. the Unified Medical Language System [70]) to assist pre-processing before being used in analyses or model development. Within psychiatry and mental health the number of clinical notes for any individual can be very large and written in a narrative but terminologically dense manner and often contain a high proportion of redundant text [71]. Further, unlike other medical and surgical specialities (that can utilise EMR-based sources of routinely collected structured data), psychiatry is far more reliant on clinical information such as symptoms, behaviour and clinical assessments within the unstructured notes. The major task is to represent this clinical text in a useful way for both algorithms and clinicians alike.
The computational processing and analysis of human language found in the unstructured text (clinical notes) falls under the broad field of natural language processing (NLP), which pertains to the statistical [72, 73] and deterministic (e.g. rule-based) representation and processing of language. NLP seeks to represent words, sentences, paragraphs and sometimes, the entire text corpus in such a way that algorithms can be deployed to automate task-specific analyses of the text. Contemporary NLP usually combines rule-based methods with statistical (usually machine and deep learning methods) to represent written and spoken language. The current state of the art for NLP focuses on pre-trained language models (PLMs, very-large deep learning NLP networks trained using a language modelling objective) like BERT [74] and GPT-3 [75]. PLMs better capture semantic nuances contained in sequences of text and have seen state-of-the-art performance in a considerable number of domains e.g. finance, internet of things, biomedical [75–77]. Most impactful applications of PLMs to EMR-free text have focused on Information Extraction, e.g. named entity recognition. This has spawned a number of tools to create structured representations of that free text to aid clinical decision support, such as MedCAT [78], NeuroBlu [3], and Med-7 [79]. However, the research into the representation of clinical notes in psychiatry as a whole is still relatively limited, especially in relation to the latest trends in NLP.
A concrete example of a challenge in NLP applied to narrative EMR data in psychiatry concerns the vernacular use of diagnostic terminology; for example, a healthcare professional might summarily describe their impression that “the patient seems depressed”. In isolation, this statement might refer to signs (observations by the professional), symptoms (difficulties reported by the patient) or a summary diagnosis (the signs and symptoms observed in this clinical encounter meet diagnostic criteria for a depressive disorder or episode). Similarly, a recording of clinical state might read “Mood: normal” and could refer to the patient’s mood being normal for them (referencing a previously observed clinical state), a normative assessment representing a lack of pathology (where the clinician’s recording references their own experience of the population of people with “abnormal” mood) or could represent a change over time (i.e. that the patient’s mood has returned to some baseline). Resolving these different interpretations remains difficult using data-driven lexical or statistical analyses of language and necessarily, resource-intensive expert human annotation is required.
Resource challenges using machine learning-based NLP within psychiatry
Contemporary neural networks (NN) are computationally expensive when compared to other mature machine- and statistical-learning methods. Practical development of NN models requires parallel processing using Graphical Processing Units (GPU) that are costly. The last few years have seen neural networks reaching the size of hundreds of billions of parameters, and the amount of data used to train them is usually comparably vast. A prominent large language model, GPT-3 [75], has ~175 billion model parameters (by comparison, the human brain has ~86 billion neurons). Commercial interests often obfuscate accurate costing, but speculative estimates are of the order of several million US dollars to train models of this magnitude [80, 81]. This trend of increasing performance through scaling of model size/complexity is problematic for resource-constrained environments such as publicly-funded hospitals (i.e. the UK’s National Health Service).
A crucial component of any AI/ML-driven algorithm or tool is that it is trusted and usable by human clinicians and patients; Critically, imbuing trust in a model requires that the algorithm deliver outputs that include justifications or reasons for reaching a given output or decision, sometimes referred to as XAI (eXplainable AI). Many ML methods (and especially deep learning neural networks) are opaque or “black-box” models, where the computational processes that intervene between input and output are too complex to be easily understood by any human user. There is an active research field dedicated to illuminating the machinery of such models, although the concept of what constitutes an explainable or interpretable model remains controversial [82]. If clinicians and patients are to trust an AI/ML model, they will likely favour model transparency and simplicity—often described as intrinsically interpretable models [83]—over the often modest performance gains given by complex DL models [84]. Free text data in sensitive (and, for psychiatry, often stigmatising) settings carries serious privacy risks due to the difficulty in adequately anonymising data and removing personally identifiable content [69]. For this reason, these data are often warehoused with strict data access regulations that necessarily inhibit reproducibility and replicability efforts.
Problems with data linkages and selection bias in EMR
Linking together information about the same individual across multiple data sources can further enhance existing data [85, 86], improve the quality of information, and offer a relatively quick and low-cost means to exploit existing data sources. One benefit of data linkage in psychiatry is it can provide additional information on other non-psychiatric conditions and medications [87], allowing more detailed information about patient’s medical history, which can be used to reduce biases in research studies. Although data linkages can improve knowledge about psychiatric research [87, 88], there are limitations. Errors in the data linkage process can introduce bias of unknown size and direction, which could feed through into final research results, leading to overestimating or underestimating results [89]. Missingness of different participant characteristics in EMR, such as age, gender and race, can also lead to systematic bias and issues with the validity and generalisability of research results [90, 91].
Selection bias is a common problem in observational research and occurs when characteristics influence whether a person is included in a group. For example, in psychiatry, only those with extreme mental health conditions enter secondary care due to the different priorities of healthcare providers and government funds. Therefore, any research studies using EMR in secondary care will differ from the general population [92]. Furthermore, selection biases can exacerbate existing disparities, such as those relating to ethnicity, sexual orientation and socioeconomic status, that can lead to inequalities in treatment and healthcare [93–97]. Findings from psychiatric research conducted in selected groups should be interpreted with great caution unless selection bias has been explicitly addressed.
Phenotyping in psychiatric research
Phenotyping is the process of identifying specific patients with a clinical condition or characteristic(s) based on information in their EMR [98]. It can involve combining different types of data such as diagnosis codes, procedures, medication data, laboratory and test results, and unstructured text [99] with growing interest in using data from smartphones and other digital wearables [15, 100]. Phenotypes can be derived using algorithms that use filters and rule-based algorithms or machine learning methods based on structured data [101, 102]. The Electronic MEdical Records and GEnomics (eMERGE) Network [103] and CALIBER [104] have both shown that phenotypes can be identified and validated and consequently used in research. Patients identified with a specific phenotype can be included in cohort studies in order for further study of risk factors or drug safety surveillance, genetic studies as well as recruitment for clinical trials [105–110]. The psychiatry field presents a unique challenge for phenotyping as the majority of psychiatric diagnoses typically rely on self-reported symptoms, behaviour and clinical judgement, meaning a combination of structured and unstructured text has been shown to give rise to more accurate phenotypes with less misclassification of cases [111, 112]. Problems arise with phenotyping when there is no consistency in the phenotyping process, only using structured data may not accurately represent the disease status of the patient, what types or combinations of data could be used from different healthcare datasets and the lack of translation of phenotypes to different health care settings and countries [113]. As phenotyping is a dynamic process, it requires clinical expertise and multiple cycles of review and can take many months of development [114]. Once a phenotype has been derived, validation of the phenotype is a critical process [115]. A phenotype must have high sensitivity and specificity, limiting both false positives and false negatives. Validation can be done using a variety of different approaches [104], such as by cross-referencing different data EMR sources and case note reviews by clinical experts to confirm a diagnosis based on the phenotype developed. Accuracy measures can then dictate how useful a phenotype will be for use in further research [106, 116, 117].
Future considerations for optimising the use of electronic medical records in psychiatry
There are many examples of the use of EMR to generate evidence in psychiatric research. However, to aid in the improvement and research applications of EMR we discuss future considerations which could optimise the use of EMR.
Design, statistical techniques to address biases and reporting in observational psychiatric research
The design, analysis and reporting are vital components for optimising the use of EMR. Observational research using EMR is utilised because RCTs that would answer causal questions are sometimes not feasible, unethical and take too long. By applying the study design principles of RCTs to observational studies, the causal effect of an intervention [118, 119] can be estimated, and this helps avoid biases such as selection and immortal time biases [120]. This approach, called “target trial emulation” [118], uses EMR to emulate a clinical trial—the target trial—that would answer the causal research question. If target trial emulation is successful the results from observational data can yield similar results to the RCT [121–125]. Target trial emulation is now being used for a wide variety of conditions, such as showing potential beneficial effects of statins with dementia risk [126] and harmful effects of protein pump inhibitors with dementia risk [127]. Other applications include determining optimal drug plasma concentrations in bipolar disorder [128] and establishing the risk of diabetes with anticonvulsant mood stabilisers [129]. Target trial emulation cannot remove bias due to the lack of randomisation of observational data [118]. However, methods to address this, such as propensity scores, can be applied to reduce this confounding [130, 131].
Clinical decision support tools for psychiatry could include identifying or detecting those at risk of certain disorders, illness progression/prognosis and using treatment response data to improve personalised care. However, specifically for predictive models, research has shown that over 90% were at high risk of bias [53]. Therefore, in order to optimise the use of EMR for developing clinical decision support, we require careful attention to model development, sample sizes [132], internal and external validation, including calibration and assessment of clinical utility and generalisability should be adopted [59, 133].
Studies using EMR can be prone to publication bias and reporting bias [134, 135]. On top of this, published research often omits important information or the information is unclear and very often, the nature of EMR data means it cannot be shared for interrogation, reproducibility and replication studies. These biases are a concern because they undermine the validity of studies. Study analysis plans and study results should be reported transparently, including what was planned, what was carried out, what was found, and what conclusions were drawn. Researchers can now register statistical analysis plans for a study prior to analysis (e.g. clinicaltrials.gov, researchregistry.com, encepp.eu) and the STROBE [136] and TRIPOD [137] guidelines offer a checklist of items that should be addressed in articles reporting studies to increase transparency [138, 139]. Furthermore, in order to improve reproducibility and ambiguity, analytical code should also be freely available [140].
Precision medicine to provide individualised healthcare
With the increase in the availability of accurate deep phenotyping information from unstructured text researchers will be able to make more precise insights about disease outcomes from clinical information. This has expanded the scope of evidence-based prediction and tools designed to triangulate evidence from multiple sources are now being developed for applications in precision medicine. For example, the Petrushka [105] web-based tool uses data from multiple sources, including QResearch (primary care), EMR (secondary care) and available literature to make personalised medication recommendations in individuals with unipolar depression. Other projects seek to incorporate other data modalities, such as wearables to give a more detailed digital phenotype [141]. However, further validation is needed to convince clinicians of the benefits of supported clinical decision-making.
Triangulation of evidence from multimodal data for large-scale psychiatry research
There is a vast array of data acquired in research and healthcare which covers a variety of different modalities. These different modalities, such as omics, histology, imaging, clinical and smart technology, can help researchers unveil novel mechanistic insights to help understand crucial information about the complexity of mental health and neurological conditions. Triangulation of evidence is an approach where one can obtain more confidence in results by carrying out analyses integrating different statistical methodologies and/or data modalities [142, 143]. The key is that each analysis has different sources of potential bias that may be unrelated to each other. If the results from each different analysis point to the same conclusion, this strengthens the confidence in the findings obtained. Examples of this triangulation approach in mental health research include assessing the relationship between cultural engagement and depression, where the authors used three different statistical methodologies with different strengths and weaknesses to show lower cultural engagement is associated with depression outcomes [144]. Other examples used observational data and genetic data to triangulate evidence between smoking and suicide ideation and attempts [145], and anxiety disorders and anorexia nervosa [146]; however, the triangulated results were inconsistent with each other, potentially questioning the causal relationships established using any of the sources. For psychiatry, triangulation could be used by applying different statistical approaches, using different EMRs across different countries and healthcare settings and/or integrating other non-EMR data as discussed below to help provide further understanding regarding causality and optimise big data in psychiatric research.
Incorporating biomarkers in EMR phenotyping
Biomarkers are biological measures utilised to better diagnose, track or predict psychiatric disorders. These can range from clinical assays and brain imaging to digital biomarkers from wearables. In EMR research, they can be used to help define disease phenotypes or better understand outcomes and applications in precision medicine. In Alzheimer’s Disease, fluid biomarkers (cerebrospinal fluid and blood plasma) for the tau protein are used to determine disease pathology to aid in trial recruitment. Biomarkers of neurodegeneration have been successful, neurofilament light measured in blood or CSF can be used to assess axon damage [147]. Measures of inflammation, such as C-reactive protein, have applications in many psychiatric disorders. Disorder-specific markers have been identified and replicated in meta-analyses for Vitamin B6 in schizophrenia and basal cortisol awakening in bipolar disorder [148]. However, in many psychiatric disorders, translation to clinical applications is limited [148, 149] and further work will be necessary to validate these potential candidates in suitable cohorts.
The suitability of the marker modality should be considered when selecting a biomarker. In mental health conditions, the development of a digital marker captured by remote monitoring might aid in diagnosis by adding information to the self-reporting of symptoms from a patient, for example, if the marker can act as a proxy for behavioural signs of mental illness that cannot be captured by a single measurement of clinical state when consulting a clinician. The development of phone-based applications allows clinicians to collect data on changes through a series of symptom-based questions [150]. However, the future of biomarker discovery is likely the ability to measure, compare and combine multiple variables and here, resources are key. The Penn Medicine Biobank [151] includes genetics and biomarkers alongside EMR to enable precision medicine and the discovery of new phenotypes.
Interrogation of EMRs has revealed the potential value of routinely recorded data to identify and validate the use of existing and exploratory biomarkers. For example, in a study of sepsis, biomarkers were used alongside EMR to study progression [152]. Biomarkers were employed to study different time periods whereby early-life mental health impacts midlife using a panel of markers [153]. Elsewhere the combination of biomarkers and EMR has been recommended for aiding risk reduction profiles and identifying new clinical biomarkers [154].
The use of genetics for large-scale psychiatry research
The observational nature of many findings precludes drawing firm conclusions about causality due to residual confounding and reverse causation. With the recent explosion of large-scale genetic data available, methods using this data, such as polygenetic risk scores (PRS) and Mendelian randomisation (MR) allow the elucidation of causal relationships between psychiatric disorders, risk factors and drug treatments. Leveraging genetic data using PRS and MR offers a cost-effective approach with the future potential to embed genetic data into healthcare settings to help improve patient care [155] and could provide complementary evidence as part of the triangulation process.
PRS are weighted sums of genetic variants associated with a particular condition [156]. Therefore, PRS can estimate the genetic risk of an individual for a disease or trait. Due to the complex polygenic nature of many conditions (including mental health and neurological conditions [157–161], PRS can only capture a fraction of the overall risk, with clinical and demographic factors usually explaining most variance. This means that on their own, PRS is unlikely to definitively predict future diagnoses in a healthcare setting [162, 163]. However, PRS could be included with other measures to predict future risk and may show promise in aiding clinical decision-making. Adding PRS to risk prediction models alongside other clinical factors such as age, gender and family history has been shown to improve model performance for predicting the risk of conditions such as dementia [164] and certain mental health conditions [162, 165, 166]. On top of this, PRS may have some potential to inform treatment response if polygenetic complex traits can be predicted from an individual’s genetics [167] and those traits are robustly associated with treatment response. PRS can be used in conjunction with other methods, such as Mendelian randomisation [168], to uncover casual insights between complex psychiatric traits and treatments.
Mendelian randomisation (MR) is a statistical approach that uses genetic variants to provide evidence that an exposure has a causal effect on an outcome [169–173] (Fig. 2). A genetic variant (or variants) is used which is associated with an exposure (e.g., insomnia) but not associated with any other risk factors which affect the outcome (e.g., depression). By doing so, any association of the genetic variant(s) with the outcome must act via the variants’ association with the exposure and imply a causal effect of the exposure on the outcome. As genetic variants are inherited randomly at conception, genetic variants are not susceptible to reverse causation and confounding, like observational studies using EMR. Results from MR can help support results from EMR by using data triangulation, as discussed previously.
Fig. 2. The concept of Mendelian randomisation (MR) and its assumptions.
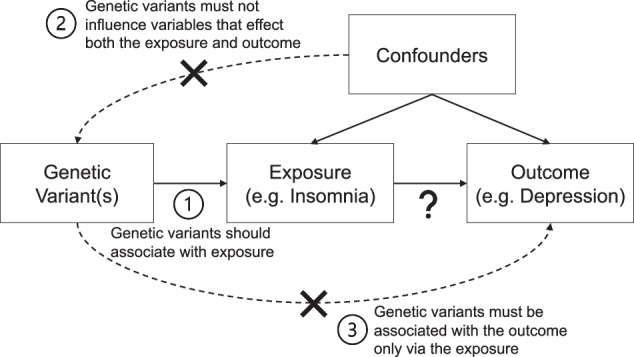
There are a growing number of MR studies being published that show causal effects related to disease-disease associations and drug-disease associations for mental health and old age psychiatry [179–186] with extensions to traditional MR approaches, which could offer further insights [187–189].
Conclusion
Large-scale research approaches are at the forefront of EMR use in psychiatry. With the advances in interpretation using NLP and access to diverse data resources, the scope of research questions is rapidly expanding. However, care is needed to make sure that potential biases are considered. Not considering limitations with big data can lead to incorrect inferences about a population which could mean poorer care for high-risk populations such as those with mental health conditions and neurodegenerative conditions.
In order to optimise EMR for psychiatry a clear understanding of such biases in the data is vital. A researcher must carefully consider if the research question can be answered in the data source they want to use and develop the best study design and statistical analysis. By cautiously incorporating the strengths of the EMR format it will be possible to make exciting contributions to mental health and neurological research.
Acknowledgements
LW is supported by an Alzheimer’s Research UK fellowship (ARUK-RF2020A-005) and funding from Virtual Brain Cloud from the European Commission [grant number H2020-SC1-DTH-2018-1] and Rosetrees Trust (M937). NT is supported by the EPSRC Center for Doctoral Training in Health Data Science (EP/S02428X/1). DWJ is supported in part by the NIHR AI Award for Health and Social Care (NIHR-AI-AWARD0-2183). DWJ is supported by the NIHR Oxford Health Biomedical Research Centre (grant BRC-1215-20005). The views expressed are those of the authors and not necessarily those of the UK National Health Service, the NIHR, the UK Department of Health, or the University of Oxford.
Author contributions
DN, NT, and LW drafted the manuscript. LW and DWJ critically revised and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Koning NR, Büchner FL, van den Berg AW, Choi SYA, Leeuwenburgh NA, Paijmans IJM, et al. The usefulness of electronic health records from preventive youth healthcare in the recognition of child mental health problems. Front Public Health 2021;9. 10.3389/fpubh.2021.658240. [DOI] [PMC free article] [PubMed]
- 2.Werbeloff N, Osborn DPJ, Patel R, Taylor M, Stewart R, Broadbent M, et al. The Camden & Islington Research Database: using electronic mental health records for research. PLoS ONE. 2018;13:e0190703. doi: 10.1371/journal.pone.0190703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patel R, Wee SN, Ramaswamy R, Thadani S, Tandi J, Garg R, et al. NeuroBlu, an electronic health record (EHR) trusted research environment (TRE) to support mental healthcare analytics with real-world data. BMJ Open. 2022;12:e057227. doi: 10.1136/bmjopen-2021-057227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castillo EG, Olfson M, Pincus HA, Vawdrey D, Stroup TS. Electronic health records in mental health research: a framework for developing valid research methods. Psychiatr Serv. 2015;66:193–6. doi: 10.1176/appi.ps.201400200. [DOI] [PubMed] [Google Scholar]
- 5.Power MC, Engelman BC, Wei J, Glymour MM. Closing the gap between observational research and randomized controlled trials for prevention of Alzheimer disease and dementia. Epidemiol Rev. 2022;44:17–28. doi: 10.1093/epirev/mxac002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jankovic J, Parsons J, Jovanović N, Berrisford G, Copello A, Fazil Q, et al. Differences in access and utilisation of mental health services in the perinatal period for women from ethnic minorities—a population-based study. BMC Med. 2020;18:245. doi: 10.1186/s12916-020-01711-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dennis M, Shine L, John A, Marchant A, McGregor J, Lyons RA, et al. Risk of adverse outcomes for older people with dementia prescribed antipsychotic medication: a population based e-cohort study. Neurol Ther. 2017;6:57–77. doi: 10.1007/s40120-016-0060-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heikal SA, Salama M, Richard Y, Moustafa AA, Lawlor B. The impact of disease registries on advancing knowledge and understanding of dementia globally. Front Aging Neurosci 2022;14. https://www.frontiersin.org/articles/10.3389/fnagi.2022.774005. [DOI] [PMC free article] [PubMed]
- 9.Hoque DME, Kumari V, Hoque M, Ruseckaite R, Romero L, Evans SM. Impact of clinical registries on quality of patient care and clinical outcomes: a systematic review. PLoS ONE. 2017;12:e0183667. doi: 10.1371/journal.pone.0183667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berger K, Rietschel M, Rujescu D. The value of ‘mega cohorts’ for psychiatric research. World J Biol Psychiatry. 2023;24:860–4. [DOI] [PubMed]
- 11.Davis K, Hotopf M. Mental health phenotyping in UK Biobank. Prog Neurol Psychiatry. 2019;23:4–7. doi: 10.1002/pnp.522. [DOI] [Google Scholar]
- 12.Fry A, Littlejohns TJ, Sudlow C, Doherty N, Adamska L, Sprosen T, et al. Comparison of sociodemographic and health-related characteristics of UK Biobank participants with those of the general population. Am J Epidemiol. 2017;186:1026–34. doi: 10.1093/aje/kwx246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis KAS, Coleman JRI, Adams M, Allen N, Breen G, Cullen B, et al. Mental health in UK Biobank—development, implementation and results from an online questionnaire completed by 157 366 participants: a reanalysis. BJPsych Open. 2020;6:e18. doi: 10.1192/bjo.2019.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bauer M, Glenn T, Geddes J, Gitlin M, Grof P, Kessing LV, et al. Smartphones in mental health: a critical review of background issues, current status and future concerns. Int J Bipolar Disord. 2020;8:2. doi: 10.1186/s40345-019-0164-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brietzke E, Hawken ER, Idzikowski M, Pong J, Kennedy SH, Soares CN. Integrating digital phenotyping in clinical characterization of individuals with mood disorders. Neurosci Biobehav Rev. 2019;104:223–30. doi: 10.1016/j.neubiorev.2019.07.009. [DOI] [PubMed] [Google Scholar]
- 16.Rykov Y, Thach T-Q, Bojic I, Christopoulos G, Car J. Digital biomarkers for depression screening with wearable devices: cross-sectional study with machine learning modeling. JMIR MHealth UHealth. 2021;9:e24872. doi: 10.2196/24872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee S, Kim H, Park MJ, Jeon HJ. Current advances in wearable devices and their sensors in patients with depression. Front Psychiatry 2021;12. https://www.frontiersin.org/articles/10.3389/fpsyt.2021.672347. [DOI] [PMC free article] [PubMed]
- 18.Torous J, Bucci S, Bell IH, Kessing LV, Faurholt‐Jepsen M, Whelan P, et al. The growing field of digital psychiatry: current evidence and the future of apps, social media, chatbots, and virtual reality. World Psychiatry. 2021;20:318–35. doi: 10.1002/wps.20883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merkow RP, Schwartz TA, Nathens AB. Practical guide to comparative effectiveness research using observational data. JAMA Surg. 2020;155:349–50. doi: 10.1001/jamasurg.2019.4395. [DOI] [PubMed] [Google Scholar]
- 20.Berger ML, Dreyer N, Anderson F, Towse A, Sedrakyan A, Normand S-L. Prospective observational studies to assess comparative effectiveness: the ISPOR Good Research Practices Task Force Report. Value Health. 2012;15:217–30. doi: 10.1016/j.jval.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 21.Hammond GC, Croudace TJ, Radhakrishnan M, Lafortune L, Watson A, McMillan-Shields F, et al. Comparative effectiveness of cognitive therapies delivered face-to-face or over the telephone: an observational study using propensity methods. PLoS ONE. 2012;7:e42916. doi: 10.1371/journal.pone.0042916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crescenzo FD, Garriga C, Tomlinson A, Coupland C, Efthimiou O, Fazel S, et al. Real-world effect of antidepressants for depressive disorder in primary care: protocol of a population-based cohort study. BMJ Ment Health. 2020;23:122–6. doi: 10.1136/ebmental-2020-300149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alemi F, Min H, Yousefi M, Becker LK, Hane CA, Nori VS et al. Effectiveness of common antidepressants: a post market release study. eClinicalMedicine 2021;41. 10.1016/j.eclinm.2021.101171. [DOI] [PMC free article] [PubMed]
- 24.Centorrino F, Meyers AL, Ahl J, Cincotta SL, Zun L, Gulliver AH, et al. An observational study of the effectiveness and safety of intramuscular olanzapine in the treatment of acute agitation in patients with bipolar mania or schizophrenia/schizoaffective disorder. Hum Psychopharmacol Clin Exp. 2007;22:455–62. doi: 10.1002/hup.870. [DOI] [PubMed] [Google Scholar]
- 25.Singh B, Kung S, Pazdernik V, Schak KM, Geske J, Schulte PJ, et al. Comparative effectiveness of intravenous ketamine and intranasal esketamine in clinical practice among patients with treatment-refractory depression: an Observational Study. J Clin Psychiatry. 2023;84:45331. doi: 10.4088/JCP.22m14548. [DOI] [PubMed] [Google Scholar]
- 26.Touriño AG, Feixas G, Medina JC, Paz C, Evans C. Effectiveness of integrated treatment for eating disorders in Spain: protocol for a multicentre, naturalistic, observational study. BMJ Open. 2021;11:e043152. doi: 10.1136/bmjopen-2020-043152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ayton A, Ibrahim A, Dugan J, Galvin E, Wright OW. Ultra-processed foods and binge eating: a retrospective observational study. Nutrition. 2021;84:111023. doi: 10.1016/j.nut.2020.111023. [DOI] [PubMed] [Google Scholar]
- 28.Molero Y, Kaddoura S, Kuja-Halkola R, Larsson H, Lichtenstein P, D’Onofrio BM, et al. Associations between β-blockers and psychiatric and behavioural outcomes: a population-based cohort study of 1.4 million individuals in Sweden. PLoS Med. 2023;20:e1004164. doi: 10.1371/journal.pmed.1004164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newby D, Linden AB, Fernandes M, Molero Y, Winchester L, Sproviero W, et al. Comparative effect of metformin versus sulfonylureas with dementia and Parkinson’s disease risk in US patients over 50 with type 2 diabetes mellitus. BMJ Open Diabetes Res Care. 2022;10:e003036. doi: 10.1136/bmjdrc-2022-003036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newby D, Prieto-Alhambra D, Duarte-Salles T, Ansell D, Pedersen L, van der Lei J, et al. Methotrexate and relative risk of dementia amongst patients with rheumatoid arthritis: a multi-national multi-database case-control study. Alzheimers Res Ther. 2020;12:38. doi: 10.1186/s13195-020-00606-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Desai RJ, Mahesri M, Lee SB, Varma VR, Loeffler T, Schilcher I, et al. No association between initiation of phosphodiesterase-5 inhibitors and risk of incident Alzheimer’s disease and related dementia: results from the Drug Repurposing for Effective Alzheimer’s Medicines study. Brain Commun. 2022;4:fcac247. doi: 10.1093/braincomms/fcac247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Desai RJ, Varma VR, Gerhard T, Segal J, Mahesri M, Chin K, et al. Comparative risk of Alzheimer disease and related dementia among medicare beneficiaries with rheumatoid arthritis treated with targeted disease-modifying antirheumatic agents. JAMA Netw Open. 2022;5:e226567. doi: 10.1001/jamanetworkopen.2022.6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ding J, Davis-Plourde KL, Sedaghat S, Tully PJ, Wang W, Phillips C, et al. Antihypertensive medications and risk for incident dementia and Alzheimer’s disease: a meta-analysis of individual participant data from prospective cohort studies. Lancet Neurol. 2020;19:61–70. doi: 10.1016/S1474-4422(19)30393-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ballard C, Aarsland D, Cummings J, O’Brien J, Mills R, Molinuevo JL, et al. Drug repositioning and repurposing for Alzheimer disease. Nat Rev Neurol. 2020;16:661–73. doi: 10.1038/s41582-020-0397-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livingston G, Huntley J, Sommerlad A, Ames D, Ballard C, Banerjee S, et al. Dementia prevention, intervention, and care: 2020 report of the Lancet Commission. The Lancet. 2020;396:413–46. doi: 10.1016/S0140-6736(20)30367-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coupland CAC, Hill T, Dening T, Morriss R, Moore M, Hippisley-Cox J. Anticholinergic drug exposure and the risk of dementia: a Nested Case-Control Study. JAMA Intern Med. 2019;179:1084–93. doi: 10.1001/jamainternmed.2019.0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Group BMJP. Anticholinergic drugs and risk of dementia: case-control study. BMJ. 2019;367:l6213. doi: 10.1136/bmj.l6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joyce G, Ferido P, Thunell J, Tysinger B, Zissimopoulos J. Benzodiazepine use and the risk of dementia. Alzheimers Dement Transl Res Clin Interv. 2022;8:e12309. doi: 10.1002/trc2.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He Q, Chen X, Wu T, Li L, Fei X. Risk of dementia in long-term benzodiazepine users: evidence from a meta-analysis of observational studies. J Clin Neurol Seoul Korea. 2019;15:9–19. doi: 10.3988/jcn.2019.15.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fox MP, Murray EJ, Lesko CR, Sealy-Jefferson S. On the need to revitalize descriptive epidemiology. Am J Epidemiol. 2022;191:1174–9. doi: 10.1093/aje/kwac056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lesko CR, Fox MP, Edwards JK. A framework for descriptive epidemiology. Am J Epidemiol. 2022;191:2063–70. doi: 10.1093/aje/kwac115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moreno-Agostino D, Wu Y-T, Daskalopoulou C, Hasan MT, Huisman M, Prina M. Global trends in the prevalence and incidence of depression: a systematic review and meta-analysis. J Affect Disord. 2021;281:235–43. doi: 10.1016/j.jad.2020.12.035. [DOI] [PubMed] [Google Scholar]
- 43.Kessler RC, Wang PS. The descriptive epidemiology of commonly occurring mental disorders in the United States. Annu Rev Public Health. 2008;29:115–29. doi: 10.1146/annurev.publhealth.29.020907.090847. [DOI] [PubMed] [Google Scholar]
- 44.Stauffacher MWD, Stiefel F, Dorogi Y, Michaud L. Observational study of suicide in Switzerland: comparison between psychiatric in- and outpatients. Swiss Med Wkly. 2022;152:w30140. doi: 10.4414/SMW.2022.w30140. [DOI] [PubMed] [Google Scholar]
- 45.Mejareh ZN, Abdollahi B, Hoseinipalangi Z, Jeze MS, Hosseinifard H, Rafiei S, et al. Global, regional, and national prevalence of depression among cancer patients: a systematic review and meta-analysis. Indian J Psychiatry. 2021;63:527–35. doi: 10.4103/indianjpsychiatry.indianjpsychiatry_77_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santomauro DF, Herrera AMM, Shadid J, Zheng P, Ashbaugh C, Pigott DM, et al. Global prevalence and burden of depressive and anxiety disorders in 204 countries and territories in 2020 due to the COVID-19 pandemic. The Lancet. 2021;398:1700–12. doi: 10.1016/S0140-6736(21)02143-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raventós B, Pistillo A, Reyes C, Fernández-Bertolín S, Aragón M, Berenguera A, et al. Impact of the COVID-19 pandemic on diagnoses of common mental health disorders in adults in Catalonia, Spain: a population-based cohort study. BMJ Open. 2022;12:e057866. doi: 10.1136/bmjopen-2021-057866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walby FA, Myhre MØ, Mehlum L. Suicide among users of mental health and addiction services in the first 10 months of the COVID-19 pandemic: observational study using national registry data. BJPsych Open. 2022;8:e111. doi: 10.1192/bjo.2022.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doherty AS, Shahid F, Moriarty F, Boland F, Clyne B, Dreischulte T, et al. Prescribing cascades in community‐dwelling adults: a systematic review. Pharmacol Res Perspect. 2022;10:e01008. doi: 10.1002/prp2.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo Y, Kataoka Y, Ostinelli EG, Cipriani A, Furukawa TA. National prescription patterns of antidepressants in the treatment of adults with major depression in the US between 1996 and 2015: a population representative survey based analysis. Front Psychiatry. 2020;11:35. doi: 10.3389/fpsyt.2020.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCarthy LM, Savage R, Dalton K, Mason R, Li J, Lawson A, et al. ThinkCascades: a tool for identifying clinically important prescribing cascades affecting older people. Drugs Aging. 2022;39:829–40. doi: 10.1007/s40266-022-00964-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trenaman SC, Bowles SK, Kirkland S, Andrew MK. An examination of three prescribing cascades in a cohort of older adults with dementia. BMC Geriatr. 2021;21:297. doi: 10.1186/s12877-021-02246-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meehan AJ, Lewis SJ, Fazel S, Fusar-Poli P, Steyerberg EW, Stahl D, et al. Clinical prediction models in psychiatry: a systematic review of two decades of progress and challenges. Mol Psychiatry. 2022;27:2700–8. doi: 10.1038/s41380-022-01528-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salazar de Pablo G, Studerus E, Vaquerizo-Serrano J, Irving J, Catalan A, Oliver D, et al. Implementing precision psychiatry: a systematic review of individualized prediction models for clinical practice. Schizophr Bull. 2021;47:284–97. doi: 10.1093/schbul/sbaa120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gradus JL, Rosellini AJ, Horváth-Puhó E, Jiang T, Street AE, Galatzer-Levy I, et al. Predicting sex-specific nonfatal suicide attempt risk using machine learning and data from Danish National Registries. Am J Epidemiol. 2021;190:2517–27. doi: 10.1093/aje/kwab112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jørgensen TSH, Osler M, Jorgensen MB, Jorgensen A. Mapping diagnostic trajectories from the first hospital diagnosis of a psychiatric disorder: a Danish nationwide cohort study using sequence analysis. Lancet Psychiatry. 2023;10:12–20. doi: 10.1016/S2215-0366(22)00367-4. [DOI] [PubMed] [Google Scholar]
- 57.Sajjadian M, Lam RW, Milev R, Rotzinger S, Frey BN, Soares CN, et al. Machine learning in the prediction of depression treatment outcomes: a systematic review and meta-analysis. Psychol Med. 2021;51:2742–51. doi: 10.1017/S0033291721003871. [DOI] [PubMed] [Google Scholar]
- 58.Vyas A, Aisopos F, Vidal M-E, Garrard P, Paliouras G. Identifying the presence and severity of dementia by applying interpretable machine learning techniques on structured clinical records. BMC Med Inform Decis Mak. 2022;22:271. doi: 10.1186/s12911-022-02004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y, Wang S, Hermann A, Joly R, Pathak J. Development and validation of a machine learning algorithm for predicting the risk of postpartum depression among pregnant women. J Affect Disord. 2021;279:1–8. doi: 10.1016/j.jad.2020.09.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang SH, LePendu P, Iyer SV, Tai-Seale M, Carrell D, Shah NH. Toward personalizing treatment for depression: predicting diagnosis and severity. J Am Med Inform Assoc JAMIA. 2014;21:1069–75. doi: 10.1136/amiajnl-2014-002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bayramli I, Castro V, Barak-Corren Y, Madsen EM, Nock MK, Smoller JW, et al. Predictive structured-unstructured interactions in EHR models: a case study of suicide prediction. NPJ Digit Med. 2022;5:15. doi: 10.1038/s41746-022-00558-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.De Mauro A, Greco M, Grimaldi M. A formal definition of Big Data based on its essential features. Libr Rev. 2016;65:122–35. doi: 10.1108/LR-06-2015-0061. [DOI] [Google Scholar]
- 63.Pastorino R, De Vito C, Migliara G, Glocker K, Binenbaum I, Ricciardi W, et al. Benefits and challenges of Big Data in healthcare: an overview of the European initiatives. Eur J Public Health. 2019;29:23–27. doi: 10.1093/eurpub/ckz168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Monteith S, Glenn T, Geddes J, Bauer M. Big data are coming to psychiatry: a general introduction. Int J Bipolar Disord. 2015;3:21. doi: 10.1186/s40345-015-0038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meng X-L. Statistical paradises and paradoxes in big data (I): law of large populations, big data paradox, and the 2016 US presidential election. Ann Appl Stat. 2018;12:685–726. doi: 10.1214/18-AOAS1161SF. [DOI] [Google Scholar]
- 66.Msaouel P. The big data paradox in clinical practice. Cancer Investig. 2022;40:567–76. doi: 10.1080/07357907.2022.2084621. [DOI] [PubMed] [Google Scholar]
- 67.Lazer D, Kennedy R, King G, Vespignani A. The parable of Google flu: traps in big data analysis. Science. 2014;343:1203–5. doi: 10.1126/science.1248506. [DOI] [PubMed] [Google Scholar]
- 68.Bradley VC, Kuriwaki S, Isakov M, Sejdinovic D, Meng X-L, Flaxman S. Unrepresentative big surveys significantly overestimated US vaccine uptake. Nature. 2021;600:695–700. doi: 10.1038/s41586-021-04198-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tayefi M, Ngo P, Chomutare T, Dalianis H, Salvi E, Budrionis A, et al. Challenges and opportunities beyond structured data in analysis of electronic health records. WIREs Comput Stat 2021;13. 10.1002/wics.1549.
- 70.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32:D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Searle T, Ibrahim Z, Teo J, Dobson R. Estimating redundancy in clinical text. J Biomed Inform. 2021;124:103938. doi: 10.1016/j.jbi.2021.103938. [DOI] [PubMed] [Google Scholar]
- 72.Charniak E. Statistical language learning (language, speech, and communication). (The MIT Press; 1996).
- 73.Jones KS. Natural language processing: a historical review. In: Zampolli A, Calzolari N, Palmer M, editors. Current issues in computational linguistics: in honour of Don Walker. Springer Netherlands: Dordrecht; 1994. pp. 3–16.
- 74.Devlin J, Chang M-W, Lee K, Toutanova K. “BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding.” North American Chapter of the Association for Computational Linguistics. 2019.
- 75.Brown TB, Mann B, Ryder N, Subbiah M, Kaplan J, Dhariwal P et al. Language models are few-shot learners. [Preprint]. 2020. 10.48550/arXiv.2005.14165.
- 76.Chan JY-L, Bea KT, Leow SMH, Phoong SW, Cheng WK. State of the art: a review of sentiment analysis based on sequential transfer learning. Artif Intell Rev. 2023;56:749–80. doi: 10.1007/s10462-022-10183-8. [DOI] [Google Scholar]
- 77.Mars M. From word embeddings to pre-trained language models: a state-of-the-art walkthrough. Appl Sci. 2022;12:8805. doi: 10.3390/app12178805. [DOI] [Google Scholar]
- 78.Kraljevic Z, Searle T, Shek A, Roguski L, Noor K, Bean D et al. Multi-domain clinical natural language processing with MedCAT: the Medical Concept Annotation Toolkit. [Preprint] 2021. 10.48550/arXiv.2010.01165. [DOI] [PubMed]
- 79.Kormilitzin A, Vaci N, Liu Q, Nevado-Holgado A. Med7: a transferable clinical natural language processing model for electronic health records. Artif Intell Med. 2021;118:102086. doi: 10.1016/j.artmed.2021.102086. [DOI] [PubMed] [Google Scholar]
- 80.Bender EM, Gebru T, McMillan-Major A, Shmitchell S. On the dangers of stochastic parrots: can language models be too big? In Conference on Fairness, Accountability, and Transparency(FAccT ’21), March 3–10, 2021, Virtual Event, Canada. 14 pages (ACM, New York, NY, USA, 2021). 10.1145/3442188.3445922.
- 81.Sharir O, Peleg B, Shoham Y. The cost of training NLP models: a concise overview. [Preprint] 2020. http://arxiv.org/abs/2004.08900.
- 82.Joyce DW, Kormilitzin A, Smith KA, Cipriani A. Explainable artificial intelligence for mental health through transparency and interpretability for understandability. Npj Digit Med. 2023;6:1–7. doi: 10.1038/s41746-023-00751-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rudin C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat Mach Intell. 2019;1:206–15. doi: 10.1038/s42256-019-0048-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li B, Qi P, Liu B, Di S, Liu J, Pei J, et al. Trustworthy AI: from principles to practices. ACM Comput Surv. 2023;55:177. doi: 10.1145/3555803. [DOI] [Google Scholar]
- 85.Gilbert R, Lafferty R, Hagger-Johnson G, Harron K, Zhang L-C, Smith P, et al. GUILD: GUidance for Information about Linking Data sets†. J Public Health. 2018;40:191–8. doi: 10.1093/pubmed/fdx037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Green E, Ritchie F, Mytton J, Webber DJ, Deave T, Montgomery A, et al. Enabling data linkage to maximise the value of public health research data: summary report. 2015. https://uwe-repository.worktribe.com/output/836851/enabling-data-linkage-to-maximise-the-value-of-public-health-research-data-summary-report.
- 87.Perera G, Broadbent M, Callard F, Chang C-K, Downs J, Dutta R, et al. Cohort profile of the South London and Maudsley NHS Foundation Trust Biomedical Research Centre (SLaM BRC) case register: current status and recent enhancement of an electronic mental health record-derived data resource. BMJ Open. 2016;6:e008721. doi: 10.1136/bmjopen-2015-008721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Carson L, Jewell A, Downs J, Stewart R. Multisite data linkage projects in mental health research. Lancet Psychiatry. 2020;7:e61. doi: 10.1016/S2215-0366(20)30375-8. [DOI] [PubMed] [Google Scholar]
- 89.Hagger-Johnson G, Harron K, Fleming T, Gilbert R, Goldstein H, Landy R, et al. Data linkage errors in hospital administrative data when applying a pseudonymisation algorithm to paediatric intensive care records. BMJ Open. 2015;5:e008118. doi: 10.1136/bmjopen-2015-008118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bohensky MA, Jolley D, Sundararajan V, Evans S, Pilcher DV, Scott I, et al. Data Linkage: a powerful research tool with potential problems. BMC Health Serv Res. 2010;10:346. doi: 10.1186/1472-6963-10-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Grath-Lone LM, Libuy N, Etoori D, Blackburn R, Gilbert R, Harron K. Ethnic bias in data linkage. Lancet Digit Health. 2021;3:e339. doi: 10.1016/S2589-7500(21)00081-9. [DOI] [PubMed] [Google Scholar]
- 92.Patten SB. Selection bias in studies of major depression using clinical subjects. J Clin Epidemiol. 2000;53:351–7. doi: 10.1016/S0895-4356(99)00215-2. [DOI] [PubMed] [Google Scholar]
- 93.Yu S. Uncovering the hidden impacts of inequality on mental health: a global study. Transl Psychiatry. 2018;8:98. doi: 10.1038/s41398-018-0148-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Semlyen J, King M, Varney J, Hagger-Johnson G. Sexual orientation and symptoms of common mental disorder or low wellbeing: combined meta-analysis of 12 UK population health surveys. BMC Psychiatry. 2016;16:67. doi: 10.1186/s12888-016-0767-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Reiss F, Meyrose A-K, Otto C, Lampert T, Klasen F, Ravens-Sieberer U. Socioeconomic status, stressful life situations and mental health problems in children and adolescents: results of the German BELLA cohort-study. PLoS ONE. 2019;14:e0213700. doi: 10.1371/journal.pone.0213700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Howe CJ, Robinson WR. Survival-related selection bias in studies of racial health disparities: the importance of the target population and study design. Epidemiol Camb Mass. 2018;29:521–4. doi: 10.1097/EDE.0000000000000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Thornton J. Ethnic minority patients receive worse mental healthcare than white patients, review finds. BMJ. 2020;368:m1058. doi: 10.1136/bmj.m1058. [DOI] [PubMed] [Google Scholar]
- 98.Pendergrass SA, Crawford DC. Using electronic health records to generate phenotypes for research. Curr Protoc Hum Genet. 2019;100:e80. doi: 10.1002/cphg.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hemingway H, Asselbergs FW, Danesh J, Dobson R, Maniadakis N, Maggioni A, et al. Big data from electronic health records for early and late translational cardiovascular research: challenges and potential. Eur Heart J. 2018;39:1481–95. doi: 10.1093/eurheartj/ehx487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wisniewski H, Henson P, Torous J. Using a smartphone app to identify clinically relevant behavior trends via symptom report, cognition scores, and exercise levels: a case series. Front Psychiatry. 2019;10:652. doi: 10.3389/fpsyt.2019.00652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang S, Varghese P, Stephenson E, Tu K, Gronsbell J. Machine learning approaches for electronic health records phenotyping: a methodical review. J Am Med Inform Assoc JAMIA. 2023;30:367–81. doi: 10.1093/jamia/ocac216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Banda JM, Seneviratne M, Hernandez-Boussard T, Shah NH. Advances in electronic phenotyping: from rule-based definitions to machine learning models. Annu Rev Biomed Data Sci. 2018;1:53. doi: 10.1146/annurev-biodatasci-080917-013315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gottesman O, Kuivaniemi H, Tromp G, Faucett WA, Li R, Manolio TA, et al. The Electronic Medical Records and Genomics (eMERGE) network: past, present, and future. Genet Med. 2013;15:761–71. doi: 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Denaxas S, Gonzalez-Izquierdo A, Direk K, Fitzpatrick NK, Fatemifar G, Banerjee A, et al. UK phenomics platform for developing and validating electronic health record phenotypes: CALIBER. J Am Med Inform Assoc. 2019;26:1545–59. doi: 10.1093/jamia/ocz105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tomlinson A, Furukawa TA, Efthimiou O, Salanti G, Crescenzo FD, Singh I, et al. Personalise antidepressant treatment for unipolar depression combining individual choices, risks and big data (PETRUSHKA): rationale and protocol. BMJ Ment Health. 2020;23:52–56. doi: 10.1136/ebmental-2019-300118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ingram WM, Baker AM, Bauer CR, Brown JP, Goes FS, Larson S, et al. Defining major depressive disorder cohorts using the EHR: multiple phenotypes based on ICD-9 codes and medication orders. Neurol Psychiatry Brain Res. 2020;36:18–26. doi: 10.1016/j.npbr.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mayer MA, Gutierrez-Sacristan A, Leis A, De La Peña S, Sanz F, Furlong LI. Using electronic health records to assess depression and cancer comorbidities. Stud Health Technol Inform. 2017;235:236–40. [PubMed] [Google Scholar]
- 108.Deferio JJ, Levin TT, Cukor J, Banerjee S, Abdulrahman R, Sheth A, et al. Using electronic health records to characterize prescription patterns: focus on antidepressants in nonpsychiatric outpatient settings. JAMIA Open. 2018;1:233. doi: 10.1093/jamiaopen/ooy037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.James G, Collin E, Lawrance M, Mueller A, Podhorna J, Zaremba-Pechmann L, et al. Treatment pathway analysis of newly diagnosed dementia patients in four electronic health record databases in Europe. Soc Psychiatry Psychiatr Epidemiol. 2021;56:409–16. doi: 10.1007/s00127-020-01872-2. [DOI] [PubMed] [Google Scholar]
- 110.Smoller JW. The use of electronic health records for psychiatric phenotyping and genomics. Am J Med Genet Part B. 2018;177:601. doi: 10.1002/ajmg.b.32548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ford E, Carroll JA, Smith HE, Scott D, Cassell JA. Extracting information from the text of electronic medical records to improve case detection: a systematic review. J Am Med Inform Assoc JAMIA. 2016;23:1007–15. doi: 10.1093/jamia/ocv180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Moldwin A, Demner-Fushman D, Goodwin TR. Empirical findings on the role of structured data, unstructured data, and their combination for automatic clinical phenotyping. AMIA Summits Transl Sci Proc. 2021;2021:445–54. [PMC free article] [PubMed] [Google Scholar]
- 113.Morley KI, Wallace J, Denaxas SC, Hunter RJ, Patel RS, Perel P, et al. Defining disease phenotypes using national linked electronic health records: a case study of atrial fibrillation. PLoS ONE. 2014;9:e110900. doi: 10.1371/journal.pone.0110900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kirby JC, Speltz P, Rasmussen LV, Basford M, Gottesman O, Peissig PL, et al. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. J Am Med Inform Assoc JAMIA. 2016;23:1046–52. doi: 10.1093/jamia/ocv202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Newton KM, Peissig PL, Kho AN, Bielinski SJ, Berg RL, Choudhary V, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc JAMIA. 2013;20:e147–154. doi: 10.1136/amiajnl-2012-000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.McGuinness LA, Warren‐Gash C, Moorhouse LR, Thomas SL. The validity of dementia diagnoses in routinely collected electronic health records in the United Kingdom: a systematic review. Pharmacoepidemiol Drug Saf. 2019;28:244–55. doi: 10.1002/pds.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bellows BK, LaFleur J, Kamauu AWC, Ginter T, Forbush TB, Agbor S, et al. Automated identification of patients with a diagnosis of binge eating disorder from narrative electronic health records. J Am Med Inform Assoc JAMIA. 2014;21:e163–e168. doi: 10.1136/amiajnl-2013-001859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hernán MA, Wang W, Leaf DE. Target trial emulation: a framework for causal inference from observational data. JAMA. 2022;328:2446–7. doi: 10.1001/jama.2022.21383. [DOI] [PubMed] [Google Scholar]
- 119.Hernán MA, Robins JM. Using big data to emulate a target trial when a randomized trial is not available. Am J Epidemiol. 2016;183:758–64. doi: 10.1093/aje/kwv254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hernán MA, Sauer BC, Hernández-Díaz S, Platt R, Shrier I. Specifying a target trial prevents immortal time bias and other self-inflicted injuries in observational analyses. J Clin Epidemiol. 2016;79:70–75. doi: 10.1016/j.jclinepi.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Admon AJ, Donnelly JP, Casey JD, Janz DR, Russell DW, Joffe AM, et al. Emulating a novel clinical trial using existing observational data. predicting results of the prevent study. Ann Am Thorac Soc. 2019;16:998–1007. doi: 10.1513/AnnalsATS.201903-241OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dickerman BA, García-Albéniz X, Logan RW, Denaxas S, Hernán MA. Avoidable flaws in observational analyses: an application to statins and cancer. Nat Med. 2019;25:1601–6. doi: 10.1038/s41591-019-0597-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Boyne DJ, Cheung WY, Hilsden RJ, Sajobi TT, Batra A, Friedenreich CM, et al. Association of a shortened duration of adjuvant chemotherapy with overall survival among individuals with Stage III colon cancer. JAMA Netw Open. 2021;4:e213587. doi: 10.1001/jamanetworkopen.2021.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Matthews AA, Szummer K, Dahabreh IJ, Lindahl B, Erlinge D, Feychting M, et al. Comparing effect estimates in randomized trials and observational studies from the same population: an application to percutaneous coronary intervention. J Am Heart Assoc. 2021;10:e020357. doi: 10.1161/JAHA.120.020357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hoffman KL, Schenck EJ, Satlin MJ, Whalen W, Pan D, Williams N, et al. Comparison of a target trial emulation framework vs. Cox regression to estimate the association of corticosteroids with COVID-19 mortality. JAMA Netw Open. 2022;5:e2234425. doi: 10.1001/jamanetworkopen.2022.34425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Caniglia EC, Rojas-Saunero LP, Hilal S, Licher S, Logan R, Stricker B, et al. Emulating a target trial of statin use and risk of dementia using cohort data. Neurology. 2020;95:e1322–e1332. doi: 10.1212/WNL.0000000000010433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ahn N, Nolde M, Günter A, Güntner F, Gerlach R, Tauscher M, et al. Emulating a target trial of proton pump inhibitors and dementia risk using claims data. Eur J Neurol. 2022;29:1335–43. doi: 10.1111/ene.15284. [DOI] [PubMed] [Google Scholar]
- 128.Chen Y-CB, Liang C-S, Wang L-J, Hung K-C, Carvalho AF, Solmi M, et al. Comparative effectiveness of valproic acid in different serum concentrations for maintenance treatment of bipolar disorder: a retrospective cohort study using target trial emulation framework. EClinicalMedicine. 2022;54:101678. doi: 10.1016/j.eclinm.2022.101678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sun JW, Young JG, Sarvet AL, Bailey LC, Heerman WJ, Janicke DM, et al. Comparison of rates of type 2 diabetes in adults and children treated with anticonvulsant mood stabilizers. JAMA Netw Open. 2022;5:e226484. doi: 10.1001/jamanetworkopen.2022.6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tian Y, Schuemie MJ, Suchard MA. Evaluating large-scale propensity score performance through real-world and synthetic data experiments. Int J Epidemiol. 2018;47:2005–14. doi: 10.1093/ije/dyy120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Austin PC. An Introduction to propensity score methods for reducing the effects of confounding in observational studies. Multivar Behav Res. 2011;46:399–424. doi: 10.1080/00273171.2011.568786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Riley RD, Ensor J, Snell KIE, Harrell FE, Martin GP, Reitsma JB, et al. Calculating the sample size required for developing a clinical prediction model. BMJ. 2020;368:m441. doi: 10.1136/bmj.m441. [DOI] [PubMed] [Google Scholar]
- 133.Steyerberg EW. Clinical prediction models: a practical approach to development, validation, and updating. Springer International Publishing: Cham; 2019.
- 134.Dalton JE, Bolen SD, Mascha EJ. Publication bias: the elephant in the review. Anesth Analg. 2016;123:812–3. doi: 10.1213/ANE.0000000000001596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nguyen VT, Engleton M, Davison M, Ravaud P, Porcher R, Boutron I. Risk of bias in observational studies using routinely collected data of comparative effectiveness research: a meta-research study. BMC Med. 2021;19:279. doi: 10.1186/s12916-021-02151-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.von Elm E, Altman DG, Egger M, Pocock SJ, Gøtzsche PC, Vandenbroucke JP, et al. The strengthening the reporting of observational studies in epidemiology (STROBE) statement: guidelines for reporting observational studies. Lancet Lond Engl. 2007;370:1453–7. doi: 10.1016/S0140-6736(07)61602-X. [DOI] [PubMed] [Google Scholar]
- 137.Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD Statement. BMC Med. 2015;13:1. doi: 10.1186/s12916-014-0241-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Williams RJ, Tse T, Harlan WR, Zarin DA. Registration of observational studies: Is it time? Can Med Assoc J. 2010;182:1638–42. doi: 10.1503/cmaj.092252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Thor M, Oh JH, Apte AP, Deasy JO. Registering study analysis plans (SAPs) before dissecting your data—updating and standardizing outcome modeling. Front Oncol. 2020;10:978. doi: 10.3389/fonc.2020.00978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Goldacre B, Morton CE, DeVito NJ. Why researchers should share their analytic code. BMJ. 2019;367:l6365. doi: 10.1136/bmj.l6365. [DOI] [PubMed] [Google Scholar]
- 141.Koutsouleris N, Hauser TU, Skvortsova V, Choudhury MD. From promise to practice: towards the realisation of AI-informed mental health care. Lancet Digit Health. 2022;4:e829–e840. doi: 10.1016/S2589-7500(22)00153-4. [DOI] [PubMed] [Google Scholar]
- 142.Lawlor DA, Tilling K, Davey Smith G. Triangulation in aetiological epidemiology. Int J Epidemiol. 2016;45:1866–86. doi: 10.1093/ije/dyw314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hammerton G, Munafò MR. Causal inference with observational data: the need for triangulation of evidence. Psychol Med. 2021;51:563–78. doi: 10.1017/S0033291720005127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Fancourt D, Steptoe A. Cultural engagement and mental health: does socio-economic status explain the association? Soc Sci Med 1982. 2019;236:112425. doi: 10.1016/j.socscimed.2019.112425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Harrison R, Munafò MR, Davey Smith G, Wootton RE. Examining the effect of smoking on suicidal ideation and attempts: triangulation of epidemiological approaches. Br J Psychiatry J Ment Sci. 2020;217:701–7. doi: 10.1192/bjp.2020.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lloyd EC, Sallis HM, Verplanken B, Haase AM, Munafò MR. Understanding the nature of association between anxiety phenotypes and anorexia nervosa: a triangulation approach. BMC Psychiatry. 2020;20:495. doi: 10.1186/s12888-020-02883-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gaetani L, Blennow K, Calabresi P, Filippo MD, Parnetti L, Zetterberg H. Neurofilament light chain as a biomarker in neurological disorders. J Neurol Neurosurg Psychiatry. 2019;90:870–81. doi: 10.1136/jnnp-2018-320106. [DOI] [PubMed] [Google Scholar]
- 148.Carvalho AF, Solmi M, Sanches M, Machado MO, Stubbs B, Ajnakina O, et al. Evidence-based umbrella review of 162 peripheral biomarkers for major mental disorders. Transl Psychiatry. 2020;10:1–13. doi: 10.1038/s41398-020-0835-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.García-Gutiérrez MS, Navarrete F, Sala F, Gasparyan A, Austrich-Olivares A, Manzanares J. Biomarkers in psychiatry: concept, definition, types and relevance to the clinical reality. Front Psychiatry 2020;11. https://www.frontiersin.org/articles/10.3389/fpsyt.2020.00432. [DOI] [PMC free article] [PubMed]
- 150.Goodday SM, Atkinson L, Goodwin G, Saunders K, South M, Mackay C, et al. The true colours remote symptom monitoring system: a decade of evolution. J Med Internet Res. 2020;22:e15188. doi: 10.2196/15188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Verma A, Damrauer SM, Naseer N, Weaver J, Kripke CM, Guare L, et al. The Penn Medicine BioBank: towards a genomics-enabled learning healthcare system to accelerate precision medicine in a diverse population. J Pers Med. 2022;12:1974. doi: 10.3390/jpm12121974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Taneja I, Reddy B, Damhorst G, Dave Zhao S, Hassan U, Price Z, et al. Combining biomarkers with EMR data to identify patients in different phases of sepsis. Sci Rep. 2017;7:10800. doi: 10.1038/s41598-017-09766-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ploubidis GB, Batty GD, Patalay P, Bann D, Goodman A. Association of early-life mental health with biomarkers in midlife and premature mortality: evidence from the 1958 British Birth Cohort. JAMA Psychiatry. 2021;78:38–46. doi: 10.1001/jamapsychiatry.2020.2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ryan D, Blakey J, Chisholm A, Price D, Thomas M, Ställberg B, et al. Use of electronic medical records and biomarkers to manage risk and resource efficiencies. Eur Clin Respir J. 2017;4:1293386. doi: 10.1080/20018525.2017.1293386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Stark Z, Dolman L, Manolio TA, Ozenberger B, Hill SL, Caulfied MJ, et al. Integrating genomics into healthcare: a global responsibility. Am J Hum Genet. 2019;104:13–20. doi: 10.1016/j.ajhg.2018.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wray NR, Lin T, Austin J, McGrath JJ, Hickie IB, Murray GK, et al. From basic science to clinical application of polygenic risk scores: a primer. JAMA Psychiatry. 2021;78:101–9. doi: 10.1001/jamapsychiatry.2020.3049. [DOI] [PubMed] [Google Scholar]
- 157.Levine ME, Crimmins EM, Prescott CA, Phillips D, Arpawong TE, Lee J. A polygenic risk score associated with measures of depressive symptoms among older adults. Biodemography Soc Biol. 2014;60:199–211. doi: 10.1080/19485565.2014.952705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Baker E, Escott-Price V. Polygenic risk scores in Alzheimer’s disease: current applications and future directions. Front Digit Health 2020;2. https://www.frontiersin.org/articles/10.3389/fdgth.2020.00014. [DOI] [PMC free article] [PubMed]
- 159.Escudero I, Johnstone M. Genetics of schizophrenia. Curr Psychiatry Rep. 2014;16:502. doi: 10.1007/s11920-014-0502-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Smeland OB, Frei O, Dale AM, Andreassen OA. The polygenic architecture of schizophrenia—rethinking pathogenesis and nosology. Nat Rev Neurol. 2020;16:366–79. doi: 10.1038/s41582-020-0364-0. [DOI] [PubMed] [Google Scholar]
- 161.Lopes FL, Zhu K, Purves KL, Song C, Ahn K, Hou L, et al. Polygenic risk for anxiety influences anxiety comorbidity and suicidal behavior in bipolar disorder. Transl Psychiatry. 2020;10:1–7. doi: 10.1038/s41398-020-00981-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Murray GK, Lin T, Austin J, McGrath JJ, Hickie IB, Wray NR. Could polygenic risk scores be useful in psychiatry?: a review. JAMA Psychiatry. 2021;78:210–9. doi: 10.1001/jamapsychiatry.2020.3042. [DOI] [PubMed] [Google Scholar]
- 163.Kember RL, Merikangas AK, Verma SS, Verma A, Judy R, Abecasis G, et al. Polygenic risk of psychiatric disorders exhibits cross-trait associations in electronic health record data from European ancestry individuals. Biol Psychiatry. 2021;89:236–45. doi: 10.1016/j.biopsych.2020.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gao XR, Chiariglione M, Qin K, Nuytemans K, Scharre DW, Li Y-J, et al. Explainable machine learning aggregates polygenic risk scores and electronic health records for Alzheimer’s disease prediction. Sci Rep. 2023;13:450. doi: 10.1038/s41598-023-27551-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wang M, Hu K, Fan L, Yan H, Li P, Jiang T et al. Predicting treatment response in schizophrenia with magnetic resonance imaging and polygenic risk score. Front Genet 2022;13. https://www.frontiersin.org/articles/10.3389/fgene.2022.848205. [DOI] [PMC free article] [PubMed]
- 166.Landi I, Kaji DA, Cotter L, Van Vleck T, Belbin G, Preuss M, et al. Prognostic value of polygenic risk scores for adults with psychosis. Nat Med. 2021;27:1576–81. doi: 10.1038/s41591-021-01475-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Meerman JJ, ter Hark SE, Janzing JGE, Coenen MJH. The potential of polygenic risk scores to predict antidepressant treatment response in major depression: a systematic review. J Affect Disord. 2022;304:1–11. doi: 10.1016/j.jad.2022.02.015. [DOI] [PubMed] [Google Scholar]
- 168.Burgess S, Thompson SG. Use of allele scores as instrumental variables for Mendelian randomization. Int J Epidemiol. 2013;42:1134–44. doi: 10.1093/ije/dyt093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wootton RE, Jones HJ, Sallis HM. Mendelian randomisation for psychiatry: how does it work, and what can it tell us? Mol Psychiatry. 2022;27:53–57. doi: 10.1038/s41380-021-01173-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Davies NM, Holmes MV, Smith GD. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018;362:k601. doi: 10.1136/bmj.k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Cai L, Bao Y, Fu X, Cao H, Baranova A, Zhang X, et al. Causal links between major depressive disorder and insomnia: a Mendelian randomisation study. Gene. 2021;768:145271. doi: 10.1016/j.gene.2020.145271. [DOI] [PubMed] [Google Scholar]
- 172.Nassan M, Daghlas I, Winkelman JW, Dashti HS, Saxena R. Genetic evidence for a potential causal relationship between insomnia symptoms and suicidal behavior: a Mendelian randomization study. Neuropsychopharmacology. 2022;47:1672–9. doi: 10.1038/s41386-022-01319-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Sun X, Liu B, Liu S, Wu DJH, Wang J, Qian Y, et al. Sleep disturbance and psychiatric disorders: a bidirectional Mendelian randomisation study. Epidemiol Psychiatr Sci. 2022;31:e26. doi: 10.1017/S2045796021000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Herrett E, Gallagher AM, Bhaskaran K, Forbes H, Mathur R, van Staa T, et al. Data resource profile: Clinical Practice Research Datalink (CPRD) Int J Epidemiol. 2015;44:827–36. doi: 10.1093/ije/dyv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Davies MR, Kalsi G, Armour C, Jones IR, McIntosh AM, Smith DJ, et al. The Genetic Links to Anxiety and Depression (GLAD) Study: online recruitment into the largest recontactable study of depression and anxiety. Behav Res Ther. 2019;123:103503. doi: 10.1016/j.brat.2019.103503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Religa D, Fereshtehnejad S-M, Cermakova P, Edlund A-K, Garcia-Ptacek S, Granqvist N, et al. SveDem, the Swedish Dementia Registry—a tool for improving the quality of diagnostics, treatment and care of dementia patients in clinical practice. PLoS ONE. 2015;10:e0116538. doi: 10.1371/journal.pone.0116538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.German National Cohort (GNC) Consortium. The German National Cohort: aims, study design and organization. Eur J Epidemiol. 2014;29:371–82. doi: 10.1007/s10654-014-9890-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Choi KW, Stein MB, Nishimi KM, Ge T, Coleman JRI, Chen C-Y, et al. An exposure-wide and Mendelian randomization approach to identifying modifiable factors for the prevention of depression. Am J Psychiatry. 2020;177:944–54. doi: 10.1176/appi.ajp.2020.19111158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Choi KW, Chen C-Y, Stein MB, Klimentidis YC, Wang M-J, Koenen KC, et al. Assessment of bidirectional relationships between physical activity and depression among adults. JAMA Psychiatry. 2019;76:399–408. doi: 10.1001/jamapsychiatry.2018.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Jefsen OH, Speed M, Speed D, Østergaard SD. Bipolar disorder and cannabis use: A bidirectional two-sample Mendelian randomization study. Addict Biol. 2021;26:e13030. doi: 10.1111/adb.13030. [DOI] [PubMed] [Google Scholar]
- 182.Huang P, Zou Y, Zhang X, Ye X, Wang Y, Yu R, et al. The causal effects of insomnia on bipolar disorder, depression, and schizophrenia: a two-sample Mendelian Randomization Study. Front Genet 2021;12. https://www.frontiersin.org/articles/10.3389/fgene.2021.763259. [DOI] [PMC free article] [PubMed]
- 183.Yang J, Fan Y, Yan B, Zhao B, Qian L, Gao F, et al. Mendelian randomization analyses reveal novel drug targets for anorexia nervosa. Prog Neuropsychopharmacol Biol Psychiatry. 2022;112:110427. doi: 10.1016/j.pnpbp.2021.110427. [DOI] [PubMed] [Google Scholar]
- 184.Sproviero W, Winchester L, Newby D, Fernandes M, Shi L, Goodday SM, et al. High blood pressure and risk of dementia: a two-sample Mendelian randomization study in the UK Biobank. Biol Psychiatry. 2021;89:817–24. doi: 10.1016/j.biopsych.2020.12.015. [DOI] [PubMed] [Google Scholar]
- 185.Baird DA, Liu JZ, Zheng J, Sieberts SK, Perumal T, Elsworth B, et al. Identifying drug targets for neurological and psychiatric disease via genetics and the brain transcriptome. PLoS Genet. 2021;17:e1009224. doi: 10.1371/journal.pgen.1009224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ryan DK, Karhunen V, Su B, Traylor M, Richardson TG, Burgess S, et al. Genetic evidence for protective effects of angiotensin-converting enzyme against Alzheimer disease but not other neurodegenerative diseases in European populations. Neurol Genet. 2022;8:e200014. doi: 10.1212/NXG.0000000000200014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Burgess S, Thompson SG. Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol. 2015;181:251–60. doi: 10.1093/aje/kwu283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Burgess S, Davies NM, Thompson SG, Consortium on behalf of E-I. Instrumental variable analysis with a nonlinear exposure–outcome relationship. Epidemiology. 2014;25:877. doi: 10.1097/EDE.0000000000000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Rees JMB, Foley CN, Burgess S. Factorial Mendelian randomization: using genetic variants to assess interactions. Int J Epidemiol. 2019;49:1147–58. doi: 10.1093/ije/dyz161. [DOI] [PMC free article] [PubMed] [Google Scholar]


