Abstract
Mature human immunodeficiency virus type 1 (HIV-1) particles contain a cone-shaped core structure consisting of the internal ribonucleoprotein complex encased in a proteinaceous shell derived from the viral capsid protein. Because of their very low stability after membrane removal, HIV-1 cores have not been purified in quantities sufficient for structural and biochemical analysis. Based on our in vitro assembly experiments, we have developed a novel method for isolation of intact mature HIV-1 cores. Concentrated virus suspensions were briefly treated with nonionic detergent and immediately centrifuged in a microcentrifuge for short periods of time. The resuspended pellet was subsequently analyzed by negative-stain and thin-section electron microscopy and by immunoelectron microscopy. Abundant cone-shaped cores as well as tubular and aberrant structures were observed. Stereo images showed that core structures preserved their three-dimensional architecture and exhibited a regular substructure. Detailed analysis of 155 cores revealed an average length of ca. 103 nm, an average diameter at the base of ca. 52 nm, and an average angle of 21.3°. There was significant variability in all parameters, indicating that HIV cores are not homogeneous. Immunoblot analysis of core preparations allowed semiquantitative estimation of the relative amounts of viral and cellular proteins inside the HIV-1 core, yielding a model for the topology of various proteins inside the virion.
Retroviruses contain a RNA genome that is reverse transcribed and integrated into the host cell genome in the early phase of infection, before viral proteins are expressed. This mode of replication requires the presence of all necessary components of the replication and integration complexes either within the infecting virion or in the target cell. In contrast to many other enveloped viruses, retrovirus entry does not require a pH-dependent step (25), and removal of the membrane may suffice to trigger uncoating of the genome. It appears likely, therefore, that retroviruses contain a metastable internal structure which is stable in the enveloped virion but readily disintegrates upon fusion of the viral and target cell membranes. Electron microscopic (EM) analysis revealed an internal electron-dense structure, termed the core, which has a characteristic morphology for various groups of retroviruses (reviewed in references 13 and 36). This core consists of the viral genome and replication proteins encased in a proteinaceous shell. Human immunodeficiency virus type 1 (HIV-1) is a typical lentivirus with a characteristic cone-shaped core, while other retroviruses have spherical or tubular cores (36). Isolation and analysis of murine and avian retroviral cores were performed about 25 years ago (3, 30, 48, 49), while lentiviral cores, with the exception of that of equine infectious anemia virus (EIAV) (42), appear to be unstable and are rapidly disintegrated upon membrane removal (27, 32, 62). Consequently, purification and structural and biochemical analysis of intact HIV-1 cores has not been possible.
Some information on the molecular organization of HIV-1 can be derived from knowledge of the assembly process. Retrovirus particles bud from the plasma membrane and are released as immature noninfectious virus, containing a spherical electron-dense shell underneath the membrane, instead of the mature core (reviewed in reference 13). The immature core is stable and can be easily recovered upon delipidation of the virion (39, 43, 46, 57). Extracellular maturation of the infectious virion requires proteolytic cleavage of viral polyproteins (Gag and Gag-Pol) by the viral proteinase (PR) and leads to morphological condensation of the core (reviewed in reference 51). This mode of assembly allows initial formation of a stable structure which is subsequently destabilized by proteolysis, once all components are confined in the budding virion. Proteolytic maturation, therefore, switches from the assembly to the disassembly mode and prepares the virus for the subsequent entry step.
The dry mass of retroviral particles consists of ca. 60% protein, ca. 2 to 3% nucleic acid (genomic RNA, tRNA, and other small RNAs), and ca. 30% lipid (53). The main structural proteins of the virion are derived from the Gag polyprotein, Pr55 in the case of HIV-1. Gag alone is both necessary and sufficient for formation of extracellular particles with immature morphology (reviewed in reference 52). The Pr55gag polyprotein consists of the matrix (MA), capsid (CA), nucleocapsid (NC), and p6 domains as well as some small spacer peptides. In the immature virion, Gag polyproteins arrange radially, with the N-terminal membrane-binding MA domain closely apposed to the virion membrane and the C-terminal NC and p6 domains pointing toward the center (10, 35, 58). Interestingly, individual domains are separated by stretches of extended protein chain which contain the PR cleavage sites (10, 58). Proteolysis of the Gag polyprotein occurs in a sequential ordered manner, and individual cleavages are likely to be important for defined steps of maturation (1, 57). In the mature virion, the order of Gag domains is largely preserved, with MA forming a thin layer underneath the virion membrane, CA corresponding to the core shell, and NC being part of the internal ribonucleoprotein (RNP) complex (14). The localization of p6, which is involved in the late stages of virus release, is not known. The viral replication proteins are synthesized as parts of a Gag-Pol fusion protein and are also proteolytically released in the maturing virion (reviewed in reference 51). PR itself is encoded on the Gag-Pol polyprotein and appears to be activated in the budding process. In the immature virion, the Pol-derived proteins PR, reverse transcriptase (RT), and integrase (IN) are probably localized toward the center since they are encoded C terminally of Gag. Following maturation, at least the nucleic acid binding proteins RT and IN are likely to be part of the internal core and to be retained with the genome upon target cell entry. The viral surface and transmembrane glycoproteins gp120 and gp41 are also synthesized as polyproteins but are transported and processed via the vesicular route and are acquired by the budding virion at the plasma membrane (25).
HIV-1 encodes a number of regulatory proteins besides the prototypic Gag, Pol, and Env proteins, and some of these are also incorporated into the virion (reviewed in reference 6). Most notable is Vpr, which is a major constituent of the virion and is packaged via interaction with p6 (28, 40). In addition, a small amount of the HIV-1 Nef protein is incorporated into HIV-1 particles and cleaved by the viral PR (38, 56). The situation is more controversial for Vif, which was reported to be packaged into the virion (33), while more recent data suggest that it is mostly present in cell-derived vesicles copurifying with HIV-1 (7). Finally, the cellular chaperone cyclophilin A (9, 50) and the cytoskeletal protein actin (37, 58) are also packaged in substantial amounts into HIV-1 virions, most likely by interaction with the CA and NC domains of Gag, respectively. However, except for cellular glycoproteins incorporated into the virion membrane (14, 34), very little is known about the topology and function of the viral and cellular accessory proteins in the HIV-1 virion.
Most of our current understanding of the molecular organization of HIV-1 is derived from immuno-EM studies of complete virions (8, 14, 18, 34, 54), from biochemical fractionation of subviral components (4, 26, 33), and by analogy with other members of the family (reviewed in reference 52). Because of the inherent instability of the HIV-1 core, detailed characterization of its morphology and molecular architecture has not been performed. Here, we report a new procedure for the rapid isolation of intact HIV-1 cores and the structural characterization of these core particles by negative-stain and thin-section EM as well as biochemical analysis of their protein composition.
MATERIALS AND METHODS
Cell culture and virus preparation.
MT-4 (21) and C8166 (44) cells were maintained at 37°C and 5% CO2 in RPMI 1640 supplemented with 10% heat-inactivated fetal calf serum, penicillin (100 U/ml), streptomycin (100 μg/ml), 4 mM glutamine, and 5 mM HEPES. Stocks of HIV-1 strain NL4-3 (2) were produced by transfection of HeLa cells. MT-4 cells were initially infected with cell-free virus, and infected cultures were subsequently expanded by cocultivation. Uninfected cells were diluted to 5 × 105 cells per ml 4 h prior to infection and added to an infected culture at a ratio of 10:1 followed by vigorous mixing. Virus preparations for isolation of HIV-1 cores were harvested before pronounced cytopathic effects were observed (24 to 32 h postinfection). Virus-containing supernatants were cleared by low-speed centrifugation, filtered through 0.45-μm-pore-size cellulose-acetate filters (Schleicher & Schuell), and analyzed for antigen content by a CA-specific enzyme-linked immunosorbent assay. Infectious titers were determined as 50% tissue culture infectious dose by endpoint titration using serial 10-fold dilutions of virus on octuplicate cultures of C8166 cells.
Isolation of HIV-1 cores.
Virus particles were concentrated from cleared culture medium by centrifugation through a cushion of 20% (wt/wt) sucrose in phosphate-buffered saline (PBS) at 130,000 × g for 2 h at 4°C. The pellet was slowly resuspended in PBS (ca. 3.5 μl per ml of initial culture volume). Subsequently, 40 μl of fresh virus suspension was mixed with an equal volume of 200 mM NaCl–100 mM morpholinepropanesulfonic acid (MOPS; pH 7.0), and virions were lysed for 2 min at room temperature by adding Triton X-100 to a final concentration of 0.5%. HIV-1 cores were recovered by centrifugation in a microcentrifuge at full speed (13,800 × g) for 8 min at 4°C. The pellets were washed twice with 100 mM NaCl–50 mM MOPS (pH 7.0) and resuspended in 8 μl of the same buffer. Core suspensions were processed immediately for further analysis.
Preparation of HIV-1 cores was also attempted by ultracentrifugation through a detergent cushion (57). In this case, concentrated virus was layered on top of a sucrose step gradient containing or lacking detergent. The step gradient consisted of a layer of 2 ml of 20% (wt/vol) sucrose, a 1.5-ml layer of 15% (wt/vol) sucrose with or without 0.5% Triton X-100, a 1.5-ml layer of 10% (wt/vol) sucrose without detergent, and PBS. Gradients were centrifuged at 220,000 × g for 2 h at 4°C. Pellets were resuspended in 100 mM NaCl–50 mM MOPS (pH 7.0) and processed immediately for EM analysis.
EM.
For negative staining, 8-μl samples of HIV-1 core suspensions were applied on Parafilm and covered with a UV-irradiated Formvar-carbon-coated grid (mesh size, 200) for 5 min. After binding, grids were washed four times with 100 mM NaCl–50 mM MOPS (pH 7.0) and stained with 2% uranyl acetate for 5 min. Excess stain was soaked off by touching the grid to a filter paper. After staining, the grid was air dried.
For thin-section analysis of HIV-1 virions, freshly infected MT-4 cells were drawn into cellulose tubes by capillary action (24), and tubes were immersed overnight in fresh medium to allow in situ virus production. The detailed procedure for analysis of virus particles will be described elsewhere. Briefly, the capillary tubes were washed in PBS 1 day after infection, and cells were fixed for 1 h with 2.5% glutaraldehyde in PBS. Subsequently, tubes were washed with PBS and cells were postfixed within the tubes for 30 min with 1% OsO4 in PBS, washed with water, stained for 30 min with 1% uranyl acetate in water, and dehydrated in a graded series of ethanol. Capillary tubes were embedded in ERL resin for sectioning. Ultrathin sections were counterstained with 2% uranyl acetate and lead citrate. For immunolabeling of ERL-embedded HIV-1 virions, antigens were exposed by etching ultrathin sections for 5 min with 1% sodium periodate in 0.5% acetic acid. Subsequently, sections were incubated overnight with polyclonal antiserum against CA (dilution of 1:500) at 4°C, and immune complexes were detected with protein A conjugated to 10-nm gold particles.
Immunolabeling of isolated cores was performed by aspirating core suspensions into cellulose tubes and fixing the cells with 2.5% paraformaldehyde in PBS for 30 min. Tubes were washed with PBS, stained for 30 min with 1% OsO4 in PBS, washed with water, and embedded in ERL resin for sectioning. Ultrathin sections were blocked with 1% bovine serum albumin in PBS, and antigens were detected as described above. All electron micrographs were taken with a Philips CM120 transmission electron microscope at 80 kV.
For morphometric analysis of negatively stained cores, images were taken at an initial magnification of ×65,000 to ×125,000, and high-resolution prints at a final magnification of ×165,000 to ×397,000-fold were digitized. Trace measurements of length, diameter, and angle were performed on structurally well preserved cores, using the SigmaScan software package (Jandel Scientific/SPSS Inc.). Statistical analysis was performed with the SPSS software package or Microsoft Excel.
Analysis of protein composition of HIV-1 virions and core particles.
Virion or core particle extracts were separated by polyacrylamide gel electrophoresis (PAGE) on sodium dodecyl sulfate (SDS)-polyacrylamide gels containing 17.5 or 10% polyacrylamide and were either stained with Coomassie blue or silver (23) or used for immunoblot analysis. Smaller proteins were analyzed on Tris-Tricine gels (45). For immunoblot analysis, proteins were transferred to nitrocellulose membranes (Schleicher & Schuell) by electroblotting and reacted with specific polyclonal antisera. Peroxidase-conjugated anti-rabbit serum (dilution of 1:10,000; Jackson Immunochemicals Inc.) was used as the secondary antibody, and immune complexes were visualized by enhanced chemiluminescence (Amersham) according to the manufacturer's instructions. In some cases, blots were stripped and reprobed as recommended by the manufacturer. Rabbit polyclonal antisera against MA, CA, NC, PR, IN, and Nef had been raised in our laboratory, using purified bacterially expressed proteins. Rabbit polyclonal antisera against p6, RT, Vpr, cyclophilin A, actin, and gp120 were kind gifts of S. Campbell, R. Goody, U. Schubert, U. von Schwedler, G. Rutter, and V. Bosch, respectively.
RESULTS
Production of high-titered HIV-1 preparations and isolation of viral cores.
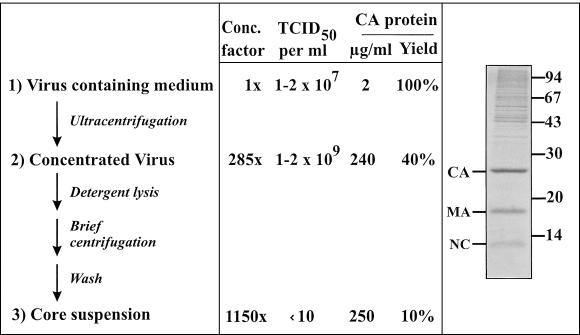
For isolation of intact viral cores, it is important to obtain large amounts of pure and highly infectious HIV-1 particles. Retroviruses are generally rather unstable, and the half-life of infectious HIV-1 (strain NL4-3) in tissue culture is on the order of 4 to 6 h at 37°C (data not shown). Therefore, harvesting virus preparations late at the time of peak antigen concentration results in a large excess of noninfectious particles in the culture medium and a very low infectious unit-to-particle ratio. Furthermore, HIV-1 infection leads to pronounced cytopathic effects in the later stages of infection, causing release of cellular vesicles which tend to copurify with virions (16, 37). For preparation of high-titered HIV-1 stocks devoid of major cellular contaminants, it would therefore appear optimal to harvest virus from a synchronized infection of highly productive cells at an early time point, well before cell lysis occurs. To this end, we infected rapidly growing MT-4 cells by coculture, with essentially all cells positive by immunofluorescence at 16 h postinfection and peak virus production at 20 to 30 h postinfection (data not shown). HIV-1 particles were harvested 24 to 32 h after infection, before pronounced cytopathic effects were observed. Under these conditions, virus preparations contained at least 1 to 2 μg of CA per ml and routinely had a titer of >107 infectious units per ml (Fig. 1). Assuming ca. 1,800 Gag molecules per virion (53), this corresponds to a particle-to-infectious unit ratio of approximately 1,000:1. Concentrating HIV-1 particles by ultracentrifugation through a sucrose cushion led to recovery of 40 to 60% of input CA antigen at a similar particle-to-infectious unit ratio, leading to a final infectious titer of at least 109 per ml (Fig. 1). Analysis of virion preparations showed that viral Gag proteins were the major constituents, with comparatively little contamination by cellular proteins (Fig. 1).
FIG. 1.
Flow chart outlining the preparation of HIV-1 cores. Concentration of particles, relative infectivity (50% tissue culture infectious dose [TCID50]), and HIV-1 CA protein recovery relative to the amount present in the culture medium are shown for various steps of a representative experiment. Comparable results were obtained in five experiments independently performed by three members of the laboratory. The insert shows a Coomassie blue-stained gel of concentrated virus used for core preparation. HIV-1 structural proteins are identified on the left; positions of molecular mass standards (in kilodaltons) are shown on the right.
When HIV-1 particles were lysed with nonionic detergents and subjected to ultracentrifugation, no intact cone-shaped cores were observed on negative-stain EM analysis (data not shown). Previous experiments had indicated that CA protein is partially retained when HIV-1 is centrifuged through a layer of detergent (57), and we therefore analyzed the pellet fraction of sucrose step gradients containing detergent layers for the presence of intact cores. However, only very few cone-shaped particles which appeared damaged on negative-stain EM were observed with a large excess of unstructured aggregates, probably corresponding to disintegrated viruses (Fig. 2A). In our in vitro assembly experiments, we recently observed that core-like particles tend to aggregate and can be recovered by short centrifugation in a microcentrifuge (20a). Assuming that this may also apply to mature HIV-1 cores, we briefly incubated concentrated HIV-1 particles with 0.5% Triton X-100, followed by centrifugation in the microcentrifuge for 8 min. This procedure led to recovery of 5 to 10% of input CA antigen and virtually complete loss of infectivity (Fig. 1), with numerous intact cone-shaped cores detected by negative-stain EM (Fig. 2B). The residual infectivity may be due either to incomplete virus lysis or to a very low infectivity of delipidated viral cores which might be internalized by endocytotic uptake.
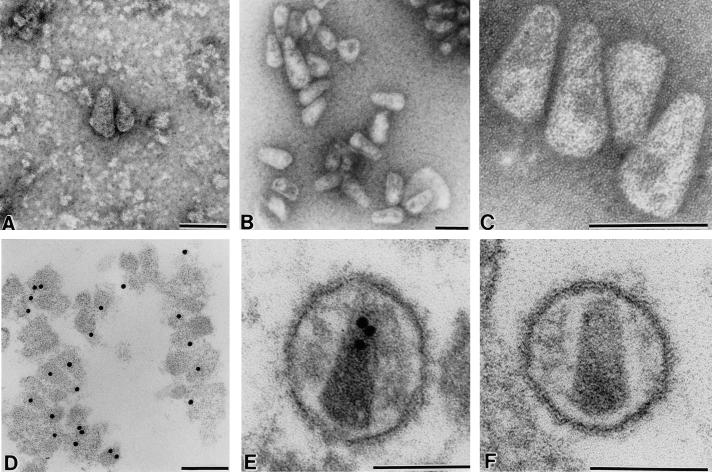
FIG. 2.
EM analysis of isolated HIV-1 cores (A to D) or mature virions (E and F). Particles were visualized either by negative staining (A to C) or on ultrathin sections (D to F). In panel A, cores were prepared by ultracentrifugation through a detergent cushion; in panels B to D, cores were prepared by low-speed centrifugation following detergent stripping. Postembedding immunolabeling of HIV-1 cores (D) and virions (E) was performed with polyclonal antiserum against HIV-1 CA and detection with protein A coupled to 10-nm gold particles. Size bars, 100 nm.
The yield of intact HIV-1 cores depended primarily on the virus concentration before stripping. Virtually no CA antigen and no core particles were recovered when 10-fold less virus was used in this procedure. In addition, recovery of intact cores was affected by the pH and salt concentration of the buffer, with both, low, and high salt concentrations causing a decrease of cone-shaped particles in negative-stain EM. Thorough resuspension of the input virus concentrate was also important for core recovery and was best performed in PBS.
EM analysis of HIV-1 core particles.
Analysis of HIV-1 core preparations by negative-stain EM revealed numerous ordered particles which were mostly cone shaped and tended to form loose aggregates (Fig. 2B). The tendency to aggregate was more obvious when core preparations were analyzed by thin-section EM (Fig. 2D and data not shown). Images taken from negatively stained preparations at higher magnification (Fig. 2C) indicated that most cores were intact and had a continuous wall with a diameter of 5 to 6 nm. Cores appeared to be capped on both sides. The particles closely resembled the internal structure of mature infectious HIV-1 virions as observed by thin-section EM (Fig. 2F). Isolated cores as well as the inner capsid structure of the virion could be labeled in immuno-EM with antiserum against CA (Fig. 2D and E), while no reactivity of these structures with antiserum against MA was detected (data not shown). Besides intact cores, we also observed damaged spherical structures and unstructured aggregates in core preparations which may correspond to the remnants of lysed virions. Some of these structures could be labeled in immuno-EM with antiserum against MA (data not shown).
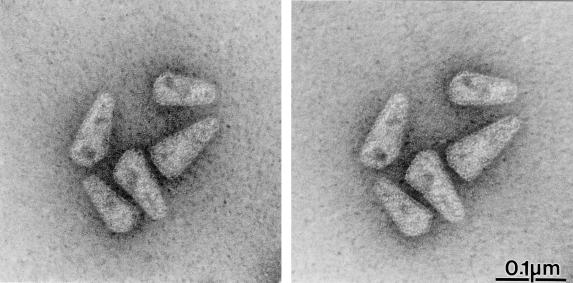
Thin-section EM analysis of intact HIV-1 reveals the prototypic lentiviral cone-shaped core (Fig. 2F), but the appearance of this structure varies depending on the plane of section (reviewed in reference 13). Negative-stain EM analysis of isolated intact cores, on the other hand, produces a projection of the three-dimensional structure into a photographic plane and should therefore allow a more thorough determination of core geometry, provided the cores are not collapsed. In stereoscopic image pairs of negatively stained cores tilted by ±6° in the 0° plane, cores projected from the grid into different directions and were clearly visible as three-dimensional objects without any apparent compression (Fig. 3). Image pairs tilted in the 30° plane yielded a similar result (data not shown), providing further evidence that the three-dimensional architecture of the cores was preserved during preparation. In addition, the walls of core particles exhibited a regular pattern similar to that observed for in vitro-assembled cylindrical particles derived from HIV-1 CA protein (20). Most core particles contained internal structures of higher density which localized toward the basis of the core and most likely correspond to the internal ribonucleoprotein complex (Fig. 2 to 4).
FIG. 3.
Stereo image of negatively stained HIV-1 cores. Images were taken at an angle of −6° and +6°.
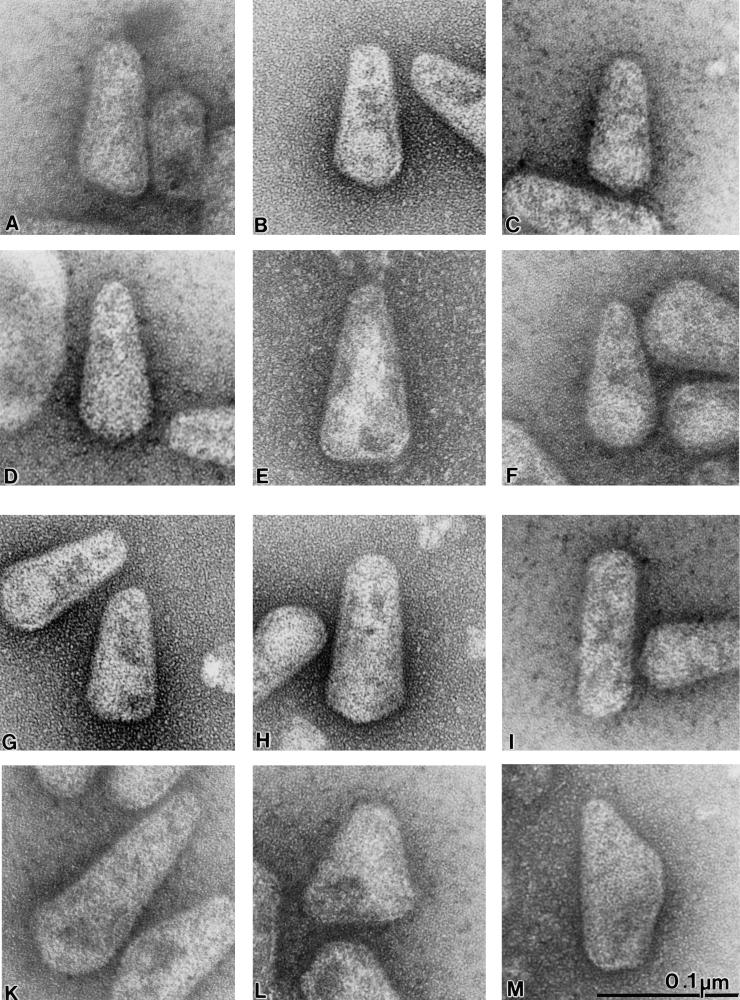
FIG. 4.
Gallery of isolated HIV-1 cores. Typical examples of negatively stained cores selected from four independent preparations are shown at the same magnification. The size bar is indicated in panel M.
Analysis of several hundred isolated HIV-1 cores revealed considerable heterogeneity in size and shape. Most particles exhibited a cone-shaped morphology, but cylindrical (Fig. 4I) and aberrantly shaped (Fig. 4L and M) particles were also observed. Significant differences were particularly obvious at the base of the cone, with the majority of particles showing a tip-like projection (Fig. 4A to D). Considering the three-dimensional architecture of a cone-shaped structure with rotational symmetry along a central longitudinal axis, random absorption of core structures to the grid would be expected. If the cone base is oblique and not perpendicular to the central axis of rotation, the two-dimensional projection of a given core can appear (i) more triangular (Fig. 4A), (ii) with a tip between the edge and the center (Fig. 4B and C), or (iii) like an arrowhead where the tip projects exactly onto the central symmetry axis (Fig. 4D). More than 70% of cores exhibited tips in an intermediate position, while the other two cases were rarely observed, as would be expected in the case of random adsorption. A few particles exhibited a base that appeared to be perpendicular to the central longitudinal axis (Fig. 4E) or a round base (Fig. 4F). We also observed variation regarding the angle at the tip of the cone (see below), yielding roughly triangular shapes (Fig. 4A to G), bullet-shaped variants (Fig. 4H), and, at a frequency of 5 to 10%, cylindrical isoforms (Fig. 4I). Rarely cores of very aberrant length (Fig. 4K) or morphology (Fig. 4L and M) were observed.
Analysis of HIV-1 core particle geometry.
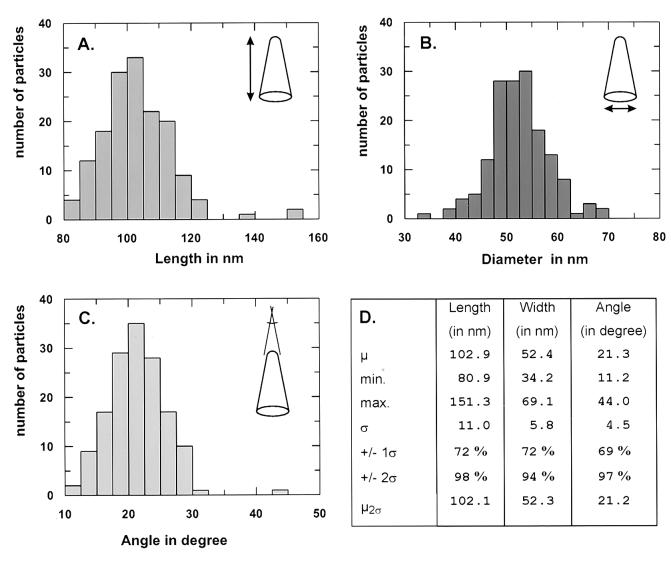
To describe the variation in particle size and morphology more quantitatively, we measured defined geometric parameters for 155 negatively stained HIV-1 cores. Morphometric analysis of overall length and diameter and of the angle at the narrow end was performed on digitized images, using the SigmaScan software package (Fig. 5). The maximal particle length was measured along the central axis of symmetry, and the maximal diameter was measured perpendicular to this axis (Fig. 5A and B). For a graphic representation, we defined arbitrary classes for each parameter and plotted the number of cores found in each class. As shown in Fig. 5 and confirmed by the Kolmogorov-Smirnov test for goodness of fit (data not shown), all three parameters followed a Gaussian distribution and length and diameter showed some skewing to the right.
FIG. 5.
Size and geometric variation of HIV-1 cores. Histograms show the distribution of length (A), diameter (B), and angle at the narrow end (C) of HIV-1 cores. Measurements were performed on digitized images taken of 155 individual cores after negative staining. Images were photographed in 26 microscopic fields and are derived from five independent core preparations. Length and diameter were measured once on each core, while the mean of three independent measurements was used in case of the angle. For each parameter, arbitrary size classes were defined and the number of cores found for each class was plotted. (D) Tabulation of the results. Abbreviations: μ, mean; min., minimal value; max., maximal value; ς, standard deviation; +/− 1ς or +/− 2ς, fraction of particles within the range of 1 or 2 standard deviations from the mean; μ2ς, mean value calculated for those particles within the range of 2 standard deviations from the overall mean.
Particle length and diameter varied around means of 103 and 52 nm, respectively (Fig. 5A and B; μ in Fig. 5D). The standard deviation in both cases was approximately 10% of the respective mean (Fig. 5D, ς); more than two-thirds of the particles were found within 1 standard deviation and ca. 95% were found within 2 standard deviations from the mean (Fig. 5D, +/− 1ς or 2ς). Measurement of the angle at the tip of the core gave a mean of 21.3° (Fig. 5C) with a larger degree of variation (standard deviation ca. 20% of the mean; Fig. 5D). Again, more than two-thirds of particles and more than 95% of particles were found within 1 and 2 standard deviations from the mean, respectively (Fig. 5D). No significant differences were observed when the mean was calculated only for those particles within 2 standard deviations from the overall mean (Fig. 5D, μ2ς), indicating that the result was not biased by particles of aberrant dimensions. Linear regression analysis of parameters revealed positive correlations between diameter and length and between angle and diameter and an inverse correlation between length and angle (data not shown).
Comparative protein analysis of virion and core preparations.
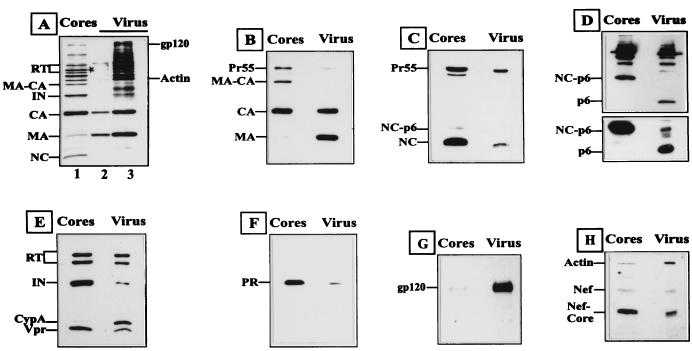
To determine the protein composition of HIV-1 cores, we performed a comparative immunoblot analysis of core preparations and the virus concentrate from which they were derived (Fig. 6). For a semiquantitative estimation of the relative amounts of specific viral and cellular proteins, the amount of core or virus extracts analyzed in immunoblots was normalized for CA (Fig. 6B to H). In Fig. 6A, core (lane 1) and virus (lanes 2 and 3) extracts corresponding to 1.5 μg (lanes 1 and 3) and 0.3 μg (lane 2) of CA, respectively, were analyzed on a silver-stained gel. Most HIV-1 Gag, Pol, and Env proteins can be clearly identified in these extracts. As expected, the HIV-1 surface glycoprotein gp120 (Fig. 6G) and the membrane-associated MA protein were substantially depleted in the core preparation, with the amount of MA corresponding to <5 to 10% of that found in the virus concentrate (Fig. 6A and B). EM analysis had shown that core preparations were not completely pure; therefore, it is not possible, to discriminate between a small amount of core-associated MA or a contamination with fragmented virions as suggested by the presence of residual gp120. Pr55 and incompletely processed Gag precursors, on the other hand, were enriched in the core preparation (Fig. 4A to D), possibly due to a small population of immature virions with a stable spherical shell. The nucleic acid binding proteins NC (Fig. 6A and C), IN, and RT (Fig. 6A and E) were also substantially enriched in the core preparation. Since the extracts loaded had been normalized for CA, this means an enrichment relative to CA which forms the core shell.
FIG. 6.
Analysis of protein composition of HIV-1 virions and cores obtained by brief centrifugation. Extracts from HIV-1 cores or the concentrated virus preparation they were derived from were separated by SDS-PAGE and analyzed by silver staining (A) or immunoblotting (B to H). In panel A, the amount of extract loaded in lanes 1 (cores) and 3 (virus) was adjusted to ca. 1.5 μg of CA; fivefold less virus was loaded in lane 2. Note that NC stains very poorly with silver but was clearly detected on staining of virus concentrate with Coomassie blue (Fig. 1). The asterisk denotes the position of Pr55 in the core preparation. In panels B to H, core (left lanes) and virus (right lanes) extracts were normalized for comparable amounts of CA protein. Western blots were probed with the following polyclonal rabbit antisera against viral and cellular proteins: (B) cocktail of anti-MA and anti-CA; (C) anti-NC; (D) anti-p6 (extracts were separated on regular SDS-gels [lower panel] or on Tris-Tricine gels [upper panel] for better resolution of small proteins); (E) cocktail of anti-RT, anti-IN, anti-Vpr, and anti-cyclophilin A (CypA); (F) anti-PR; (G) anti-gp120; (H) cocktail of anti-Nef and anti-actin. The additional band migrating between Nef and actin in panel H probably corresponds to IN and has been observed previously for this antiserum (55). Specific viral and cellular proteins are identified on the left of each panel.
In contrast, the cellular protein cyclophilin A, which binds to CA (Fig. 6E), and the viral p6 protein, which corresponds to the C-terminal domain of Gag (Fig. 6D), were essentially lost during core preparation. Loss of p6 was unexpected because the C-terminal end of Gag would be presumed to be inside the maturing core after cleavage. In addition, p6 is responsible for incorporating Vpr into the virion (reviewed in reference 6), and Vpr is clearly enriched in the core preparation (Fig. 6E). A p6-reactive protein probably corresponding to NC-p6 (Fig. 6C and D) was enriched in the core preparation, most likely due to the nucleic acid binding capacity of the NC domain. It should be noted, however, that the p6 antiserum used exhibits a stronger reactivity toward precursor forms (compare the relative intensity of Gag and intermediate cleavage products in Fig. 6D), and NC-p6 is not as abundant as it appears in Fig. 6D (see NC-p6 in Fig. 6C). Unexpectedly, HIV-1 PR was substantially enriched in the core preparation (Fig. 6F). Furthermore, the proteolytically cleaved core fragment of Nef was also enriched in the core preparation, while actin was clearly depleted (Fig. 6H).
DISCUSSION
We have developed a simple strategy to isolate intact mature cores from infectious HIV-1 in quantities sufficient for structural and biochemical analysis. So far this has not been possible for HIV-1 because of the notorious instability of its core structure after removal of the lipid membrane. The low stability of the mature core in the absence of the viral membrane is common to all retroviruses but appears to be more pronounced for HIV-1 (reviewed in reference 52). Accordingly, several groups reported isolation of avian and murine retroviral cores about 25 years ago (3, 30, 48, 49), but applying the same methods to HIV-1 did not yield sufficient intact cone-shaped cores.
Our method relies on three basic parameters: (i) use of a concentrated virus preparation of high biological activity as starting material, (ii) brief exposure of the virus to nonionic detergent, and (iii) gently and rapidly collecting the cores from the medium, using their intrinsic ability to aggregate into large complexes. The idea to collect the cores by brief centrifugation was based on our in vitro assembly experiments which had shown that spherical assembly products were almost quantitatively sedimented upon brief centrifugation in the microcentrifuge (20a). Since the g force and time of centrifugation applied are far from sufficient for sedimentation of single particles of this size, it is clear that the purification method must rely on aggregation. Accordingly, thin-section EM of in vitro-assembled spheres (20a) or of stripped cone-shaped cores recovered by brief centrifugation showed aggregates which were apparently dissociated in negatively stained preparations. Recovery of in vitro assembly products was lost when assembly was performed at protein concentrations below 300 μg per ml (M. Grättinger and H.-G. Kräusslich, unpublished data), suggesting that there is a critical threshold concentration for aggregation. The concentrated HIV-1 suspension used for core preparation usually had a CA concentration of 200 to 500 μg per ml, and approximately 10% of this antigen was recovered in the core fraction, similar to previously described preparation methods for other retroviruses (3, 30, 42, 48, 49). If the concentration of the HIV-1 suspension was below 100 μg of CA per ml, however, virtually no cores were recovered by the described method.
Besides protein concentration, the recovery of core particles was also influenced by the pH and salt concentration of the buffer and by the time of detergent treatment, while the effects of different detergents or variation of detergent concentrations were not analyzed in this study. In 1972, the relative effectivities of 61 detergents for the isolation of cores from avian myeloblastosis virus (AMV) had been determined, indicating that nonionic detergents like Triton X-100 are quite effective and were surpassed only by detergents of the polyoxyethylene alcohol class (47). While this report was under review, Kotov et al. (29) also reported isolation of intact HIV-1 cores by density gradient centrifugation through a detergent-containing layer. The success of this procedure appeared to depend on a high virus concentration in the starting material and very brief exposure to detergent as well.
The core preparations that we obtained contained numerous intact cone-shaped particles, but cores were not pure and remnants of lysed virions were also detected. Therefore, the results of comparative immunoblot analysis can provide only an estimate of the relative presence or absence of certain proteins in the core preparation; they cannot rule out the possibility that small amounts of a specific protein are present within the cores as has been suggested for the HIV-1 MA protein (4, 11, 26). Trace amounts of MA and of the viral glycoproteins were previously observed in core preparations from AMV, feline leukemia virus, mouse mammary tumor virus, EIAV, and simian immunodeficiency virus (SIV) that had been obtained by density gradient fractionation (3, 42, 47–49, 62), suggesting that it may be difficult to separate all envelope components from the retroviral core.
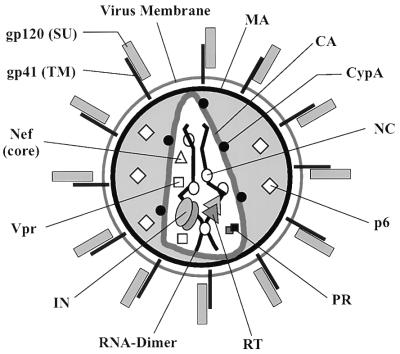
The localization of the main structural proteins within the HIV-1 virion has previously been studied by immuno-EM (reviewed in references 13 and 52) and the immunoblot analysis of our core preparation yielded largely similar results. A model of the topology of viral and cellular proteins in mature HIV-1 particles is presented in Fig. 7. CA and NC were the main components of the cores, while MA and gp120 were almost completely depleted. Semiquantitative comparison of the core preparation and the virus suspension from which it was derived showed that NC as well as the replication proteins RT and IN and the accessory Vpr protein were significantly enriched relative to CA in the core preparation. This result could be due to partial disintegration of the cores causing loss of their CA protein shell and sedimentation of the internal RNP complex. Alternatively, not all of the CA protein in the virion would be part of the core shell.
FIG. 7.
Model of HIV-1 showing the topology of individual proteins within the mature virion. Note that the number of symbols shown for each protein does not reflect the stoichiometry of the respective protein in the virion. Abbreviations for retroviral proteins (31) are given in the text. CypA, cyclophilin A.
Besides IN and RT, we also detected a significant enrichment of HIV-1 PR in the core preparation. PR is a very soluble protein and does not bind to nucleic acid, indicating that this result reflects its true association with the HIV-1 core. PR was also found in core preparations of AMV and EIAV (42, 48) and may thus be a constituent of all retroviral cores. Since some PR was detected in preintegration complexes isolated from early HIV-1-infected cells (26), one may speculate on a PR function in the early steps of the retroviral life cycle (41), besides its essential role in maturation. The accessory protein Vpr was found significantly enriched in core preparations, supporting its postulated role in transport of the HIV-1 preintegration complex to the nucleus (4, 22, 26). Using immuno-EM analysis, Wang et al. (54) detected Vpr primarily underneath the membrane and not in the core of HIV-1. However, these virions were mostly immature, and the quality of the immuno-EM images is insufficient to determine the precise localization of the protein. The situation is controversial for the homologous Vpx proteins of HIV-2 and SIV. Vpx from SIVmac was reported to localize primarily outside the core (32, 62), while HIV-2 Vpx was found to cofractionate with cores in sucrose density gradients (27). Additional support for Vpr being a constituent of the core comes from the observation that Vpr fusions which package heterologous proteins into HIV-1 particles were often degraded by PR inside the virion (59). This is not surprising, considering that both PR and Vpr are enriched within the same virion compartment. The same may be true for HIV-1 Nef, which is packaged into HIV-1 particles, where it is cleaved by HIV-1 PR (38, 56). The apparent association of the larger Nef fragment with the HIV-1 core, which was also observed by Kotov et al. (29), may indicate that virion-associated Nef plays a role in the early phase of infection (17, 29, 38, 55, 56). Similar to MA, the C-terminal p6 product of Gag was completely solubilized during core preparation, suggesting that it is not a component of the core. The corresponding p9 protein of EIAV is also located outside the core (42). The cellular chaperone cyclophilin A, which is packaged by interaction with the CA domain of Gag, is depleted almost quantitatively during core preparation, which correlates with its weak affinity for CA (61) and is consistent with its proposed localization on the outer surface of the core shell (19). Actin, which appears to be incorporated through the NC domain of Gag (58) was also depleted in the cores, indicating that it is probably not a structural constituent of the core and is unlikely to be associated with NC in the mature virion.
EM analysis of negatively stained isolated HIV-1 cores revealed that they were not collapsed but exhibited a defined three-dimensional architecture. Both ends of the core appeared to be capped. Some cores showed an ordered wall pattern, similar to that found for in vitro-assembled CA-derived cylinders (20). However, a more detailed analysis of their structure will require high-resolution cryo-EM with image analysis; these experiments are currently in progress. The majority of cores were cone shaped, resembling isolated cores from HIV-2, SIV, and EIAV (5, 27, 42, 62). Cylindrical isoforms were observed at a frequency of 5 to 10%; this result supports the hypothesis that the organization of in vitro-assembled CA-derived cylinders may be similar to that of the mature core shell. Interestingly, the diameters at the narrow and wide ends of the mature cores were similar to the minimal and maximal diameters of CA-derived in vitro-assembled cylinders (ca. 25 to 65 nm) (D. Thomas, T. Wilk, T. Rutten, I. Gross, H.-G. Kräusslich, and S. Fuller, unpublished data). In vitro-assembled cylinders exhibit a helical arrangement of CA proteins, Thomas et al., unpublished data), and it is conceivable that CA can form helices of only a certain range of diameters. This might suggest that the mature cone-shaped core shell corresponds to a helix of continuously changing diameters; some support for this hypothesis can be derived from recent cryo-EM analysis of in vitro assembly products (Thomas et al., unpublished data).
Despite their overall similarity, a large degree of variation in size and shape of isolated cores was observed. Size variation was not unexpected since intact retroviruses are also not uniform and exhibit a wide range of sizes (10, 15, 53, 60). The average diameter of HIV-1 virions has been determined by various EM methods to be between 120 and 160 nm (10, 14, 15). Since cryo-EM is least likely to cause shrinkage of the virion, such data may be most accurate. Recently, Fuller et al. (10) reported that immature HIV-1 particles ranged in diameter from 120 to 260 nm, with a mean of 160 nm. A similar size variation was reported for immature and mature murine leukemia virus particles (60) and was also found for mature HIV-1 (T. Wilk, R. Welker, H.-G. Kräusslich, and S. Fuller, unpublished data). Using a precise determination of the mass of Rous sarcoma virus by scanning transmission EM, Vogt and Simon recently showed a wide variation, with one- to two-thirds of the virions deviating from the mean by more than 10% (53). These differences are probably due to variable numbers of Gag molecules used to assemble the immature virion, suggesting that retroviruses tolerate remarkable variations in their assembly process. It appears likely that the observed variation in core length and width reflects the differences in size of the respective immature virions. Since the negative staining technique leads to loss of water and shrinking of the structures, it is likely that the true values for length and diameter are somewhat higher than the reported values of 103 and 52 nm, respectively. A higher degree of variability was observed for the angle at the narrow end of the cone, with a standard deviation of 20%. This higher variability may be partly due to measurement error but also reflects the true geometric variability, as is evident when one inspects the gallery of cores in Fig. 4. The mean angle of 21.3° is slightly different from the 19° angle recently reported for in vitro-assembled structures (12). The significant variation in angles at the narrow end of isolated cores which exhibited a Gaussian distribution between 11° and 32° in addition to the fact that cylindrical and bullet-shaped isoforms were detected suggests a significant degree of freedom in capsid assembly. However, all retroviral preparations contain a large excess of noninfectious particles; although this may be due to various defects, it is likely that some of the observed core structures are aberrant and therefore noninfectious.
ACKNOWLEDGMENTS
We are grateful to B. Cardel for excellent technical assistance, I. Gross for providing the basis for this approach, R. Schwarz for help with statistical analysis, and I. Ellhof for photography. We thank S. Fuller, D. Thomas, and T. Wilk for many important suggestions and comments. We are also grateful to K. Wiegers for enzyme-linked immunosorbent assay measurements, for discussion, and for critically reading the manuscript and to G. Rutter for suggestions and discussion.
This work was supported in part by grants from the German Ministry for Education and Research and the Deutsche Forschungsgemeinschaft to H.-G.K.
REFERENCES
- 1.Accola M A, Hoglund S, Göttlinger H G. A putative alpha-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J Virol. 1998;72:2072–2078. doi: 10.1128/jvi.72.3.2072-2078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolognesi D P, Luftig R, Shaper J H. Localization of RNA tumor virus polypeptides. I. Isolation of further virus substructures. Virology. 1973;56:549–564. doi: 10.1016/0042-6822(73)90057-3. [DOI] [PubMed] [Google Scholar]
- 4.Bukrinsky M I, Sharova N, McDonald T L, Pushkarskaya T, Tarpley W G, Stevenson M. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci USA. 1993;90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chrystie I L, Almeida J D. The morphology of human immunodeficiency virus (HIV) by negative staining. J Med Virol. 1988;25:281–288. doi: 10.1002/jmv.1890250305. [DOI] [PubMed] [Google Scholar]
- 6.Cohen E A, Subbramanian R A, Göttlinger H G. Role of auxiliary proteins in retroviral morphogenesis. Curr Top Microbiol Immunol. 1996;214:219–235. doi: 10.1007/978-3-642-80145-7_7. [DOI] [PubMed] [Google Scholar]
- 7.Dettenhofer M, Yu X F. Highly purified human immunodeficiency virus type 1 reveals a virtual absence of Vif in virions. J Virol. 1999;73:1460–1467. doi: 10.1128/jvi.73.2.1460-1467.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Escaig-Haye F, Grigoriev V, Sharova I, Rudneva V, Buckrinskaya A, Fournier J G. Ultrastructural localization of HIV-1 RNA and core proteins. Simultaneous visualization using double immunogold labelling after in situ hybridization and immunocytochemistry. J Submicrosc Cytol Pathol. 1992;24:437–443. [PubMed] [Google Scholar]
- 9.Franke E K, Yuan H E, Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372:359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- 10.Fuller S D, Wilk T, Gowen B E, Kräusslich H G, Vogt V M. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr Biol. 1997;7:729–738. doi: 10.1016/s0960-9822(06)00331-9. [DOI] [PubMed] [Google Scholar]
- 11.Gallay P, Swingler S, Aiken C, Trono D. HIV-1 infection of nondividing cells: C-terminal tyrosine phosphorylation of the viral matrix protein is a key regulator. Cell. 1995;80:379–388. doi: 10.1016/0092-8674(95)90488-3. [DOI] [PubMed] [Google Scholar]
- 12.Ganser B K, Li S, Klishko V Y, Finch J T, Sundquist W I. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 13.Gelderblom H R. Assembly and morphology of HIV: potential effect of structure on viral function. AIDS. 1991;5:617–637. [PubMed] [Google Scholar]
- 14.Gelderblom H R, Hausmann E H, Özel M, Pauli G, Koch M A. Fine structure of human immunodeficiency virus (HIV) and immunolocalization of structural proteins. Virology. 1987;156:171–176. doi: 10.1016/0042-6822(87)90449-1. [DOI] [PubMed] [Google Scholar]
- 15.Gentile M, Adrian T, Scheidler A, Ewald M, Dianzani F, Pauli G, Gelderblom H R. Determination of the size of HIV using adenovirus type 2 as an internal length marker. J Virol Methods. 1994;48:43–52. doi: 10.1016/0166-0934(94)90087-6. [DOI] [PubMed] [Google Scholar]
- 16.Gluschankof P, Mondor I, Gelderblom H R, Sattentau Q J. Cell membrane vesicles are a major contaminant of gradient-enriched human immunodeficiency virus type-1 preparations. Virology. 1997;230:125–133. doi: 10.1006/viro.1997.8453. [DOI] [PubMed] [Google Scholar]
- 17.Goldsmith M A, Warmerdam M T, Atchison R E, Miller M D, Greene W C. Dissociation of the CD4 downregulation and viral infectivity enhancement functions of human immunodeficiency virus type 1 Nef. J Virol. 1995;69:4112–4121. doi: 10.1128/jvi.69.7.4112-4121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goto T, Ikuta K, Zhang J J, Morita C, Sano K, Komatsu M, Fujita H, Kato S, Nakai M. The budding of defective human immunodeficiency virus type 1 (HIV-1) particles from cell clones persistently infected with HIV-1. Arch Virol. 1990;111:87–101. doi: 10.1007/BF01310507. [DOI] [PubMed] [Google Scholar]
- 19.Grättinger M, Hohenberg H, Thomas D, Wilk T, Müller B, Kräusslich H G. In vitro assembly properties of wild-type and cyclophilin-binding defective human immunodeficiency virus capsid proteins in the presence and absence of cyclophilin A. Virology. 1999;257:247–260. doi: 10.1006/viro.1999.9668. [DOI] [PubMed] [Google Scholar]
- 20.Gross I, Hohenberg H, Kräusslich H G. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur J Biochem. 1997;249:592–600. doi: 10.1111/j.1432-1033.1997.t01-1-00592.x. [DOI] [PubMed] [Google Scholar]
- 20a.Gross, I., H. Hohenberg, T. Wilk, K. Wiegers, M. Grättinger, B. Müller, S. Fuller, and H.-G. Kräusslich. A conformational switch controlling HIV-1 morphogenesis. EMBO J., in press. [DOI] [PMC free article] [PubMed]
- 21.Harada S, Koyanagi Y, Yamamoto N. Infection of HTLV-III/LAV in HTLV-I-carrying cells MT-2 and MT-4 and application in a plaque assay. Science. 1985;229:563–566. doi: 10.1126/science.2992081. [DOI] [PubMed] [Google Scholar]
- 22.Heinzinger N K, Bukinsky M I, Haggerty S A, Ragland A M, Kewalramani V, Lee M A, Gendelman H E, Ratner L, Stevenson M, Emerman M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heukeshoven J, Dernick R. Improved silver staining procedure for fast staining in PhastSystem Development Unit. I. Staining of sodium dodecyl sulfate gels. Electrophoresis. 1988;9:28–32. doi: 10.1002/elps.1150090106. [DOI] [PubMed] [Google Scholar]
- 24.Hohenberg H, Mannweiler K, Müller M. High-pressure freezing of cell suspensions in cellulose capillary tubes. J Microsc. 1994;175:34–43. doi: 10.1111/j.1365-2818.1994.tb04785.x. [DOI] [PubMed] [Google Scholar]
- 25.Hunter E. Viral entry and receptors. In: Coffin J M, Hughes S, Varmus H, editors. Retroviruses. Cold Spring Harbor, N.Y: CSHL Press; 1997. pp. 71–120. [PubMed] [Google Scholar]
- 26.Karageorgos L, Li P, Burrell C. Characterization of HIV replication complexes early after cell-to-cell infection. AIDS Res Hum Retroviruses. 1993;9:817–823. doi: 10.1089/aid.1993.9.817. [DOI] [PubMed] [Google Scholar]
- 27.Kewalramani V N, Emerman M. Vpx association with mature core structures of HIV-2. Virology. 1996;218:159–168. doi: 10.1006/viro.1996.0176. [DOI] [PubMed] [Google Scholar]
- 28.Kondo E, Mammano F, Cohen E A, Göttlinger H G. The p6gag domain of human immunodeficiency virus type 1 is sufficient for the incorporation of Vpr into heterologous viral particles. J Virol. 1995;69:2759–2764. doi: 10.1128/jvi.69.5.2759-2764.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kotov A, Zhou J, Flicker P, Aiken C. Association of Nef with the human immunodeficiency virus type 1 core. J Virol. 1999;73:8824–8830. doi: 10.1128/jvi.73.10.8824-8830.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lange J, Frank H, Hunsmann G, Moennig V, Wollmann R, Schäfer W. Properties of mouse leukemia viruses. VI. The core of Friend virus; isolation and constituents. Virology. 1973;53:457–462. doi: 10.1016/0042-6822(73)90225-0. [DOI] [PubMed] [Google Scholar]
- 31.Leis J, Baltimore D, Bishop J M, Coffin J, Fleissner E, Goff S P, Oroszlan S, Robinson H, Skalka A M, Temin H M, et al. Standardized and simplified nomenclature for proteins common to all retroviruses. J Virol. 1988;62:1808–1809. doi: 10.1128/jvi.62.5.1808-1809.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liska V, Spehner D, Mehtali M, Schmitt D, Kirn A, Aubertin A M. Localization of viral protein X in simian immunodeficiency virus macaque strain and analysis of its packaging requirements. J Gen Virol. 1994;75:2955–2962. doi: 10.1099/0022-1317-75-11-2955. [DOI] [PubMed] [Google Scholar]
- 33.Liu H, Wu X, Newman M, Shaw G M, Hahn B H, Kappes J C. The Vif protein of human and simian immunodeficiency viruses is packaged into virions and associates with viral core structures. J Virol. 1995;69:7630–7638. doi: 10.1128/jvi.69.12.7630-7638.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meerloo T, Sheikh M A, Bloem A C, de Ronde A, Schutten M, van Els C A, Roholl P J, Joling P, Goudsmit J, Schuurman H J. Host cell membrane proteins on human immunodeficiency virus type 1 after in vitro infection of H9 cells and blood mononuclear cells. An immuno-electron microscopic study. J Gen Virol. 1993;74:129–135. doi: 10.1099/0022-1317-74-1-129. [DOI] [PubMed] [Google Scholar]
- 35.Menendez-Arias L, Risco C, Pinto da Silva P, Oroszlan S. Purification of immature cores of mouse mammary tumor virus and immunolocalization of protein domains. J Virol. 1992;66:5615–5620. doi: 10.1128/jvi.66.9.5615-5620.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nermut M V, Hockley D J. Comparative morphology and structural classification of retroviruses. Curr Top Microbiol Immunol. 1996;214:1–24. doi: 10.1007/978-3-642-80145-7_1. [DOI] [PubMed] [Google Scholar]
- 37.Ott D E, Coren L V, Kane B P, Busch L K, Johnson D G, Sowder II R C, Chertova E N, Arthur L O, Henderson L E. Cytoskeletal proteins inside human immunodeficiency virus type 1 virions. J Virol. 1996;70:7734–7743. doi: 10.1128/jvi.70.11.7734-7743.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pandori M W, Fitch N J, Craig H M, Richman D D, Spina C A, Guatelli J C. Producer-cell modification of human immunodeficiency virus type 1: Nef is a virion protein. J Virol. 1996;70:4283–4290. doi: 10.1128/jvi.70.7.4283-4290.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park J, Morrow C D. Mutations in the protease gene of human immunodeficiency virus type 1 affect release and stability of virus particles. Virology. 1993;194:843–850. doi: 10.1006/viro.1993.1328. [DOI] [PubMed] [Google Scholar]
- 40.Paxton W, Connor R I, Landau N R. Incorporation of Vpr into human immunodeficiency virus type 1 virions: requirement for the p6 region of Gag and mutational analysis. J Virol. 1993;67:7229–7237. doi: 10.1128/jvi.67.12.7229-7237.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roberts M M, Copeland T D, Oroszlan S. In situ processing of a retroviral nucleocapsid protein by the viral proteinase. Protein Eng. 1991;4:695–700. doi: 10.1093/protein/4.6.695. [DOI] [PubMed] [Google Scholar]
- 42.Roberts M M, Oroszlan S. The preparation and biochemical characterization of intact capsids of equine infectious anemia virus. Biochem Biophys Res Commun. 1989;160:486–494. doi: 10.1016/0006-291x(89)92459-5. [DOI] [PubMed] [Google Scholar]
- 43.Rose J R, Babe L M, Craik C S. Defining the level of human immunodeficiency virus type 1 (HIV-1) protease activity required for HIV-1 particle maturation and infectivity. J Virol. 1995;69:2751–2758. doi: 10.1128/jvi.69.5.2751-2758.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salahuddin S Z, Markham P D, Wong-Staal F, Franchini G, Kalyanaraman V S, Gallo R C. Restricted expression of human T-cell leukemia-lymphoma virus (HTLV) in transformed human umbilical cord blood lymphocytes. Virology. 1983;129:51–64. doi: 10.1016/0042-6822(83)90395-1. [DOI] [PubMed] [Google Scholar]
- 45.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 46.Stewart L, Schatz G, Vogt V M. Properties of avian retrovirus particles defective in viral protease. J Virol. 1990;64:5076–5092. doi: 10.1128/jvi.64.10.5076-5092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stromberg K. Surface-active agents for isolation of the core component of avian myeloblastosis virus. J Virol. 1972;9:684–697. doi: 10.1128/jvi.9.4.684-697.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stromberg K, Hurley N E, Davis N L, Rueckert R R, Fleissner E. Structural studies of avian myeloblastosis virus: comparison of polypeptides in virion and core component by dodecyl sulfate-polyacrylamide gel electrophoresis. J Virol. 1974;13:513–528. doi: 10.1128/jvi.13.2.513-528.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teramoto Y A, Cardiff R D, Lund J K. The structure of the mouse mammary tumor virus: isolation and characterization of the core. Virology. 1977;77:135–148. doi: 10.1016/0042-6822(77)90413-5. [DOI] [PubMed] [Google Scholar]
- 50.Thali M, Bukovsky A, Kondo E, Rosenwirth B, Walsh C T, Sodroski J, Göttlinger H G. Functional association of cyclophilin A with HIV-1 virions. Nature. 1994;372:363–365. doi: 10.1038/372363a0. [DOI] [PubMed] [Google Scholar]
- 51.Vogt V M. Proteolytic processing and particle maturation. Curr Top Microbiol Immunol. 1996;214:95–131. doi: 10.1007/978-3-642-80145-7_4. [DOI] [PubMed] [Google Scholar]
- 52.Vogt V M. Retroviral virions and genomes. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: CSHL Press; 1997. pp. 27–69. [PubMed] [Google Scholar]
- 53.Vogt V M, Simon M N. Mass determination of Rous sarcoma virus virions by scanning transmission electron microscopy. J Virol. 1999;73:7050–7055. doi: 10.1128/jvi.73.8.7050-7055.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang J J, Lu Y, Ratner L. Particle assembly and Vpr expression in human immunodeficiency virus type 1-infected cells demonstrated by immunoelectron microscopy. J Gen Virol. 1994;75:2607–2614. doi: 10.1099/0022-1317-75-10-2607. [DOI] [PubMed] [Google Scholar]
- 55.Welker R, Harris M, Cardel B, Kräusslich H G. Virion incorporation of human immunodeficiency virus type 1 Nef is mediated by a bipartite membrane-targeting signal: analysis of its role in enhancement of viral infectivity. J Virol. 1998;72:8833–8840. doi: 10.1128/jvi.72.11.8833-8840.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Welker R, Kottler H, Kalbitzer H R, Kräusslich H G. Human immunodeficiency virus type 1 Nef protein is incorporated into virus particles and specifically cleaved by the viral proteinase. Virology. 1996;219:228–236. doi: 10.1006/viro.1996.0240. [DOI] [PubMed] [Google Scholar]
- 57.Wiegers K, Rutter G, Kottler H, Tessmer U, Hohenberg H, Kräusslich H G. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J Virol. 1998;72:2846–2854. doi: 10.1128/jvi.72.4.2846-2854.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilk T, Gowen B, Fuller S D. Actin associates with the nucleocapsid domain of the human immunodeficiency virus Gag polyprotein. J Virol. 1999;73:1931–1940. doi: 10.1128/jvi.73.3.1931-1940.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu X, Liu H, Xiao H, Kim J, Seshaiah P, Natsoulis G, Boeke J D, Hahn B H, Kappes J C. Targeting foreign proteins to human immunodeficiency virus particles via fusion with Vpr and Vpx. J Virol. 1995;69:3389–3398. doi: 10.1128/jvi.69.6.3389-3398.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yeager M, Wilson-Kubalek E M, Weiner S G, Brown P O, Rein A. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: implications for retroviral assembly mechanisms. Proc Natl Acad Sci USA. 1998;95:7299–7304. doi: 10.1073/pnas.95.13.7299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoo S, Myszka D G, Yeh C, McMurray M, Hill C P, Sundquist W I. Molecular recognition in the HIV-1 capsid/cyclophilin A complex. J Mol Biol. 1997;269:780–795. doi: 10.1006/jmbi.1997.1051. [DOI] [PubMed] [Google Scholar]
- 62.Yu X, Matsuda Z, Yu Q C, Lee T H, Essex M. Vpx of simian immunodeficiency virus is localized primarily outside the virus core in mature virions. J Virol. 1993;67:4386–4390. doi: 10.1128/jvi.67.7.4386-4390.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]