Summary
Canonical splice site variants (CSSVs) are often presumed to cause loss-of-function (LoF) and are assigned very strong evidence of pathogenicity (according to American College of Medical Genetics/Association for Molecular Pathology criterion PVS1). The exact nature and predictability of splicing effects of unselected rare CSSVs in blood-expressed genes are poorly understood. We identified 168 rare CSSVs in blood-expressed genes in 112 individuals using genome sequencing, and studied their impact on splicing using RNA sequencing (RNA-seq). There was no evidence of a frameshift, nor of reduced expression consistent with nonsense-mediated decay, for 25.6% of CSSVs: 17.9% had wildtype splicing only and normal junction depths, 3.6% resulted in cryptic splice site usage and in-frame insertions or deletions, 3.6% resulted in full exon skipping (in frame), and 0.6% resulted in full intron inclusion (in frame). Blind to these RNA-seq data, we attempted to predict the precise impact of CSSVs by applying in silico tools and the ClinGen Sequence Variant Interpretation Working Group 2018 guidelines for applying PVS1 criterion. The predicted impact on splicing using (1) SpliceAI, (2) MaxEntScan, and (3) AutoPVS1, an automatic classification tool for PVS1 interpretation of null variants that utilizes Ensembl Variant Effect Predictor and MaxEntScan, was concordant with RNA-seq analyses for 65%, 63%, and 61% of CSSVs, respectively. In summary, approximately one in four rare CSSVs did not show evidence for LoF based on analysis of RNA-seq data. Predictions from in silico methods were often discordant with findings from RNA-seq. More caution may be warranted in applying PVS1-level evidence to CSSVs in the absence of functional data.
Keywords: transcriptomics, RNA sequencing, splicing, genetic testing, variant interpretation, genetic counseling, genome sequencing
CSSVs are often presumed to cause LoF and are assigned very strong evidence of pathogenicity. We found that approximately one in four CSSVs may not cause LoF and that in silico predictions using established tools and published guidelines were often discordant with RNA-seq data.
Introduction
Canonical splice site variants (CSSVs) are DNA variants affecting splicing donor (+1 and +2) and acceptor (−1 and −2) sites defining exon-intron boundaries.1,2 The consensus nucleotide sequences at splicing donor and acceptor sites are GT and AG, respectively, and are essential in interacting with the U2 spliceosome to result in normal splicing and generation of wildtype (WT) transcripts.2,3,4,5 CSSVs may modify the interactions between the precursor mRNA and spliceosome complex.5,6,7 The resulting splice disruption events can include exon skipping, full intron inclusion, and alternative use of nearby cryptic splice sites resulting in insertions or deletions (indels) of nucleotides.6,7,8 These effects may or may not induce a frameshift and premature termination codon, which can then trigger nonsense-mediated RNA decay (NMD) and result in a loss-of-function (LoF) of the gene.9,10
Accurate variant interpretation is foundational to both genome diagnostics and screening.11,12,13 Rare CSSVs are typically considered under the “null variant” code and assigned very strong evidence for pathogenicity (PVS1).12 In 2018, the PVS1 guidelines were refined by the ClinGen Sequence Variant Interpretation Working Group (ClinGen SVI).11 ClinGen SVI recommended assigning PVS1 at varying evidence strengths (i.e., supporting, moderate, strong, very strong, or not at all) after directly inspecting the genomic region to predict the impact of the CSSV on splicing and the overall reading frame.11 In silico tools were recognized as a valuable but imperfect adjunct method for predicting the impact of CSSVs.11 Advances since 2018 include the emergence of SpliceAI as a widely used and powerful method for annotating genetic variants with their predicted effect on splicing.14,15
To our knowledge, there have been no systematic attempts to catalog the precise consequences of rare CSSVs on splicing via cDNA sequencing. The degree to which the impact(s) of CSSVs are predictable via inspection of the genomic region and application of in silico tools is also unclear. Here, we analyzed all rare CSSVs identified by genome sequencing (GS) from children and parents, in blood-expressed but otherwise unselected genes, using genome-wide RNA sequencing (RNA-seq). We hypothesized that a significant minority of CSSVs would show no evidence of a frameshift nor of reduced expression consistent with NMD, recognizing that approximately one-third of inserted or deleted DNA segments would be expected to have a size divisible by 3.16 We determined the proportions of various splicing outcomes, and we compared the results with outcomes predicted in a blinded fashion by three in silico tools: SpliceAI, MaxEntScan, and AutoPVS1.12,13,14 Our results revealed a previously underappreciated complexity in CSSV impact prediction and underscore the value of functional data in the interpretation of rare CSSVs.
Subjects, material, and methods
Identification of rare CSSVs expressed in blood
In this study, we performed GS and RNA-seq from blood for 112 total individuals.17,18
The study was approved by the Research Ethics Board of the Hospital for Sick Children and conducted in accordance with the Declaration of Helsinki. Written informed consent was obtained for genetic analysis and publication of clinical details. Demographic details of our cohort are described in Table S1. Detailed GS methods, and a subset of the GS data, were published previously.17,18,19,20,21 All variants identified were aligned to the Genome Reference Consortium Human Build 37 (GRCh37). DNA variants from GS files were filtered according to the following criteria: (1) single nucleotide substitution in a canonical splice site identified in a MANE Select or Ensembl Canonical transcript, (2) allele frequency of less than 0.05 (per 1,000 Genomes, NHLBI-ESP, and ExAC/gnomAD; cut-off selected because of the original stand-alone evidence of benign impact criterion12), (3) at least 99% Genotype Quality Score, and (4) gene possibly detected in whole blood (according to the Genotype-Tissue Expression, V8, transcripts per million [TPM] > 0.05) and detected in our internal cohorts.19,22 If no MANE Select or Ensembl Canonical transcript was available, then we selected the blood-expressed transcript in which the variant was in a canonical splice site for a coding exon (Table S2). Variants in untranslated regions (UTRs) were excluded;23 results for n = 16 CSSVs flanking non-coding exons are available upon request. All 168 CSSVs included in this study were identified in the heterozygous state (including the variant in an X chromosome gene) (Table S2). Overall, 45 of the 164 genes were known or suspected to be associated with a germline Mendelian disease in the Online Mendelian Inheritance in Man (OMIM; searched winter 2023): 41 via either a (suspected or confirmed) mono-allelic and/or bi-allelic LoF mechanism, 2 via a (suspected or confirmed) dominant negative mechanism, and 2 via a gain-of-function mechanism (Table S2). Only 2 of the CSSVs were considered diagnostic for the phenotype(s) that prompted GS, and 40 additional probands had a non-CSSV molecular diagnosis on GS.17,18,19,20,21,24
Analysis of splicing impact of CSSVs using RNA-seq data
We analyzed the impact of CSSVs on splicing in the canonical transcript using the accompanying short-read blood-derived RNA-seq data. RNA extraction, sequencing, and data processing methods were previously described in full.19,25 The median sample sequencing depth was 112.82 million read pairs (interquartile range 26.80), and the median number of genes detected at 1 or more TPM was 11,438.5 (interquartile range 1,993). Each splicing junction was manually inspected using the Integrated Genome Visualizer by two independent evaluators (R.Y.O. and A.A.). For every CSSV, an average of five random, age-range matched controls (i.e., with normal DNA sequence at the affected CSS) from this cohort were used to identify the WT splicing event(s) and provide a reference on junction read depths to account for any possible fluctuations. Sex-matched controls were specifically used for one CSSV located on the X chromosome. Only junctions with more than five uniquely mapped reads, which is a low cut-off, were considered in the analysis. The junction with the highest read depth was considered the predominant splicing outcome. Splicing outcome categories are as follows.
-
(A)
Presumed NMD (if the raw WT splicing junction coverage was ≥20% less than control individuals but no aberrant splicing events were captured),26
-
(B)
NMD not detected (if there was comparable [≤20% difference] junction depth between individuals and no aberrant splicing events were captured),
-
(C)
Exon skipping leading to frameshift deletion,
-
(D)
Exon skipping leading to in-frame deletion,
-
(E)
Full intron inclusion leading to frameshift insertion,
-
(F)
Full intron inclusion leading to in-frame insertion,
-
(G)
Activation of a cryptic or non-canonical splice site leading to frameshift indel,
-
(H)
Activation of a cryptic or non-canonical splice site leading to in-frame indel.
Evidence supporting a known or presumed frameshift effect (i.e., splicing outcomes A, C, E, and G) led to the assignment of a CSSV to the frameshift group. The remaining variants were assigned to the non-frameshift group.
Prediction of splicing outcomes using in silico tools
Masked to the RNA-seq data, two independent assessors (D.C. and G.G.) attempted to predict the precise impact of CSSVs using in silico tools and ClinGen SVI recommendations.11 To identify alternative splice site usage outside of the canonical splice site directly impacted by the CSSV, standard score thresholds were used for MaxEntScan (>3) and for SpliceAI (delta score >0.2, for high sensitivity). For each CSSV, the splice junction was manually inspected using Alamut Visual Plus (version v1.7, SOPHiA GENETICS) with a ±20-bp window for a cryptic splice site. All CSSVs predicted to result in frameshift were presumed to undergo NMD, unless the outcome is predicted to result in a premature termination codon in the last exon or within 50 bp of the 3′ end of the penultimate exon of the gene (in which case NMD may not occur; results in a higher likelihood of an expressed protein).26,27 In the case of two or more possible cryptic splice sites in the same region, we assumed usage of the splice site with the highest or strongest in silico score. An alternative approach of assuming a non-NMD outcome whenever the use of any of the splice sites was predicted to result in an in-frame indel yielded similar results (data not shown). In the absence of a cryptic splice site in the neighboring region, for acceptor losses we predicted exon skipping and for donor losses we predicted full intron inclusion. The length of the exon or intron, respectively, was then used to predict whether the impact would be in frame or out of frame. While the ClinGen SVI recommendations specify use of a ±20-bp window, we conducted additional exploratory analyses using an expanded SpliceAI window of ±5,000 bp (“SpliceAI_expanded”). In the case where the extended window revealed the predicted loss of two consecutive CSSVs (i.e., the acceptor and donor sites flanking an exon, or the donor and acceptor sites flanking an intron), we predicted exon skipping or intron inclusion, respectively. Concordance of outcomes was then calculated if the frameshift/non-frameshift outcome predicted by the in silico tools matched the results from RNA-seq.
AutoPVS1, an automatic classification tool for PVS1 interpretation of null variants, was also used to predict the impact of CSSVs on splicing.28 This algorithm uses Variant Effect Predictor for annotation of variants and MaxEntScan to predict the use of cryptic splice sites and resulting impact on splicing.28 The three possible outcomes of AutoPVS1 are (1) exon skipping or cryptic splice site usage that leads to a frameshift and NMD, (2) exon skipping or cryptic splice site usage that leads to a frameshift without NMD, or (3) exon skipping or cryptic splice site usage that preserves the reading frame.
Results
Comparison of frameshift outcomes of CSSVs using RNA-seq vs. in silico predictions
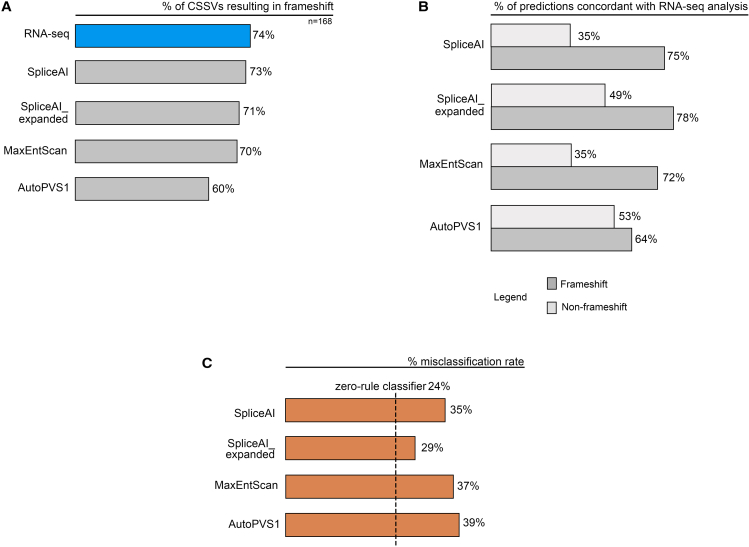
We assessed a total of 168 rare, blood-expressed CSSVs in 164 otherwise unselected genes. By RNA-seq, 26% of these CSSVs did not result in a frameshift or in reduced expression, consistent with NMD (Figure 1A). There was no apparent difference in the patterns of variant location and specific nucleotide substitution of CSSVs by frameshift/non-frameshift outcomes (Figures S1 and S2). Considering the n = 30 CSSVs that showed only WT splicing and with comparable read depth to controls (outcome category B), 18 CSSVs were in the donor splice site including three GT>GC variants29 (Figure S3). Most CSSVs (9/11) flanking a penultimate coding exon of a gene demonstrated evidence for NMD/frameshift by RNA-seq, with conflicting results predicted by in silico tools (Table S2). There was no significant difference in the median CADD Phred score between the CSSVs that did and did not show evidence of NMD/frameshift by RNA-seq (33 vs. 33; Mann-Whitney U = 3552, p = 0.53) (Table S2). There was no apparent difference in gnomAD allele frequency between the rare CSSVs that resulted in frameshift/NMD and the rare CSSVs that resulted in no frameshift/no NMD (Figure S4); most variants (161/168) had an allele frequency of less than 0.001 (Table S2).
Figure 1.
General categories of CSSV splicing outcomes
(A) A comparison of the proportion of CSSVs resulting in a frameshift according to RNA-seq analysis vs. in silico predictions.
(B) Concordance between RNA-seq and in silico predictions of the impact of CSSVs on splicing. Concordant outcomes are defined as the RNA-seq and respective in silico tool identifying the same outcome (frameshift/NMD [n = 125 by RNA-seq] or non-frameshift/no NMD [n = 43] by RNA-seq).
(C) Misclassification rates of in silico tools compared with a zero-rule classifier.
For the 168 CSSVs, blinded in silico methods predicted non-frameshift outcomes in 27% (SpliceAI), 29% (SpliceAI_expanded), 30% (MaxEntScan), and 40% (AutoPVS1) (Figure 1A). For CSSVs resulting in a frameshift/NMD per RNA-seq, SpliceAI_expanded had the greatest pairwise concordance (78%), followed by SpliceAI (75%), MaxENTScan (72%), and AutoPVS1 (64%) (Figure 1B). For CSSVs not resulting in a frameshift per RNA-seq, SpliceAI was concordant in 35%, SpliceAI_expanded in 49%, MaxENTScan in 35%, and AutoPVS1 in 53% (Figure 1B). Across all 168 variants, SpliceAI_expanded had the greatest pairwise concordance with RNA-seq with respect to the frameshift vs. non-frameshift outcome, at 71%. To assess the performance of each in silico method, we calculated the misclassification rates from each technique and contextualized these results by comparing to a zero-rule classifier (a non-recommended approach that would predict that every CSSV causes frameshift/NMD) (Figure 1C). Reasons for discordant results were often unclear, even after detailed post hoc review. For example, 27 CSSVs had a frameshift/NMD outcome per RNA-seq and a predicted non-frameshift outcome per SpliceAI (using the expanded ±5,000-bp window). In only one instance were we able to resolve this discrepancy via additional in silico review: A rare CSSV in the gene ITSN2 (MIM: 604464) was predicted to result in in-frame intron inclusion, but the insertion would include a premature stop codon.
Comparison of specific splicing outcomes of CSSVs using RNA-seq vs. in silico predictions
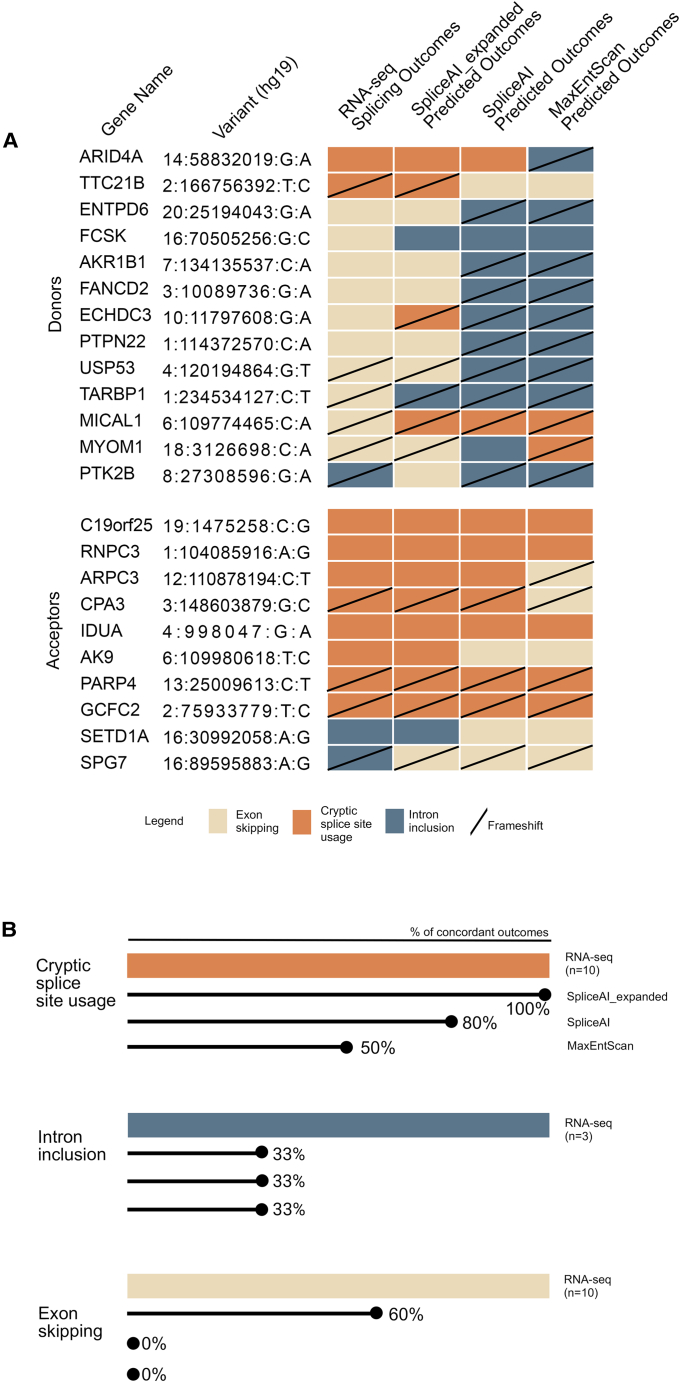
Next, we compared the specific splicing outcome of CSSVs (cryptic splice site usage, exon skipping, or intron inclusion; categories C–H above) between RNA-seq and in silico predictions, for the n = 23 variants where this could be determined from RNA-seq (Figure 2A). Total pairwise concordance was 74% (17/23) for SpliceAI_expanded (improved from 39% [9/23] for SpliceAI) and 26% (6/23) for MaxENTScan; AutoPVS1 does not provide such predictions. The performance of both in silico methods seemed to vary by outcome category, e.g., with SpliceAI_expanded correctly predicting all uses of cryptic splice sites (10/10), some of the exon skipping events (6/10), and only one of the intron inclusion events (1/3) (Figures 2A and 2B). Two selected donor CSSVs are used to illustrate that in silico tools were often correct in predicting frameshift vs. no frameshift outcomes, however, for incorrect and/or incomplete mechanisms of abnormal splicing (Figures 3 and 4).
Figure 2.
Specific categories of CSSV splicing outcomes
(A) Selected variants (n = 23) with specific splicing outcomes in RNA-seq (exon skipping, cryptic splice site use, and intron inclusion) compared with splicing outcome predictions from in silico tools. Total pairwise concordance in specific splicing outcomes was 74% (17/23) for SpliceAI_expanded, 39% (9/23) for SpliceAI, and 26% (6/23) for MaxENTScan.
(B) Concordance of specific splicing outcomes in RNA-seq vs. in silico predictions.
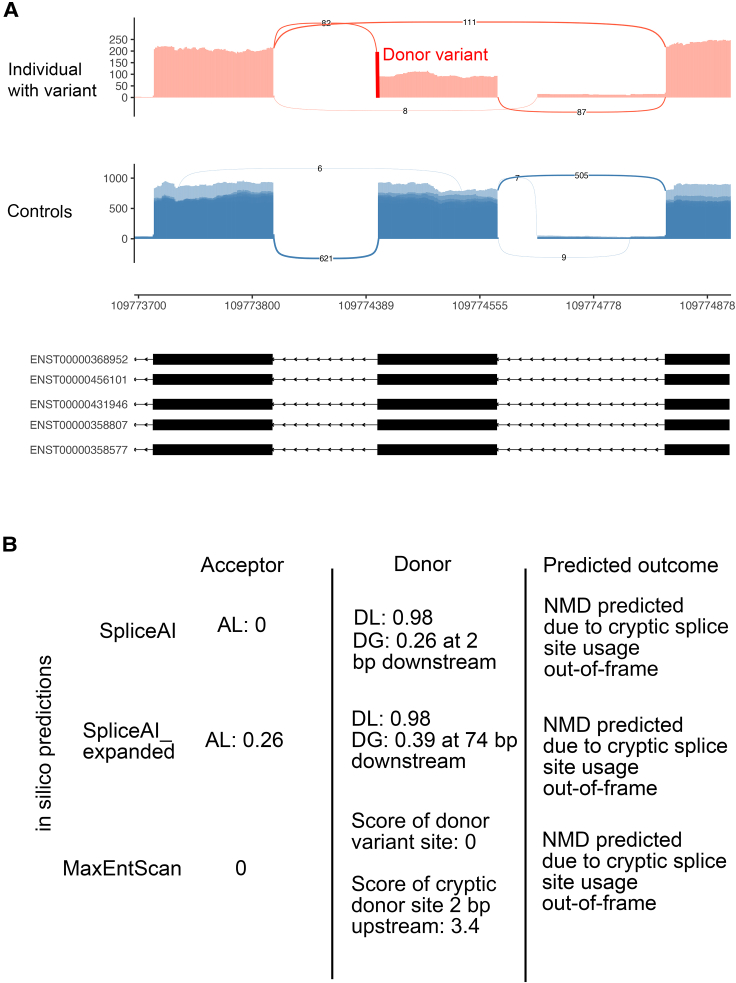
Figure 3.
Example of a donor CSSV showing discordant splicing events in blood RNA-seq vs. in silico predictions but overall correct outcome (frameshift vs. no frameshift)
(A) The sashimi plot from RNA-seq demonstrates that a rare CSSV (NM_022765.4:c.571+1G>T) in MICAL1 (MIM: 607129) results in exon skipping leading to a frameshift.
(B) SpliceAI, SpliceAI_expanded, and MaxEntScan all predicted activation of a cryptic splice site resulting in frameshift.
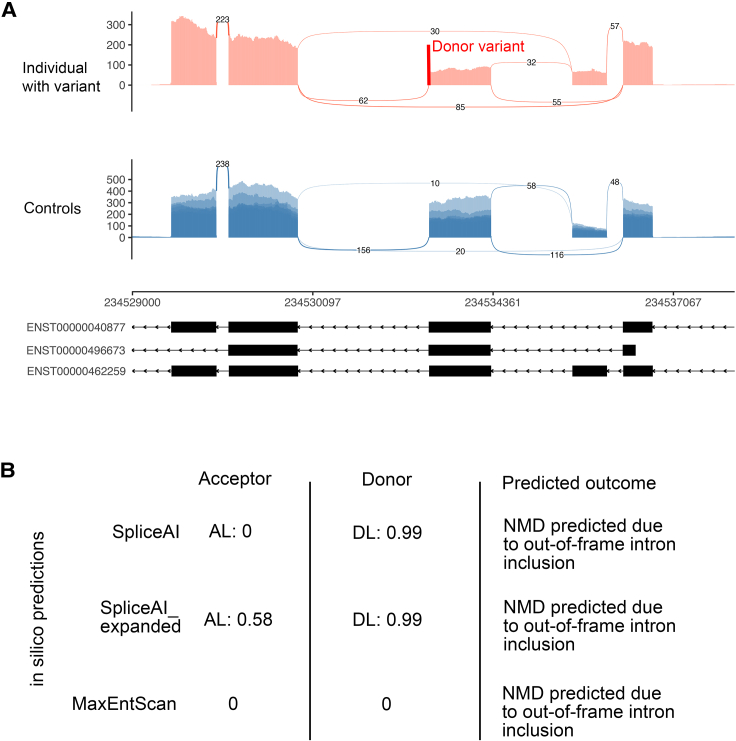
Figure 4.
Example of a donor CSSV showing discordant splicing events in blood RNA-seq vs. in silico predictions but overall correct outcome (frameshift vs. no frameshift)
(A) The sashimi plot from RNA-seq demonstrates that a rare CSSV (NM_005646.4:c.4243+1G>A) in TARBP1 (MIM: 605052) results in exon skipping, leading to a frameshift.
(B) SpliceAI, SpliceAI_expanded, and MaxEntScan all predicted intron inclusion resulting in frameshift.
Discussion
Challenging common assumptions and interpretation heuristics for CSSVs
Most human genomes contain one or more rare CSSVs, as seen in our study cohort for rare CSSVs in blood-expressed genes. Although there is widespread recognition that a rare CSSV need not necessarily result in LoF, experiences from decades of using focused clinical genetic testing (with a resulting ascertainment bias) may have contributed to a misconception that CSSVs are comparable with nonsense and frameshift variants. We present a systematic assessment of the impact of rare “unselected” CSSVs in blood-expressed genes, using RNA-seq. We found that nearly one in four CSSVs may not cause LoF, and that in silico predictions using established tools and published guidelines were often discordant with RNA-seq data.
In recent years, there have been numerous computational tools developed to predict the location of novel splice sites and, thus, the impact of DNA variants on splicing.12,14 These tools were validated using none to a limited number of CSSVs (e.g., near intronic variants ≥3 nucleotides from a canonical exon boundary in SpliceAI-10k; n = 55 in 300K-RNA Top-4) and were not created to predict specific CSSV outcomes like exon skipping or intron inclusion.14,15,30 Another tool called MutSpliceDB has been developed to facilitate interpretation of non-coding variants that affect splicing; however, it lacks data for many CSSVs (included in a list of 341 variants with no interpretations available are mostly variants in the ±1 and 2 sites) and being derived from RNA-seq data using samples from cancer cell lines, may have a bias for pathogenic variants in cancer-related genes.31 A prior report found that intron inclusion was poorly predicted using SpliceAI when compared with transcriptome sequencing data.32 AutoPVS1 does not list intron inclusion as a specific outcome, nor does it distinguish which variants result in exon skipping versus cryptic splice site usage.28 For now, no in silico methods seem to predict the precise impact of CSSVs with the sensitivity and specificity needed for clinical diagnostics.33 A recent study has proposed using the American College of Medical Genetics (ACMG)/Association for Molecular Pathology (AMP) PM4 (moderate evidence of pathogenicity) criterion for CSSVs that are predicted to result in intron inclusion and three or more in-frame events as predicted by 300K-RNA Top-4.15 Relative weighting of biological function or evolutionary conservation of the affected gene region(s) may need to be considered in addition to the length of the in-frame disruption.15 We propose, in addition, that future updates to published guidelines on the use of PVS1 should consider the use of SpliceAI with an expanded window of ±5,000 bp.
The impact of CSSVs on splicing can be complex. Multiple and/or partial effects on splicing have been observed in the past (e.g., with some aberrantly spliced transcripts resulting in LoF and others showing no apparent impact or producing a functional transcript).23 Surprisingly, some CSSVs in our data showed no direct (aberrant/non-WT splice junctions) or indirect (reduced read depth, compatible with NMD) impact on splicing. The ACMG/AMP rule, BS3, may be applied to CSSVs with evidence of normal splicing patterns demonstrated by RNA-seq fulfilling specific criteria.23,34 A recent study using cell culture-based full-length gene splicing assays demonstrated that specific nucleotide substitutions (GT>GC in the donor splice site) can generate WT transcript levels in an estimated 15%–18% of cases; no other nucleotide substitutions in the +2 donor splice site were able to generate WT transcripts.29 Moreover, WT splicing as a result of the 5′ splice site GT>GC substitutions was not accurately predicted by in silico tools.29 Our results showed that WT transcripts can be generated with diverse nucleotide substitutions, in no consistent ranking order in the 5′ donor splice site as recently described (although this study performed RNA-seq in fibroblast samples and also included common variants), affecting ±1 and 2 canonical splice sites, highlighting in vitro assays’ inability to capture the full complexity of splicing in human cells (Figure S3).35 Of note, the sensitivity of our RNA-seq methods for detecting evidence of NMD will be less than 100%. Long-read RNA-seq might have facilitated more robust quantitation of transcript isoforms, assessment for allele-specific expression in the setting of apparently normal splice junction depth (outcome category B; see Subjects, material, and methods), and revealed additional splicing outcomes.
Our study has several additional limitations. First, we recognize that in-frame indels can still result in non-functional proteins (e.g., through disruption of an essential protein domain) and that protein function cannot be inferred completely from RNA-seq. In the absence of any evidence-based guidance, we assumed for all in silico predictions that exon skipping would be the typical impact of an acceptor site loss and that intron retention/inclusion would be the typical impact of a donor site loss, whenever an alternative/cryptic splice site was not present. There is growing appreciation that this reasoning is overly simplistic.32,36 The landscape of impacts of rare CSSVs may change based on age, sex, genetic ancestry, and/or environment.37,38,39 Assessing abnormal splicing in blood may not be representative for all tissue types due to alternative splicing, resulting in differential expression of certain transcripts and ultimately tissue-specific gene expression. As blood remains the most clinically accessible tissue, we restricted our analyses to blood-expressed genes. Replicating our findings in additional tissues beyond whole blood warrants future consideration. We acknowledge that there may also be underlying ascertainment biases related to our selection of rare, unselected blood-expressed CSSVs from our study cohort, which was family based, with children presenting with medical complexity as probands for GS, resulting in the majority of individuals in our study being under the age of 19 years.40 Based on our review of detailed phenotype data and GS data, in only two instances was the participant’s initial recruitment into the study driven by a CSSV. Confirming our findings in additional cohorts will be important, although with the recognition that all cohorts will have ascertainment biases. Last, we were underpowered in this study to identify substitution- and location- or motif-specific predictors of splicing outcomes, and to explore how allele frequency/variant rarity may be correlated with CSSV impact.
Implications for diagnostics, predictive testing, and screening
These data reinforce prior expert consensus recommendations that cautioned against applying PVS1 to CSSVs in the absence of additional supportive evidence.23 Our findings are further supported by a recent pre-print publication that recommended against assigning PVS1 evidence strength (“PVS1_N/A”) for those CSSVs resulting in a functional donor/acceptor site and, thereby, WT transcripts in RNA-seq.41 We agree with their recommendations to annotate physiologically occurring alternative splicing events (or leaky splicing events, as we have also noted in some CSSVs) as candidate rescue transcripts. Consideration of gene-specific details (the location or involvement of critical region in a gene) and guidance on applying various lines of evidence (computational and in vitro assays to assess impact on splicing) will become increasingly important in navigating equivocal clinical scenarios requiring interpretation of CSSVs.41 Our findings demonstrate that in silico approaches are relatively conservative in their assignation of a frameshift/NMD outcome, relative to a zero-rule classifier, and we suggest that this conservativeness is appropriate in clinical practice while functional assays remain difficult to access.
The advantages and limitations of RNA-seq, which can be done high throughput, should be weighed against a targeted approach like RT-PCR; the latter may have greater sensitivity in detecting mis-spliced reads of low read depth as a result of low gene expression in whole blood and being able to distinguish partial from complete abnormal splicing.34 Although a majority of CSSVs result in a LoF, this assumption should be questioned when genome-wide sequencing identifies novel rare CSSVs in genes associated with ultra-rare, poorly characterized conditions with non-specific phenotypes (such as autism or developmental delay in neurodevelopmental disorders) and appropriate clinical evaluation should be undertaken.42 The nuances of CSSV interpretation take on added importance when the pre-test probability is low and phenotypes are absent or variable, as is the case with most secondary findings and in newborn genomic screening programs.33,40,43
Data and code availability
The CSSVs analyzed in this study have been uploaded to dbSNP (Handle: COSTAINLABORATORY; Batch: CSSV_HGGAdvances). RNA-seq analysis and in silico predictions data can be found in Table S2. The complete genome-wide DNA and RNA-seq datasets were not consented to be deposited in a public repository, but are available from the corresponding author on request.
Acknowledgments
We gratefully acknowledge all the individuals and their families who participated in this study. We thank the many health care providers involved in the diagnosis and care of these study participants. Special thanks to all staff affiliated with the Complex Care Program and The Center for Applied Genomics. M.D.W. was supported by the Canada Research Chairs Program. Funding was provided by Genome Canada (OGI-158; M.D.W., A.S., and J.J.D.), the SickKids Centre for Genetic Medicine and Translational Genomics Node, the Sickkids Research Institute, the Canadian Institutes of Health Research (Funding Reference Number: PJT186240), and the University of Toronto McLaughlin Centre.
Author contributions
Conceptualization: G.C., R.Y.O., A.A., D.C., G.G., S.W., H.H., and K.Y.; data curation: R.Y.O., A.A., D.C., G.G., B.H., and G.C.; formal analysis: R.Y.O., A.A., D.C., G.G., and G.C.; funding acquisition: G.C., M.D.W., A.S., and J.J.D.; investigation: R.Y.O., A.A., D.C., and G.C.; methodology: R.Y.O., A.A., D.C., G.G., H.H., K.Y., S.W., and G.C.; project administration: R.Y.O., A.A., T.K., B.T., and G.C.; resources: G.C., C.M., M.D.W., and J.J.D.; software: R.Y.O., A.A., D.C., G.G., H.H., K.Y., and G.C.; visualization: R.Y.O., A.A., D.C., H.H., and G.C.; writing – original draft: R.Y.O., A.A., and G.C.; writing – review and editing: all other authors.
Declaration of interests
S.W. is an employee of Genomics England Limited.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2024.100299.
Web resources
dbSNP: https://www.ncbi.nlm.nih.gov/snp/
Genotype-Tissue Expression (GTEx) Portal: https://gtexportal.org/home/
Online Mendelian Inheritance in Man: https://www.omim.org/
Supplemental information
References
- 1.Krawczak M., Reiss J., Cooper D.N. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum. Genet. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- 2.Burset M., Seledtsov I.A., Solovyev V.V. Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res. 2000;28:4364–4375. doi: 10.1093/nar/28.21.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hastings M.L., Krainer A.R. Pre-mRNA splicing in the new millennium. Curr. Opin. Cell Biol. 2001;13:302–309. doi: 10.1016/s0955-0674(00)00212-x. [DOI] [PubMed] [Google Scholar]
- 4.Matera A.G., Wang Z. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014;15:108–121. doi: 10.1038/nrm3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rogalska M.E., Vivori C., Valcárcel J. Regulation of pre-mRNA splicing: roles in physiology and disease, and therapeutic prospects. Nat. Rev. Genet. 2023;24:251–269. doi: 10.1038/s41576-022-00556-8. [DOI] [PubMed] [Google Scholar]
- 6.Krawczak M., Thomas N.S.T., Hundrieser B., Mort M., Wittig M., Hampe J., Cooper D.N. Single base-pair substitutions in exon–intron junctions of human genes: nature, distribution, and consequences for mRNA splicing. Hum. Mutat. 2007;28:150–158. doi: 10.1002/humu.20400. [DOI] [PubMed] [Google Scholar]
- 7.Ward A.J., Cooper T.A. The pathobiology of splicing. J. Pathol. 2010;220:152–163. doi: 10.1002/path.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dufner-Almeida L.G., do Carmo R.T., Masotti C., Haddad L.A. Chapter Two - Understanding human DNA variants affecting pre-mRNA splicing in the NGS era. Kumar D., editor. Adv. Genet. 2019;103:39–90. doi: 10.1016/bs.adgen.2018.09.002. Academic Press. [DOI] [PubMed] [Google Scholar]
- 9.Fatscher T., Boehm V., Gehring N.H. Mechanism, factors, and physiological role of nonsense-mediated mRNA decay. Cell. Mol. Life Sci. 2015;72:4523–4544. doi: 10.1007/s00018-015-2017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hug N., Longman D., Cáceres J.F. Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Res. 2016;44:1483–1495. doi: 10.1093/nar/gkw010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abou Tayoun A.N., Pesaran T., DiStefano M.T., Oza A., Rehm H.L., Biesecker L.G., Harrison S.M., ClinGen Sequence Variant Interpretation Working Group ClinGen SVI Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion. Hum. Mutat. 2018;39:1517–1524. doi: 10.1002/humu.23626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costain G., Cohn R.D., Scherer S.W., Marshall C.R. Genome sequencing as a diagnostic test. CMAJ (Can. Med. Assoc. J.) 2021;193:E1626–E1629. doi: 10.1503/cmaj.210549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaganathan K., Kyriazopoulou Panagiotopoulou S., McRae J.F., Darbandi S.F., Knowles D., Li Y.I., Kosmicki J.A., Arbelaez J., Cui W., Schwartz G.B., et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell. 2019;176:535–548.e24. doi: 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 15.Dawes R., Bournazos A.M., Bryen S.J., Bommireddipalli S., Marchant R.G., Joshi H., Cooper S.T. SpliceVault predicts the precise nature of variant-associated mis-splicing. Nat. Genet. 2023;55:324–332. doi: 10.1038/s41588-022-01293-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Resch A., Xing Y., Alekseyenko A., Modrek B., Lee C. Evidence for a subpopulation of conserved alternative splicing events under selection pressure for protein reading frame preservation. Nucleic Acids Res. 2004;32:1261–1269. doi: 10.1093/nar/gkh284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costain G., Walker S., Marano M., Veenma D., Snell M., Curtis M., Luca S., Buera J., Arje D., Reuter M.S., et al. Genome Sequencing as a Diagnostic Test in Children With Unexplained Medical Complexity. JAMA Netw. Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.18109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stavropoulos D.J., Merico D., Jobling R., Bowdin S., Monfared N., Thiruvahindrapuram B., Nalpathamkalam T., Pellecchia G., Yuen R.K.C., Szego M.J., et al. Whole Genome Sequencing Expands Diagnostic Utility and Improves Clinical Management in Pediatric Medicine. NPJ Genom. Med. 2016;1 doi: 10.1038/npjgenmed.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deshwar A.R., Yuki K.E., Hou H., Liang Y., Khan T., Celik A., Ramani A., Mendoza-Londono R., Marshall C.R., Brudno M., et al. Trio RNA sequencing in a cohort of medically complex children. Am. J. Hum. Genet. 2023;110:895–900. doi: 10.1016/j.ajhg.2023.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lionel A.C., Costain G., Monfared N., Walker S., Reuter M.S., Hosseini S.M., Thiruvahindrapuram B., Merico D., Jobling R., Nalpathamkalam T., et al. Improved diagnostic yield compared with targeted gene sequencing panels suggests a role for whole-genome sequencing as a first-tier genetic test. Genet. Med. 2018;20:435–443. doi: 10.1038/gim.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walker S., Lamoureux S., Khan T., Joynt A.C.M., Bradley M., Branson H.M., Carter M.T., Hayeems R.Z., Jagiello L., Marshall C.R., et al. Genome sequencing for detection of pathogenic deep intronic variation: A clinical case report illustrating opportunities and challenges. Am. J. Med. Genet. 2021;185:3129–3135. doi: 10.1002/ajmg.a.62389. [DOI] [PubMed] [Google Scholar]
- 22.GTEx Portal. 2023. https://gtexportal.org/home/
- 23.Ellingford J.M., Ahn J.W., Bagnall R.D., Baralle D., Barton S., Campbell C., Downes K., Ellard S., Duff-Farrier C., FitzPatrick D.R., et al. Recommendations for clinical interpretation of variants found in non-coding regions of the genome. Genome Med. 2022;14:73. doi: 10.1186/s13073-022-01073-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Costain G., Jobling R., Walker S., Reuter M.S., Snell M., Bowdin S., Cohn R.D., Dupuis L., Hewson S., Mercimek-Andrews S., et al. Periodic reanalysis of whole-genome sequencing data enhances the diagnostic advantage over standard clinical genetic testing. Eur. J. Hum. Genet. 2018;26:740–744. doi: 10.1038/s41431-018-0114-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abstracts from the 54th European Society of Human Genetics (ESHG) Conference: Oral Presentations. Eur. J. Hum. Genet. 2022;30:3–87. doi: 10.1038/s41431-021-01025-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang Y.F., Imam J.S., Wilkinson M.F. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 27.Lewis B.P., Green R.E., Brenner S.E. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl. Acad. Sci. USA. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiang J., Peng J., Baxter S., Peng Z. AutoPVS1: An automatic classification tool for PVS1 interpretation of null variants. Hum. Mutat. 2020;41:1488–1498. doi: 10.1002/humu.24051. [DOI] [PubMed] [Google Scholar]
- 29.Lin J.H., Tang X.Y., Boulling A., Zou W.B., Masson E., Fichou Y., Raud L., Le Tertre M., Deng S.J., Berlivet I., et al. First estimate of the scale of canonical 5’ splice site GT>GC variants capable of generating wild-type transcripts. Hum. Mutat. 2019;40:1856–1873. doi: 10.1002/humu.23821. [DOI] [PubMed] [Google Scholar]
- 30.de Sainte Agathe J.M., Filser M., Isidor B., Besnard T., Gueguen P., Perrin A., Van Goethem C., Verebi C., Masingue M., Rendu J., et al. SpliceAI-visual: a free online tool to improve SpliceAI splicing variant interpretation. Hum. Genom. 2023;17:7. doi: 10.1186/s40246-023-00451-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palmisano A., Vural S., Zhao Y., Sonkin D. MutSpliceDB: A database of splice sites variants with RNA-seq based evidence on effects on splicing. Hum. Mutat. 2021;42:342–345. doi: 10.1002/humu.24185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shiraishi Y., Okada A., Chiba K., Kawachi A., Omori I., Mateos R.N., Iida N., Yamauchi H., Kosaki K., Yoshimi A. Systematic identification of intron retention associated variants from massive publicly available transcriptome sequencing data. Nat. Commun. 2022;13:5357. doi: 10.1038/s41467-022-32887-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haque B., Cheerie D., Birkadze S., Xu A.L., Nalpathamkalam T., Thiruvahindrapuram B., Walker S., Costain G. Estimating the proportion of nonsense variants undergoing the newly described phenomenon of manufactured splice rescue. Eur. J. Hum. Genet. 2024;32:238–242. doi: 10.1038/s41431-023-01495-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bournazos A.M., Riley L.G., Bommireddipalli S., Ades L., Akesson L.S., Al-Shinnag M., Alexander S.I., Archibald A.D., Balasubramaniam S., Berman Y., et al. Standardized practices for RNA diagnostics using clinically accessible specimens reclassifies 75% of putative splicing variants. Genet. Med. 2022;24:130–145. doi: 10.1016/j.gim.2021.09.001. [DOI] [PubMed] [Google Scholar]
- 35.Erkelenz S., Theiss S., Kaisers W., Ptok J., Walotka L., Müller L., Hillebrand F., Brillen A.L., Sladek M., Schaal H. Ranking noncanonical 5’ splice site usage by genome-wide RNA-seq analysis and splicing reporter assays. Genome Res. 2018;28:1826–1840. doi: 10.1101/gr.235861.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rivas M.A., Pirinen M., Conrad D.F., Lek M., Tsang E.K., Karczewski K.J., Maller J.B., Kukurba K.R., DeLuca D.S., Fromer M., et al. Impact of predicted protein-truncating genetic variants on the human transcriptome. Science. 2015;348:666–669. doi: 10.1126/science.1261877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aicher J.K., Jewell P., Vaquero-Garcia J., Barash Y., Bhoj E.J. Mapping RNA splicing variations in clinically accessible and nonaccessible tissues to facilitate Mendelian disease diagnosis using RNA-seq. Genet. Med. 2020;22:1181–1190. doi: 10.1038/s41436-020-0780-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Basu M., Wang K., Ruppin E., Hannenhalli S. Predicting tissue-specific gene expression from whole blood transcriptome. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abd6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.García-Pérez R., Ramirez J.M., Ripoll-Cladellas A., Chazarra-Gil R., Oliveros W., Soldatkina O., Bosio M., Rognon P.J., Capella-Gutierrez S., Calvo M., et al. The landscape of expression and alternative splicing variation across human traits. Cell Genom. 2023;3 doi: 10.1016/j.xgen.2022.100244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paterson A.D., Seok S.C., Vieland V.J. The effect of ascertainment on penetrance estimates for rare variants: implications for establishing pathogenicity and for genetic counselling. PLoS One. 2023;18 doi: 10.1371/journal.pone.0290336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walker L.C., Hoya M.d.L., Wiggins G.A.R., Lindy A., Vincent L.M., Parsons M.T., Canson D.M., Bis-Brewer D., Cass A., Tchourbanov A., et al. Using the ACMG/AMP framework to capture evidence related to predicted and observed impact on splicing: Recommendations from the ClinGen SVI Splicing Subgroup. Am. J. Hum. Genet. 2023;110:1046–1067. doi: 10.1016/j.ajhg.2023.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanders S.J., Schwartz G.B., Farh K.K.H. Clinical impact of splicing in neurodevelopmental disorders. Genome Med. 2020;12:36. doi: 10.1186/s13073-020-00737-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Forrest I.S., Chaudhary K., Vy H.M.T., Petrazzini B.O., Bafna S., Jordan D.M., Rocheleau G., Loos R.J.F., Nadkarni G.N., Cho J.H., Do R. Population-Based Penetrance of Deleterious Clinical Variants. JAMA. 2022;327:350–359. doi: 10.1001/jama.2021.23686. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The CSSVs analyzed in this study have been uploaded to dbSNP (Handle: COSTAINLABORATORY; Batch: CSSV_HGGAdvances). RNA-seq analysis and in silico predictions data can be found in Table S2. The complete genome-wide DNA and RNA-seq datasets were not consented to be deposited in a public repository, but are available from the corresponding author on request.