Abstract
Kolmioviridae is a family for negative-sense RNA viruses with circular, viroid-like genomes of about 1.5–1.7 kb that are maintained in mammals, amphibians, birds, fish, insects and reptiles. Deltaviruses, for instance, can cause severe hepatitis in humans. Kolmiovirids encode delta antigen (DAg) and replicate using host-cell DNA-directed RNA polymerase II and ribozymes encoded in their genome and antigenome. They require evolutionary unrelated helper viruses to provide envelopes and incorporate helper virus proteins for infectious particle formation. This is a summary of the International Committee on Taxonomy of Viruses (ICTV) Report on the family Kolmioviridae, which is available at ictv.global/report/kolmioviridae.
Keywords: deltavirus, hepatitis D virus, ICTV Report, Kolmioviridae, taxonomy
Virion
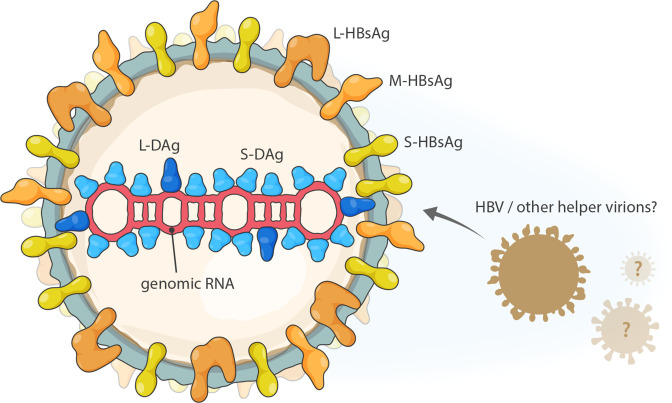
Kolmiovirions are spherical with a lipid envelope containing envelope proteins obtained from an evolutionarily unrelated helper virus (Table 1, Fig. 1). Virions contain a ribonucleoprotein (RNP) complex consisting of distinct isoforms of delta antigen (DAg) that are encoded in and closely associate with deltavirus genomic and antigenomic RNAs [1,2].
Table 1. Characteristics of members of the family Kolmioviridae.
| Example: | hepatitis D virus 1 (AF104263), species Deltavirus italiense, genus Deltavirus |
| Virion | Spherical virions (36–43 nm in diameter) with an outer envelope containing envelope proteins derived from a helper virus and an inner ribonucleoprotein consisting of genomic RNA and a nucleocapsid protein (delta antigen; DAg) that is encoded by the genomic RNA and may occur in two isoforms |
| Genome | Rod-like ribozyme-containing negative-sense, covalently closed, circular RNA (cccRNA) of about 1.5–1.7 kb |
| Replication | RNA-directed RNA synthesis by host-cell DNA-directed RNA polymerase II through a double rolling circle mechanism, and autocatalytic cleavage/ligation via encoded genomic and antigenomic ribozymes and re-cyclization in the host-cell nucleus |
| Translation | mRNA-based translation of DAg (in some cases including an isoform thereof) |
| Host range | Mammals, amphibians, birds, fish, insects and reptiles |
| Taxonomy | Realm Ribozyviria; the family includes >7 genera and >14 species |
Fig. 1. Schematic of hepatitis D virus 1 particles. L-DAg, S-DAg: large and small delta antigen; L-HBsAg, M-HBsAg, S-HBsAg: large, middle and small HBV surface antigen; HBV: hepatitis B virus.
Genome
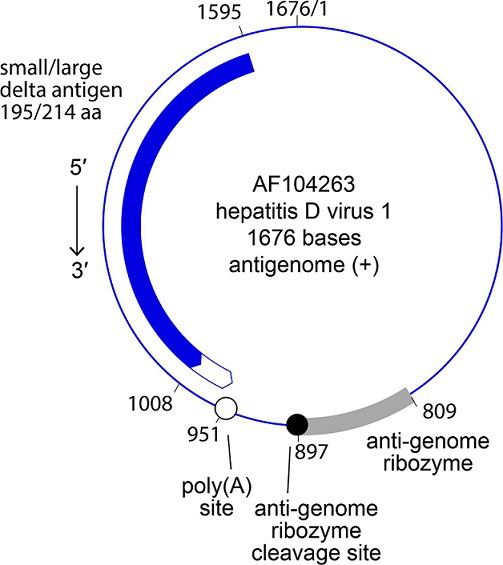
Kolmiovirids have a viroid-like, ribozyme-containing, negative-sense, covalently closed circular RNA (cccRNA) genome of about 1.5–1.7 kb (Fig. 2) that forms a rod-like structure through its high degree of self-complementarity. The genome encodes a single protein, DAg, which in some cases occurs in two isoforms [1,8].
Fig. 2. Antigenome organization of hepatitis D virus 1. Nucleotide numbering according to that for the (complementary) genome.
Replication
Kolmiovirion cell entry is mediated by interaction with cell-surface receptors and envelope proteins that are obtained from helper viruses. Membrane fusion releases the RNP into the cytosol, from where it migrates to the cell nucleus where RNA-directed RNA genome and antigenome synthesis is mediated by host-cell DNA-directed RNA polymerase II through a double rolling circle mechanism; genome- and antigenome-encoded ribozymes catalyse autocatalytic cleavage of concatenated progeny genomes and ligation/recyclization in the nucleus. DAg isoforms serve as nucleoproteins and regulate replication and packaging [1,2].
Pathogenicity
Deltaviruses cause disease in humans, typically in association with the evolutionarily unrelated hepatitis B virus (HBV, family Hepadnaviridae). Coinfection causes hepatitis D, the most severe of WHO-classified viral hepatitides [1,2].
Taxonomy
Current taxonomy: ictv.global/taxonomy. Kolmiovirids (realm Ribozyviria) are most closely related to viroids and related mobile genetic elements. Kolmiovirids share at least two of the following characteristics: (i) enveloped virions; (ii) circular negative-sense RNA genome containing ribozymes and encoding a DAg homolog; (iii) require an evolutionarily unrelated helper virus for assembly of infectious virions.
Resources
Full ICTV Report on the family Kolmioviridae: www.ictv.global/report/kolmioviridae.
Acknowledgements
We thank Evelien Adriaenssens, Holly Hughes, Elliot J. Lefkowitz, Sead Sabanadzovic, Peter Simmonds, Dann Turner, F. Murilo Zerbini, Luisa Rubino, Arvind Varsani (ICTV Report Editors) and Donald B. Smith (Managing Editor, ICTV Report). We also thank Anya Crane and Jiro Wada (Integrated Research Facility at Fort Detrick, Frederick, MD, USA) for critically editing the text and preparing figures.
Abbreviations
- cccRNA
covalently closed, circular RNA
- DAg
delta antigen
- HBV
hepatitis B virus
- L-DAg, S-DAg
large and small delta antigen
- L-HBsAg, M-HBsAg, S-HBsAg
large, middle and small hepatitis B virus surface antigen
- RNP
ribonucleoprotein
Footnotes
Funding: Production of this Profile, the ICTV Report, and associated resources was supported by the Microbiology Society. This work was supported in part through the Laulima Government Solutions, LLC, prime contract with the U.S. National Institute of Allergy and Infectious Diseases (NIAID) under Contract No. HHSN272201800013C. J.H.K. performed this work as an employee of Tunnell Government Services (TGS), a subcontractor of Laulima Government Solutions, LLC, under Contract No. HHSN272201800013C. The content of this publication should not be interpreted as necessarily representing the official policies, either expressed or implied, of the U.S. Department of Health and Human Services or of the institutions and companies affiliated with the authors.
Contributor Information
Jens H. Kuhn, Email: kuhnjens@mail.nih.gov.
Artem Babaian, Email: artem.babaian@gmail.com.
Laura M. Bergner, Email: Laura.Bergner@glasgow.ac.uk.
Paul Dény, Email: paul.deny@inserm.fr.
Dieter Glebe, Email: Dieter.Glebe@viro.med.uni-giessen.de.
Eugene V. Koonin, Email: koonin@ncbi.nlm.nih.gov.
Mart Krupovic, Email: mart.krupovic@pasteur.fr.
Marcos de la Peña, Email: rivero@ibmcp.upv.es.
Teemu Smura, Email: teemu.smura@helsinki.fi.
Jussi Hepojoki, Email: jussi.hepojoki@helsinki.fi.
References
- 1.Asselah T, Rizzetto M. Hepatitis D virus infection. N Engl J Med. 2023;389:58–70. doi: 10.1056/NEJMra2212151. [DOI] [PubMed] [Google Scholar]
- 2.Dandri M, Volmari A, Lütgehetmann M. The hepatitis delta virus and chronic hepatitis D. J Hepatol. 2022;77:1448–1450. doi: 10.1016/j.jhep.2022.05.022. [DOI] [PubMed] [Google Scholar]
- 3.Chang W-S, Pettersson JH-O, Le Lay C, Shi M, Lo N, et al. Novel hepatitis D-like agents in vertebrates and invertebrates. Virus Evol. 2019;5:vez021. doi: 10.1093/ve/vez021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hetzel U, Szirovicza L, Smura T, Prähauser B, Vapalahti O, et al. Identification of a novel deltavirus in boa constrictors. mBio. 2019;10:00014–00019. doi: 10.1128/mBio.00014-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wille M, Netter HJ, Littlejohn M, Yuen L, Shi M, et al. A divergent hepatitis D-like agent in birds. Viruses. 2018;10:e0720. doi: 10.3390/v10120720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paraskevopoulou S, Pirzer F, Goldmann N, Schmid J, Corman VM, et al. Mammalian deltavirus without hepadnavirus coinfection in the neotropical rodent Proechimys semispinosus. Proc Natl Acad Sci U S A. 2020;117:17977–17983. doi: 10.1073/pnas.2006750117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergner LM, Orton RJ, Broos A, Tello C, Becker DJ, et al. Diversification of mammalian deltaviruses by host shifting. Proc Natl Acad Sci U S A. 2021;118:e2019907118. doi: 10.1073/pnas.2019907118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lucifora J, Delphin M. Current knowledge on hepatitis delta virus replication. Antiviral Res. 2020;179:104812. doi: 10.1016/j.antiviral.2020.104812. [DOI] [PubMed] [Google Scholar]