Abstract
Objective
In this study, we aimed to determine whether proly4-hydroxylase-III (P4HA3) could be used as a biomarker for the diagnosis of colorectal cancer (CRC) as well as for determining prognosis.
Methods
We used The Cancer Genome Atlas (TCGA) database to analyze P4HA3 expression in CRC and further investigated the association between P4HA3 and clinicopathological parameters, immune infiltration, and prognosis of patients with CRC. Enrichment analysis was conducted to investigate the potential biological role of P4HA3 in CRC. To verify the results of TCGA analysis, we performed immunohistochemical staining of 180 clinical CRC tissue samples to probe into the relationship of P4HA3 expression with lymphocyte infiltration and immune checkpoints expression.
Results
The expression of P4HA3 was significantly higher in CRC tissues and associated with a higher degree of malignancy and poorer prognosis in CRC. The results of enrichment analysis indicated that P4HA3 may be associated with the epithelial-mesenchymal transition process and the immune response. Immunohistochemical staining results showed that high P4HA3 expression was associated with high infiltration levels of CD8+ and Foxp3+ TILs and high PD-1/PD- L1 expression. Lastly, patients with CRC co-expressing P4HA3 and PD-1 had a significantly worse prognosis.
Conclusion
High expression of P4HA3 is associated with adverse clinical features and immune cell infiltration in CRC, and has the potential to serve as a biomarker for predicting CRC prognosis.
Keywords: Colorectal cancer, P4HA3, Tumor microenvironment, Immune cell infiltration, Prognosis
Highlights
-
•
P4HA3 expression is upregulated in CRC and correlates with CRC malignant progression.
-
•
In CRC, P4HA3 expression was associated with infiltration of CD8+ T cells and Foxp3+ T cells.
-
•
P4HA3 expression correlates with PD-1 and PD-L1 expression, and co-expression with PD-1 is linked to a worse prognosis.
Abbreviations
- AUC
area under the curve
- CRC
colorectal cancer
- DSS
disease-specific survival
- EMT
epithelial-mesenchymal transition
- GO
gene ontology
- GSEA
gene set enrichment analysis
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- OS
overall survival
- P4HA3
proly4-hydroxylase-III
- PD-1
programmed cell death receptor-1
- PD-L1
programmed cell death receptor-1 ligand
- PFI
progression-free interval
- ROC
receiver operator characteristic curve
- TCGA
The Cancer Genome Atlas
- TILs
tumor-infiltrating lymphocytes
- TMA
tissue microarray
- TME
tumor microenvironment
1. Introduction
Colorectal cancer (CRC) ranks as the third most prevalent cancer globally [1,2]. In China, CRC has the second-highest incidence and fourth-highest number of deaths. By 2030, the global incidence of new cases and deaths from CRC is expected to reach 2.45 million and 1.22 million, respectively. Recently, there has been a global rise in the occurrence of early-onset CRC [3,4]. Despite the increasing research on the comprehensive treatment of CRC, most cancer patients do not receive proper or timely diagnosis or treatment because of the absence of visible signs in the early stages. Therefore, identifying new biomarkers to increase early detection and improve prognosis has become imperative to reduce CRC mortality.
As the critical component in the tumor matrix, the extracellular matrix is vital in the initiation and progression of diverse malignancies, such as CRC; it also promotes tumor cell metastasis. Collagen, an important structural protein of the extracellular matrix, can obstruct the production of chemokines in the tumor microenvironment (TME), thus hindering the anti-tumor immune response [5].
As a member of the collagen proly4-hydroxylase family in the endoplasmic reticulum, Proly4-hydroxylase-III (P4HA3) is a critical enzyme for collagen synthesis. Notably, multiple studies have provided evidence indicating the significant impact of P4HA3 expression on the progression of various cancer types such as colon cancer [6], head and neck squamous cell carcinoma [7], gastric cancer [8], lung cancer [9], and pituitary adenoma [10]. In the study of CRC, although the literature has reported that P4HA3 shows certain potential in predicting patient prognosis and influencing the progression of epithelial-mesenchymal transition (EMT), the expression of P4HA3 in tumor tissues, its association with patient clinical characteristics, and its relationship with immune cell infiltration have not been thoroughly investigated.
Consequently, this study performed a multilevel analysis of P4HA3 expression in CRC. We utilized The Cancer Genome Atlas (TCGA) database and clinical tissue microarray (TMA) to investigate the association between P4HA3 and clinicopathological characteristics, as well as prognosis in CRC patients. Furthermore, we predicted the biological function of P4HA3 and examined its correlation with tumor immune infiltration and immune checkpoints. Immunohistochemical staining was conducted on 180 clinical CRC tissues and their corresponding adjacent normal tissues to detect the expression of P4HA3, programmed cell death receptor-1 (PD-1), PD-1 ligand (PD-L1), and the infiltration of tumor-infiltrating lymphocytes (TILs; CD4+ TILs, CD8+ TILs, and Foxp3+ TILs) to verify the relationship between P4HA3 and immune cell infiltration. Finally, retrospective clinical data were analyzed to verify whether P4HA3 influences the prognosis of CRC patients. Our findings elucidate the significance of P4HA3 in the progression of CRC and indicate P4HA3 to be a novel prognostic factor for CRC.
2. Material and methods
2.1. Data sources
RNA-seq data of tissue from 647 CRC cases and 51 cases of normal paracancerous tissue and clinical information data were downloaded and organized from the TCGA database (https://portal.gdc.cancer.gov/). GSE9348 and GSE21815 datasets were obtained from the geographic database (https://www.Ncbi.nlm.nih.gov/geo/). The RNA-seq data format was Transcripts Per Million (TPM). A log2(TPM+1) transformation was performed on the TPM values, and the RNA-seq data were normalized by the trimmed mean of M-values (TMM) method using the calcNormFactors function in the edgeR package. Batch effects were removed using the ComBat function of the sva package. P4HA3 expression was categorized into low- and high-expression groups based on the median.
2.2. CRC tissue sample collection and patient information
The 180 CRC and corresponding adjacent normal tissues in this study were obtained from general surgery surgical resection specimens at the Zhejiang Provincial People's Hospital. In addition, the general clinical information of these 180 patients from which the samples were taken including factors such as sex, age, telephone number, operation time, lesion size, site, pathologic type, infiltration of lymphatic and neural tissues, Tumor-Node-Metastasis (TNM) stage, pathological stage, and degree of differentiation, were collected.
2.3. TMA construction
The TMA construction method was performed as described in Ref. [11]. All sample tissues were stained with hematoxylin and eosin (HE). The typical tumor tissues or normal tissues in the paraffin specimens were marked based on the HE-stained sections. Using a tissue arrayer, tissue cores with a diameter of 1.5 mm were punched out from the marked areas on the donor paraffin blocks. The tissue cores were transferred to recipient paraffin blocks with a spacing of 0.3 mm between each tissue core. The tissue cores in the paraffin blocks were incubated at 37 °C for 30 min. Subsequently, the tissue array blocks were sectioned at a speed of 4 μm per revolution using an automated tissue slicer, and the sections were mounted on glass slides with anti-detachment treatment. Each chip contained a grid of 18 by 10 tissue spots.
2.4. Immunohistochemical staining
Immunohistochemistry was employed to detect P4HA3, CD4, CD8, PD-1, PD-L1, and Foxp3 expression of TMA samples. After baking, deparaffinization, hydration, and antigen repair of the tissues, the tissues were treated with hydrogen peroxide and phosphate buffer to disrupt endogenous peroxidase. The tissues were then incubated overnight at 4 °C with anti-PD-1 (1:100, GT228107, GeneTech, Shanghai, China), anti-PD-L1 (1:100, GT228007, GeneTech, Shanghai, China), anti-Foxp3 (1:100, ab215206, Abcam, Cambridge, UK), anti-CD4 (1:1000, ab183685, Abcam, Cambridge, UK), anti-CD8 (1:2000, ab217344, Abcam, Cambridge, UK), and anti-P4HA3 (1:100, 23185-1-AP, Proteintech, Chicago, USA). The tissues were successively incubated with secondary antibody and diaminobenzidine chromogen solution. Finally, counterstaining of the slides with hematoxylin was done, followed by dehydration and mounting. In each experiment, both negative (omission of the primary antibody) and positive controls (with IgG-matched serum) were included.
2.5. Immunohistochemical scoring criteria
P4HA3 was evaluated using the histochemistry score (H-score) grading system. H-score = ∑(I × Pi), I = intensity of staining (categorized as 0, 1, 2, 3 based on staining depth), and Pi = percentage of positive cells. The H-score is between 0 and 300, with larger values indicating a stronger combined positive intensity. As has been previously described [12], the evaluation of five immune indicators (PD-1, PD-L1, CD4, CD8, and Foxp3) was conducted by determining the number of positive cells based on randomly selected 5 high-power fields (HPFs) at 200 × magnification and calculating the average level.
2.6. Gene expression and clinicopathologic features
The expression of P4HA3 was evaluated in CRC and adjacent normal tissue samples with multiple data, including TCGA-COADREAD data, GSE9348, GSE21815 datasets, and TMA. Patients’ clinical information in TCGA-COADREAD and TMA was organized and grouped, and the relationship between P4HA3 expression and each clinicopathological parameter was analyzed.
2.7. Prognostic value analysis
P4HA3 expression data and patient survival data from TCGA-COADREAD and TMA were integrated, and the impact of P4HA3 on patient survival status was assessed using the survival package. Cox regression analyses were also performed for P4HA3 and several clinical variables. The pROC package was employed to obtain receiver operator characteristic (ROC) curves, which evaluated the model's accuracy.
2.8. Functional concentration analysis
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG): single-gene correlation analysis was performed by the Spearman statistical method, and molecules with correlation with P4HA3 were screened according to the threshold (|Cor|> 0.5 and p < 0.05). Gene Set Enrichment Analysis (GSEA): the DESeq2 package was used to analyze the changes in gene expression levels between high and low expression groups of P4HA3, and obtain the log2FoldChange values. GO, KEGG, and GSEA analysis was then performed using the clusterProfiler package. Predefined gene sets were obtained from the MsigDB database (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp).
2.9. Immune analysis
The enrichment scores of various immune cell subsets in each sample were calculated using the ssGSEA algorithm provided in the R package – GSVA, enabling the inference of immune cell infiltration levels within individual samples. An analysis was performed to examine the association between P4HA3 expression and immune cell infiltration, as well as the expression levels of immunostimulatory and immunosuppressive molecules.
2.10. Statistical analysis
T-test/Wilcoxon rank sum test was chosen for the analysis of differences between any two groups. Spearman was chosen for correlation analysis. The chi-square test/Yates' correction was used to assess whether there was a difference in the constitutive ratio of clinical variables/immune-related protein expression between high and low P4HA3 expression groups. In the survival analysis, Cox regression was used for testing when variables met the assumption of equal proportional risk; otherwise, the log-rank test was chosen. All statistical analyses in this study were performed using R version [4.2.1]. Data were analyzed using stats [4.2.1], car [3.1–0], survival [3.3.1], pROC [1.18.0], rms [6.3–0], GSVA [1.46.0], DESeq2 [1.36.0], sva [3.44.0], clusterProfiler [4.4.4]. Data visualizations were created by the ggplot [3.3.6]. Significance was set at a bilateral p-value <0.05. *(P < 0.05), **(P < 0.01), ***(P < 0.001), ns(P ≥ 0.05).
3. Results
3.1. Analysis of results based on public database data
3.1.1. P4HA3 expression, correlation with clinicopathologic features, and prognostic value in TCGA COAD-READ and Gene Expression Omnibus databases
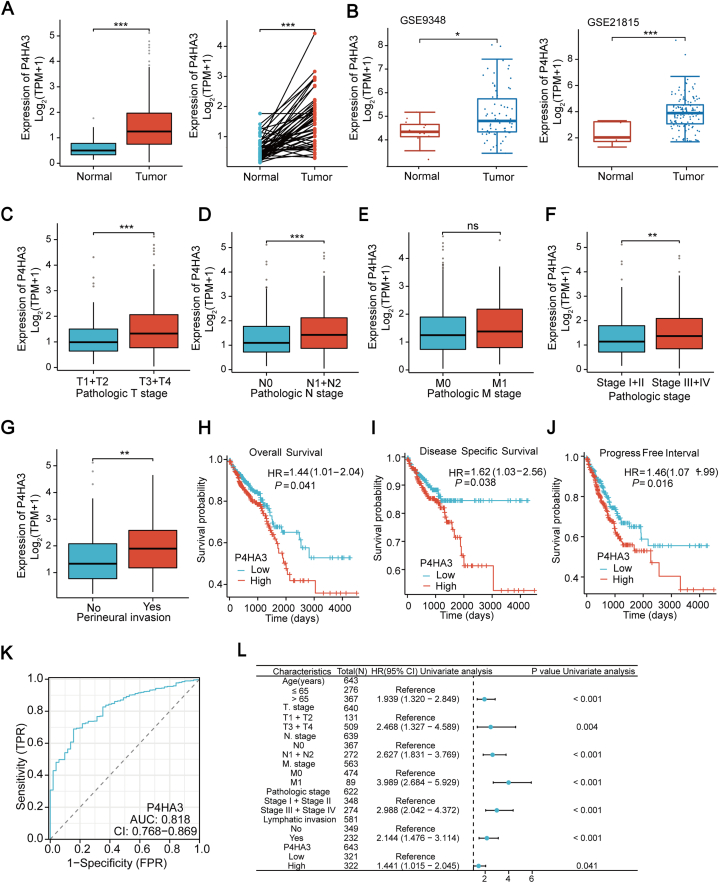
TCGA database and two Gene Expression Omnibus (GEO) Series (GSE) were analyzed, and the results showed that P4HA3 expression was markedly elevated in CRC tissues than in the adjacent normal tissues (Fig. 1A and B). Subsequently, evaluation of the expression profile of P4HA3 in association with the degree of tumor malignancy revealed that P4HA3 expression was significantly higher in T stage (T3 and T4), N stage (N1 and N2), pathological stages (stages III and stages IV), and perineural invasion, but there was no difference in P4HA3 expression in M stages (Fig. 1C–G). Therefore, the assumption that P4HA3 is associated with a higher degree of tumor malignancy is reasonable. The overall survival (OS), disease-specific survival (DSS), and progression-free interval (PFI) were characterized using Kaplan–Meier curves for patients with low and high P4HA3 expression, aiming to investigate the prognostic significance of P4HA3 in predicting the prognosis of CRC patients. The results showed that the OS, DSS, and PFI of patients with CRC with high P4HA3 expression were significantly lower than those with the low P4HA3 expression group (Fig. 1H–J). Moreover, the ROC curve revealed high diagnostic accuracy of P4HA3 for CRC (Fig. 1K; AUC = 0.818; confidence interval = 0.768–0.869). A one-way Cox analysis was conducted to ascertain whether P4HA3 is a prognostic factor for survival outcomes. Based on reports in the literature and conjunction with the clinical information contained in the TCGA database, several clinical variables that may be associated with CRC prognosis were selected for Cox regression analysis along with P4HA3 (Table S1) [13]. The analysis revealed that age, T-stage, N-stage, M-stage, pathological stage, lymphatic invasion, and P4HA3 all affected the survival time of patients (Fig. 1L).
Fig. 1.
P4HA3 expression in CRC was analyzed using TCGA COAD-READ and GEO databases, and the correlation of P4HA3 with clinicopathological features and its prognostic value were assessed using TCGA COAD-READ. (A) P4HA3 expression in unpaired (left) and paired samples (right) in TCGA. (B) P4HA3 expression between normal and tumor samples in the GSE9348 and GSE21815 datasets. (C–G) Differential expression of P4HA3 in TNM staging, pathologic staging, and perineural invasion. Survival curves showing (H) OS, (I) DSS, and (J) PFI between patients with low and high P4HA3 expression. (K) P4HA3 diagnostic ROC curves for CRC models. (L) The Forest plot representing a one-way Cox univariate regression analysis.
3.1.2. Functional enrichment of P4HA3 based on TCGA COAD-READ
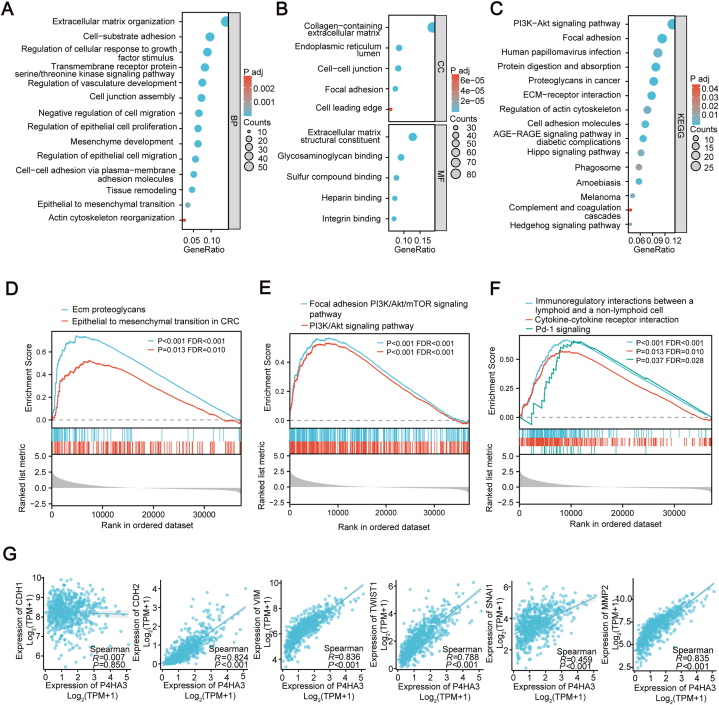
To investigate the potential biological functions and associated pathways linked to P4HA3 expression, we conducted GO and KEGG enrichment analyses on molecules pertinent to P4HA3 expression. Our analysis revealed that biological processes were predominantly enriched in cell-matrix adhesion, epithelial-to-mesenchymal cell transition, and epithelial cell migration (Fig. 2A). Regarding cellular components, enrichment was primarily observed in the collagen-containing extracellular matrix. Furthermore, functional molecular analyses highlighted a significant enhancement in structural components of the extracellular matrix (Fig. 2B). KEGG analysis indicated notable enrichment in phosphoinositide-3-kinase/Akt (PI3K/Akt) signaling and extracellular matrix-receptor interactions (Fig. 2C). Subsequently, we analyzed the differential expression patterns between high and low P4HA3 expression groups and conducted GSEA on the list of differentially expressed genes. GSEA analysis revealed that the EMT process (Fig. 2D) and PI3K-Akt signaling were significantly enriched (Fig. 2E). Interestingly, cytokine interactions and PD-1 signaling were also significantly different (Fig. 2F). Thus, we further analyzed the correlation between P4HA3 and EMT-related markers. We observed that P4HA3 expression was positively correlated with a variety of EMT molecular markers, including N-cadherin, vimentin, snai1, twist1, and MMP2 (Fig. 2G). Based on these enrichment results, we speculated that high P4HA3 expression might be associated with the EMT process and immune response.
Fig. 2.
Enrichment analysis of the P4HA3 gene. (A–C) GO and KEGG enrichment analysis of P4HA3. (D–F) GSEA analysis of P4HA3. (G) Correlation analysis of P4HA3 and EMT markers.
3.1.3. Correlation between P4HA3 expression and immune cell infiltration and immune checkpoints in TCGA COAD-READ
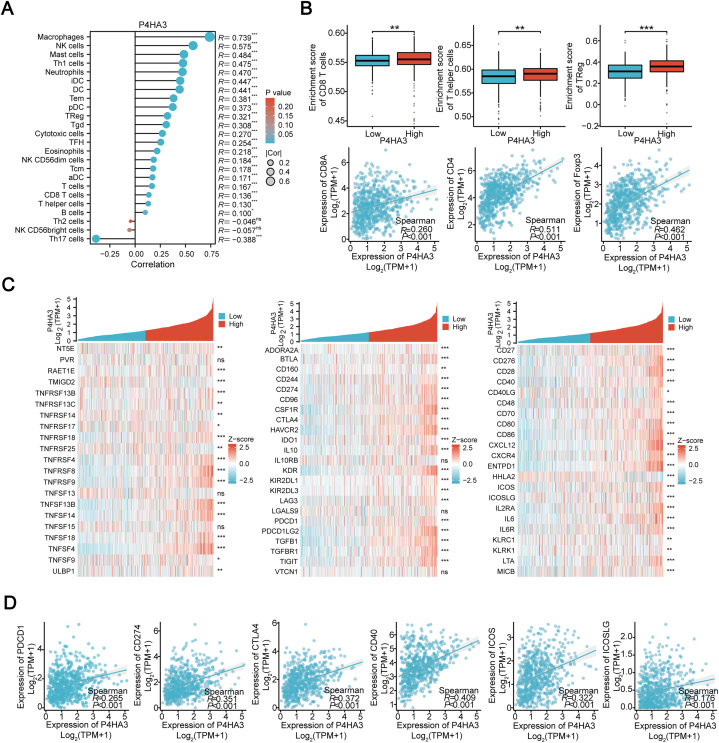
The findings from the enrichment analysis indicated a potential association of P4HA3 with immune response. Further analysis was conducted using data from the TCGA to explore the relevance of P4HA3 to immune cell infiltration and immune checkpoints. The correlation analysis results between P4HA3 and immune cell score were visualized using a lollipop plot (Fig. 3A). Results showed that P4HA3 correlated with 22 TILs, including Th1 cells, Tem cells, Regulatory T (Treg) cells, Tgd cells, Tcm cells, CD8 T cells, and Th17 cells. Individual analysis revealed that the P4HA3 high-expression group exhibited significantly higher enrichment scores for CD8 T cells, T helper cells, and Treg cells than did the P4HA3 low-expression group. In addition, P4HA3 expression was linked to the expression of CD8A, CD4, and Foxp3 (Fig. 3B). Besides, it was observed that P4HA3 expression was positively correlated with the expression of most immunosuppressive and immunostimulatory checkpoints (Fig. 3C), such as the immunosuppressive checkpoints PDCD1 (PD-1), CD274 (PD-L1), and CTLA-4 and the immunostimulatory checkpoints CD40, ICOS, and ICOSLG (Fig. 3D). Thus, P4HA3 may have a relationship with the immune response to CRC.
Fig. 3.
Correlation between P4HA3, immune cells, and immune checkpoint markers in patients with CRC. (A) Analysis of molecular and immune infiltration score correlation. In this lollipop plot, the circle's size and the bar's length indicate the correlation's strength. The shade of the color reflects the significance level represented. (B) Correlation between P4HA3 expression and CD8 T cells, T helper cells, Treg infiltration, and their cell marker expression. (C) Heatmap of P4HA3 co-expression with immunostimulatory and immunosuppressive molecules. (D) Correlation between P4HA3 expression and immunostimulatory molecules ICOS, ICOSLG, CD40, and inhibitory molecules PDCD1, CD274, and CTLA4 in patients with CRC.
3.2. Analysis of results based on clinical samples
3.2.1. P4HA3 expression in CRC tissues and normal paracancerous tissues in clinical tissue samples
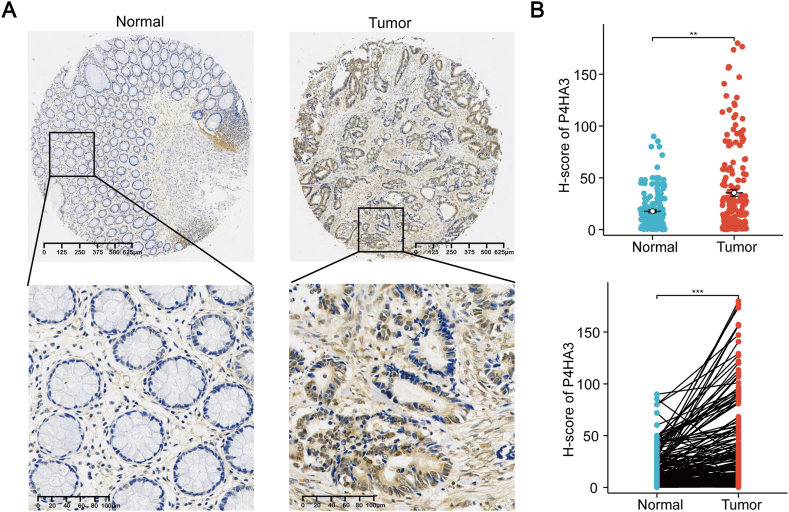
The expression of P4HA3 in 180 CRC tissues and their paired normal paracancerous tissues was examined by immunohistochemistry to verify the high expression of P4HA3 in patients with CRC as shown in TCGA and GSE datasets (Fig. 4A). Evaluation of the H-score of P4HA3 revealed a significant upregulation of P4HA3 expression in patients with CRC in paired and unpaired comparisons (Fig. 4B). This observation aligns with the outcomes obtained from the analysis conducted on public databases.
Fig. 4.
P4HA3 expression in clinical CRC tissues. (A) Representative immunohistochemical images of P4HA3 in CRC tissues and normal paracancerous tissues. Scale bar = 625 μm for the whole tissue image, and scale bar = 100 μm for the regional magnification image. (B) P4HA3 expression in clinical CRC tissue and normal paracancerous tissues.
3.2.2. Correlation between P4HA3 expression and clinicopathologic parameters in clinical tissue samples
Further correlation analysis of P4HA3 expression with clinicopathological parameters was performed based on the histochemical scores of P4HA3 in 179 CRC tissues (1 sample had missing clinical information). P4HA3 expression was classified using the mean histochemical score as the threshold, designating P4HA3 scores below the mean as low expression (n = 130) and P4HA3 scores equal to or exceeding the mean as high expression (n = 49). The results showed that the older the patients (>65 years), the higher the P4HA3 expression. In addition, patients with high P4HA3 expression exhibited higher T stages, N stages, pathological stages, and fewer differentiated tumor cells. Notably, high P4HA3 expression was also associated with lymphatic infiltration. In contrast, P4HA3 expression was not associated with sex, M stage, tumor site, tumor histological typing, vascular involvement, or neurological involvement (Table 1). These results were consistent with those in TCGA. Based on this differential expression of P4HA3 in patients with CRC and clinicopathological correlation results, we hypothesized that P4HA3 may correlate with an increased malignancy level in patients with CRC.
Table 1.
Correlation between P4HA3 expression and clinicopathological parameters.
| Characteristic | P4HA3low | P4HA3high | P value |
|---|---|---|---|
| N | 131 | 49 | |
| Gender, n (%) | 0.642 | ||
| Male | 80 (71.4 %) | 32 (28.6 %) | |
| Female | 50 (74.6 %) | 17 (25.4 %) | |
| Age (years), n (%) | 0.019 | ||
| ≤65 | 75 (80.6 %) | 18 (19.4 %) | |
| >65 | 56 (65.1 %) | 30 (34.9 %) | |
| T-stage, n (%) | 0.008 | ||
| T1 | 8 (100 %) | 0 (0 %) | |
| T2 | 27 (90 %) | 3 (10 %) | |
| T3 | 76 (71.7 %) | 30 (28.3 %) | |
| T4 | 20 (57.1 %) | 15 (42.9 %) | |
| N-stage, n (%) | < 0.001 | ||
| N0 | 76 (80.9 %) | 18 (19.1 %) | |
| N1 | 37 (78.7 %) | 10 (21.3 %) | |
| N2 | 18 (47.4 %) | 20 (52.6 %) | |
| M-stage, n (%) | 0.235 | ||
| M0 | 129 (74.1 %) | 45 (25.9 %) | |
| M1 | 2 (40 %) | 3 (60 %) | |
| Pathologic stage, n (%) | 0.018 | ||
| stageⅠ | 7 (100 %) | 0 (0 %) | |
| stage Ⅱ | 69 (80.2 %) | 17 (19.8 %) | |
| stage Ⅲ | 53 (65.4 %) | 28 (34.6 %) | |
| stage Ⅳ | 2 (40 %) | 3 (60 %) | |
| Tumor site, n (%) | 0.394 | ||
| Rectum | 65 (75.6 %) | 21 (24.4 %) | |
| Colon | 65 (69.9 %) | 28 (30.1 %) | |
| Histological type, n (%) | 0.221 | ||
| Adenocarcinoma | 123 (71.5 %) | 49 (28.5 %) | |
| Mucinous adenocarcinoma | 7 (100 %) | 0 (0 %) | |
| Degree of differentiation, n (%) | 0.040 | ||
| Moderately differentiated | 120 (76.9 %) | 36 (23.1 %) | |
| Poorly differentiated | 8 (50 %) | 8 (50 %) | |
| Vascular recidivism, n (%) | 0.356 | ||
| No | 116 (74.4 %) | 40 (25.6 %) | |
| Yes | 15 (65.2 %) | 8 (34.8 %) | |
| Lymphatic recidivism, n (%) | 0.013 | ||
| No | 101 (78.3 %) | 28 (21.7 %) | |
| Yes | 30 (60 %) | 20 (40 %) | |
| Neurorecidivism, n (%) | 0.134 | ||
| No | 121 (75.2 %) | 40 (24.8 %) | |
| Yes | 10 (55.6 %) | 8 (44.4 %) |
3.2.3. Correlation of P4HA3 expression with immune infiltration and immune checkpoints in clinical tissue samples
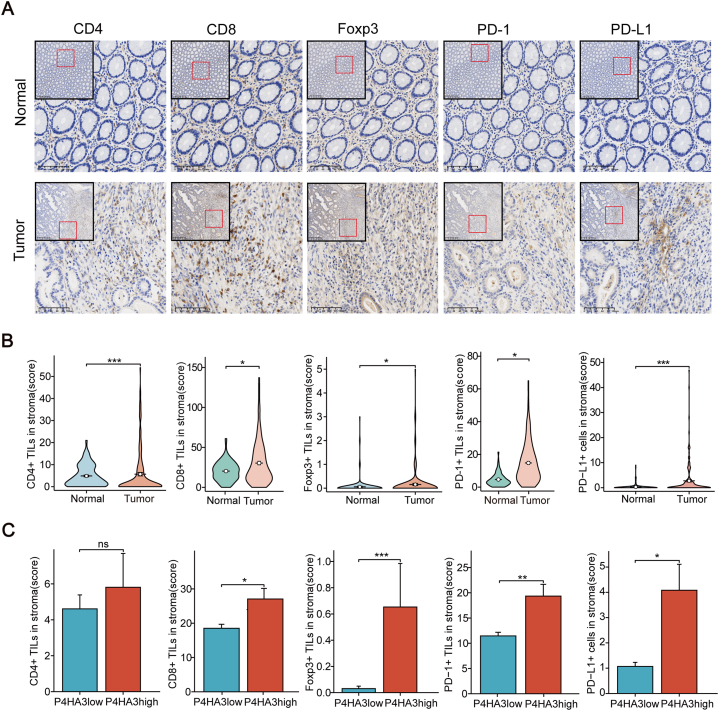
We further explored the relationship between P4HA3 and immune cell infiltration as well as immune checkpoint expression in the tumor microenvironment. According to the literature, in the tumor microenvironment of CRC, the expression of immune-related proteins in the tumor stroma is more clinically significant [[14], [15], [16]]. Therefore, we investigated the expression of P4HA3 in CRC as well as the expression of CD4, CD8, Foxp3, PD-1, and PD-L1 in CRC stroma using immunohistochemical staining (Fig. 5A). We observed a significant increase in the expression of CD4+, CD8+, and Foxp3+ TILs within the tumor stroma. In addition, the expression of PD-1 and PD-L1 was also elevated in the tumor stroma (Fig. 5B). We employed the average stain scores of P4HA3 and five other immune-related indicators as thresholds to categorize the expression levels of each indicator in the tissue samples as either high or low expression for subsequent correlation analysis. The correlation analysis showed that P4HA3 expression was correlated with CD8, Foxp3, PD-1, and PD-L1 expression in the tumor stroma. Nonetheless, there was no significant correlation observed between P4HA3 expression and CD4 expression in the tumor stroma (Table 2, Fig. S1). Simultaneously, we observed a marked increase in CD8+, Foxp3+, PD-1+, and PD-L1+ cells in the tumor stroma in the high P4HA3 expression group compared with that in the low P4HA3 expression group. Nevertheless, the high P4HA3 expression group exhibited no notable disparity in the number of CD4+ TILs compared with the low P4HA3 expression group (Fig. 5C). Based on these results, it is reasonable to speculate that P4HA3 may affect immune cell infiltration in the TME of CRC and may be associated with immunosuppression.
Fig. 5.
Correlation between P4HA3 expression and immunity in clinical tissue samples. (A) Representative images of CD4, CD8, Foxp3, PD-1, and PD-L1 staining in normal and tumor tissues. Low magnification field of view: scale bar = 300 μm. High magnification field of view: scale bar = 100 μm. (B) CD4+TILs, CD8+TILs, Foxp3+TILs, PD-1+TILs, and PD-L1+cells infiltration between normal and tumor tissues. (C) CD4+TILs, CD8+TILs, Foxp3+TILs, PD-1+TILs, and PD-L1+cells infiltration between P4HA3 high and low expression groups.
Table 2.
Correlation analysis of P4HA3 expression with the expression of immune-related markers in the CRC stroma.
| Characteristic | P4HA3low | P4HA3high | P value |
|---|---|---|---|
| n | 131 | 49 | |
| CD4, n (%) | 0.926 | ||
| CD4low | 93 (73.2 %) | 34 (26.8 %) | |
| CD4high | 37 (72.5 %) | 14 (27.5 %) | |
| CD8, n (%) | 0.033 | ||
| CD8low | 87 (78.4 %) | 24 (21.6 %) | |
| CD8high | 42 (63.6 %) | 24 (36.4 %) | |
| FOXP3, n (%) | 0.002 | ||
| FOXP3low | 128 (75.7 %) | 41 (24.3 %) | |
| FOXP3high | 3 (27.3 %) | 8 (72.7 %) | |
| PD-1, n (%) | 0.006 | ||
| PD-1low | 91 (80.5 %) | 22 (19.5 %) | |
| PD-1high | 40 (61.5 %) | 25 (38.5 %) | |
| PD-L1, n (%) | 0.016 | ||
| PD-L1low | 99 (78 %) | 28 (22 %) | |
| PD-L1high | 32 (60.4 %) | 21 (39.6 %) |
3.2.4. High P4HA3 expression predicts poor prognosis of clinical patients with CRC
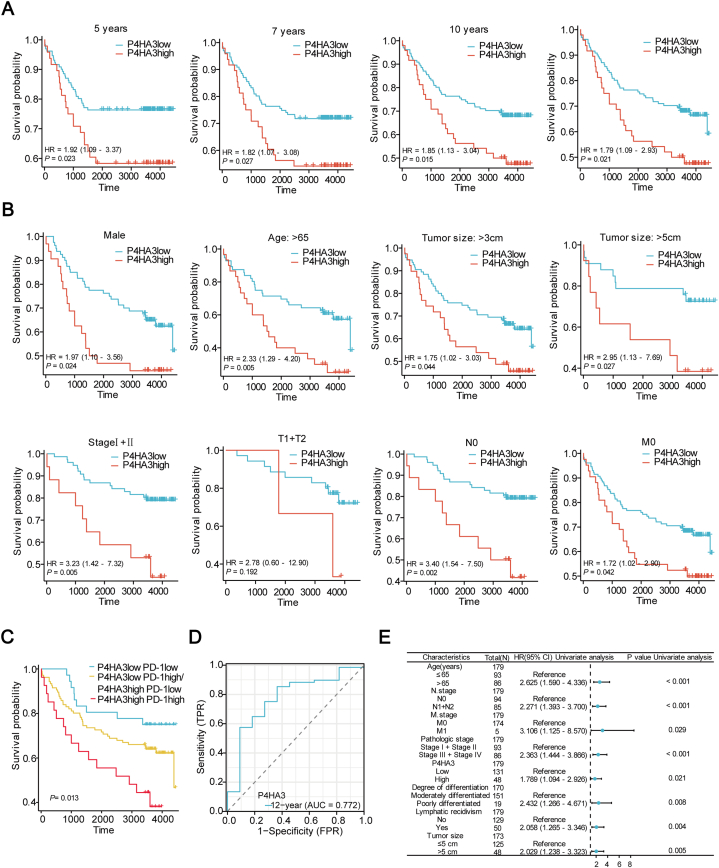
We obtained prognostic information to further analyze whether P4HA3 expression affects the prognostic situation of patients through multiple follow-ups of patients with clinical CRC. First, we analyzed the OS of patients at years 5, 7, 10, and at the end of follow-up. Our findings revealed that patients exhibiting high expression of P4HA3 had a lower OS (Fig. 6A). Subsequently, we divided patients with CRC into subgroups based on their clinicopathological features. Subgroup survival analysis showed that high P4HA3 expression was associated with clinical prognosis for most variables: male sex, age >65 years, tumor size >3 cm, tumor size >5 cm, stage I + II, N0 stage, and M0 stage (Fig. 6B). In addition, we categorized P4HA3 and PD-1 expression into high-risk (P4HA3 high/PD-1 high), intermediate-risk (P4HA3 high/PD-1 low, P4HA3 low/PD-1 high), and low-risk groups (P4HA3 low/PD-1 low). Kaplan–Meier survival analysis demonstrated a notable divergence in OS across the three groups (P = 0.013; Fig. 6C). The prognosis was the poorest in the high-risk group. The time-dependent ROC curve (AUC, 0.772) indicated that P4HA3 exhibited prognostic efficacy (Fig. 6D). Furthermore, we also performed a one-way Cox regression analysis of multiple clinical variables and P4HA3 (Table S2). The results showed that high age, high N stage, high M stage, high pathological stage, high P4HA3 expression, low degree of differentiation, lymphatic infiltration, and tumor size (>5 cm) were associated with poor prognosis in CRC patients (Fig. 6E). These data indicate that P4HA3 is an unfavorable prognostic factor for CRC patients.
Fig. 6.
Prognostic value of P4HA3 expression in clinical CRC patients. (A) OS of patients with CRC with low and high P4HA3 expression in TMA after 5, 7, and 10 years, and at the end of follow-up. (B) The OS of patients with CRC with low and high P4HA3 expression within each subgroup. (C) Prognostic impact of P4HA3 and PD-1 co-expression. (D) Time-related ROC. (E) Results of a Cox one-way univariate analysis in the form of a forest plot.
4. Discussion
P4HA3, an indispensable enzyme in the biosynthesis of collagen, its high expression is closely associated with cancer. Collagen forms the skeletal structure of the extracellular matrix in the TME, imparting rigidity and tensile strength and influencing cell proliferation, invasion, and migration [17]. Collagen has been reported to be over-deposited in tumor tissues [18]. When collagen-encoding genes are highly expressed, patients with CRC exhibit worse OS [19]. Moreover, higher collagen levels have been observed in patients with CRC developing liver metastases [20]. Through bioinformatics analysis and molecular mechanism studies, some researchers have found that P4HA3 overexpression promotes the malignant progression of melanoma, which can be reversed by inhibiting P4HA3 expression [21]. In addition, Zhang et al. [22] and Long et al. [10] found that P4HA3 may modulate the PI3K-Akt pathway to promote the progression of clear-cell kidney cancer and pituitary adenoma growth and metastasis. Notably, Zhou et al. [6] and Nakasuka et al. [9] suggested that P4HA3 accelerates the developmental process of colon cancer and non-small cell lung cancer by promoting EMT through the TGF-β pathway. Recently, the outcomes of a comprehensive analysis across various human cancers revealed that P4HA3 expression level is more closely related to immune cells [23]. Moreover, in a study on gastric cancer, P4HA3 promoted immune cell infiltration, highlighting its potential to be a therapeutic target for immunotherapy in gastric cancer [8].
However, the specific contribution of P4HA3 to the progression of CRC remains uncertain. Therefore, we investigated the biological significance of P4HA3 in CRC progression from multiple perspectives. Our findings showed a prominent upregulation of P4HA3 expression in patients with CRC, which correlated significantly with various clinicopathological parameters in patients with CRC. Moreover, the higher the T-stage, N-stage, and pathological stages, the higher the expression level of P4HA3, indicating a potential role of P4HA3 in promoting the malignant progression of CRC. Notably, GO, KEGG, and GSEA analyses revealed that P4HA3 may be associated with the EMT process. EMT promotes tumor immunosuppression and immune escape because tumor cells with a mesenchymal phenotype are less susceptible to attack by autoimmune cells [24,25]. Moreover, various cytokines released by immune cells can in turn promote EMT, which further promotes the formation of immunosuppressive TME [26]. Interestingly, a robust correlation also exists between EMT and PD-L1 [27,28]. The rate of PD-L1 positivity is much higher in patients with the mesenchymal phenotype than in those with the epithelial phenotype [29].
What's more, to investigate the relationship of P4HA3 with the immune response, we performed an analysis using the TCGA database. Notably, our findings demonstrated that P4HA3 was positively correlated with immune cell infiltration, immunosuppressive molecules, and immunostimulatory molecule expression. GSEA showed that P4HA3 was associated with the PD-1 signaling pathway. In addition, immunohistochemical staining showed increased CD8+ TIL infiltration into the stroma of tumors in the high P4HA3 expression group. Therefore, we hypothesized that high P4HA3 expression might have an effect on immune cell infiltration in TME of CRC. The well-established understanding within the scientific community affirms the immuno-suppressive functions of PD-1+ TILs, PD-L1+ cells, and Tregs in the TME, helping tumor cells evade T cell attacks. Our study confirmed that increased numbers of Tregs in the stroma and higher protein expression of PD-1 and PD-L1 are associated with elevated P4HA3 expression. These observations suggest that high P4HA3 expression may be associated with the formation of an immunosuppressive TME.
Our evaluation of survival in patients with CRC stratified into high and low P4HA3 expression groups revealed that patients with high P4HA3 expression had significantly poorer prognoses. Additionally, in the pathological stages I + II, N0, M0, and moderately differentiated subgroups, patients exhibiting high P4HA3 expression had significantly shorter survival times. This finding suggests that P4HA3 is an ideal prognostic marker for early CRC. Notably, we found that patients had a significantly worse prognosis when P4HA3 was co-overexpressed with PD-1. This introduces a novel perspective regarding the impact of P4HA3 on patient prognosis. This study showed that P4HA3 is a CRC prognostic factor closely linked to the immune response.
Nevertheless, this study has some limitations. First, the investigation was primarily restricted to bioinformatics analysis and clinical sample studies and lacked the validation of basic experiments. Second, although it was found that high P4HA3 expression may be associated with EMT and immune cell infiltration, the exact mechanism of action remains unclear. Therefore, additional comprehensive studies are warranted to validate and elucidate these aspects.
5. Conclusions
The findings of this study established a strong correlation between elevated P4HA3 expression and increased malignancy as well as unfavorable prognosis among patients with CRC. P4HA3 may be linked to immune cell infiltration in the tumor stroma. The results also showed that patients with CRC had a significantly poorer prognosis when P4HA3 and PD-1 were co-expressed. Therefore, P4HA3 could be a novel biomarker for CRC that correlates with prognosis and immune infiltration.
Consent for publication
All the authors have provided consent for the publication.
Data availability statement
The TCGA and GEO data used in this study are available in public databases (relevant web pages are provided in the article). Clinical patient data from this study can be obtained by contacting the corresponding author.
Ethics statement
This research was conducted in compliance with the principles outlined in the Declaration of Helsinki and received ethical approval from the Institutional Review Board of Zhejiang Provincial People's Hospital under protocol number QT2023190. Due to the anonymity of patient data, the need for informed consent was waived.
Funding
This work was supported by the National Natural Science Foundation of China (8217082371); the Key Project of Science and Technology of Zhejiang Province in China (2022C03186); the Key Projects by the Province of Zhejiang Medical and Health Science and Technology Project (WKJ-ZJ-2101) and the Zhejiang Provincial Science and Technology Program of Traditional Medicine (no. 2022ZA016).
CRediT authorship contribution statement
Xiaohuan Guo: Writing – original draft, Investigation, Data curation, Conceptualization. Yu Zhang: Writing – review & editing, Resources, Methodology. Lina Peng: Visualization, Validation. Yaling Wang: Visualization, Validation. Cheng-Wen He: Writing – review & editing, Data curation. Kaixuan Li: Validation, Methodology. Ke Hao: Resources. Kaiqiang Li: Methodology. Zhen Wang: Project administration, Funding acquisition. Haishan Huang: Supervision. Xiaolin Miao: Supervision, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors would like to thank Xin Zhang for her technical support for this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e31695.
Contributor Information
Xiaohuan Guo, Email: gxiaohuan22@163.com.
Yu Zhang, Email: 1013950738@qq.com.
Lina Peng, Email: plnpenglina@163.com.
Yaling Wang, Email: wangyay23@163.com.
Cheng-Wen He, Email: hcw_7_14@163.com.
Kaixuan Li, Email: 1072124370@qq.com.
Ke Hao, Email: haoke@hmc.edu.cn.
Kaiqiang Li, Email: likaiqiang@hmc.edu.cn.
Zhen Wang, Email: wangzhen@hmc.edu.cn.
Haishan Huang, Email: haishan_333@163.com.
Xiaolin Miao, Email: mxl@eye.ac.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Xi Y., Xu P. Global colorectal cancer burden in 2020 and projections to 2040. Transl Oncol. 2021;14 doi: 10.1016/j.tranon.2021.101174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rho Y.S., Gilabert M., Polom K., Aladashvili A., Kopeckova K., Megdanova V., Coleman N., Greally M., Marrelli D., Roviello F., et al. Comparing clinical characteristics and outcomes of young-onset and late-onset colorectal cancer: an international collaborative study. Clin. Colorectal Cancer. 2017;16:334–342. doi: 10.1016/j.clcc.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 4.Scott R.B., Rangel L.E., Osler T.M., Hyman N.H. Rectal cancer in patients under the age of 50 years: the delayed diagnosis. Am. J. Surg. 2016;211:1014–1018. doi: 10.1016/j.amjsurg.2015.08.031. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y., Kim J., Yang S., Wang H., Wu C.J., Sugimoto H., Lebleu V.S., Kalluri R. Type I collagen deletion in αSMA+ myofibroblasts augments immune suppression and accelerates progression of pancreatic cancer. Cancer Cell. 2021;39:548–565.e6. doi: 10.1016/j.ccell.2021.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou H., Zou J., Shao C., Zhou A., Yu J., Chen S., Xu C. Prolyl 4-hydroxylase subunit alpha 3 facilitates human colon cancer growth and metastasis through the TGF-β/Smad signaling pathway. Pathol. Res. Pract. 2022;230 doi: 10.1016/j.prp.2021.153749. [DOI] [PubMed] [Google Scholar]
- 7.Wang T., Wang Y.X., Dong Y.Q., Yu Y.L., Ma K. Prolyl 4-hydroxylase subunit alpha 3 presents a cancer promotive function in head and neck squamous cell carcinoma via regulating epithelial-mesenchymal transition. Arch. Oral Biol. 2020;113 doi: 10.1016/j.archoralbio.2020.104711. [DOI] [PubMed] [Google Scholar]
- 8.Niu X., Ren L., Wang S., Gao D., Ma M., Hu A., Qi H., Zhang S. High prolyl 4-hydroxylase subunit alpha 3 expression as an independent prognostic biomarker and correlated with immune infiltration in gastric cancer. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.952335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakasuka F., Tabata S., Sakamoto T., Hirayama A., Ebi H., Yamada T., Umetsu K., Ohishi M., Ueno A., Goto H., et al. TGF-β-dependent reprogramming of amino acid metabolism induces epithelial-mesenchymal transition in non-small cell lung cancers. Commun. Biol. 2021;4:782. doi: 10.1038/s42003-021-02323-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Long R., Liu Z., Li J., Yu H. COL6A6 interacted with P4HA3 to suppress the growth and metastasis of pituitary adenoma via blocking PI3K-Akt pathway. Aging. 2019;11:8845–8859. doi: 10.18632/aging.102300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glinsmann-Gibson B., Wisner L., Stanton M., Larsen B., Rimsza L., Maguire A. Recommendations for tissue microarray construction and quality assurance. Appl. Immunohistochem. Mol. Morphol. 2020;28:325–330. doi: 10.1097/PAI.0000000000000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsuchiya K., Yoshimura K., Inoue Y., et al. YTHDF1 and YTHDF2 are associated with better patient survival and an inflamed tumor-immune microenvironment in non-small-cell lung cancer. OncoImmunology. 2021;10(1) doi: 10.1080/2162402X.2021.1962656. Published 2021 Aug 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marks K.M., West N.P., Morris E., Quirke P. Clinicopathological, genomic and immunological factors in colorectal cancer prognosis. Br. J. Surg. 2018;105(2):e99–e109. doi: 10.1002/bjs.10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Z., Li Y., Wang Y., et al. A deep learning quantified stroma-immune score to predict survival of patients with stage II-III colorectal cancer. Cancer Cell Int. 2021;21(1):585. doi: 10.1186/s12935-021-02297-w. Published 2021 Oct 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kather J.N., Krisam J., Charoentong P., et al. Predicting survival from colorectal cancer histology slides using deep learning: a retrospective multicenter study. PLoS Med. 2019;16(1) doi: 10.1371/journal.pmed.1002730. Published 2019 Jan 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calon A., Lonardo E., Berenguer-Llergo A., et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat. Genet. 2015;47(4):320–329. doi: 10.1038/ng.3225. [DOI] [PubMed] [Google Scholar]
- 17.Pozzi A., Wary K.K., Giancotti F.G., Gardner H.A. Integrin alpha1beta1 mediates a unique collagen-dependent proliferation pathway in vivo. J. Cell Biol. 1998;142:587–594. doi: 10.1083/jcb.142.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brauchle E., Kasper J., Daum R., Schierbaum N., Falch C., Kirschniak A., Schäffer T.E., Schenke-Layland K. Biomechanical and biomolecular characterization of extracellular matrix structures in human colon carcinomas. Matrix Biol. 2018;68–69:180–193. doi: 10.1016/j.matbio.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 19.Liang Y., Lv Z., Huang G., Qin J., Li H., Nong F., Wen B. Prognostic significance of abnormal matrix collagen remodeling in colorectal cancer based on histologic and bioinformatics analysis. Oncol. Rep. 2020;44:1671–1685. doi: 10.3892/or.2020.7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lalmahomed Z.S., Bröker M.E., van Huizen N.A., Coebergh van den Braak R.R., Dekker L.J., Rizopoulos D., Verhoef C., Steyerberg E.W., Luider T.M., Ijzermans J.N. Hydroxylated collagen peptide in urine as biomarker for detecting colorectal liver metastases. Am. J. Cancer Res. 2016;6:321–330. [PMC free article] [PubMed] [Google Scholar]
- 21.Atkinson A., Renziehausen A., Wang H., Lo Nigro C., Lattanzio L., Merlano M., Rao B., Weir L., Evans A., Matin R., et al. Collagen prolyl hydroxylases are bifunctional growth regulators in melanoma. J. Invest. Dermatol. 2019;139:1118–1126. doi: 10.1016/j.jid.2018.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Z., Zhang Y., Zhang R. P4HA3 promotes clear cell renal cell carcinoma progression via the PI3K/AKT/GSK3β pathway. Med. Oncol. 2023;40:70. doi: 10.1007/s12032-022-01926-2. [DOI] [PubMed] [Google Scholar]
- 23.Wu Y., Zhang B., Nong J., Rodrìguez R.A., Guo W., Liu Y., Zhao S., Wei R. Systematic pan-cancer analysis of the potential tumor diagnosis and prognosis biomarker P4HA3. Front. Genet. 2023;14 doi: 10.3389/fgene.2023.1045061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nieto M.A., Huang R.Y., Jackson R.A., Thiery J.P. EMT. EMT: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 25.Dongre A., Rashidian M., Reinhardt F., Bagnato A., Keckesova Z., Ploegh H.L., Weinberg R.A. Epithelial-to-mesenchymal transition contributes to immunosuppression in breast carcinomas. Cancer Res. 2017;77:3982–3989. doi: 10.1158/0008-5472.CAN-16-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma H.Y., Liu X.Z., Liang C.M. Inflammatory microenvironment contributes to epithelial-mesenchymal transition in gastric cancer. World J. Gastroenterol. 2016;22:6619–6628. doi: 10.3748/wjg.v22.i29.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alsuliman A., Colak D., Al-Harazi O., Fitwi H., Tulbah A., Al-Tweigeri T., Al-Alwan M., Ghebeh H. Bidirectional crosstalk between PD-L1 expression and epithelial to mesenchymal transition: significance in claudin-low breast cancer cells. Mol. Cancer. 2015;14:149. doi: 10.1186/s12943-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang Y., Zhan H. Communication between EMT and PD-L1 signaling: new insights into tumor immune evasion. Cancer Lett. 2020;468:72–81. doi: 10.1016/j.canlet.2019.10.013. [DOI] [PubMed] [Google Scholar]
- 29.Kim S., Koh J., Kim M.Y., Kwon D., Go H., Kim Y.A., Jeon Y.K., Chung D.H. PD-L1 expression is associated with epithelial-to-mesenchymal transition in adenocarcinoma of the lung. Hum. Pathol. 2016;58:7–14. doi: 10.1016/j.humpath.2016.07.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The TCGA and GEO data used in this study are available in public databases (relevant web pages are provided in the article). Clinical patient data from this study can be obtained by contacting the corresponding author.