Abstract
Overcoming resistance to immune checkpoint inhibitors is an important issue in patients with non‐small‐cell lung cancer (NSCLC). Transcriptome analysis shows that adenocarcinoma can be divided into three molecular subtypes: terminal respiratory unit (TRU), proximal proliferative (PP), and proximal inflammatory (PI), and squamous cell carcinoma (LUSQ) into four. However, the immunological characteristics of these subtypes are not fully understood. In this study, we investigated the immune landscape of NSCLC tissues in molecular subtypes using a multi‐omics dataset, including tumor‐infiltrating leukocytes (TILs) analyzed using flow cytometry, RNA sequences, whole exome sequences, metabolomic analysis, and clinicopathologic findings. In the PI subtype, the number of TILs increased and the immune response in the tumor microenvironment (TME) was activated, as indicated by high levels of tertiary lymphoid structures, and high cytotoxic marker levels. Patient prognosis was worse in the PP subtype than in other adenocarcinoma subtypes. Glucose transporter 1 (GLUT1) expression levels were upregulated and lactate accumulated in the TME of the PP subtype. This could lead to the formation of an immunosuppressive TME, including the inactivation of antigen‐presenting cells. The TRU subtype had low biological malignancy and “cold” tumor‐immune phenotypes. Squamous cell carcinoma (LUSQ) did not show distinct immunological characteristics in its respective subtypes. Elucidation of the immune characteristics of molecular subtypes could lead to the development of personalized immune therapy for lung cancer. Immune checkpoint inhibitors could be an effective treatment for the PI subtype. Glycolysis is a potential target for converting an immunosuppressive TME into an antitumorigenic TME in the PP subtype.
Keywords: immunotherapy, lung cancer, molecular subtype, tumor microenvironment, tumor‐infiltrating lymphocyte
Our study shows the immune tumor microenvironment in different lung adenocarcinoma molecular subtypes: terminal respiratory unit and proximal proliferative had immunosuppressive microenvironment but differ in phenotypes, while the proximal inflammatory subtype has upregulated immune response. Understanding these traits may enable personalized immune therapy for lung cancer.
Abbreviations
- CDR
complementarity determining region
- CTLA‐4
cytotoxic T lymphocyte‐associated protein 4
- CXCL
chemokine (C‐X‐C motif) ligand
- DC
dendritic cell
- EFS
event‐free survival
- FCM
flow cytometry
- GSEA
Gene Set Enrichment Analysis
- ICI
immune checkpoint inhibitor
- IHC
immunohictochemistry
- LUAD
lung adenocarcinoma
- LUSQ
lung squamous cell carcinoma
- MFI
mean fluorescence intensity
- NK
natural killer
- NSCLC
non‐small‐cell lung cancer
- PD‐1
programmed death receptor 1
- PD‐L1
PD‐1 ligand 1
- PI
proximal inflammatory
- PP
proximal proliferative
- RNA‐seq
RNA sequencing
- TCM
central memory T cell
- TCR
T‐cell receptor
- TIL
tumor‐infiltrating leukocyte
- TLS
tertiary lymphoid structure
- TMB
tumor mutation burden
- TME
tumor microenvironment
- TPM
transcripts per million
- TRU
terminal respiratory unit
- UMI
unique molecular identifier
- WES
whole exome sequencing
1. INTRODUCTION
Lung cancer is the leading cause of cancer‐related deaths worldwide. 1 , 2 The emergence of ICIs has revolutionized the treatment of several cancers, including NSCLC. While some patients respond dramatically to treatment, most do not respond to ICI therapy; the response rate is only 20%–30%. 3 , 4 The main reasons for ICI‐refractory cancers and relapse of ICI‐sensitive cancer are an immunosuppressive TME caused by a variety of factors, including impaired intratumoral immune infiltration, immunosuppressive cell infiltration, and immunosuppressive cytokines. 5 As the characteristics of an immunosuppressive TME differ in each case, 6 it is important to develop specialized immunotherapies that are appropriate for each type of immune susceptibility.
There is an urgent need to identify reliable prognostic biomarkers that can predict the response to immunotherapy and consequently enhance patient outcomes in NSCLC. While PD‐L1 is among the extensively studied biomarkers using IHC staining, conflicting findings have emerged due to the inconsistent application of cut‐off values and the absence of standardized methods in its assessment. 3 , 4 Studies have indicated that in histological classification, the predominance of a solid or micropapillary pattern in adenocarcinoma (LUAD) shows higher PD‐L1 expression compared to other subtypes. 7 , 8 However, the correlation between distinct histological subtypes and their responsiveness to immunotherapy remains unclear.
Non‐small‐cell carcinoma is a heterogeneous disease categorized into two major types, LUAD and LUSQ. Several molecular subtypes within LUAD and LUSQ tumors have been identified that differ in their prognosis, underlying genomic alterations, and potential response to treatment. 9 , 10 , 11 There are three LUAD subtypes, TRU (formerly bronchoid), PP (formerly magnoid), and PI (formerly squamoid). 10 The four LUSQ subtypes are primitive, classical, basal, and secretory. 11
Molecular subtyping, as it reflects intrinsic biological characteristics, is anticipated to correlate with the immune condition of the TME and serve as predictive indicators for immune‐based therapies. 11 , 12 Previous studies have highlighted the value of molecular subtyping in identifying patients who are likely to benefit from immunotherapy, exemplified in gastric adenocarcinomas 13 and urothelial cancer. 14 Similarly, in cases of LUAD and LUSQ, specific molecular subtypes have shown a TME characterized by an activated immune response, suggesting their potential as immunotherapy biomarkers. 12 , 15 The assessment of the immune cell content within the TME of molecular subtypes has primarily relied on a combination of immune‐related gene expressions 12 , 15 ; however, it remains uncertain whether this gene expression‐based immune profile accurately mirrors the immune conditions within the TME.
Comprehensive immune TME profiling using multi‐omics analysis is useful to analyze an immunosuppressive TME for a variety of cancer types. 16 , 17 To decipher the immune TME, we constructed a database using FCM, RNA‐seq, WES, TCR repertoires, and metabolomic analyses, accompanied by clinicopathologic findings. In this study, we used this database to elucidate the immunological, transcriptional, and genomic characteristics of LUAD and LUSQ molecular subtypes. Converting the immunosuppressive TME into an antitumorigenic state might be possible by regulating the immune subtype‐specific pathways and responsible gene.
2. MATERIALS AND METHODS
2.1. Patients and samples
The patients' characteristics were described previously. 18 This study enrolled 217 patients with NSCLC (LUAD, n = 142; LUSQ, n = 75) who underwent surgical resection at the National Cancer Center Hospital, Japan between 2017 and 2019. Single‐cell RNA‐seq analysis was undertaken on the combination of six NSCLC tissue samples. Patient characteristics are summarized in Table S1.
2.2. Tissue preparation and FCM analysis
Flow cytometry analysis was carried out as previously reported. 18 Briefly, tumors were received within 2 h of resection, immediately processed for single‐cell dissociation, and stored at −150°C. The cells were incubated with Fixable Viability Dye eFluor 506 (Thermo Fisher Scientific) and FcR blocking reagent (Miltenyi Biotec), followed by staining with mAbs. The cells were fixed and permeabilized for intracellular protein staining using a FOXP3 transcription factor buffer set (Thermo Fisher Scientific) and stained with mAbs. Data were acquired on a FACSymphony A5 instrument (BD Biosciences) and analyzed using the FlowJo software (BD Biosciences). The staining panel and gating strategy used were derived from a previously published paper. 18
2.3. Immune profiling data clustering
To compare TIL‐based molecular subtypes, reflecting the quantity and frequency of each immune cell type, the cell proportion per CD45+ cells (%CD45) and log10 transformed cell count per gram of tumor +1 (cell density) were scaled across patients.
2.4. RNA sequencing
RNA sequencing was carried out as previously described. 18 Complementary DNA was prepared from the isolated RNA from the tissues. The prepared RNA‐seq libraries were analyzed using the Agilent 4200 TapeStation (Agilent Technologies), then subjected to next‐generation sequencing of 125 bp paired‐end reads using the Hiseq PE Cluster Kit version 4 cBot and Hiseq SBS Kit version 4 with a Hiseq2500 platform (Illumina). Raw data from the Hiseq system were converted to the FASTQ format with bcl2fastq (Illumina) and cleaned using the QCleaner software (Amelieff) and the Resequence analysis pipeline (Amelieff). To undertake expression profiling with RNA‐seq data, paired‐end reads were aligned to the hg38 human genome assembly using STAR; count values were then calculated using the STAR quant mode. Following this, we used RSEM with bowtie mapping to determine the TPM value for each RefSeq transcript.
2.5. Expression analysis and GSEA
Expression analysis and GSEA were carried out as described previously. 18 To determine the molecular subtype of each sample, we carried out the following steps: TPM + 1 values were log‐transformed and gene median‐centered, then we calculated the Pearson correlations between each sample and the molecular subtype predictor centroids for LUAD 10 and LUSQ. 11 The molecular subtype was predicted based on the centroid with the highest correlation value.
2.6. Immunohistochemical staining
The IHC staining was carried out as described previously. 18 Immunostaining for CD8 was undertaken on 4 μm‐thick tissue microarray sections using the autostainer DAKO LINK48 (Agilent Technologies). The digital images were then analyzed using the CytoNuclear Algorithm version 2.0.5 for immune cell markers (CD8) with the HALO system (version 3.1; Indica Labs).
2.7. Whole exome sequencing and mutation analysis
Sequencing libraries were prepared for WES, and the adaptor‐ligated samples were amplified as previously described. 18 Massively parallel sequencing of the isolated fragments was carried out using a HiSeq2500 platform (Illumina). Mutation calling was undertaken using the best practice workflow in GATK version 4.0.8.1. Paired‐end WES reads were independently aligned to the human reference genome (hg38) using BWA‐MEM. Mutect2 was used to identify somatic mutations. Gene mutations were annotated using SnpEff. 19 CNVkit was used to estimate the log2 copy ratio 20 in the R 3.6.1 environment.
2.8. T‐cell receptor repertoire analysis
The TCR immune repertoire sequencing libraries were prepared from tumor RNA as previously described. 18 The TCR cDNA libraries were sequenced using MiSeq sequencer (Illumina) in paired‐ end mode with 251 base reads. IMSEQ was used to analyze the sequencing data. Reads were aligned to the reference V, D, and J regions of the TCRα, TCRβ, TCRδ, and TCRγ genes. CDR3 gene regions were then extracted from the aligned reads. To minimize false‐positive CDR3 clonotype calls, a minimum of seven supporting UMIs were established. Shannon entropy for TCRα, β, δ, and γ‐chain sequences were calculated using the R “vegan” package based on UMI counts for each patient.
2.9. Metabolome analysis
Metabolomic analysis was undertaken as described previously. 18 , 21 Briefly, frozen tissue samples (~10 mg) were immersed in methanol containing internal standards. After homogenization, both chloroform and Milli‐Q water were added to the sample, and the resulting solution was thoroughly mixed. To eliminate proteins, the upper aqueous fraction was filtered, and the filtrate was dried using an evacuated centrifuge and then dissolved in Milli‐Q water containing reference compounds (3‐aminopyrrolidine and trimesic acid) prior to capillary electrophoresis–mass spectrometry analysis, which was carried out and analyzed as previously described. 21
2.10. Single‐cell RNA‐seq using 10X chromium
Single‐cell RNA‐seq was carried out using the Chromium Next GEM Single Cell 3′ GEM, Library & Gel Bead Kit version 3.1 (10X Genomics), following the manufacturer's instructions. Each well was targeted for a total of 10,000 cells. The libraries were sequenced on the NovaSeq 6000 platform (Illumina) with pair‐ended sequencing and dual indexing, running 28, 8, and 91 cycles for Read 1, i7 index, and Read 2, respectively.
2.11. Statistical analysis
Statistical analysis was undertaken using R version 4.0.1 (R Foundation for Statistical Computing). The relationships between parameters were examined using the Mann–Whitney U‐test, Kruskal–Wallis test, and Fisher's exact test, and Spearman's correlation. The event‐free survival differences among groups were analyzed using the Kaplan–Meier method and log‐rank test. The level of statistical significance was set at p < 0.05.
3. RESULTS
3.1. Clinicopathologic features of lung cancer molecular subtypes
To comprehensively analyze the features of NSCLC molecular subtypes, we used a multi‐omics dataset of surgically resected tumor tissues in 142 treatment‐naive patients with LUAD and 75 patients with LUSQ. Adenocarcinoma was divided into the TRU (n = 49; 35%), PP (n = 47; 33%), and PI subtypes (n = 46; 32%), while LUSQ was divided into the primitive (n = 10; 13%), classical (n = 26; 35%), secretory (n = 20; 27%), and basal subtypes (n = 19; 25%) by RNA‐seq data (Figure 1A) as previously reported. 10 , 11 To show the clinical significance of subtyping, we examined the patient outcomes (EFS) of each molecular subtype. The EFS varied significantly across subtypes in LUAD (Figure 1B). Subsequently, we undertook an EFS analysis to compare each subtype. Consistent with previous reports, the PP subtype showed a notably poorer prognosis than TRU and tended to be far worse than that of the PI subtype (Figure S1a). 10 , 22 The primitive subtype prognosis tended to be poorer than that for the other LUSQ subtypes; however, this difference was not statistically significant (Figure 1B).
FIGURE 1.
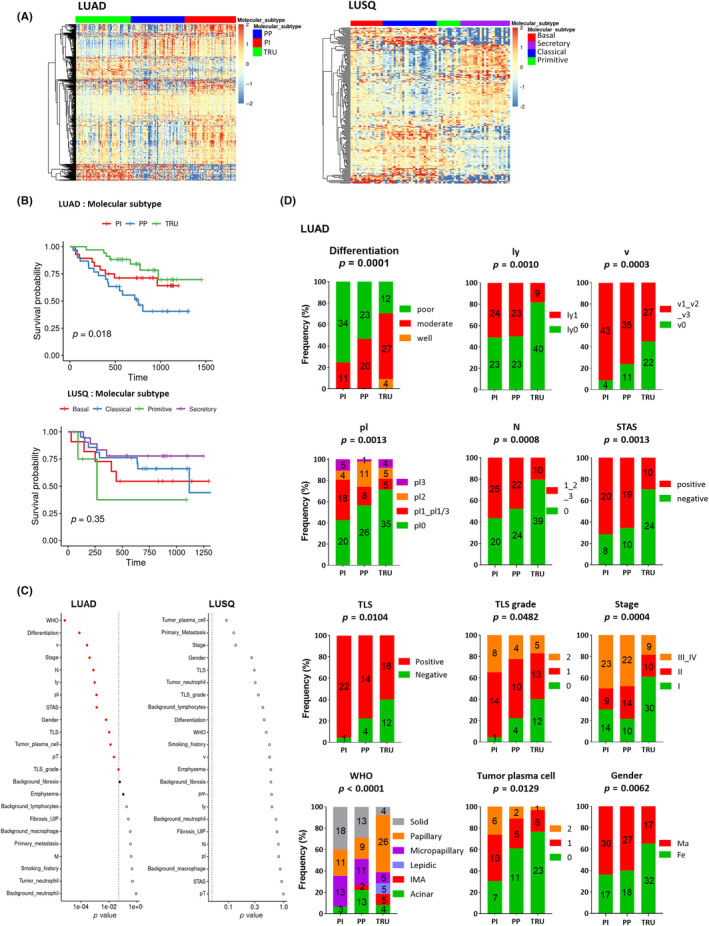
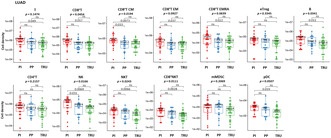
Clinicopathologic features of lung cancer molecular subtypes. (A) Heatmap of mRNAs in different subtypes of lung adenocarcinoma (LUAD; n = 142) (proximal inflammatory [PI], n = 46; proximal proliferative [PP], n = 47; terminal respiratory unit [TRU], n = 49) (left) and lung squamous cell carcinoma (LUSQ; n = 75) (basal, n = 19; classical, n = 26, primitive, n = 10; secretory, n = 20) (right) by RNA sequencing. (B) Kaplan–Meier event‐free survival curves for each molecular subtype of LUAD (upper) and LUSQ (lower) in days. (C) Differences in clinical variables for molecular subtypes of LUAD (left) and LUSQ (right). p values were determined using Fisher's exact test; in the plots, red circles indicate p ≤ 0.05, and filled red circles indicate Holm‐adjusted p ≤ 0.2. (D) Frequencies of each classification of LUAD molecular subtype. Fe, female; IMA, invasive mucinous adenocarcinoma; ly, lymphatic invasion; M, distant metastasis; Ma, male; N, lymph node metastasis; pl, pleural involvement; Pm, intrapulmonary metastasis; pT, tumor size; STAS, spared through air spaces; TLS, tertiary lymphoid structure; V, vascular invasion.
Based on the significant difference in stage among the subtypes of LUAD (Figure 1C,D), we divided LUAD cases into early (stage I and II) and advanced stages (stage III and IV), and compared the EFS of subtypes. No significant differences were observed in advanced stages, whereas EFS was significantly different among three subtypes in the cases at the early stage (Figure S1b).
We then examined the relationship between molecular subtypes and clinical variables. Different molecular subtypes were significantly associated with various clinicopathologic factors in LUAD but not in LUSQ (Figure 1C). Specifically, in LUAD, the TRU subtype was associated with fewer plasma cells in the tumor, more histologically well‐differentiated tumors, low clinical stage, and no vascular invasion, lymphatic invasion, or lymph node metastasis (Figure 1C,D). Tumors spreading through air spaces, which is an independent unfavorable prognostic factor in lung cancer, 23 was less frequent in the TRU subtype than in other subtypes (Figure 1D). Moreover, the TLS, which is associated with positive immunogenicity and favorable prognosis, 24 and grade were fewer in the TRU subtype than in other LUAD subtypes, indicating that the TRU subtype might be associated with a nonimmunogenic TME (Figure 1D). Together, these data suggested that the TRU subtype has lower biological malignancy and lower invasive potential, as well as a nonimmunogenic TME, compared to the other two LUAD subtypes.
3.2. Immune profile of lung cancer molecular subtypes
To elucidate the immunological characteristics of the TME of different LUAD and LUSQ molecular subtypes, we analyzed the TIL profile in cancer tissue. Multicolor FCM showed no association between immune cell fractions and LUSQ subtypes, whereas various immune cell fractions were associated with different LUAD subtypes (Figures 2A and S2b). First, we compared the number of immune cell types per gram of tumor tissue (cell density) (Figure 2B). In the PI subtype, the number of effector cells, including NK and CD8+ T cells, tended to be higher than in the other two subtypes (Figure 2B). High CD8+ T cell frequency was confirmed with IHC CD8 staining (Figure 2C,D). Inversely, in the TRU subtype, the number of TILs, including T, CD8+ T, CD8+ TCM, CD8+ effector memory T, CD8+ terminally differentiated effector memory T, B, NK, NKT, CD8+ NKT, and plasmacytoid dendritic cells, was lower than that in the PI subtype (Figure 2B). In the PP subtype, the number of CD8+ TCM and NK cells was significantly lower than that in the PI subtype. These results suggest that the immune response was less activated in the TRU and PP subtypes than in the PI subtype. No association between the percentages of immune cell type per CD45+ cells (%CD45) in molecular subtypes was detected in LUAD or LUSQ (Figure S2A,C). We analyzed the TIL profile in LUAD, categorizing cases into early and advanced stages. We observed differences in the frequencies of immune cells that remained consistent across disease stages (Figure S3).
FIGURE 2.
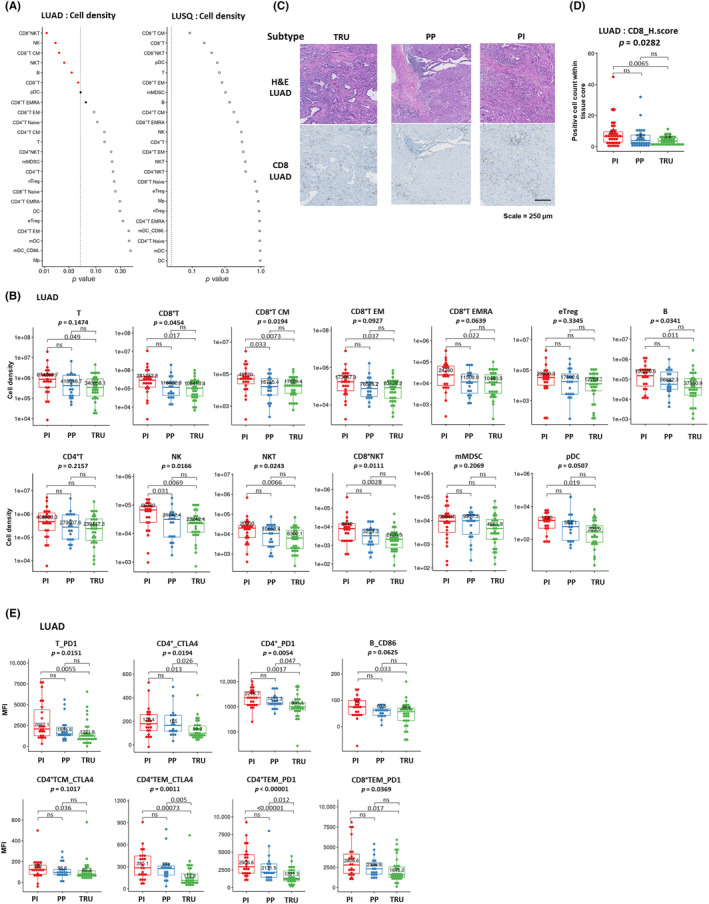
Immune profile of lung cancer molecular subtypes. (A) Relationships between cell density for each immune cell fraction infiltrated in tumors and molecular subtypes of lung adenocarcinoma (LUAD; left) and lung squamous cell carcinoma (LUSQ; right). (B) Cell densities of immune cell fractions in each LUAD molecular subtype were analyzed using flow cytometry (FCM) (proximal inflammatory [PI], n = 23; proximal proliferative [PP], n = 19; terminal respiratory unit [TRU], n = 30). (C) H&E and immunohistochemical (IHC) staining for CD8 in a representative case of each LUAD molecular subtype (PI, n = 35; PP, n = 34; TRU, n = 37). The positivity of each marker is given as the number of total positive cells within each tissue core. (D) Immune‐cell count by IHC staining in lung cancer tissues in each LUAD molecular subtype. The positivity of each marker is given as the number of total positive cells within each tissue core. (E) Mean fluorescence intensity (MFI) of functional molecules in immune cell fractions according to LUAD molecular subtype by FCM. CM, central memory; DC, dendritic cell; EM, effector memory; EMRA, effector memory cells re‐expressing CD45RA; mMDSC, monocytic myeloid‐derived suppressor cell, NK, natural killer; ns, not significant.
The definition of molecular subtypes in LUAD encompassed 507 genes, while in LUSQ, it encompassed 208 genes. 10 , 11 As several immune reaction‐related genes, such as CCL4, CCL5, CCL8, CCL13, CCL18, CCL19, CXCL9, CXCL10, HLA‐DMA, HLA‐DRA, GZMB, CD1c, CD4, CD14, and CD163, meet the criteria for molecular subtype classification, 10 , 11 we examined the association between the expression of various immune‐related genes and the number of immune cells. We found that the expression of CD8+ T cell‐related genes such as CCL4, CCL5, CXCL9, CXCL10, and GZMB was, to some degree, positively correlated with the number of CD8+ T cells analyzed by FCM (Figure S4A). However, there was no significant correlation between the number of other immune cells such as macrophages, dendritic cells, and CD4+ T cells and their related gene expressions (Figure S4A). Additionally, we estimated the immune cell profile within the TME by calculating an immune score using transcriptome data using the xCell software 25 (Figure S4B). The TRU subtype had a moderate immune score, which was consistent with previous reports. 12 , 15
To further analyze the characteristics of each immune cell type in different molecular subtypes, we examined the MFI of functional molecules, such as PD‐1, CTLA‐4, PD‐L1, and Ki‐67 on immune cells using FCM. Following analysis of immune cell fractions, we found that MFIs of functional molecules were associated with the LUAD subtype (Figure S5). The MFI of CTLA‐4 in CD4+ T cells and of PD‐1 in CD4+ T and CD8+ T cells, which are associated with ICI efficacy in clinical settings, 26 , 27 were downregulated in the TRU subtype (Figure 2E). Together, these results suggest that the PI subtype was immunogenic, which was compatible with the immune cell profile (Figure 2B), while the TRU subtype was nonimmunogenic.
3.3. Genomic alterations and immune‐related gene expression in molecular subtypes of lung cancer
We then examined the relationship between molecular subtypes and genomic alterations using WES data from tumor tissues. Only MUC3A mutation was associated with molecular subtypes in LUSQ, whereas EGFR and TP53 mutations were significantly related to the LUAD subtype (Figure 3A). The rate of TP53 mutation in the PP subtype was higher than that in other LUAD subtypes (Figures 3B and S6A). The rate of EGFR mutation in the TRU subtype was higher than that in other LUAD subtypes (Figures 3B and S6B). The EGFR mutation was associated with a noninflamed TME in LUAD through IRF1 downregulation, 28 which is consistent with our finding that the TRU subtype had lower TIL levels in our LUAD cohort (Figure 2B). RNA sequencing data showed that, in the TRU and PP subtypes, IRF1 expression was downregulated and CD8+ T cell‐recruiting chemokine expression levels, including CXCL9, CXCL10, and CXCL11, 29 were decreased (Figure 3C). As the CXCL9 and CXCL10 genes were used for subtype discrimination, the increased expression of CXCL9 and CXCL10 in the PI subtype of LUAD appears reasonable and expected. Moreover, we confirmed that the differences in the expression of these chemokine and IRF genes were consistent across disease stages, as depicted in Figure S6B.
FIGURE 3.
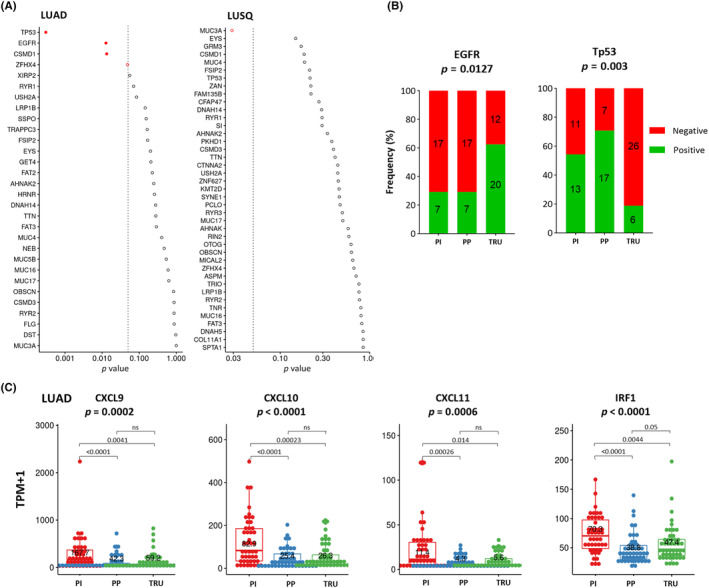
Genomic alterations and immune‐related gene expression in molecular subtypes of lung cancer. (A) Differences in gene alterations for molecular subtypes in lung adenocarcinoma (LUAD; left) and lung squamous cell carcinoma (LUSQ; right). (B) Frequencies of gene alterations in each molecular subtype of LUAD. (C) Expression of CD8+ T cell‐attractant chemokines (chemokine [C‐X‐C motif] ligand [CXCL]9, CXCL10, and CXCL11) and IRF1 in each molecular subtype of LUAD. EGFR, epidermal growth factor receptor; ns, not significant; PI, proximal inflammatory; PP, proximal proliferative; TPM, transcripts per million; TRU, terminal respiratory unit.
3.4. Characteristics of predictive biomarkers for ICIs in different molecular subtypes
Immune checkpoint inhibitors, such as CTLA‐4, PD‐1, and its ligand PD‐L1 Abs, have changed cancer treatment. These treatments can lead to complete, durable responses in some patients with NSCLC. 3 , 30 , 31 However, despite these promising results, predicting who will benefit from ICI treatment is difficult. Most clinically used ICI response biomarkers available in pretreatment settings are a high frequency of CD8+ T cell infiltration in the tumor, intratumoral PD‐L1 expression, and high TMB levels. Thus, we examined these ICI biomarkers in the respective molecular subtypes. As shown in Figure 2B, the number of infiltrated CD8+ T cells in the PI subtype was higher than in the other two LUAD subtypes, which was confirmed by IHC staining of CD8+ cells (Figure 2D). In the PI subtype TME, the immune response was actively induced, which was reflected by increased cytotoxicity marker expression levels (interferon‐gamma, perforin, and granzyme B), compared to the other LUAD subtypes (Figure 4A). In addition, our RNA‐seq data showed that PD‐L1 expression was significantly higher in the PI subtype than in the other LUAD subtypes (Figure 4B). The general trends in cytotoxicity markers and PD‐L1 expression remained consistent in both early and advanced stages, mirroring those observed in the analysis encompassing all stages (Figure S7).
FIGURE 4.
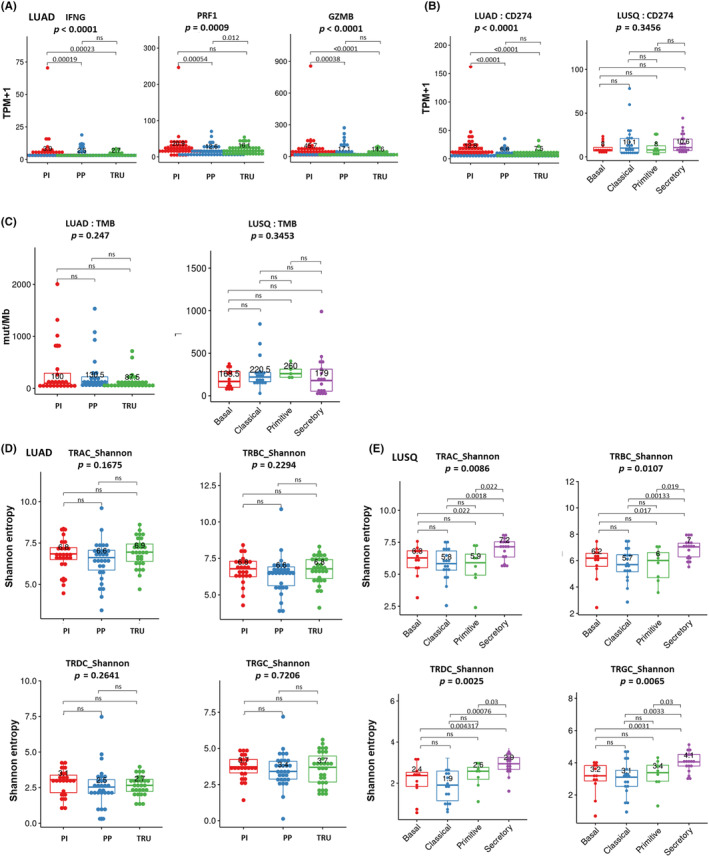
Characteristics of predictive biomarkers for immune checkpoint inhibitors in different molecular subtypes of lung cancer. (A) Expression of cytotoxic activity markers (interferon‐gamma [IFNG], perforin [PRF1], and granzyme B [GZMB]) in each molecular subtype of lung adenocarcinoma (LUAD). (B) Expression of immune checkpoint molecules in each molecular subtype of LUAD (left) and lung squamous cell carcinoma (LUSQ; right). CD274 expression levels were obtained from RNA sequencing. (C) Distribution difference of tumor mutational burden (TMB) in each molecular subtype of LUAD (proximal inflammatory [PI], n = 24; proximal proliferative [PP], n = 24; terminal respiratory unit [TRU], n = 30) (left) and LUSQ (basal, n = 12; classical, n = 20, primitive, n = 5; secretory, n = 17) (right). (D, E) Boxplots showing Shannon entropies of canonical T‐cell receptor (TCR) repertoires (TCRα, TCRβ, TCRγ, and TCRδ) in each molecular subtype of (D) LUAD (PI, n = 25; PP, n = 30; TRU, n = 31) and (E) LUSQ (basal, n = 11; classical, n = 15, primitive, n = 8; secretory, n = 18). CTLA4, cytotoxic T lymphocyte‐associated protein 4; mut, mutation; ns, not significant; PD‐1, programmed death receptor‐1; TCM, central memory T cell; TEM, effector memory T cell.
Tumor mutational burden did not differ significantly among the LUAD subtypes (Figure 4C). Overall, the PI subtype reflected ICI‐responsive characteristics, while the TRU and PP subtypes did not. In LUSQ, CD8+ T cell infiltration, PD‐L1 expression, and TMB were not associated with molecular subtype (Figures 4B,C and S2B,C). In addition, it was reported that the clonality of the TCR repertoire, which was calculated by next‐generation deep sequencing of TCR β‐chain complementarity determining regions (CDR3s), was associated with PD‐1 therapy response in some cancers, including lung carcinoma. 32 , 33 In our cohort, the molecular subtype was associated with TCR clonality in LUSQ, but not in LUAD (Figure 4D,E).
3.5. Immunological characteristics of TME in PP subtype
As shown in Figure 1B, patient prognosis in the PP subtype was significantly poorer than in other LUAD subtypes. To explore the reason for this worse prognosis, we investigated the activated or suppressed signaling pathways in the TME of the PP subtype. The GSEA with a Gene Ontology (GO) set clarified that immune reaction‐related signatures were significantly downregulated in the PP subtype compared to the other subtypes (Figures 5A and S8–, S10). This was compatible with the reduced number of effector cells in the PP subtype (Figure 2B). We then focused on DCs as they act as the conductor of the orchestra of immunological responses. Flow cytometry showed that the activation status of DCs in the PP subtype tended to be decreased compared to the other two types (Figure 5B). The immunogram score on the cancer‐immunity cycle 34 based on RNA‐seq data confirmed that the DC status was suppressed in the PP subtype (Figure 5C). The downregulated DC status might explain the immunosuppressive TME and poor prognosis in the PP subtype. Next, we examined the cause of the downregulated DC status in the PP subtype. We found that lactate levels, which reportedly inhibit DC activation, 35 , 36 tended to be elevated in the PP subtype (Figure 5D). We then examined glucose–lactose‐related gene expression. The RNA‐seq data from LUAD showed that glucose metabolism‐related gene expression, including TPI1, LDHA, and GLUT1, was higher in the PP subtype than in the other subtypes (Figure 5E). In the three‐group comparison, no statistically significant differences were detected regarding the activation status of DC and the level of lactate (Figure 5B,D), possibly due to the small sample size and the potential influence of multiple comparisons. When comparing the PP with the other two subtypes, glycolysis was significantly increased and DC activity was reduced in the PP subtype (Figure S11A–C). Single‐cell RNA‐seq data showed GLUT1 upregulation in malignant cells (Figure 5F), thus confirming the cell types responsible for GLUT1 expression. These results suggest that in the PP subtype, glycolysis was promoted in malignant cells and lactate production was upregulated, followed by DC inactivation in the TME of the PP subtype. The overarching trends regarding the activation status of DC, lactate concentration, and glycolysis‐related genes in both early and advanced stages were consistent with those observed in the analysis encompassing all stages (Figure S11D–F).
FIGURE 5.
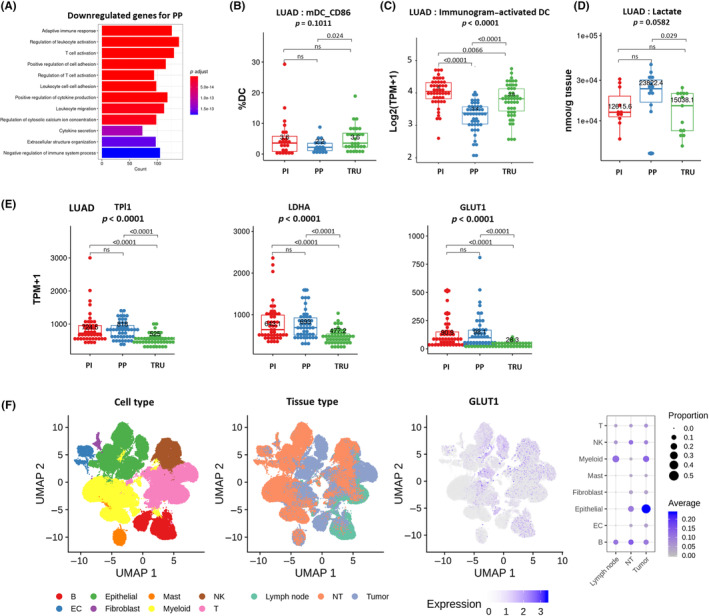
Immunological characteristics of the tumor microenvironment in the proximal proliferative (PP) subtype of lung adenocarcinoma (LUAD). (A) Suppressed pathways in the PP subtype were identified by Gene Set Enrichment Analysis using the Gene Ontology gene set. (B) Proportion of CD86+ dendritic cells (DCs) in each molecular subtype of LUAD. (C) Immunogram (activated DC) of each molecular subtype of LUAD. (D) Lactate content in each molecular subtype of LUAD. (E) Expression of glucose metabolism‐related genes (TPI1, LDHA, and GLUT1) in each molecular subtype of LUAD. (F) Expression of GLUT1 determined by single‐cell RNA sequencing in multiple cell types within tumor and nontumor (NT) sites. Dot plot heatmap showing average scaled expression (color) and percentage of cells (dot size) expressing each gene. EC, enterochromaffin cell; NK, natural killer; ns, not significant; PI, proximal inflammatory; TPM, transcripts per million; TRU, terminal respiratory unit; UMAP, uniform manifold approximation and projection.
4. DISCUSSION
Through the integration of multi‐omics data, our study sheds light on the distinct immune TME across various NSCLC molecular subtypes. Table 1 summarizes the clinicopathologic, immunologic, and multi‐omics characteristics of the LUAD subtypes. Although TRU and PP had an immunosuppressive microenvironment, they had different microenvironmental phenotypes, while the PI subtype TME had an upregulated immune response. We highlight the following subtype characteristics that might contribute to immune escape: defects in immune cell migration to the tumor site in the TRU subtype, antigen‐presenting cell inactivation in the PP subtype, and high immune checkpoint molecule expression levels in the PI subtype.
TABLE 1.
Summary of clinicopathologic, immunologic, and multi‐omics characteristics of lung adenocarcinoma subtypes.
| Molecular subtype | |||
|---|---|---|---|
| PI | PP | TRU | |
| Clinicopathologic features | |||
| Prognosis | Favorable | Poor | Favorable |
| Stage | High | High | Low |
| Differentiation | Poor | Moderate | Well |
| Lymphatic invasion | High | High | Low |
| Vascular invasion | High | Moderate | Low |
| Lymph node metastasis | High | Moderate | Low |
| Spread through air spaces | High | Moderate | Low |
| Tertiary lymphoid structures | High | Moderate | Low |
| TIL profile | |||
| CD8+ T cells | High | Moderate | Low |
| B cells | High | Moderate | Low |
| NK cells | High | Low | Low |
| pDC | High | Moderate | Low |
| PD‐1+ CD4 | High | Moderate | Low |
| Activation status of DC | High | Low | High |
| Genomic mutations | |||
| EGFR | Low | Low | High |
| TP53 | Moderate | High | Low |
| Glycolysis‐related molecules | |||
| GLUT1 expression | Moderate | High | Low |
| LDHA expression | Moderate | High | Low |
| Lactate | Low | High | Low |
Abbreviations: DC, dendritic cell; EGFR, epidermal growth factor receptor; GLUT1, glucose transporter 1; LDHA, lactate dehydrogenase A; NK, natural killer; PD‐1, programmed cell death 1; PI, proximal inflammatory; PP, proximal proliferative; TIL, tumor‐infiltrating leukocyte; TRU, terminal respiratory unit.
Based on the correlation observed between LUAD subtypes and stage (Figure 1D), we categorized LUAD cases into an early stage (stages I and II) and an advanced stage (stages III and IV). We then compared several factors, including EFS, the TIL profile, gene expression (chemokines, cytotoxic markers, CD274, and glycolysis‐related genes), and lactate concentration in LUAD. While some distinctions identified in the analysis across all stages did not attain statistical significance, likely due to the limited number of cases, the overall trends remained consistent with those observed across all stages. This suggests that the impact of the stage on the analysis of immunological aspects might not be substantially influential.
The definition of molecular subtypes comprises several immune reaction‐related genes. 10 , 11 This suggests that molecular subtyping might reflect the immune condition of the TME. However, the expression of immune genes was not necessarily correlated with the immune cell density (Figure S4A,B). Therefore, it is important to note that we utilized a database encompassing FCM, TCR repertoire, and metabolome analyses to assess the immune TME. This multifaceted approach is distinct from analyses based solely on transcriptome data, minimizing the likelihood of our study being confounded by transcriptome analysis alone. Detailed immunological analyses would be valuable in comprehending the immune condition specific to molecular subtypes within the TME. This understanding could significantly contribute to developing personalized immune therapy for lung cancer.
We believe the results of this study have important implications for clinical applications as our results may contribute to the selection of appropriate patients for ICI treatment. In addition to earlier findings about high levels of immune checkpoint molecules (CTLA‐4 and PD‐L1) and adaptive immune cell infiltration, the PI subtype showed high TLS expression levels, high cytotoxic marker levels, and upregulated immune reaction‐related signatures. These results reveal that the PI subtype had a TME wherein the immune response was activated; they also demonstrate that high immune checkpoint molecule expression levels might lead to immune escape. Patients with the PI subtype might be the best candidates for immunotherapy and eligible for preoperative and postoperative immune adjuvant therapy.
Despite previous reports suggesting an immune‐active TME in the TRU subtype akin to the PI subtype analyzed through transcriptome analysis 12 , 15 (Figure S4B), our multi‐omics analysis revealed that the TRU subtype harbors a nonimmunogenic TME characterized by low immune cell infiltration, reduced TLS presence, and diminished cytotoxic marker levels. This discrepancy highlights the importance of integrating various analyses beyond transcriptome analysis alone for a more precise understanding of the immune TME. Incorporating immunostaining and FCM alongside transcriptome analysis provides a more comprehensive and accurate assessment of the immune microenvironment. Despite the nonimmunogenic TME in the TRU subtype, patient prognosis was better than in the PP subtype. This discrepancy could be explained because the TRU subtype has low biological malignancy with low frequencies of lymph node metastasis, lymphatic invasion, vascular invasion, and advanced stage cases. The diminished presence of immune cells within the TRU subtype suggests a potential limitation in the efficacy of ICIs. However, preclinical models have indicated that the EGFR inhibitor changed the immune status and that the combination of ICI and EGFR inhibitor was effective in EGFR‐mutant lung cancers, representing a significant proportion of this subtype. 28 The combination of ICIs with EGFR inhibitors holds promise as a potential therapeutic approach for TRU subtype lung cancer.
Previous findings of low overall immune activity were confirmed in the PP subtype 12 , 15 ; immune‐related pathway signatures were downregulated compared to other subtypes, according to GSEA (Figure 5A). The PP subtype has an intriguing relationship between the immunological status and DC downregulation. Mechanistically, tumors significantly increase glucose metabolism‐related gene expression levels (Figure 5E), and as a result, lactate, the end product of glycolysis, accumulates in the TME of the PP subtype. Tumor‐derived lactate promotes a tolerogenic phenotype in differentiating human DCs and inhibits the ability of DCs to present antigens to T cells, 37 which allows tumors to evade the immune response. Moreover, studies have indicated that a highly glycolytic TME amplifies PD‐1 expression and augments the suppressive function of regulatory T cells. Kumagai et al. have proposed that interventions targeting glycolysis could potentially restore immune activation and bolster the efficacy of immunotherapy. 38 These results may underscore the significance of targeting the highly glycolytic TME, and ICI could be effective after the change of an immune suppressive TME to an antitumor TME by suppressing glucose metabolism‐related gene expression and lactate accumulation. However, it was also reported that the glycolytic pathway plays a pivotal role in T cell activation. 39 Therefore, careful investigation is imperative to ascertain whether targeting glycolysis can induce an effective antitumor immune response in animal models and clinical settings.
Prior studies based on transcriptome data highlighted the secretory subtype in LUSQ as showing an upregulated immune response pathway 11 and a high immune score. 15 However, our observations did not reveal significant immunological distinctions among the subtypes, including the TIL profile and clinicopathologic features. Our analysis suggests that molecular subtyping might not be linked to immunological aspects in LUSQ. This implies the need for alternative classification criteria to tailor precise immunotherapy strategies for LUSQ. The TCR repertoire did not differ in LUAD subtypes, but notably, the secretory subtype in LUSQ showed a significantly higher TCR repertoire compared to the other three subtypes. This suggests that the secretory subtype could shape a distinct immune TME in LUSQ.
Our research has some limitations; for example, our immunogenomic analysis cannot confirm causal relationships. Additionally, the impact of molecular subtypes on ICI treatment outcomes in LUAD remains incompletely understood. Limited cases received ICI treatment, leaving the role of molecular subtypes in predicting immunotherapy outcomes uncertain. Investigating the correlation between LUAD molecular subtypes and ICI efficacy is crucial for future studies in comprehending their influence on treatment outcomes. Potential target pathways, such as glycolysis, require further functional validation. However, our multi‐omics and immunophenotyping data of clinical samples are among the first in the line of investigation and could present valuable information for planning and focusing future research by the scientific community.
AUTHOR CONTRIBUTIONS
Hironori Fukuda: Writing – original draft. Kosuke Arai: Conceptualization; writing – original draft; writing – review and editing. Hideaki Mizuno: Data curation; formal analysis; validation; visualization. Yukari Nishito: Data curation; formal analysis. Noriko Motoi: Formal analysis; investigation. Yasuhito Arai: Investigation. Nobuyoshi Hiraoka: Investigation. Tatsuhiro Shibata: Investigation. Yukiko Sonobe: Data curation; formal analysis. Yoko Kayukawa: Data curation; formal analysis. Eri Hashimoto: Data curation; formal analysis. Mina Takahashi: Data curation; formal analysis. Etsuko Fujii: Data curation; formal analysis. Toru Maruyama: Data curation; formal analysis; visualization. Kenta Kuwabara: Data curation; formal analysis; visualization. Takashi Nishizawa: Data curation; formal analysis. Yukihiro Mizoguchi: Data curation; investigation. Yukihiro Yoshida: Resources. Shun‐ichi Watanabe: Resources. Makiko Yamashita: Investigation. Shigehisa Kitano: Investigation. Hiromi Sakamoto: Investigation. Yuki Nagata: Resources. Risa Mitsumori: Resources. Kouichi Ozaki: Resources. Shumpei Niida: Resources. Yae Kanai: Investigation. Akiyoshi Hirayama: Investigation. Tomoyoshi Soga: Investigation. Keisuke Tsukada: Data curation; formal analysis. Nami Yabuki: Data curation; formal analysis. Mei Shimada: Data curation; formal analysis. Takehisa Kitazawa: Data curation; formal analysis. Osamu Natori: Data curation; formal analysis. Noriaki Sawada: Data curation; formal analysis. Atsuhiko Kato: Data curation; formal analysis. Teruhiko Yoshida: Investigation. Kazuki Yasuda: Investigation. Atsushi Ochiai: Funding acquisition; project administration. Hiroyuki Tsunoda: Data curation; formal analysis; supervision. Kazunori Aoki: Funding acquisition; project administration; supervision; writing – original draft; writing – review and editing.
CONFLICT OF INTEREST STATEMENT
T. Shibata, Y. Kanai, T. Soga, T. Yoshida, A. Ochiai, and K. Aoki are associate editors of Cancer Science. H. Mizuno, Y. Nishito, Y. Sonobe, Y. Kayukawa, E. Hashimoto, M. Takahashi, E. Fujii, T. Nishizawa, T. Maruyama, K. Kuwabara, K. Tsukada, N. Yabuki, M. Shimada, T. Kitazawa, O. Natori, N. Sawada, A. Kato, and H. Tsunoda are employees of Chugai Pharmaceutical Co., Ltd. H. Tsunoda reports personal remuneration from Chugai Pharmaceutical Co., Ltd. The other authors have no conflicts of interest.
FUNDING INFORMATION
This work was supported by Funding for Research to Expedite Effective Drug Discovery by Government, Academia and Private Partnership (GAPFREE) under Grant Numbers JP19ak0101044h0104 and JP19ak0101043h0105A. It was also supported by a grant‐in‐aid for Practical Research for Innovative Cancer Control under Grant Numbers 21ck0106532h0002 and 21ck010640h0002 from the Japan Agency for Medical Research and Development (AMED), and by the National Cancer Center Research and Development Fund (2020‐J‐2), a Grant‐in‐Aid for Scientific Research C (18K07036, 21K06900, 21K07951).
ETHICS STATEMENTS
Approval of the research protocol by an institutional review board: This study was approved (2016‐124) by the National Cancer Center Ethics Committee.
Informed consent: All patients provided written informed consent before sampling, according to the Declaration of Helsinki.
Registry and the registration no. of the study/trial: N/A
Animal studies: N/A.
Supporting information
Figure S1.
Figure S2.
Figure S3.
Figure S4.
Figure S5.
Figure S6.
Figure S7.
Figure S8.
Figure S9.
Figure S10.
Figure S11.
Table S1.
ACKNOWLEDGMENTS
We are deeply grateful to Professor Takao Shimizu for his excellent advice.
Fukuda H, Arai K, Mizuno H, et al. Molecular subtypes of lung adenocarcinoma present distinct immune tumor microenvironments. Cancer Sci. 2024;115:1763‐1777. doi: 10.1111/cas.16154
Hironori Fukuda and Kosuke Arai contributed equally as co‐first authors.
Contributor Information
Hiroyuki Tsunoda, Email: tsunodahry@chugai-pharm.co.jp.
Kazunori Aoki, Email: kaoki@ncc.go.jp.
REFERENCES
- 1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7‐33. doi: 10.3322/caac.21654 [DOI] [PubMed] [Google Scholar]
- 2. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209‐249. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 3. Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus docetaxel in advanced squamous‐cell non‐small‐cell lung cancer. N Engl J Med. 2015;373(2):123‐135. doi: 10.1056/NEJMoa1504627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Herbst RS, Baas P, Kim DW, et al. Pembrolizumab versus docetaxel for previously treated, PD‐L1‐positive, advanced non‐small‐cell lung cancer (KEYNOTE‐010): a randomised controlled trial. Lancet. 2016;387(10027):1540‐1550. doi: 10.1016/S0140-6736(15)01281-7 [DOI] [PubMed] [Google Scholar]
- 5. Jenkins RW, Barbie DA, Flaherty KT. Mechanisms of resistance to immune checkpoint inhibitors. Br J Cancer. 2018;118(1):9‐16. doi: 10.1038/bjc.2017.434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen DS, Mellman I. Elements of cancer immunity and the cancer–immune set point. Nature. 2017;541(7637):321‐330. doi: 10.1038/nature21349 [DOI] [PubMed] [Google Scholar]
- 7. Tancoš V, Grendár M, Farkašová A, et al. Programmed death ligand 1 protein expression, histological tumour differentiation and intratumoural heterogeneity in pulmonary adenocarcinoma. Pathology. 2020;52(5):538‐545. doi: 10.1016/j.pathol.2020.03.012 [DOI] [PubMed] [Google Scholar]
- 8. Miyazawa T, Marushima H, Saji H, et al. PD‐L1 expression in non‐small‐cell lung cancer including various adenocarcinoma subtypes. Ann Thorac Cardiovasc Surg. 2019;25(1):1‐9. doi: 10.5761/atcs.oa.18-00163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cancer Genome Atlas Research Network . Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543‐550. doi: 10.1038/nature13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hayes DN, Monti S, Parmigiani G, et al. Gene expression profiling reveals reproducible human lung adenocarcinoma subtypes in multiple independent patient cohorts. J Clin Oncol. 2006;24(31):5079‐5090. doi: 10.1200/JCO.2005.05.1748 [DOI] [PubMed] [Google Scholar]
- 11. Wilkerson MD, Yin X, Hoadley KA, et al. Lung squamous cell carcinoma mRNA expression subtypes are reproducible, clinically important, and correspond to normal cell types. Clin Cancer Res. 2010;16(19):4864‐4875. doi: 10.1158/1078-0432.CCR-10-0199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Faruki H, Mayhew GM, Serody JS, Hayes DN, Perou CM, Lai‐Goldman M. Lung adenocarcinoma and squamous cell carcinoma gene expression subtypes demonstrate significant differences in tumor immune landscape. J Thorac Oncol. 2017;12(6):943‐953. doi: 10.1016/j.jtho.2017.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu X, Ye Y, Vega KJ, Yao J. Consensus molecular subtypes efficiently classify gastric adenocarcinomas and predict the response to anti‐PD‐1 immunotherapy. Cancers (Basel). 2022;14(15):3740. doi: 10.3390/cancers14153740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim J, Kwiatkowski D, McConkey DJ, et al. The cancer genome atlas expression subtypes stratify response to checkpoint inhibition in advanced urothelial cancer and identify a subset of patients with high survival probability. Eur Urol. 2019;75(6):961‐964. doi: 10.1016/j.eururo.2019.02.017 [DOI] [PubMed] [Google Scholar]
- 15. Öjlert ÅK, Halvorsen AR, Nebdal D, et al. The immune microenvironment in non‐small cell lung cancer is predictive of prognosis after surgery. Mol Oncol. 2019;13(5):1166‐1179. doi: 10.1002/1878-0261.12475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tada Y, Togashi Y, Kotani D, et al. Targeting VEGFR2 with ramucirumab strongly impacts effector/activated regulatory T cells and CD8+ T cells in the tumor microenvironment. J Immunother Cancer. 2018;6(1):106. doi: 10.1186/s40425-018-0403-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kumagai S, Togashi Y, Sakai C, et al. An oncogenic alteration creates a microenvironment that promotes tumor progression by conferring a metabolic advantage to regulatory T cells. Immunity. 2020;53(1):187‐203.e8. doi: 10.1016/j.immuni.2020.06.016 [DOI] [PubMed] [Google Scholar]
- 18. Aoki K, Nishito Y, Motoi N, et al. Tumor‐infiltrating leukocyte profiling defines three immune subtypes of NSCLC with distinct signaling pathways and genetic alterations. Cancer Res Commun. 2023;3(6):1026‐1040. doi: 10.1158/2767-9764.CRC-22-0415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cingolani P, Platts A, Wang LL, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso‐2; iso‐3. Fly. 2012;6(2):80‐92. doi: 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: genome‐wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol. 2016;12(4):e1004873. doi: 10.1371/journal.pcbi.1004873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hirayama A, Kami K, Sugimoto M, et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time‐of‐flight mass spectrometry. Cancer Res. 2009;69(11):4918‐4925. doi: 10.1158/0008-5472.CAN-08-4806 [DOI] [PubMed] [Google Scholar]
- 22. Wilkerson MD, Yin X, Walter V, et al. Differential pathogenesis of lung adenocarcinoma subtypes involving sequence mutations, copy number, chromosomal instability, and methylation. PLoS One. 2012;7(5):e36530. doi: 10.1371/journal.pone.0036530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Han YB, Kim H, Mino‐Kenudson M, et al. Tumor spread through air spaces (STAS): prognostic significance of grading in non‐small cell lung cancer. Mod Pathol. 2021;34(3):549‐561. doi: 10.1038/s41379-020-00709-2 [DOI] [PubMed] [Google Scholar]
- 24. Dieu‐Nosjean MC, Antoine M, Danel C, et al. Long‐term survival for patients with non–small‐cell lung cancer with intratumoral lymphoid structures. J Clin Oncol. 2008;26(27):4410‐4417. doi: 10.1200/JCO.2007.15.0284 [DOI] [PubMed] [Google Scholar]
- 25. Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18(1):220. doi: 10.1186/s13059-017-1349-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kumagai S, Togashi Y, Kamada T, et al. The PD‐1 expression balance between effector and regulatory T cells predicts the clinical efficacy of PD‐1 blockade therapies. Nat Immunol. 2020;21(11):1346‐1358. doi: 10.1038/s41590-020-0769-3 [DOI] [PubMed] [Google Scholar]
- 27. Inomata M, Kado T, Okazawa S, et al. Peripheral PD1‐positive CD4 T‐lymphocyte count can predict progression‐free survival in patients with non‐small cell lung cancer receiving immune checkpoint inhibitor. Anticancer Res. 2019;39(12):6887‐6893. doi: 10.21873/anticanres.13908 [DOI] [PubMed] [Google Scholar]
- 28. Sugiyama E, Togashi Y, Takeuchi Y, et al. Blockade of EGFR improves responsiveness to PD‐1 blockade in EGFR‐mutated non‐small cell lung cancer. Sci Immunol. 2020;5(43):eaav3937. doi: 10.1126/sciimmunol.aav3937 [DOI] [PubMed] [Google Scholar]
- 29. Nagarsheth N, Wicha MS, Zou W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat Rev Immunol. 2017;17(9):559‐572. doi: 10.1038/nri.2017.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hellmann MD, Paz‐Ares L, Bernabe Caro R, et al. Nivolumab plus ipilimumab in advanced non–small‐cell lung cancer. N Engl J Med. 2019;381(21):2020‐2031. doi: 10.1056/NEJMoa1910231 [DOI] [PubMed] [Google Scholar]
- 31. Reck M, Rodríguez‐Abreu D, Robinson AG, et al. Pembrolizumab versus chemotherapy for PD‐L1‐positive non‐small‐cell lung cancer. N Engl J Med. 2016;375(19):1823‐1833. doi: 10.1056/NEJMoa1606774 [DOI] [PubMed] [Google Scholar]
- 32. Riaz N, Havel JJ, Makarov V, et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell. 2017;171(4):934‐949.e16. doi: 10.1016/j.cell.2017.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Forde PM, Chaft JE, Smith KN, et al. Neoadjuvant PD‐1 blockade in resectable lung cancer. N Engl J Med. 2018;378(21):1976‐1986. doi: 10.1056/NEJMoa1716078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Karasaki T, Nagayama K, Kuwano H, et al. An immunogram for the cancer‐immunity cycle: towards personalized immunotherapy of lung cancer. J Thorac Oncol. 2017;12(5):791‐803. doi: 10.1016/j.jtho.2017.01.005 [DOI] [PubMed] [Google Scholar]
- 35. Peng X, He Y, Huang J, Tao Y, Liu S. Metabolism of dendritic cells in tumor microenvironment: for immunotherapy. Front Immunol. 2021;12:613492. doi: 10.3389/fimmu.2021.613492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ding J, Karp JE, Emadi A. Elevated lactate dehydrogenase (LDH) can be a marker of immune suppression in cancer: interplay between hematologic and solid neoplastic clones and their microenvironments. Cancer Biomark. 2017;19(4):353‐363. doi: 10.3233/CBM-160336 [DOI] [PubMed] [Google Scholar]
- 37. Gottfried E, Kunz‐Schughart LA, Ebner S, et al. Tumor‐derived lactic acid modulates dendritic cell activation and antigen expression. Blood. 2006;107(5):2013‐2021. doi: 10.1182/blood-2005-05-1795 [DOI] [PubMed] [Google Scholar]
- 38. Kumagai S, Koyama S, Itahashi K, et al. Lactic acid promotes PD‐1 expression in regulatory T cells in highly glycolytic tumor microenvironments. Cancer Cell. 2022;40(2):201‐218.e9. doi: 10.1016/j.ccell.2022.01.001 [DOI] [PubMed] [Google Scholar]
- 39. Chang CH, Qiu J, O'Sullivan D, et al. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell. 2015;162(6):1229‐1241. doi: 10.1016/j.cell.2015.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1.
Figure S2.
Figure S3.
Figure S4.
Figure S5.
Figure S6.
Figure S7.
Figure S8.
Figure S9.
Figure S10.
Figure S11.
Table S1.


