Abstract
The EICP0 protein of equine herpesvirus 1 (EHV-1) is an early, viral regulatory protein that independently trans-activates EHV-1 immediate-early (IE), early, γ1 late, and γ2 late promoters. To assess whether this powerful trans-activator functions in conjunction with three other EHV-1 regulatory proteins to activate expression of the various classes of viral promoters, transient cotransfection assays were performed in which effector plasmids expressing the EICP22, EICP27, and IE proteins were used either singly or in combination with an EICP0 effector construct. These analyses revealed that (i) independently, the EICP0 and IE proteins are powerful trans-activators but do not function synergistically, (ii) the IE protein inhibits the ability of the EICP0 protein to trans-activate the IE, γ1 late, and γ2 late promoters, (iii) the EICP22 and EICP0 proteins do not function together to significantly trans-activate any EHV-1 promoter, and (iv) the EICP27 and EICP0 proteins function synergistically to trans-activate the early and γ1 late promoters. A panel of EICP0 truncation and deletion mutant plasmids was generated and used in experiments to define the domains of the 419-amino-acid (aa) EICP0 protein that are important for the trans-activation of each class of EHV-1 promoters. These studies revealed that (i) carboxy-terminal truncation mutants of the EICP0 protein exhibited a progressive loss of trans-activating ability as increasing portions of the carboxy terminus were removed, (ii) the amino terminus of the EICP0 protein containing the RING finger (aa 8 to 46) and the acidic region (aa 71 to 84) was necessary but not sufficient for activation of all classes of EHV-1 promoters, (iii) the RING finger was absolutely essential for activation of EHV-1 promoters, since deletion of the entire RING finger motif (aa 8 to 46) or a portion of it (aa 19 to 30) completely abrogated the ability of these mutants to activate any promoter tested, (iv) the acidic region contributed to the ability of the EICP0 protein to activate the early and γ1 late promoters, and deletion of the acidic region enhanced the ability of this mutant to activate the IE promoter, (v) the carboxy terminus (aa 325 to 419), which is rich in glutamine residues, was dispensable for the EICP0 trans-activation function, (vi) a motif resembling a nuclear localization signal (aa 289 to 293) was unnecessary for the EICP0 protein to trans-activate promoters of any temporal class, and (vii) the EICP0 protein was phosphorylated during infection, and deletion of the serine-rich region (aa 210 to 217), a potential site for phosphorylation, reduced by more than 70% the ability of the EICP0 protein to activate the γ2 late class of promoters.
Equine herpesvirus 1 (EHV-1), an important pathogen of the horse, causes rhinopneumonitis, spontaneous abortions in pregnant mares, and myeloencephalitis (1, 35). The 77 genes of EHV-1 are transcribed in an ordered and temporally controlled cascade termed immediate-early (IE), early (E), and late (L) (4, 17, 18). Five EHV-1 regulatory proteins (EICP22, EICP27, EICP0, IE, and E-TIF [EHV-1 α-trans-inducing protein]) are known to control the specific transcription of viral genes (2, 20, 22, 26, 27–29, 35, 40–43, 47).
The sole IE gene, the first gene to be transcribed during infection, encodes the key regulatory protein of EHV-1 (3, 19, 40). IE gene transcription occurs in the absence of viral protein synthesis and is induced by E-TIF (7, 28, 29, 37). The IE protein activates expression of the early promoters, autoregulates expression of its own promoter (40), and is essential for EHV-1 replication, since deletion of both copies of the IE gene renders the virus unable to grow on noncomplementing cells (16).
The second set of genes to be transcribed during an EHV-1 infection are early genes, which encode additional viral regulatory proteins (EICP0, EICP22, and EICP27), as well as proteins involved in the replication of the viral genome (2, 4, 20, 21, 46). Early gene transcription requires the presence of the IE protein and occurs before the initiation of viral DNA synthesis. Late genes are the last set to be transcribed, and the majority encode viral structural proteins (4). The late genes can be further divided into two subclasses: (i) γ1 or “leaky late” genes, whose expression begins prior to viral DNA replication but does not reach maximal levels until after the onset of viral DNA replication and (ii) γ2 or “true late” genes, whose synthesis is totally dependent on viral DNA replication.
The EICP0 protein is the only EHV-1 regulatory protein that independently activates all classes of viral promoters (IE, early, γ1 late, and γ2 late) in transient transfection chloramphenicol acetyltransferase (CAT) assays (2). Our present understanding of EHV-1 gene regulation suggests that the EICP0 protein is important in the switch from early to late gene expression. The relative amounts of the IE and EICP0 proteins appear to be a factor in this temporal switch, since these two regulatory proteins have an antagonistic relationship, as we previously reported (25). The goals of the studies presented here were to determine whether the EICP0 protein functions in conjunction with other EHV-1 regulatory proteins (IE, EICP22, and EICP27) and to define the domains of the EICP0 protein that are important for its trans-activation function. Synergy among some of the EHV-1 regulatory proteins has been described previously. For example, the IE protein functions in conjunction with the EICP22 or EICP27 early regulatory proteins to up-regulate expression from early and γ1 late promoters (22, 42, 47). The EICP22 protein increases the ability of the IE protein to bind to its cognate DNA binding site, thereby enhancing the ability of the IE protein to activate transcription from different promoters (26, 27). The precise mechanism by which the EICP27 accessory protein functions to increase IE protein trans-activation is unknown and is currently under investigation.
Studies presented here indicate that the IE and EICP0 proteins do not function synergistically but instead function antagonistically. The EICP27 protein, but not the EICP22 protein, has the ability to enhance EICP0 trans-activation of early and γ1 late promoters. In addition, mutational analyses of the EHV-1 EICP0 protein indicated that several regions of this protein are important for its trans-activation function, including the RING finger, the acidic, and the serine-rich regions.
MATERIALS AND METHODS
Cells and virus.
Cultures of L-M (murine fibroblast) cells were maintained as described previously (40, 43). The Kentucky A (KyA) strain of EHV-1 was propagated in L-M suspension culture at a low multiplicity of infection (MOI) of 0.01 PFU per cell and was assayed for infectivity by plaque titration (3, 4, 17).
Plasmids.
The generation of the effector constructs pSVIE, pSVUL3, pCDR4, and pSVICP0K and the reporter constructs pIE-CAT, pTK2-CAT, pIR5-CAT, and pgK-CAT has been described previously (2, 22, 40, 43, 47). These effector constructs express the EHV-1 IE (pSVIE), EICP27 (pSVUL3), EICP22 (pCDR4), and EICP0 (pSVICP0K) proteins under the control of either the simian virus 40 promoter (SVIE, SVUL3, and SVICP0K) or the cytomegalovirus promoter (pCDR4). To construct carboxy-terminal truncation mutants n163 and n213, pSVICP0K was partially digested with Ecl136II, and singly cut DNA fragments were purified by using the Gene Clean kit (Bio 101, Vista, Calif.). The purified fragments were ligated in the presence of an XbaI linker (CTAGTCTAGACTAG; New England Biolabs, Beverly, Mass.) that contains a nonsense codon in all three open reading frames (6). To generate carboxy-terminal truncation mutants n227 and n325, pSVICP0K was partially digested with HincII, and purified fragments were ligated as described above. To create carboxy-terminal truncation mutants n135, n141, and n282, pSVICP0K was partially digested with AluI, and singly cut DNA fragments were ligated to the XbaI linker as described above. The locations of the linkers were verified by restriction enzyme digestion and agarose gel electrophoresis. The n135, n141, n163, n213, n227, n282, and n325 mutants express the first 135, 141, 163, 213, 227, 282, and 325 amino acids (aa) of the EICP0 protein, respectively. To generate the d105 construct, the Sma3 clone (2) was digested with NcoI (nucleotide [nt] 572) and EcoRV (nt 1702), and the 1,130-bp fragment was filled in with Klenow fragment and cloned into a SmaI-digested pSVSPORT1 vector (Life Technologies, Gaithersburg, Md.). This clone expresses an amino-terminal-truncated form of the EICP0 protein that lacks the first 105 aa.
The ΔACID (deletion of aa 71 to 84 [Δ71-84]), ΔNLS (Δ289-293), ΔSRICH (Δ210-217), ΔRING8-46, ΔRING19-30, and DIEBS deletion mutant plasmids were generated via a “Quik Change” site-directed mutagenesis kit from Stratagene (La Jolla, Calif.) that is based on a Pfu polymerase mutagenesis protocol. To generate the ΔACID mutant, PCRs were performed with the complementary mutagenic primers ACID#1 and ACID#2 (5′ GAA ACA AAG GTG AGC GTG GGG CAA TTT TTG GCC GTG 3′ and 5′ CAC GGC CAA AAA TTG CCC CAC GCT CAC CTT TGT TTC 3′, respectively), which delete bp 257 to 298 of pSVICP0K, corresponding to aa 71 to 84 of the EICP0 protein, upon PCR amplification. An aliquot of this PCR product was digested with DpnI, leaving only the amplified mutagenized plasmids, which were then transformed into supercompetent XL-1 Blue Escherichia coli cells. The presence of the desired mutation was initially identified via restriction enzyme digestion and was confirmed by DNA sequence analyses. The ΔNLS, ΔSRICH, ΔRING19-30, ΔRING8-46, and DIEBS deletion mutant plasmids were generated essentially as described above except that different mutagenic PCR primers were used. For the ΔNLS mutant, the primers used were NLS#1 and NLS#2 (5′ GTC GGC GCA GCG CCC AGC CCA GAA CCA AC 3′ and 5′ GTT GGT TCT GGG CTG GGC GCT GCG CCG AC 3′, respectively), which deleted bp 911 to 925 of pSVICP0K, corresponding to aa 289 to 293 of the EICP0 protein. For the preparation of the ΔSRICH mutant, the mutagenic primers used were SRICH#1 and SRICH#2, which contained the sequences 5′ GAG GGG TAG AAT ACA TTG ACG AGG AAG AAA CAG ACA GC 3′ and 5′ GCT GTC TGT TTC TTC CTC GTC AAT GTA TTC TAC CCC TC 3′, respectively. Amplification of pSVICP0K with these primers would delete bp 674 to 694 of pSVICP0K, corresponding to aa 210 to 217 of the EICP0 protein. To prepare ΔRING19-30, the mutagenic primers used were RING#3 and RING#4, which contained the sequences 5′ CTG GAG GAC CCC AGC AAC TAC GTG TGT ATT ACG CGC TGG ATA 3′ and 5′ TAT CCA GCG CGT AAT ACA CAC GTA GTT GCT GGG GTC CTC CAG 3′, respectively. Amplification of pSVICP0K with these primers would delete the base pairs of pSVICP0K corresponding to aa 19 to 30 of the EICP0 protein. To generate ΔRING8-46, in which EICP0 aa 8 to 46 were deleted, PCR primers RING#1 and RING#2 were used; they contained the sequences 5′ ATG GCA ACT GTT GCA GAG CGA AAA GTG CCG GTC GAA TCT GTG 3′ and 5′ CAC AGA TTC GAC CGG CAC TTT TCG CTC TGC AAC AGT TGC CAT 3′, respectively. The DIEBS mutant plasmid is devoid of a degenerate IE binding site (ATCGc) located at bp −12 to −8 relative to the EICP0 translation initiation site. This plasmid was generated with the mutagenic primers IEBS#1 and IEBS#2, which contain the sequences 5′ TCA TTT GGA AAC TCT TCC AAG CCA CCA TGG CAA CTG TTG C 3′ and 5′ GCA ACA GTT GCC ATG GTG GCT TGG AAG AGT TTC CAA ATG A 3′, respectively.
Because this PCR-based mutagenesis protocol depended on the amplification of the entire plasmid, larger regions of the EICP0 gene corresponding to the deletions were reinserted back into the parental pSVICP0K plasmid to ensure that the only mutagenized region was the desired one. To generate the ΔNLS clone that was used in subsequent transient transfection assays, the original ΔNLS clone (described above) was digested with NgoAIV and BsmBI, and the 252-bp fragment was cloned into NgoAIV- and BsmBI-digested pSVICP0K. To generate the ΔACID, DIEBS, ΔRING19-30, and ΔRING8-46 clones later used in transient transfection assays, the original mutagenized clones (described above) were digested with EcoRI and SstII, and the 649-bp fragment from each mutant was cloned into EcoRI- and SstII-digested pSVICP0K. To generate the ΔSRICH clone for use in transient transfection assays, the original mutagenized ΔSRICH clone (described above) was digested with SstII and NgoAIV, and the 225-bp fragment was cloned into SstII- and NgoAIV-digested pSVICP0K. The authenticity of these newly generated deletion mutant clones was confirmed by DNA sequence analysis of the cloned fragments.
Transfection procedure and CAT assays.
Transient transfections were performed by combining the appropriate amounts of effector and reporter constructs in 500 μl of Eagle's minimal essential medium (EMEM) and allowing this mixture to incubate at room temperature. In a separate tube, 22 μl of Lipofectin reagent (Life Technologies) was added to 1 ml of EMEM and allowed to incubate at room temperature for 45 min for liposome formation. Following these incubations, the plasmid DNA and Lipofectin were mixed, gently swirled, and allowed to incubate for an additional 15 min at room temperature. Finally, 1.5 ml of this mixture was added to each 60-mm dish of approximately 4 × 106 L-M cells for a 5-h incubation period at 37°C under 5% CO2. The cells were harvested at 62 h posttransfection, and CAT assays were performed as described previously (2). Each CAT assay was independently repeated at least three times, and within individual experiments each sample was assayed in triplicate. Data were analyzed for statistical significance by the Student t test.
Infection, metabolic labeling, and immunoprecipitations.
To label the infected cell proteins with [32P]orthophosphate, approximately 1.5 × 107 L-M cells were washed twice, placed in phosphate-free EMEM, and infected with EHV-1 KyA at an MOI of 20 for 1.5 h. At 1.5 h postinfection, the cells were again washed and placed in phosphate-free EMEM, and [32P]orthophosphate was added at a concentration of 100 μCi/ml. Cells were harvested at 4, 5, 6, 7, and 8 h postinfection. Cells were lysed in 1 ml of radioimmunoprecipitation (RIPA) buffer (150 mM NaCl, 50 mM Tris-HCl [pH 8.0], 0.1% sodium dodecyl sulfate [SDS], 0.5% deoxycholate, 1.0% Nonidet P-40) containing protease inhibitors (aprotinin [50 μg/ml], leupeptin [50 μg/ml], and phenylmethylsulfonyl fluoride [300 μg/ml]), passed through a 21-gauge needle three times, and centrifuged, and the cell lysates were transferred to a fresh tube. Equivalent volumes of sample per time point were used for the immunoprecipitation reactions. The samples were precleared by the addition of a 1:20 dilution of preimmune rabbit serum for 1 h at 4°C with rocking. Thirty-five microliters of protein A-agarose beads (Sigma Chemical Company, St. Louis, Mo.) was added to the protein extracts, which were incubated for 1 h at 4°C with rocking. The beads were pelleted by centrifugation, and the precleared extract was placed in a fresh microcentrifuge tube. These metabolically labeled cell extracts were then subjected to immunoprecipitation at 4°C overnight using 5 μl of TrpE-ICP0 antiserum (2). Thirty-five microliters of protein A-Sepharose beads was added, and the mixture was incubated overnight with rocking. The beads were pelleted by centrifugation and washed three times in RIPA buffer, and the proteins were eluted in Laemmli sample buffer (2% SDS, 10% glycerol, 5% 2-mercaptoethanol, 0.1% bromophenol blue, 62.5 mM Tris-HCl [pH 6.8]). Samples were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE), and the proteins were visualized by autoradiography (3, 4).
RESULTS
The IE protein inhibits the ability of the EICP0 protein to activate EHV-1 promoters.
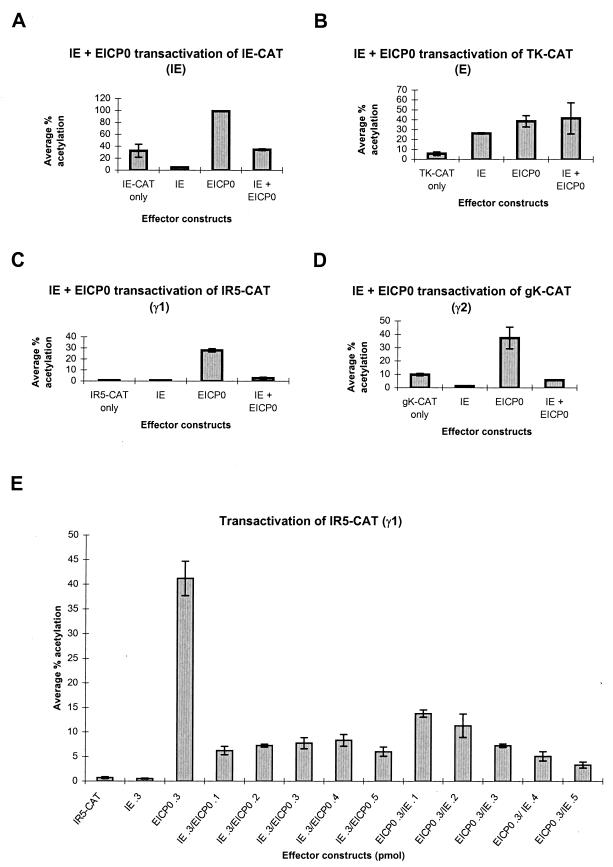
Both the IE and EICP0 proteins are potent trans-activators of EHV-1 promoters (2, 40). To assess whether these proteins function synergistically, transient cotransfection analyses were performed with EHV-1 promoters representing each kinetic class. As observed previously (Fig. 1A), the EICP0 protein is an activator of the EHV-1 IE promoter and increased its expression approximately 3.1-fold above basal levels; the IE protein down-regulates its own promoter, resulting in a 7-fold decrease from basal levels (2, 40). The presence of both the EICP0 and IE proteins resulted in a level of trans-activation approximately equal to basal levels, indicating that either the IE protein interferes with EICP0 function or EICP0 cannot completely overcome the ability of the IE protein to autoregulate its own promoter. Both the IE and EICP0 proteins independently trans-activated the EHV-1 early thymidine kinase (TK) promoter and increased expression from this promoter approximately 4.7- and 6.9-fold, respectively (Fig. 1B). However, when used together, they did not function synergistically.
FIG. 1.
trans-Activation of representative EHV-1 IE, early, γ1 late, and γ2 late promoters linked to the CAT reporter gene by effector constructs expressing the IE and/or EICP0 proteins. (A through D) L-M cells were transfected with each promoter-CAT reporter construct and 0.3 pmol of either pSVIE and/or pSVICP0K. Transfected cells were harvested, and CAT assays were performed as described previously (2). Each transfection was performed at least in triplicate, and the experiment was carried out independently at least three times. Error bars, standard deviations. Shown is trans-activation of the EHV-1 IE reporter construct (pIE-CAT; 1.4 pmol) (A), the EHV-1 early (E) TK reporter construct (pTK2-CAT; 1.4 pmol) (B), the EHV-1 γ1 late IR5 promoter (pIR5-CAT; 1.4 pmol) (C), and the EHV-1 γ2 late gK promoter (pgK-CAT; 2.0 pmol) (D). (E) Dose-dependent trans-activation of the γ1 IR5 promoter (pIR5-CAT; 1.4 pmol). The amount of each effector plasmid is given along the x axis.
The EICP0 protein activated expression of the γ1 late IR5 promoter approximately 40-fold (Fig. 1C), and, as previously reported, the IE protein did not activate this promoter (40). The IE protein significantly retarded the ability of the EICP0 protein to activate the IR5 γ1 late promoter, since CAT expression in the presence of both proteins was approximately 3.9-fold above basal levels. A similar effect was observed with the glycoprotein K (gK) γ2 late promoter (Fig. 1D). Interestingly, the IE protein also down-regulated the gK promoter, approximately 8-fold. Recent studies revealed that the IE protein binds near the transcription initiation site of the gK promoter, thereby repressing its transcription (25).
A titration experiment using the IR5 γ1 late promoter was performed to determine if the antagonism between EICP0 and IE proteins is dose dependent. As shown in Fig. 1E, the IE protein alone did not activate expression from this promoter, whereas 0.3 pmol of the EICP0 plasmid increased expression from this late promoter 59-fold. When any combination of the IE and EICP0 effector constructs was tested, the levels of trans-activation never approached that obtained when the EICP0 effector construct was transfected alone. For example, when 0.3 pmol of the IE effector construct was cotransfected with 0.1 to 0.5 pmol of the EICP0 plasmid, increased expression from the IR5 promoter ranged from 9- to 12-fold. When the amount of the EICP0 plasmid transfected was held constant at 0.3 pmol, the level of activation varied from 20- to 4.7-fold depending on the amount of the IE plasmid cotransfected. At 0.1 pmol of IE plasmid, the level of activation was 20-fold, the highest observed. When 0.5 pmol of the pSVIE plasmid was cotransfected with the EICP0 plasmid, the level of activation decreased to 4.7-fold. Therefore, increasing the amount of the IE plasmid transfected reduced the ability of the EICP0 protein to activate the IR5 γ1 late promoter. These findings indicate that the IE protein inhibits the function of the EICP0 protein at all concentrations of EICP0 plasmid tested and that the degree of inhibition is dose dependent.
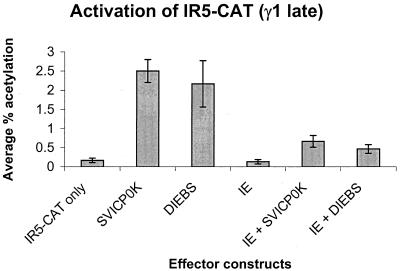
A degenerate IE binding site (ATCGc) (S. K. Kim and D. J. O'Callaghan, unpublished data) is located in the EICP0 untranslated region (bp −12 to −8 relative to the EICP0 translation initiation site). To determine whether the ability of the IE protein to bind to this site could account for the IE–EICP0 antagonism, an EICP0 expression vector (DIEBS) that lacked this IE binding site was generated. Both the wild-type EICP0 expression vector (SVICP0K) and DIEBS were able to independently activate the γ1 late IR5 promoter, 15- and 13-fold, respectively (Fig. 2), whereas the IE protein did not activate this promoter. The IE protein, however, was able to inhibit the ability of DIEBS to activate the IR5 promoter, and the level of inhibition was similar to that observed with the wild-type SVICP0K construct (fourfold activation for SVICP0K; threefold activation for DIEBS). These data suggest that a mechanism other than the binding of the IE protein to this site in the EICP0 promoter accounts for the antagonistic relationship of these two regulatory proteins.
FIG. 2.
trans-Activation of the γ1 late IR5 promoter by effector constructs expressing the IE and/or EICP0 proteins. L-M cells were transfected with 1.4 pmol of the pIR5-CAT construct and 0.3 pmol of either SVICP0K, SVIE, DIEBS, or combinations of these three effector constructs. Transfected cells were harvested, and CAT assays were performed as described previously (2). Each transfection was performed in triplicate, and the experiment was carried out independently two times. Error bars, standard deviations.
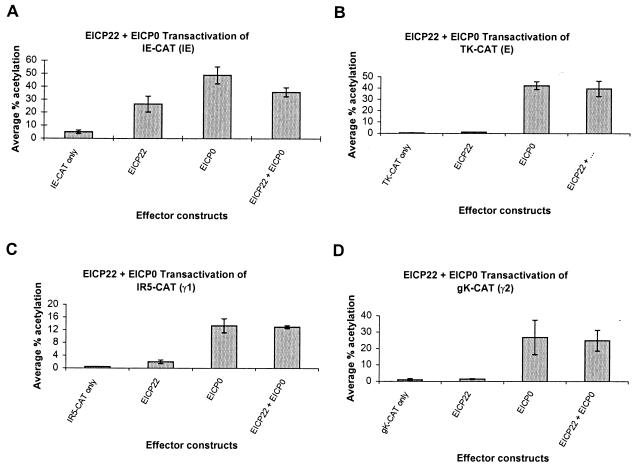
The EICP22 protein does not significantly enhance the ability of the EICP0 protein to activate EHV-1 promoters.
To determine whether the EICP0 and EICP22 proteins function synergistically, a series of transient cotransfection analyses were performed with the IE, early (TK), γ1 late (IR5), and γ2 late (gK) promoters (Fig. 3). As demonstrated in Fig. 3, the EICP22 protein was able to activate the IE, TK, IR5, and gK promoters only to minimal levels. As reported previously (2), the EICP0 protein was a very powerful activator of EHV-1 promoters, resulting in 9.8-fold activation of the IE promoter, 71-fold activation of the TK promoter, 26-fold activation of the γ1 late IR5 promoter, and 26-fold activation of the γ2 late gK promoter (Fig. 3). The presence of both the EICP22 and EICP0 proteins neither enhanced nor reduced expression of the IE, TK, IR5, or gK promoters beyond the levels obtained with the EICP0 protein alone, indicating that these two regulatory proteins do not function synergistically or antagonistically on these promoters (Fig. 3).
FIG. 3.
trans-Activation of representative EHV-1 IE, early, γ1 late, and γ2 late promoters linked to the CAT reporter gene by effector constructs expressing the EICP22 and/or EICP0 proteins. L-M cells were transfected with each promoter-CAT reporter construct and 0.3 pmol of either pCDR4 (EICP22 expression construct) and/or pSVICP0K. Transfected cells were harvested, and CAT assays were performed as described previously (2). Each transfection was performed in triplicate, and the experiment was carried out independently three times. Error bars, standard deviations. (A) trans-Activation of the EHV-1 IE reporter construct (pIE-CAT; 1.4 pmol); (B) trans-activation of the EHV-1 early (E) TK reporter construct (TK2-CAT; 1.4 pmol); (C) trans-activation of the EHV-1 γ1 late IR5 promoter (pIR5-CAT; 1.4 pmol); (D) trans-activation of the EHV-1 γ2 late gK promoter (pgK-CAT; 2.0 pmol).
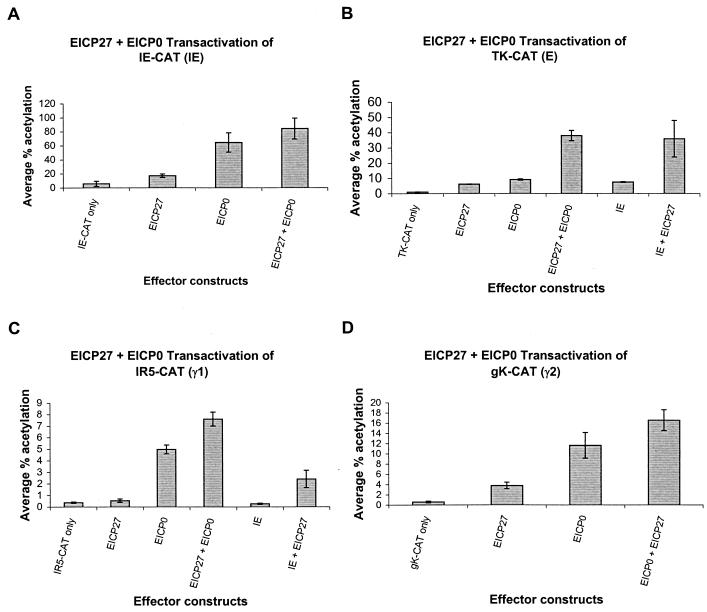
The EHV-1 EICP0 and EICP27 proteins function synergistically to activate early and γ1 late promoters.
To address whether the EICP27 and EICP0 proteins function together, cotransfection experiments were performed with reporter CAT constructs representing all classes of EHV-1 promoters. In these experiments, the EICP0 protein independently activated the IE, TK, IR5, and gK promoters 11-, 9-, 13.5-, and 19-fold, respectively, whereas the EICP27 protein activated these promoters only minimally (Fig. 4). No synergy or antagonism between the EICP0 and EICP27 proteins was observed in the case of either the IE or the gK promoter (Fig. 4A and D). However, the EICP0 and the EICP27 proteins functioned synergistically in the case of both the TK and IR5 promoters (Fig. 4B and C). As reported previously (42, 47), the IE and the EICP27 proteins function synergistically to enhance expression of both the TK and IR5 promoters (Fig. 4B and C). Results from these analyses indicate that the EICP27 protein is capable of enhancing the trans-activation function of the IE protein as well as that of the EICP0 protein with regard to expression from early and γ1 late promoters.
FIG. 4.
trans-Activation of representative EHV-1 IE, early, γ1 late, and γ2 late promoters linked to the CAT reporter gene by effector constructs expressing the IE, EICP0, and/or EICP27 proteins. L-M cells were transfected with each promoter-CAT reporter construct and 0.3 pmol of pSVIE, pSVICP0K, and/or pSVUL3 (EICP27 effector construct). Transfected cells were harvested, and CAT assays were performed as described previously (2). Each transfection was performed in triplicate, and the experiment was carried out independently three times. Error bars, standard deviations. (A) trans-Activation of the EHV-1 IE reporter construct (pIE-CAT; 1.4 pmol); (B) trans-activation of the EHV-1 early TK2 reporter construct (pTK2-CAT; 1.4 pmol); (C) trans-activation of the EHV-1 γ1 late IR5 promoter (pIR5-CAT; 1.4 pmol); (D) trans-activation of the EHV-1 γ2 late gK promoter (pgK-CAT; 2.0 pmol).
Construction and characterization of EICP0 nonsense and deletion mutant plasmids.
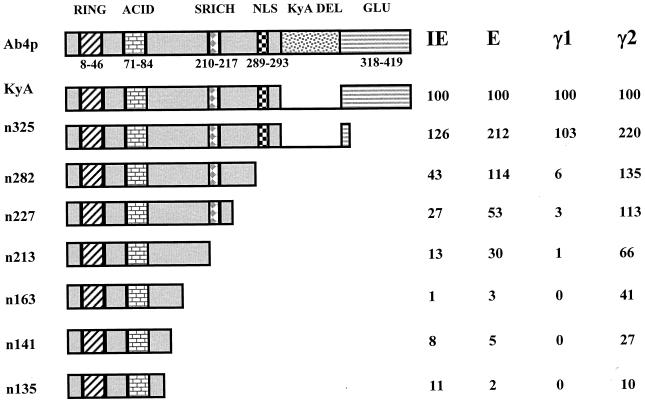
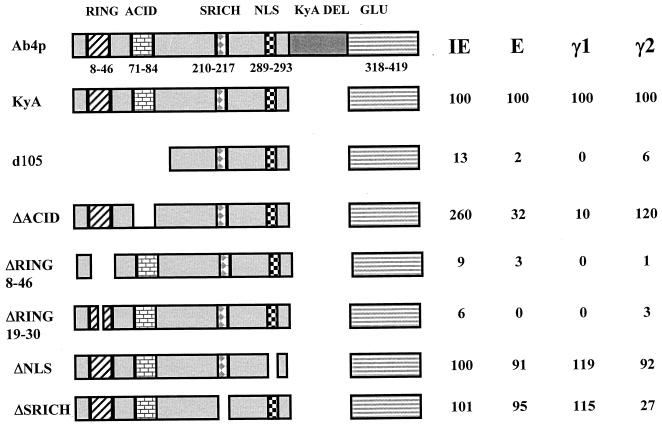
The EICP0 protein can be divided into five major regions: (i) a RING finger motif (RING; aa 8 to 46), (ii) an acidic region (ACID; aa 71 to 84), (iii) a serine-rich region (SRICH; aa 210 to 217), (iv) a putative nuclear localization signal (NLS; aa 289 to 293), and (v) a glutamine-rich region (GLU; aa 318 to 419) (Fig. 5 and 6). Our previous study indicated that the EICP0 proteins of the KyA strain and the Ab4p strain differ in size due to a 113-aa in-frame deletion in the latter (2) (Fig. 5 and 6). Both of these EICP0 proteins activated EHV-1 promoter-CAT reporter constructs identically, indicating that this 113-aa segment is not essential for the trans-activation function (reference 2 and data not shown). By using the KyA EICP0 protein, a panel of 13 EICP0 nonsense and deletion mutant plasmids that targeted these domains (RING, ACID, SRICH, NLS, and GLU) was constructed (Fig. 5 and 6). The results of restriction enzyme analyses of these mutants indicated that the desired mutation was obtained (data not shown). In addition, the authenticity of each deletion mutant clone was verified by DNA sequence analyses (data not shown).
FIG. 5.
Relative abilities of EICP0 nonsense mutants to trans-activate EHV-1 IE, early, γ1 late, and γ2 late promoters. The construction of each mutant was described in Materials and Methods. L-M cells were transfected with each promoter-CAT reporter construct (1.4 pmol of pIE-CAT, pTK2-CAT, or pIR5-CAT; 2.0 pmol of pgK-CAT) and 0.3 pmol of each effector construct. Transfected cells were harvested, and CAT assays were performed as described previously (2). Each transfection was performed in triplicate in individual experiments. Each experiment was carried out independently at least three times. The relative activity of each nonsense mutant compared to wild-type EICP0 on each class of promoters (IE, early, γ1 late, and γ2 late) is shown beside each mutant and is expressed as a percentage of the wild-type value, which was set at 100%. The Ab4p and KyA EICP0 proteins are from different EHV-1 strains and differ only by the presence or absence (Kya DEL) of a 113-aa region. Both forms of this protein function identically in CAT assays.
FIG. 6.
Relative abilities of EICP0 deletion mutants d105, ΔACID, ΔNLS, ΔSRICH, ΔRING19-39, and ΔRING8-46 to trans-activate representative EHV-1 promoters. The construction of each mutant was described in Materials and Methods. L-M cells were transfected with each promoter-CAT reporter construct (1.4 pmol of pIE-CAT, pTK2-CAT, or pIR5-CAT; 2.0 pmol of pgK-CAT) and 0.3 pmol of each effector construct. Transfected cells were harvested, and CAT assays were performed as described previously (2). Each transfection was performed in triplicate in individual experiments. Each experiment was carried out independently at least three times. Values obtained for the mutants are expressed as percentages of the wild-type value, which was set at 100%.
To confirm that each mutant clone expressed a protein of the expected size, each mutant plasmid was in vitro transcribed and translated, and the resulting proteins were subjected to either SDS-PAGE analyses followed by autoradiography or Western blot analyses with EICP0 antiserum (data not shown). The mutant proteins expressed from every construct were of the expected size with the exception of the d105 protein, which lacks both the RING finger and acidic regions and migrated more rapidly than expected. Such aberrant migration by a RING-less herpes simplex virus type 1 (HSV-1) ICP0 protein was also observed by Everett (9). In addition, every EICP0 mutant protein was shown to be expressed in mouse L-M cells and to be detectable by Western blot analyses following transfection with plasmids that express each EICP0 mutant (data not shown).
The RING finger domain is critical for activation of EHV-1 promoters by the EICP0 protein.
The trans-activating abilities of the EICP0 mutants were examined in transfection assays using the reporter plasmids IE-CAT, TK-CAT (early), IR5-CAT (γ1 late), and gK-CAT (γ2 late). With the exception of the IR5 (γ1 late) promoter, the nonsense mutants exhibited a progressive loss of function as more of the carboxy terminus of the protein was removed (Fig. 5). Activation of the IR5 promoter was severely inhibited by the loss of aa 283 to 419 (the n282 mutant). Interestingly, this mutant protein (n282) was able to activate the TK and gK promoters to levels that were at or slightly above those with the wild-type protein. Amino acids 326 to 419, which contain the glutamine-rich region, were dispensable for EICP0 function, since the n325 mutant exhibited levels of trans-activation equal to or, in some instances, greater than wild-type levels (Fig. 5).
The RING finger (aa 8 to 46) and acidic region (aa 71 to 84) were necessary but not sufficient for activation of the four promoters tested. The d105 construct, which lacks the first 105 aa, including the RING finger and acidic regions, was impaired in its ability to activate transcription from all promoters (Fig. 6). Conversely, constructs that expressed only the RING finger and the acidic regions (such as constructs n135 and n141) were not able to fully activate any promoter tested (Fig. 5). Loss of only the acidic region impaired the ability of the mutant EICP0 protein to activate the early and γ1 late promoters (Fig. 6), suggesting that the RING finger is the region most responsible for trans-activation activity. Interestingly, deletion of the acidic region did not adversely affect the ability of the EICP0 protein to activate the IE and γ2 promoters; in fact, the levels of activation were near wild-type levels or enhanced (120 to 260% of wild-type activity).
To assess the role of the RING finger directly, mutants that lacked either the entire RING finger motif (ΔRING8-46) or one-half of the RING finger motif (ΔRING19-30) were generated. In transient transfection assays, both of these RING finger mutants were severely impaired in the ability to activate all promoters (Fig. 6). These data indicate that the RING finger motif is critical for EICP0 function.
Deletion of a putative NLS located at aa 289 to 293 did not impair the ability of the mutant protein to trans-activate any promoter tested, suggesting that either this region is not the nuclear localization domain or the 50-kDa EICP0 protein is of a size sufficient to enter the nucleus without an NLS (Fig. 6).
The EICP0 protein is phosphorylated during EHV-1 infection.
The EICP0 protein may be phosphorylated at the serine-rich region (aa 210 to 217). Deletion of this region (ΔSRICH) did not affect the ability of the EICP0 protein to fully activate the IE, early, and γ1 late promoters but reduced the ability of this mutant protein to activate the γ2 late promoter to only 27% of wild-type activity. This finding suggests that the degree of phosphorylation of the EICP0 protein in infection may play an important role in its ability to activate the γ2 late class of promoters. To determine whether the EICP0 protein was phosphorylated, L-M cells were infected with the EHV-1 KyA strain in the presence of [32P]orthophosphate. Lysates of mock-infected and infected cells were prepared at 4, 5, 6, 7, and 8 h postinfection and subjected to immunoprecipitation with EICP0 antiserum. The EICP0 protein was phosphorylated by 4 h postinfection (Fig. 7, lane 2), and only one of the three forms (2) of the EICP0 protein was phosphorylated. To date, the phosphorylated form of the EICP0 protein has been detected only in infected cells (Fig. 7), since [32P]orthophosphate-labeled EICP0 protein was not detected in either EICP0-expressing cell lines or cells transiently transfected with the SVICP0K construct (data not shown). These data suggest that a viral protein may contribute to the modification of the EICP0 protein and that the level of phosphorylation of the EICP0 protein may be important in its ability to activate the γ2 late promoters.
FIG. 7.
The EICP0 protein is phosphorylated during infection. Mock-infected L-M cells (lane 1) and infected labeled L-M cells harvested at 4, 5, 6, 7, and 8 h postinfection (lanes 2 to 6, respectively) were immunoprecipitated with anti-EICP0 antiserum (2). Immunoprecipitations were analyzed by SDS-PAGE gel analyses followed by autoradiography.
DISCUSSION
In this study, evidence is presented that the EICP0 and IE proteins, the two most powerful EHV-1 trans-activators, do not function synergistically. Instead, the IE protein, an essential EHV-1 regulatory protein, interferes with the ability of the EICP0 protein to activate EHV-1 promoters. The antagonism exhibited by the IE and EICP0 proteins differs from the relationship of their counterparts in HSV-1 and varicella-zoster virus (VZV). One well-characterized feature of the HSV-1 ICP0 and ICP4 proteins (ICP4 is the HSV-1 homolog of the EHV-1 IE protein) is their ability to function synergistically (5, 10–13, 36). The VZV EICP0 protein equivalent, the ORF61 protein, can down-regulate the trans-activation functions of the VZV IE (ORF62) and EICP27 (ORF4) proteins (34).
A possible mechanism for the antagonism observed between the IE and EICP0 proteins is that the IE protein, a sequence-specific DNA binding protein, may bind to an IE consensus binding site in the pSVICP0K expression vector and thereby inhibit its transcription. Although such a binding site is present in the pSVICP0K plasmid, gel shift analyses indicate that the IE protein does not bind to this site (Kim and O'Callaghan, unpublished data). The finding that the trans-activation ability of the DIEBS EICP0 mutant is inhibited by the IE protein confirms that binding of the IE protein to the EICP0 promoter does not account for the antagonism observed between the EICP0 and IE proteins. A possible explanation for this antagonism involves a physical interaction between the two proteins. Although there is no evidence that these two EHV-1 proteins physically interact, it is documented that HSV-1 ICP0 and ICP4 physically associate and that the sequences responsible for synergy and physical interactions map to the carboxy terminus of the HSV-1 ICP0 protein (45). The n325 mutant, which lacks the carboxy terminus of the EICP0 protein, is a better trans-activator than the wild-type EICP0 protein, tempting speculation that the IE protein interacts with this region of the EICP0 protein and inhibits its function. Another possibility to explain this antagonism is that both the IE and EICP0 proteins compete for cellular transcription factors. The HSV-1 ICP4 protein interacts with TFIIB and TFIID (39), and recent data from our laboratory reveal that the IE protein interacts specifically with TFIIB (unpublished data). The HSV-1 ICP0 protein interacts with three cellular proteins, but none of these proteins is directly involved in transcription (14, 23, 24, 31, 32). However, to date there is no evidence that the EICP0 protein interacts with any cellular protein.
Previous studies from our laboratory indicated that the EICP27 protein is an early accessory regulatory protein capable of (i) independently activating the EHV-1 IE promoter and minimally activating EHV-1 early promoters, (ii) functioning synergistically with the IE protein to up-regulate expression from early and γ1 late promoters, and (iii) functioning synergistically with the EICP22 protein to trans-activate the IE promoter (22, 47). Here we report that the EICP27 protein can also function synergistically with the EICP0 protein to activate early and γ1 late promoters. The precise mechanism of EICP27 function is unknown at present, but it is interesting that EICP27 synergy with the IE protein and its synergy with the EICP0 protein involve a common set of EHV-1 promoters.
It should be noted that a trivial explanation for the findings concerning the effects of the IE, EICP22, or EICP27 protein on the transactivation activity of the EICP0 protein would be that these regulatory proteins affect the level of the EICP0 protein. However, Western blot analyses of cells cotransfected with plasmids that express these other EHV-1 regulatory proteins showed that this was not the case and that the level of the EICP0 protein was not affected by expression of these other proteins (data not shown).
Mutational analyses of the EICP0 protein indicated that several regions of this protein are important for its ability to trans-activate; the most important region maps within the amino-terminal 105 aa. This region includes a RING finger motif that binds zinc, is conserved among all ICP0 homologs, and is also found in a plethora of cellular proteins (reviewed in references 15 and 38). Mutational analyses of the VZV ORF61 and HSV-1 ICP0 proteins also indicated that the RING finger of each protein was necessary for its function (8, 33). Recently, Lium and Silverstein (30) showed that alteration of any of the cysteine or histidine residues in the C3HC4 consensus RING finger domain abolished the trans-activation ability of the HSV-1 ICP0 protein. Evidence presented here shows that deletion of the entire RING finger or a portion of the RING finger abrogated the ability of these mutant EICP0 proteins to activate any EHV-1 promoter tested.
Stevenson et al. (44) mapped the NLS of the ORF61 protein of VZV and predicted that the NLS of the EICP0 protein of EHV-1 is located at aa 289 to 293 (RLRRR). Our results indicate that deletion of these 5 aa has no effect on the activation of any of the EHV-1 promoters tested. It should be noted that the ORF61 protein comprises 467 aa and may require an NLS for entry into the nucleus. In contrast, the EICP0 protein is smaller (419 aa) and may not require a specific domain to mediate nuclear entry.
Interestingly, a serine-rich tract located at aa 210 to 217 is not necessary for EICP0 activation of IE, early, or γ1 late promoters but is required for full activation of the γ2 late gK promoter. This finding suggests that the phosphorylation state of the EICP0 protein may be important for the activation of γ2 late promoters. In this regard the EICP0 protein exists as three distinct species, only one of which is the phosphorylated form (2). Phosphorylation of EICP0 is not detected in cells transiently transfected with EICP0 expression constructs or in cell lines that constitutively express the EICP0 protein. Thus, it is likely that modification of this regulatory protein is mediated by an EHV-1 gene product.
The glutamine-rich region located between aa 318 and 419 is dispensable for EICP0 function, since the n325 mutant, which lacks a major portion of this region, still activated all EHV-1 promoters tested at wild-type or higher levels. The carboxy-terminal region of the HSV-1 ICP0 protein was not required for ICP0 to independently transactivate HSV-1 promoters but was required for synergy and physical association with the HSV-1 ICP4 protein (reviewed in reference 45). Studies are currently in progress to determine whether the IE and EICP0 proteins physically interact and, if so, whether the carboxy-terminal portion of the EICP0 protein is involved in mediating this physical association. Hopefully, these and additional experiments will contribute more insight into the role of EICP0 in the EHV-1 replication cycle. Our present understanding of EHV-1 gene regulation indicates that the EICP0 protein plays a very important role in the activation of late genes. However, the mechanism by which EICP0 can independently trans-activate EHV-1 promoters and the mechanism by which the IE protein interferes with EICP0 function are not understood and are important questions to be addressed.
ACKNOWLEDGMENTS
We thank Suzanne Zavecz for excellent technical assistance. We thank P. Smith, A. Frampton, W. Derbigny, and R. Albrecht for critical reading of the manuscript.
This investigation was supported by research grants from the National Institutes of Health (AI-22001).
REFERENCES
- 1.Allen G P, Bryans J T. Molecular epizootiology, pathogenesis, and prophylaxis of equine herpesvirus infections. Prog Vet Microbiol Immunol. 1986;2:78–144. [PubMed] [Google Scholar]
- 2.Bowles D E, Holden V R, Zhao Y, O'Callaghan D J. The ICP0 protein of equine herpesvirus 1 is an early protein that independently transactivates expression of all classes of viral promoters. J Virol. 1997;71:4904–4914. doi: 10.1128/jvi.71.7.4904-4914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caughman G B, Robertson A T, Gray W L, Sullivan D C, O'Callaghan D J. Characterization of equine herpesvirus type 1 immediate early proteins. Virology. 1988;163:563–571. doi: 10.1016/0042-6822(88)90297-8. [DOI] [PubMed] [Google Scholar]
- 4.Caughman G B, Staczek J, O'Callaghan D J. Equine herpesvirus type 1 infected cell polypeptides: evidence for immediate early/early/late regulation of viral gene expression. Virology. 1985;145:49–61. doi: 10.1016/0042-6822(85)90200-4. [DOI] [PubMed] [Google Scholar]
- 5.Chen J, Zhu X, Silverstein S. Mutational analysis of the sequence encoding ICP0 from herpes simplex virus type 1. Virology. 1991;180:207–220. doi: 10.1016/0042-6822(91)90025-7. [DOI] [PubMed] [Google Scholar]
- 6.DeLuca N A, Schaffer P A. Activities of herpes simplex virus type 1 (HSV-1) ICP4 genes specifying nonsense peptides. Nucleic Acids Res. 1987;15:4491–4511. doi: 10.1093/nar/15.11.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eliott G, O'Hare P. Equine herpesvirus 1 gene 12, the functional homologue of herpes simplex virus VP16, transactivates via octamer sequences in the equine herpesvirus IE gene promoter. Virology. 1995;213:258–262. doi: 10.1006/viro.1995.1568. [DOI] [PubMed] [Google Scholar]
- 8.Everett R, O'Hare P, O'Rourke D, Barlow P, Orr A. Point mutations in the herpes simplex virus type 1 Vmw110 RING finger helix affect activation of gene expression, viral growth, and interaction with PML-containing nuclear structures. J Virol. 1995;69:7339–7344. doi: 10.1128/jvi.69.11.7339-7344.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Everett R D. Construction and characterization of herpes simplex virus type 1 mutants with defined lesions in immediate early gene 1. J Gen Virol. 1989;70:1185–1202. doi: 10.1099/0022-1317-70-5-1185. [DOI] [PubMed] [Google Scholar]
- 10.Everett R D. A detailed mutational analysis of Vmw110, a trans-acting transcriptional activator encoded by herpes simplex virus type 1. EMBO J. 1987;6:2069–2076. doi: 10.1002/j.1460-2075.1987.tb02472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Everett R D. Functional and genetic analysis of the role of Vmw110 in herpes simplex virus replication. Boca Raton, Fla: CRC Press; 1991. [Google Scholar]
- 12.Everett R D. The products of herpes simplex virus type 1 (HSV-1) immediate early genes 1, 2, and 3 can activate HSV-1 gene expression in trans. J Gen Virol. 1986;67:2507–2513. doi: 10.1099/0022-1317-67-11-2507. [DOI] [PubMed] [Google Scholar]
- 13.Everett R D. Trans activation of transcription by herpes virus products: requirement for two HSV-1 immediate-early polypeptides for maximum activity. EMBO J. 1984;3:3135–3141. doi: 10.1002/j.1460-2075.1984.tb02270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Everett R D, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 1997;16:566–577. doi: 10.1093/emboj/16.3.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freemont P S. The RING finger. A novel protein sequence motif related to the Zinc finger. Ann N Y Acad Sci. 1993;684:174–192. doi: 10.1111/j.1749-6632.1993.tb32280.x. [DOI] [PubMed] [Google Scholar]
- 16.Garko-Buczynski K A, Smith R H, Kim S K, O'Callaghan D J. Complementation of a replication-defective mutant of equine herpesvirus type 1 by a cell line expressing the immediate-early protein. Virology. 1998;248:83–94. doi: 10.1006/viro.1998.9247. [DOI] [PubMed] [Google Scholar]
- 17.Gray W L, Baumann R P, Robertson A T, Caughman G B, O'Callaghan D J, Staczek J. Regulation of equine herpesvirus type 1 gene expression: characterization of immediate early, early, and late transcription. Virology. 1987;158:79–87. doi: 10.1016/0042-6822(87)90240-6. [DOI] [PubMed] [Google Scholar]
- 18.Gray W L, Baumann R P, Robertson A T, O'Callaghan D J, Staczek J. Characterization and mapping of equine herpesvirus type 1 immediate early, early, and late transcripts. Virus Res. 1987;8:233–244. doi: 10.1016/0168-1702(87)90018-9. [DOI] [PubMed] [Google Scholar]
- 19.Grundy F J, Baumann R P, O'Callaghan D J. DNA sequence and comparative analyses of the equine herpesvirus 1 immediate early gene. Virology. 1989;172:223–236. doi: 10.1016/0042-6822(89)90124-4. [DOI] [PubMed] [Google Scholar]
- 20.Holden V R, Caughman G B, Zhao Y, Harty R N, O'Callaghan D J. Identification and characterization of the ICP22 protein of equine herpesvirus 1. J Virol. 1994;68:4329–4340. doi: 10.1128/jvi.68.7.4329-4340.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holden V R, Yalamanchili R R, Harty R N, O'Callaghan D J. ICP22 homolog of equine herpesvirus 1: expression from early and late promoters. J Virol. 1992;66:664–673. doi: 10.1128/jvi.66.2.664-673.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holden V R, Zhao Y, Thompson Y, Caughman G B, Smith R H, O'Callaghan D J. Characterization of the regulatory function of the ICP22 protein of equine herpesvirus type 1. Virology. 1995;210:273–282. doi: 10.1006/viro.1995.1344. [DOI] [PubMed] [Google Scholar]
- 23.Kawaguchi Y, Bruni R, Roizman B. Interaction of herpes simplex virus 1 α regulatory protein ICP0 with elongation factor 1δ: ICP0 affects translational machinery. J Virol. 1997;71:1019–1024. doi: 10.1128/jvi.71.2.1019-1024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawaguchi Y, Van Sant C, Roizman B. Herpes simplex virus 1 α regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J Virol. 1997;71:7328–7336. doi: 10.1128/jvi.71.10.7328-7336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim S K, Bowles D E, O'Callaghan D J. The γ2 late glycoprotein K promoter of equine herpesvirus 1 is differentially regulated by the IE and EICP0 proteins. Virology. 1999;256:173–179. doi: 10.1006/viro.1999.9608. [DOI] [PubMed] [Google Scholar]
- 26.Kim S K, Holden V R, O'Callaghan D J. The ICP22 protein of equine herpesvirus 1 cooperates with the IE protein to regulate viral gene expression. J Virol. 1997;71:1004–1012. doi: 10.1128/jvi.71.2.1004-1012.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim S K, Smith R H, O'Callaghan D J. Characterization of DNA binding properties of the immediate-early gene product of equine herpesvirus type 1. Virology. 1995;213:46–56. doi: 10.1006/viro.1995.1545. [DOI] [PubMed] [Google Scholar]
- 28.Lewis J B, Thompson Y G, Caughman G B. Transcriptional control of the equine herpesvirus 1 immediate early gene. Virology. 1993;197:788–792. doi: 10.1006/viro.1993.1658. [DOI] [PubMed] [Google Scholar]
- 29.Lewis J B, Thompson Y G, Feng X, Holden V R, O'Callaghan D J, Caughman G B. Structural and antigenic identification of the ORF12 protein (αTIF) of equine herpesvirus 1. Virology. 1997;230:369–375. doi: 10.1006/viro.1997.8477. [DOI] [PubMed] [Google Scholar]
- 30.Lium E K, Silverstein S. Mutational analysis of the herpes simplex virus type 1 ICP0 C3HC4 zinc ring finger reveals a requirement for ICP0 in the expression of the essential α27 gene. J Virol. 1997;71:8602–8614. doi: 10.1128/jvi.71.11.8602-8614.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meredith M, Orr A, Elliott M, Everett R. Separation of sequence requirements for HSV-1 Vmw110 multimerisation and interaction with a 135-kDa cellular protein. Virology. 1995;209:174–187. doi: 10.1006/viro.1995.1241. [DOI] [PubMed] [Google Scholar]
- 32.Meredith M, Orr A, Everett R. Herpes simplex virus type 1 immediate-early protein Vmw110 binds strongly and specifically to a 135-kDa cellular protein. Virology. 1994;200:457–469. doi: 10.1006/viro.1994.1209. [DOI] [PubMed] [Google Scholar]
- 33.Moriuchi H, Moriuchi M, Cohen J I. The RING finger domain of the varicella-zoster virus open reading frame 61 protein is required for its transregulatory functions. Virology. 1994;205:238–246. doi: 10.1006/viro.1994.1639. [DOI] [PubMed] [Google Scholar]
- 34.Nagpal S, Ostrove J M. Characterization of a potent varicella-zoster virus-encoded trans-repressor. J Virol. 1991;65:5289–5296. doi: 10.1128/jvi.65.10.5289-5296.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Callaghan D J, Osterrieder N. Equine herpesviruses. In: Webster R G, Granoff A, editors. Encyclopedia of virology. 2nd ed. London, United Kingdom: Academic Press, Ltd.; 1999. [Google Scholar]
- 36.O'Hare P, Hayward G S. Evidence for a direct role for both the 175,000- and 110,000-molecular-weight immediate-early proteins of herpes simplex virus in the transactivation of delayed-early promoters. J Virol. 1985;53:751–760. doi: 10.1128/jvi.53.3.751-760.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Purewal A S, Allsopp R, Riggio M, Telford E A R, Azam S, Davison A J. Equid herpesviruses 1 and 4 encode functional homologs of the herpes simplex virus type 1 virion transactivator protein, VP16. Virology. 1994;198:385–389. doi: 10.1006/viro.1994.1047. [DOI] [PubMed] [Google Scholar]
- 38.Saurin A J, Borden K L B, Boddy M N, Freemont P S. Does this have a familiar RING? Trends Biochem Sci. 1996;21:208–214. [PubMed] [Google Scholar]
- 39.Smith C A, Bates P, Rivera-Gonzalez R, Gu B, DeLuca N A. ICP4, the major transcriptional regulatory protein of herpes simplex virus type 1, forms a tripartite complex with TATA-binding protein and TFIIB. J Virol. 1993;67:4676–4687. doi: 10.1128/jvi.67.8.4676-4687.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith R H, Caughman G B, O'Callaghan D J. Characterization of the regulatory functions of the equine herpesvirus 1 immediate-early gene product. J Virol. 1992;66:936–945. doi: 10.1128/jvi.66.2.936-945.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith R H, Holden V R, O'Callaghan D J. Nuclear localization and transcriptional activation activities of truncated versions of the immediate-early gene product of equine herpesvirus 1. J Virol. 1995;69:3857–3862. doi: 10.1128/jvi.69.6.3857-3862.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith R H, Zhao Y, O'Callaghan D J. The equine herpesvirus 1 (EHV-1) UL3 gene, an ICP27 homolog, is necessary for full activation of gene expression directed by an EHV-1 late promoter. J Virol. 1993;67:1105–1109. doi: 10.1128/jvi.67.2.1105-1109.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith R H, Zhao Y, O'Callaghan D J. The equine herpesvirus type 1 immediate-early gene product contains an acidic transcriptional activation domain. Virology. 1994;202:760–770. doi: 10.1006/viro.1994.1398. [DOI] [PubMed] [Google Scholar]
- 44.Stevenson D, Coleman K L, Davison A J. Delineation of a sequence required for nuclear localization of the protein encoded by varicella-zoster virus gene 61. J Gen Virol. 1994;75:3229–3233. doi: 10.1099/0022-1317-75-11-3229. [DOI] [PubMed] [Google Scholar]
- 45.Yao F, Schaffer P A. Physical interaction between the herpes simplex virus type 1 immediate-early regulatory proteins ICP0 and ICP4. J Virol. 1994;68:8158–8168. doi: 10.1128/jvi.68.12.8158-8168.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao Y, Holden V R, Harty R N, O'Callaghan D J. Identification and transcriptional analyses of the UL3 and UL4 genes of equine herpesvirus 1, homologs of the ICP27 and glycoprotein K genes of herpes simplex virus. J Virol. 1992;66:5363–5372. doi: 10.1128/jvi.66.9.5363-5372.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao Y, Holden V R, Smith R H, O'Callaghan D J. Regulatory function of the equine herpesvirus 1 ICP27 gene product. J Virol. 1995;69:2786–2793. doi: 10.1128/jvi.69.5.2786-2793.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]