Abstract
The study of protein-RNA interactions is critical for our understanding of cellular processes and regulatory circuits controlled by RNA binding proteins (RBPs). Recent next generation sequencing-based approaches significantly promoted our understanding of RNA biology and its importance for cell function. We present a streamlined protocol for Photoactivatable-Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation (PAR-CLIP), a technique that allows for the characterization of RBP binding sites on target RNAs at nucleotide resolution and transcriptome-wide scale. PAR-CLIP involves irreversible UV-mediated crosslinking of RNAs labeled with photoreactive nucleosides to interacting proteins, followed by stringent purification steps and the conversion of crosslinked RNA into small RNA cDNA libraries compatible with next-generation sequencing. The defining hallmark of PAR-CLIP is a diagnostic mutation at the crosslinking site that is introduced into cDNA during the library preparation process. This feature allows for efficient computational removal of contaminating sequences derived from non-crosslinked fragments of abundant cellular RNAs. In the following, we present two different step-by-step procedures for PAR-CLIP, which differ in the small RNA cDNA library preparation procedure: (1) Standard library preparation involving gel size selections after each enzymatic manipulation, and (2) A modified PAR-CLIP procedure (“on-beads” PAR-CLIP), where most RNA manipulations including the necessary adapter ligation steps are performed on the immobilized RNP. This streamlined procedure reduces the protocol preparation time by three days compared to the standard workflow.
Keywords: RNA binding protein, Posttranscriptional Gene Regulation, PAR-CLIP, Small RNA cDNA library, Next-generation sequencing
1. Introduction
Throughout their entire life-cycle cellular RNAs are associated with RNA binding proteins (RBPs) forming ribonucleoprotein particles (RNPs). In humans, more than 1500 genes are predicted as RBPs [1], and hundreds of other proteins, lacking a canonical RNA binding domain, have been suggested to function as RBPs [2]. In accordance, RBPs make up ~20% of the protein-encoding transcriptome and are expressed to higher levels than other functional groups of transcripts [1]. The basic cellular machinery, including RNA polymerases, the spliceosome and ribosomes constitute the most abundant RNPs, while the majority of characterized RBPs have been implicated in posttranscriptional gene regulatory processes. RBPs were shown to regulate aspects of genome organization, transcription, RNA splicing and maturation, nuclear export and localization of mRNAs, translation, and RNA turnover (for reviews, see [1,2]). In some cases specific RNA ligands have also been shown to regulate activity of interacting proteins [3].
In order to dissect regulatory circuits and networks that rely on RBPs it is essential to characterize their interactions with RNA ligands at high resolution. Most RBPs bind to short (4–8 nt) and often degenerate sequence or structural motifs termed RNA recognition elements (RREs) [4]. Taking into account competition between RBPs and the abundance of RREs, RBPs may populate distinct sets of their potential binding sites depending on cell type, regulatory state, or other biological contexts [5]. Therefore, experimental approaches, rather than computational ones remain necessary to comprehensively identify occupied RREs. In recent years, multiple protocols that combine next-generation sequencing (NGS) with crosslinking and immunoprecipitation (CLIP) were developed for the comprehensive identification of RREs [6–10]. For the following, we focus on the method we introduced, PAR-CLIP (Photoactivatable Ribonucleoside-enhanced Crosslinking and Immunoprecipitation) [10].
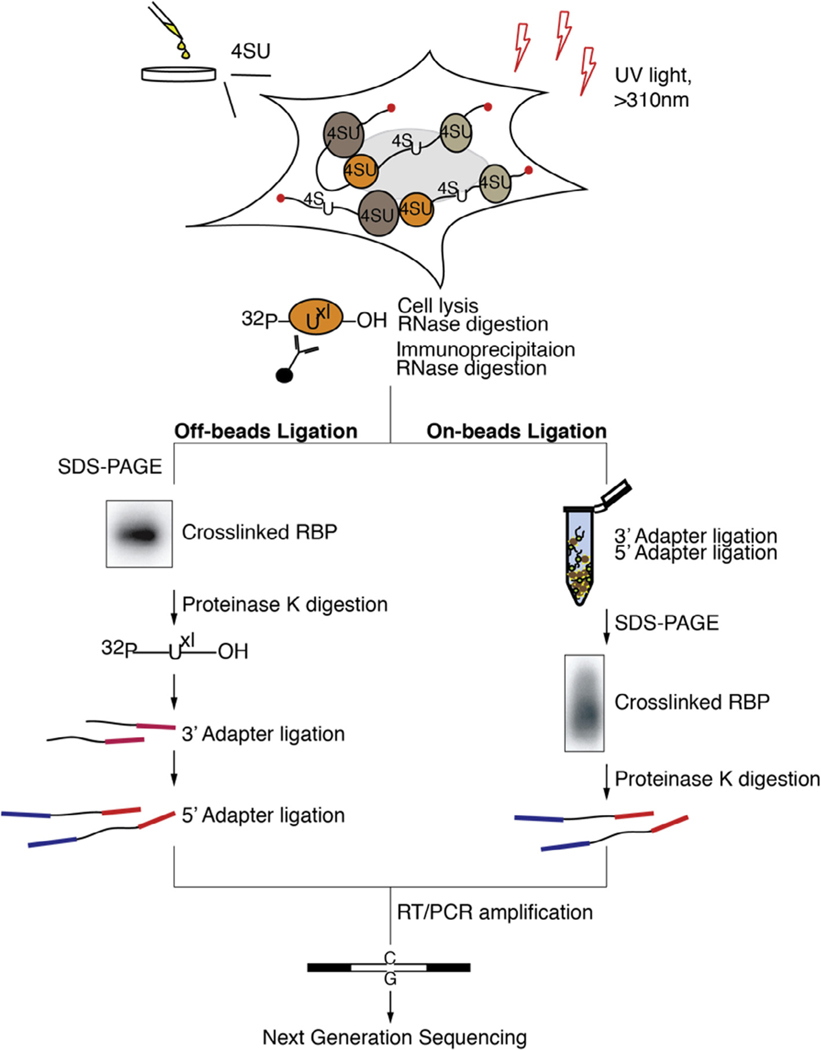
In PAR-CLIP (Fig. 1), photoactivatable ribonucleosides (4-thiouridine [4SU] or 6-thioguanine [6SG]) are incorporated into nascent RNA transcripts in living cells. Upon exposure of cells to long-wavelength ultraviolet (UV) light (>310 nm), incorporated 4SU or 6SG nucleosides form a photoadduct with reactive amino acid side chains of interacting RBPs [11,12]. This reaction proceeds at a 100–1000 fold higher efficiency compared to uridine-protein crosslinking by UV-C light (<260 nm wavelength) [10]. Importantly, upon crosslinking 4SU and 6SG undergo a structural change, which likely results in altering of the Watson-Crick face of the crosslinked nucleoside. This altered structure leads to a specific and diagnostic T-to-C, or G-to-A mutation, when using 4SU or 6SG, respectively, in the resulting cDNA. This feature enables the efficient filtration of contaminating non-crosslinked sequences derived from fragments of abundant cellular RNAs, which contribute the majority of experimental noise in CLIP procedures [13–15].
Fig. 1.
Overview of key steps in PAR-CLIP for the standard (left panel) and “on beads” (right panel) approaches. These methods differ in the small RNA cDNA library preparation steps, which are shortened in the “on beads” protocol by three days. In the latter approach, all adapter ligation steps are performed on matrix-immobilized RNPs, thus avoiding gel fractionation steps.
Following stringent purification of the crosslinked RNP, the protected RNA is recovered and converted into an NGS-compatible cDNA library [16,17]. The RNA is first dephosphorylated at its ends followed by a 5′ phosphorylation. This allows to take advantage of a 5′ phosphate dependent small RNA cDNA preparation procedure developed for sequencing of miRNAs and other 5′ phosphate containing small RNA families [16,17]. These protocols minimize undesired side-reactions during adapter ligation steps to the RNA 5′ and 3′ ends, which introduce constant sequences allowing for reverse transcription and PCR amplification. Usually, after each of the ligations, products need to be size-selected by Urea-Polyacrylamide gel electrophoresis (Urea-PAGE) in order to remove side-products and the excess of unreacted adapter molecules. After reverse transcription, PCR amplification and an additional gel fractionation step that removes PCR-byproducts, the cDNA library is ready to be sequenced.
In order to streamline and shorten the PAR-CLIP procedure, we developed a modified protocol, here called “on-beads” PAR-CLIP. Instead of recovering interacting RNA from the immunoprecipitated RNPs and carrying it through the small RNA cDNA library protocol, the RNP is kept immobilized onto the matrix. Adapters introducing constant regions are ligated onto the RNP while it is still associated to the beads allowing for efficient cleaning and buffer exchange, and dispending gel-electrophoresis based size selections. Altogether, “on-beads” PAR-CLIP saves approximately 3 days compared to the standard, “off-beads” protocol (Fig. 1). We want to note that the “on beads” protocol requires the accessibility of RNA ends in the RNP complex and thus the method may be unsuited for a number of RNP complexes. From our experience, accessible 5′ and 3′ ends are usually prevalent in RNPs with relatively long protected footprints (25–45 bases). However this needs to be tested empirically per RBP as in our hands both the “on-beads” and “off-beads” versions gave comparable results for most RBPs tested, including those that exhibited relatively short footprints (15–35 bases, data not shown). Below we present a combined point-by-point protocol for both the “off-beads” and the “on-beads” versions of PAR-CLIP.
2. PAR-CLIP protocol
2.1. Materials
2.1.1. Reagents
| 4-thiouridine (4SU) | Sigma Aldrich, T4509 |
| 6-thioguanosine (6GS) | Carbosynth, NT04480 |
| RNase T1 (1000 U/μl) | Fermentas, EN0541 |
| Calf intestinal phosphatase (CIP) | New England Biolabs (NEB), M0290 |
| T4 polynucleotide kinase (T4 PNK) | NEB, M0201 |
| γ−32P-ATP, 10 mCi/ml, concentration 1.6 μM | Perkin Elmer, NEG002Z001MC |
| Proteinase K | Roche, 03 115 879 001 |
| Glycoblue (15 mg/ml) | Ambion, AM9516 |
| Truncated and mutated RNA ligase 2, T4 Rnl2(1-249)K227Q(1 mg/ml) | NEB, M0351 |
| T4 RNA ligase, T4 Rnl1 (1 mg/ml) | ThermoFisher |
| Scientific, EL0021 | |
| Low melting point agarose | ThermoFisher |
| Scientific, 16520050 | |
| SuperScript III reverse transcriptase kit | Invitrogen, 18080-044 |
| Taq DNA polymerase | Invitrogen, 10966018 |
2.1.2. Buffers
| IP buffer | 20 mM Tris, pH 7.5 |
| 150 mM NaCl | |
| 2 mM EDTA | |
| 1% (v/v) NP40 | |
| 0.5 mM DTT (added fresh) | |
| RIPA buffer | 10 mM Tris-HCl (pH 8.0) |
| 1 mM EDTA | |
| 0.5 mM EGTA | |
| 1% Triton X-100 | |
| 0.1% sodium deoxycholate | |
| 0.1% SDS | |
| 140 mM NaCl | |
| High-salt wash buffer | IP buffer with 500 mM NaCl |
| Dephosphorylation buffer or 1× NEB Buffer 3 | 50 mM Tris-HCl, pH 7.9 |
| 100 mM NaCl | |
| 10 mM MgCl2 | |
| 1 mM DTT | |
| Polynucleotide kinase (PNK) buffer without DTT | 50 mM Tris-HCl, pH 7.5 |
| 50 mM NaCl | |
| 10 mM MgCl2 | |
| Polynucleotide kinase (PNK) buffer with DTT | 70 mM Tris-HCl, pH 7.6 |
| 10 mM MgCl2 | |
| or 1 × NEB PNK buffer | 5 mM DTT |
| Acid phenol/chloroform, pH 4.5 | Ambion, AM9720 |
| 2× Proteinase K buffer | 100 mM Tris-HCl, pH 7.5 |
| 150 mM NaCl | |
| 12.5 mM EDTA | |
| 2% (w/v) SDS | |
| 10 × RNA ligase buffer without ATP or 10× NEB RNA ligase buffer | 500 mM Tris-HCl, pH 7.6, |
| 100 mM MgCl2 | |
| 10 mM DTT | |
| 10× RNA ligase buffer with ATP or 10× Thermo scientific RNA ligase buffer |
500 mM Tris-HCl, pH 7.6 |
| 100 mM MgCl2 | |
| 100 mM DTT | |
| 10 mM ATP | |
| Formamide gel loading dye | 50 mM EDTA 0.05% (w/v) bromophenol blue formamide ad 100% |
| Transfer buffer | 25 mM Tris 190 mM Glycine 10% methanol |
| 10 × dNTP | 2 mM dATP |
| 2 mM dCTP | |
| 2 mM dGTP | |
| 2 mM dTTP |
2.1.3. Oligonucleotide sequences RNA oligonucleotides
| 19 nt RNA size marker | 5′CGUACGCGGGUUUAAACGA |
| 35 nt RNA size marker | 5′ CUCAUCUUGGUCGUACGCGGAAUAGUUUAAACUGU |
| 5′ adapter | GUUCAGAGUUCUACAGUCCGACGAUC |
|
| |
| DNA oligonucleotides | |
|
| |
| 3′ PCR primer | CAAGCAGAAGACGGCATACGA |
| 5′ PCR primer | AATGATACGGCGACCACCGACAGGTTCAGAGTTCTACAGTCCGA |
| 3′ adapters* | |
| Ad01 | App-TCACTTCGTATGCCGTCTTCTGCTTG-L |
| Ad02 | App-TCATCTCGTATGCCGTCTTCTGCTTG-L |
| Ad03 | App-TCCACTCGTATGCCGTCTTCTGCTTG-L |
| Ad04 | App-TCCGTTCGTATGCCGTCTTCTGCTTG-L |
| Ad05 | App-TCCTATCGTATGCCGTCTTCTGCTTG-L |
| Ad06 | App-TCGATTCGTATGCCGTCTTCTGCTTG-L |
| Ad07 | App-TCGCGTCGTATGCCGTCTTCTGCTTG-L |
| Ad08 | App-TCTAGTCGTATGCCGTCTTCTGCTTG-L |
| Ad09 | App-TCTCCTCGTATGCCGTCTTCTGCTTG-L |
| Ad10 | App-TCTGATCGTATGCCGTCTTCTGCTTG-L |
Listed are ten 3′ adapters we frequently use in the lab [18,19]. App, 5′ terminal adenosine residue connected via a 5′,5′-diphosphate bridge to the 5′OH of the 5′ nucleotide; L, 3′ aminohexyl blocking group. Oligonucleotides can be obtained from suppliers of customized oligonucleotides [e.g. IDT (http://www.idtdna.com) or MultiplexDX (http://www.multiplexdx.com)]. Note: to further reduce possible biases in ligation efficiency randomized nucleotides at the 5′ end of the 3′ adapter, as well as on the 3′ end of the 5′ adapter can be introduced. Nevertheless, we do not find substantial improvement in library representation by these modifications, consistent with our previous finding that secondary structure formation of RNA substrates contributed most to ligation biases [17].
2.1.4. Instrumentation and other material
| Spectrolinker XL-1500 | Spectronics Corporation |
| Sonics Vibra cell | Sonics, VCX130 |
| DNA LoBind Tube | Eppendorf, 022431021 |
| 5 μm membrane syringe filter | Pall Acrodisc |
| GeneJET Gel Extraction Kit | ThermoFisher Scientific |
| 3% Pippin gel cassettes | Sage Science, CSD3010 |
| DNA clean & concentrator-5 | Zymo Research |
| Gel breaker tubes | Fisher Scientific |
| 5 μm filter tube | Fisher Scientific |
| V16–2 Protein Gel Electrophoresis System; 17 × 15 cm Gel size | LabRepCo |
| Urea Gel | National Diagnostics, EC-829 |
2.2. Experimental procedure
2.2.1. Preparation of radiolabeled size markers
The radioactively labeled 19 nt and 35 nt RNA size marker allows for monitoring of adapter ligation efficiency, as well as for RNA size selection in the traditional small RNA cDNA library preparation protocol [18]. Note that these marker molecules are not required for the “on beads” library preparation protocol.
Radiolabel the size markers individually at 37 °C for 15 min by preparing the following reaction mix: 10 pmol RNA, 10 U T4 PNK kinase, 50 μCi γ−32P-ATP. Make up to 10 μl final volume in 1× PNK buffer with DTT.
Add 10 μl of formamide gel loading solution and denature the sample at 90 °C for 1 min.
Load samples onto a 15% denaturing Urea-PAGE, leaving one empty lane in-between samples.
Run the gel for approximately 45 min at 30 W until the bromophenol blue dye reaches the bottom of gel.
Dismantle the gel, leaving it attached on one glass plate. Mark the corners of the gel with 0.2 μl of radioactive material (preferably use waste, avoid unnecessary consumption of source material, see Section 2.2.2.4) by pushing a hole onto to the gel with the pipet tip or slicing a small “X” with a scalpel. This will facilitate superimposition of the autoradiograph image onto the gel.
Wrap the gel in plastic film/Saran wrap.
Expose the gel to a blank phosphorimager screen for 5 min and develop on a phosphorimager (longer exposure time should be performed at −20 °C to avoid diffusion).
Print the image in the original size and align it to the gel.
Cut out the sample from the lanes and collect them in gel breaker tubes. Place the gel breaker tube inside a 1.5 ml microcentrifuge tube.
Centrifuge the tubes for 1 min at 12000g.
Add 400 μl of 0.3 M NaCl to the shredded gel pieces and incubate at 60 °C for 45 min with vigorous shaking.
Place a filter tube inside a 1.5 ml microcentrifuge collection tube and add the gel suspension to the filter.
Spin at 5000g for 1 min.
Precipitate the RNA in the flow through by adding 1 μl of glycogen (10 mg/ml), mixing, followed by addition of 3 volumes of ethanol and incubation at −20 °C for 20 min.
Centrifuge the sample at >12000g for 20 min and remove all ethanol traces.
Dissolve pellet in 10 μl of water.
Combine size markers solutions to obtain the size marker cocktail.
Dilute 1/100 before using. Note: the 19/35nt size marker can be used over a period of several weeks. Before use, measure the activity of the marker mix using a Geiger counter in order to ensure sufficient detectability. Use appropriate amounts to match the intensity of your sample.
2.2.2. Step 1 – PAR-CLIP
2.2.2.1. Cell culture, extraction and crosslinking.
-
1
Culture cells to 80% confluency in appropriate culturing media. A typical experiment utilizes 10–20 15 cm plates or roughly 100–200 million cells, yielding 1–2 ml of wet cell pellet. Note: The required amount of input material depends on two factors, (1.) the expression level of the RBP of interest, and (2.) the affinity of the antibody used for immunoprecipitation. For highly expressed or tagged RBPs we frequently reduce the input material to 2–3 15 cm plates.
-
2
16 h before crosslinking, add 4-thiouridine (4SU) or 6-thioguanosine (6SG) to the cell culture media at a final concentration of 100 μM. Treatment overnight ensures largely uniform labeling of the transcriptome of typical mammalian cell lines, which exhibit a median mRNA half-life of ~8 h [20].
2.2.2.1.1. Adherent cells.
-
3
After 16 h incubation with 4SU or 6SG, wash the cells once with 1× PBS, decant the supernatant. Keep the culture plate uncovered for crosslinking.
-
4
Crosslink the cells with 0.15 J/cm2 of >300 nm UV light in a Spectrolinker XL-1500 (Spectronics Corporation) or comparable instrument. The optimal excitation wavelength for 4SU is 320 nm and we crosslink our cells at 312 nm.
-
5
Scrape the cells in 1 ml of 1× PBS per plate and pellet at 500g for 5 min at 4 °C. Discard the supernatant.
-
6
Pausing point: cell pellets can be flash frozen in liquid nitrogen and stored at —80 °C for up to 12 months.
2.2.2.1.2. Suspension cells.
-
3
After 16 h incubation with 4SU or 6SG, pellet the cells at 500g for 5 min at 4 °C.
-
4
Wash the cell pellet once with 1× PBS.
-
5
Spin down at 500g for 5 min, resuspend the cell pellet in 20 ml of 1× PBS and transfer to a 15 cm plate. Keep the culture plate uncovered for crosslinking.
-
6
Crosslink the cells with 0.2 J/cm2 of > 300 nm UV light in a Spectrolinker XL-1500 (Spectronics Corporation) or comparable instrument.
-
7
Transfer the crosslinked cells into a 50 ml centrifugation tube and pellet at 500g for 5 min. Discard the supernatant.
-
8
Pausing point: cell pellets can be flash frozen in liquid nitrogen and stored at −80 °C for up to 12 months.
2.2.2.2. Cell extraction.
Note: prior to cell extraction, couple antibody to Protein G magnetic beads outlined in Section 2.2.2.3.1.
2.2.2.2.1. Abundant and/or cytoplasmic RBP.
Resuspend the cell pellet in 3 volumes of IP buffer (e.g. for a 2 ml pellet add 6 ml of IP buffer) and incubate on ice for 10 min.
Clear the lysate by centrifugation at 13000g for 15 min. Collect the supernatant and discard the cell debris.
Optional: filter the supernatant through a 5 μm syringe filter.
Add RNase T1 to a final concentration of 1 U/μl. Incubate for 15 min at 22 °C.
Cool the reaction on ice for 5 min.
Save 50 μl of the reaction as input to control for RBP expression.
2.2.2.2.2. Nuclear associated RBP.
Resuspend the cell pellet in 3 volumes of RIPA buffer (or similar) (e.g. for a 2 ml pellet add 6 ml of RIPA buffer) and incubate on ice for 10 min.
Sonicate the cell lysate for 30 s at 60% amplitude. Cool on ice for 30 s. Repeat this step 3 times.
Clear the lysate by centrifugation at 13000g for 15 min. Collect the supernatant.
Add RNase T1 to a final concentration of 1 U/μl. Incubate for 15 min at 22 °C.
Cool the reaction on ice for 5 min.
Save 50 μl of the reaction as input to control for RBP expression.
2.2.2.3. Immunoprecipitation.
2.2.2.3.1. Coupling antibody to protein G magnetic beads.
Transfer 20 μl of Protein G magnetic bead suspension per ml of cell lysate to a 1.5 ml microcentrifuge tube (e.g. for a 6 ml cell lysate use 120 μl of bead suspension).
Wash the beads 2 times with 1 ml 1× PBS. Note: to wash beads using a magnetic separator place the 1.5 ml microcentrifuge tube in a magnetic rack and let stand until the suspension clears and the magnetic particles are collected. Aspirate the 1× PBS and add fresh 1× PBS. Remove the 1.5 ml microcentrifuge tube from the magnetic rack and suspend the beads by pipetting several times. Repeat the wash.
Resuspend beads in 1 original bead suspension of 1× PBS (e.g. 120 μl for 6 ml original cell lysate).
Add 0.25 mg of antibody per ml of original bead suspension. Note: make sure that the antibody you use is suitable for IP and optimize the amount of antibody necessary for efficient IP.
Incubate the antibody and bead mixture for 1 h at 4 °C with gentle rotation.
Wash the beads 2 times with 1 ml 1× PBS.
Resuspend beads in 1 original bead volume of 1× PBS.
2.2.2.3.2. Immunoprecipitation and RNase T1 treatment.
Incubate the antibody-coupled beads with the cell lysate for 2 h at 4 °C. Note: for weak antibody-antigen interactions the IP can be performed overnight at 4 °C with gentle rotation.
Save 50 μl of the supernatant to control for RBP depletion.
Wash the beads 3 times with 1 ml IP buffer.
Resuspend the beads in 1 bead volume of IP buffer (e.g. 120 μl).
Add RNase T1 to a final concentration of 1–100 U/μl and incubate for 15 min at 22 °C. Cool the reaction on ice for 5 min. Note: RNase T1 digestion should be optimized per RBP of interest. Over-digestion may result in short fragments that are less accessible for subsequent enzymatic manipulation. In addition, we only consider sequence reads > 19 nt during our computational analyses in order to ensure unambiguous mapping. Insufficient digestion may result in maintenance of indirect, RNA-bridged, interactions between the RBP of interest and other RBPs, as well as the recovery of RNA segments longer than protected by the RBP of interest complicating the identification of the RRE. We recommend optimizing RNase T1 concentration by testing 1, 10, 100 U/μl in a small scale PAR-CLIP experiment and determining the size of the RBP protected RNA fragment by denaturing polyacrylamide electrophoresis, followed by transfer to a nitrocellulose membrane and visualization by autoradiograph. In our experience 1 or 10 U/μl are suitable for most PAR-CLIP experiments. Also note that other RNases without sequence specificity, such as RNase I and micrococcal nuclease, can be used instead of RNAse T1. While using RNase T1 might result in an underrepresentation of G nucleotide in recovered RNAs, its characteristic signature allows for more stringent filtering during the bioinformatic analysis by scoring for the presence of a G directly upstream and at the 3′ end of the mapped sequence read. Wash the beads 3 times with 1 ml IP buffer.
Optional: wash the beads 3 times with 1 ml high salt wash buffer. Note: make sure the antibody tolerates the 500 mM NaCl concentration for efficient IP.
Resuspend the beads in 1 bead volume of dephosphorylation buffer.
Add 0.5 U/μl of CIP alkaline phosphatase and incubate for 10 min at 37 °C with shaking. Note: adjust the shaking speed on the thermomixers such that the beads do not settle.
Wash beads 2 times with 1 ml of dephosphorylation buffer.
Wash beads 2 times with 1 ml PNK buffer without DTT.
Resuspend the beads in 1 bead volume of PNK buffer containing DTT.
2.2.2.4. Radiolabeling of crosslinked RNA.
Add 0.5 μCi γ−32P-ATP final concentration and 1 U/μl of T4 PNK kinase to the bead suspension.
Incubate for 30 min at 37 °C with shaking. Note: adjust the shaking speed on the thermomixers such that the beads do not settle.
Add non-radioactive ATP to a final concentration of 100 μM and incubate for additional 5 min at 37 °C.
Wash beads 5 times with 1 ml of PNK wash buffer without DTT. Note: Exposure to >1 mM DTT for a prolonged time may damage magnetic beads and should only be used in the reaction buffer. Note: keep 50 μl of the radioactive waste, which is collected after the first bead wash, to mark the nitrocellulose membrane and/or Urea-PAGEs described below.
In the following we present two options for the preparation of the small RNA cDNA library for sequencing (see Fig. 1). For Option A, the RNA is recovered from immunoprecipitated RNPs and carried through a small RNA cDNA library preparation [10] (Section 2.2.3). This method can be used for any RNA binding protein and is recommended if no information on the length of the protected fragment of the RBP is available. Option B, relies on manipulations of the crosslinked RNA while RNPs remain immobilized on beads and should be most suitable for RNA fragment with accessible 3′ and 5′ ends [17] (Section 2.2.4).
2.2.3. Step 2 (Option A) – standard small RNA cDNA library preparation
2.2.3.1. SDS-PAGE, nitrocellulose transfer and proteinase K digestion.
Resuspend the beads in 70 μl of 2× SDS-PAGE loading buffer and incubate for 5 min at 95 °C. Pausing point: the eluted sample can be stored at −20 °C overnight.
Vortex vigorously and centrifuge briefly to collect the suspension.
Place the tube on a magnetic separator to collect magnetic beads and transfer the supernatant to a clean 1.5 ml microcentrifuge tube.
Load 65 μl of the sample into two adjacent wells, i.e. 32.5 μl per well, on a 4–12% Bis-Tris SDS-PAGE. Run the gel at 200 V for approximately 45 min until the bromophenol blue dye reaches the bottom of the gel. Note: run the remaining 5 μl of the obtained IP material on a separate SDS-PAGE, along with 10% of input material saved in Section 2.2.2.2.1 step 6 or 2.2.2.2.2 step 6 and 10% of the IP supernatant saved in Section 2.2.2.3.2 step 2 to confirm proper pull-down of the RBP of interest, as well as depletion of antigen from lysate (Fig. 2A).
Transfer the gel using a semi-dry blotting apparatus to a nitrocellulose membrane for 1 h at 17 V using standard transfer buffer containing 10% methanol.
Mark the corners of the membrane and the protein ladder with 0.2 μl of radioactive material (collected in Section 2.2.2.4) to facilitate correct alignment of an autora-diograph to the gel, as well as to relate sizes RNP band sizes. Note: we recommend to mark the corners of the membrane with a marker and spot the radioactive waste on top to facilitate finding the alignment points.
Wrap the nitrocellulose membrane with plastic film/Saran wrap.
Expose the membrane to a blank phosphorimager screen for 1 h or longer at −20 °C and develop on a phosphorimager.
Print out the image on transparent paper and align it to the radioactive marks on the nitrocellulose membrane to correctly identify and excise the protein-RNA band. Cut the membrane slice into smaller pieces to increase the efficiency of RNA recovery.
Transfer the membrane slices to a 1.5 ml microcentrifuge tube. Pausing point: you can store the membrane slices in −20 °C overnight.
- Release the RNA from the membrane slices by digesting the proteins with proteinase K in three subsequent steps, each time adding to the existing volume to make a final volume of 500 μl. Note: We observed that adding fresh proteinase K significantly increases the recovery of RNA. To measure the efficiency of RNA recovery, measure the radioactivity on the nictrocellulose membrane slice before and after proteinase K digestion. If successful, all the radioactive signal will be transferred from the nitrocellulose membrane into solution.
- add 1.2 mg/ml proteinase K in 200 μl of 1× Proteinase K buffer. Incubate the sample at 50 °C in a heat block under vigorous shaking for 30 min.
- add 0.75 mg/ml proteinase K in 150 μl of 1× Proteinase K buffer. Incubate the sample at 50 °C in a heat block under vigorous shaking for 30 min.
- add 0.75 mg/ml proteinase K in 150 μl of 1× Proteinase K buffer. Incubate the sample at 50 °C in a heat block under vigorous shaking for 30 min.
Transfer the supernatant to a new 1.5 ml microcentrifuge tube and discard the membrane slices.
Add 50 μl of 3 M NaCl and 300 μl acidic phenol–chloroform (pH 4.5) and mix by vortexing.
Centrifuge at 12000g for 2 min and transfer the aqueous phase to a new 1.5 ml microcentrifuge tube.
Add 300 μl of chloroform and mix by vortexing.
Centrifuge at 12000g for 2 min and transfer the aqueous phase to a new 1.5 ml microcentrifuge tube.
Precipitate the RNA by adding 1 μl of glycogen (10 mg/ml), mixing, followed by addition of 3 volumes of ethanol and incubation at −20 °C for at least 1 h.
Centrifuge the sample at >12000g for 20 min and remove all ethanol traces.
Dry the pellet for 5 min at room temperature.
Dissolve the pellet in 9 μl of water.
Optional: size select RNA fragments between 19 and 35 nt range by fractionating samples on a 15% denaturing Urea-PAGE. See Section 2.2.3.2 steps 8–21 for detailed protocol for gel purification procedure.
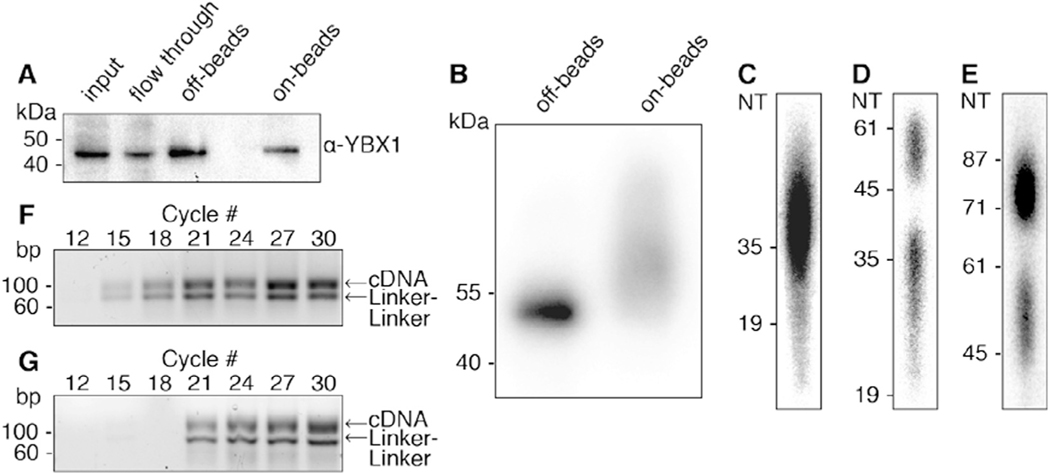
Fig. 2.
Illustration of steps during PAR-CLIP. (A) Immunoblot using antibody against the YBX1 protein. Samples from left to right: input, IP flow through, “off-beads” and “on-beads” IP elutions. (B) Autoradiograph showing the separation of radiolabeled YBX1 RNP immunoprecipitated from “off-beads” and “on-beads” procedures. Eluted RNPs are separated by SDS-PAGE and transferred to a nitrocellulose membrane. (C-E) Autoradiography of denaturing polyacrylamide gels that are used along the “off-beads” procedure to visualize and select: (C) RNA fragments extracted after Proteinase K digestion of RNPs, (D) products of 3′ adapter ligation, and (E) products of 5′ adapter ligation. (F, G) Agarose gel separation of PCR products at increasing cycle numbers for off-beads (F) and on-beads (G) procedures. For library preparation we chose 15 and 18 cycles, respectively. The expected PCR product runs at ~100 bp. Linker-linker side-products run at 71 bp.
2.2.3.2. 3′ adapter ligation.
For the following steps the 19 nt and 35 nt RNA size marker mixture will be used, generated in Section 2.2.1, allowing for the determination of the correct size and efficient ligation of the RNA.
Prepare the following reaction mixture for ligation of the 3′ adenylated adapter, one for your sample and one for the 19/35 nt RNA size marker: 2 μl RNA ligase buffer without ATP, 6 μl 50% PEG-8000, 2 μl of 50 μM adenylated 3′ adapter. Note: We recommend using two different barcoded adapters for the size marker reaction and the PAR-CLIP sample and alternating different barcoded adapters in subsequent experiments in order to track possible contaminations from previously prepared small RNA cDNA libraries.
Add 10 μl of the reaction mixture to the 9 μl sample solution.
Add 10 μl of the reaction mixture and 7 μl of water to 2 μl of 19/35 nt RNA marker mix.
Denature the RNA samples at 90 °C for 1 min.
Immediately cool the reaction on ice for 2 min.
Add 1 μl of Rnl2(1–249)K227Q ligase, mix gently and incubate for 2 h at 16 °C or overnight at 4 °C while shaking.
Add 20 μl of formamide gel loading solution and denature the sample at 90 °C for 1 min.
Load samples onto a 15% denaturing Urea-PAGE, leaving one empty lane in-between samples. Flank your samples with 20 μl of the 19/35 nt RNA size marker control reaction on both sides.
Run the gel for approximately 45 min at 30 W until the bromophenol blue dye reaches the bottom of gel.
Dismantle the gel leaving it attached on one glass plate. Mark the corners of the gel with 0.2 μl of radioactive material by pushing a whole onto to the gel with the pipet tip or slicing a small “X” with a scalpel, to facilitate superimposition of the autoradiograph onto the gel.
Wrap the gel in plastic film/Saran wrap.
Expose the wrapped gel to a blank phosphorimager screen for 1 h (or overnight) at −20 °C and develop on a phosphorimager.
Print the image in the original size and align it to the gel.
Cut out the sample from the lane in-between the 19/35 nt RNA size marker, as well as the 19/35 nt RNA size markers, and collect them in gel shredder tubes. Place the gel shredder tube inside a 1.5 ml microcentrifuge tube.
Centrifuge the tubes for 1 min at 12000g.
Add 400 μl of 0.3 M NaCl to the shredded gel pieces and incubate at 60 °C for 45 min with vigorous shaking.
Place a filter tube inside a 1.5 ml microcentrifuge collection tube and add the gel suspension to the filter.
Spin at 5000g for 1 min.
Precipitate the RNA in the flow through by adding 1 μl of glycogen (10 mg/ml), mixing, followed by addition of 3 volumes of ethanol and incubation at −20 °C for at least 1 h. Pausing point: you can store the sample in −20 °C overnight.
Centrifuge the sample at >12000g for 20 min.
Dissolve pellet in 9 μl of water.
2.2.2.3. 5′ adapter ligation.
Prepare the following reaction mixture for ligation of the 5′ adapter, one for your sample and one for the 19/35 nt RNA size marker: 2 μl RNA ligase buffer containing ATP, 6 μl 50% PEG-8000, 1 μl of 100 μM 5′ adapter.
Add 9 μl of the mixture to the samples
Denature the RNA at 90 °C for 1 min.
Immediately cool the reaction on ice for 2 min.
Add 2 μl of Rnl1, mix gently, and incubate 1 h at 37 °C while shaking.
Fractionate the sample by Urea-PAGE as outlined in Section 2.2.3.2 steps 7–21. Note: use a 12% Urea-PAGE for this step.
Dissolve pellet in 4.6 μl of water. The samples are now ready for reverse transcription. See Section 2.2.5.
2.2.4. Step 3 (Option B) – on bead ligations of 3′ and 5′ adapters
2.2.4.1. On bead 3′ adapter ligation.
Prepare the following reaction mixture for ligation of the 3′ adenylated adapter: 22.5 μl water, 5 μl 10× RNA ligase buffer without ATP, 15 μl 50% aqueous PEG-8000, 5 μl 50 μM adenylated 3′ adapter oligonucleotide, 2.5 μl Rnl2(1–249)K227Q ligase.
Wash the beads twice with 1 ml 1× RNA ligase buffer without ATP.
Add 50 μl of the reaction mixture to the beads and mix gently.
Incubate the sample for 2 h at 16 °C.
2.2.4.2. On bead 5′ adapter ligation.
Prepare the following reaction mixture for ligation of the 5′ adapter: 22.5 μl water, 5 μl 10× RNA ligase buffer without ATP, 15 μl 50% aqueous PEG-8000, 2.5 μl 100 μM 5′ adapter oligonucleotide, 5 μl Rnl1.
Wash the beads twice with 1× RNA ligase buffer with ATP.
Add 50 μl of the reaction mixture to the beads and mix gently.
Incubate the tube for 2 h at 37 °C while shaking.
Wash the beads 2 times with 1 ml of IP buffer.
2.2.4.3. SDS-PAGE, transfer to nitrocellulose and proteinase K digestion.
1. Follow the procedure outlined in Section 2.2.3.1 steps 1–19 described above.
2. Dissolve the pellet in 4.6 μl of water. The samples are now ready for reverse transcription. See Section 2.2.5.
2.2.5. Step 4 – reverse transcription and PCR amplification of product
2.2.5.1. Reverse transcription.
Prepare the following reaction mixture. 1.5 μl DTT, 3 μl 5× first strand buffer, 4.2 μl 10× dNTP, 1 μl 3′ PCR primer (100 μM stock).
Denature the recovered RNA by incubating at 90 °C for 30 s in a thermocycler.
Add 9.7 μl of the reaction mixture to your samples and incubate for 3 min at 50 °C.
Add 0.75 μl Superscript III reverse transcriptase and incubate for further 2 h at 50 °C.
Dilute the sample by adding 85 μl of water. Pausing point: you can store the sample in −20 °C.
2.2.5.2. PCR amplification.
PCR amplify your barcoded RNA segments by preparing the following reaction mixture: 64.4 μl water, 10 μl 10× dNTPs, 3 μl MgCl2 (50 mM stock), 10 μl 10× PCR buffer, 0.5 μl 5′ PCR primer (100 lM stock), 0.5 μl 3′ PCR primer (100 μM stock) and 0.4 μl Taq DNA polymerase.
Add 10 μl of cDNA to 90 μl of the above PCR master mix.
Program the thermocycler: 94 °C, 2 min; 94 °C, 45 s; 50 °C, 85 s; 72 °C, 1 min for a total of 30 cycles.
Remove a 10 μl aliquot after every three cycles between cycle number 12 and 30 (i.e. cycles 12, 15, 18, 21, 24, 27 and 30). This will help you determine the optimal PCR cycles needed for your specific sequencing library. Note: it is very important to determine the optimal cycle number for amplification of the library for sequencing to avoid PCR artifacts. The cDNA should be amplified with a cycle number where the PCR is still in the exponential amplification phase.
Analyze the PCR product on a 2.5% agarose gel in 1× TBE buffer, using a 20 bp DNA ladder. Determine the optimal PCR cycles (Fig. 2F, G). Note: the optimal PCR cycle usually ranges from cycle 12–21; generally, we use the least number of cycles resulting in visible PCR product on the ethidiumbromide stained agarose gel.
Set up three 100 μl PCR reactions with the determined optimized cycle number.
Once the PCR is done, pool the samples together and use a PCR cleanup kit to clean and concentrate your samples. Elute the samples in 30 μl of water.
- Clean up the PCR product to remove all linker-linker byproduct (Fig. 2F, G). We recommend one of the following methods:
- Run 30 μl of the PCR product on a 2.5% agarose gel, in 2 wells as to not overload the gel. Cut out the band, which correlates to the 100 bp product. The sample is extracted using a gel purification kit. Elute your sample in 30 μl of water.
- Run 30 μl PCR product on a 3% Pippin Prep, following the manufacturer’s instructions and collect PCR products from 90–106 bp.
Quantify DNA concentration by TapeStation, Picogreen, Qubit, or a similar method. It is important to quantify the concentration of the samples accurately to prevent over- or under-clustering of the Illumina sequencing flow cell. We recommend using at least 2 different methods of quantification for each sample.
The samples are now ready to be sequenced on an Illumina HiSeq machine. Note: the primers we are using are only compatible with Illumina HiSeq and not with NextSeq or MiSeq. Several biocomputational pipelines for PAR-CLIP data analysis have been made available. For a comparative list of available pipelines see [21].
3. Expected results
The above protocol details all the required steps to perform PAR-CLIP, including two methodologies for transforming isolated RNA fragments into NGS libraries. In the following, results obtained in our lab for the intensely studied RBP, YBX1, will be used to illustrate expected results of main steps along the protocol, and where relevant, discuss differences between standard and “on-beads” library preparation procedures. “On-beads” processing of RNPs accelerates the transformation of RNA footprints into sequencing libraries by streamlining adapter ligations and necessary buffer exchange, purification and size selection steps. On the other hand, while the standard “off-beads” approach requires additional time and labor, it includes size-selection gels that enable monitoring intermediate steps of the procedure at higher resolution.
The success of individual steps of the PAR-CLIP protocol, including in vivo labeling, crosslinking, lysis, RNase treatment, IP, and radiolabeling, are not controlled prior to elution of immunoprecipitated (IPed) RNPs. The IP efficiency can be quantified by sampling the eluted IP material and comparing it to input and supernatant (Fig. 2A). While the RNA covalently linked to the RBP can increase the molecular weight and thus the retention in gel electrophoresis, we find that the vast majority (~99%) of IPed protein is not cross-linked to RNA, and is thus detected at the predicted size for the RBP alone (Fig. 2A). Extended manipulation “on beads” can lead to the loss of a portion of the RBP during washing steps (Fig. 2A, compare the “off-beads” and “on-beads” elution).
Crosslinked RNPs are visualized by autoradiography (Fig. 2B). In our experience, “off-beads” PAR-CLIP results in a homogenous band on the autoradiograph, migrating close to the predicted size of the RBP (Fig. 2B, left lane). “On-beads” processing of RNPs includes ligation of adapters (26 base, 8–10 kDa, each) at both the 3′ and 5′ ends of protected RNA fragments prior to elution of RNPs. The eluted RNPs are expected to include subpopulations of non-ligated, single-ligated (at either the 3′ or 5′ end), or fully ligated RNA fragments. Adding up to 20 kDa in size distribution, autoradiography of “on-beads” ligated RNPs reflect a rather heterogeneous size distribution (Fig. 2B, right lane).
The RNA component of the immunoprecipitated RNP is released by Proteinase K digestion and an optional gel fractionation step in the “off-beads” procedure allows for size assessment and selection of the desired length of RNA fragments (Fig. 2C). Similarly, both the 3′ and the 5′ adapter ligation steps are followed by denaturing gel size selection step (Fig. 2D and E, respectively). Denaturing gel separation and visualization of the ligation products monitor the efficiency of ligation (ligated RNA fragment form a higher molecular weight band) and enable the removal of unligated RNA and excess of adapter molecules.
Once the ligated RNA fragments have been isolated the “on-beads” and “off-beads” protocols converge at the steps of reverse-transcription (RT) and PCR amplification. PCR amplification has to be optimized to avoid generation of PCR artifacts. In the example presented in Fig. 2F the optimal number of PCR cycles is between 12 and 15 cycles. Importantly, neither the “off-beads” size selecting gels, nor the extensive washing during “on-beads” procedures entirely prevent the carryover and amplification of linker-linker products (3′ adapters ligated to 5′ adapters). These products undergo RT and are amplified by PCR, yielding a 71 bp long product. Therefore, in both protocols PCR-amplified libraries have to be size selected for depletion of amplified linker-linker products before sequencing (Fig. 2F, G).
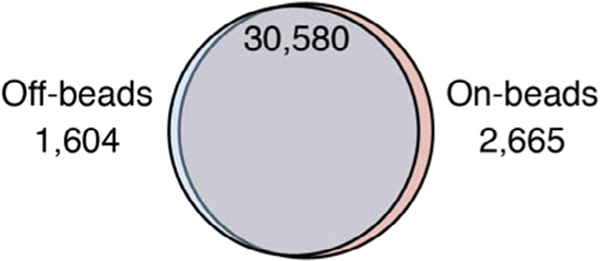
The bioinformatic analysis pipeline is identical for both approaches, and multiple computational approaches have been described [21]. We want to note again that “on-beads” PAR-CLIP requires accessibility of RNA ends within the RNP complex and therefore may technically fit some RBPs better than others. Nevertheless, we applied “on-beads” and “off-beads” PAR-CLIP to several RBPs and find good concordance between the obtained results, as demonstrated by the greater than 90% overlap of the ~30,000 identified YBX1 binding sites (Fig. 3).
Fig. 3.
Overlap of YBX1 binding sites determined by PAR-CLIP using the “off-beads” and the “on-beads” cDNA library preparation procedures. Overlap of at least one nucleotide was determined using the bedtools intersect program using the –u option. Hypergeometric test for the significance of the overlap results in P = 10E-12881.21.
Acknowledgments
D.B., H.M., A.S., and M.H. are supported by the Intramural Research Program of NIAMS. A.S. acknowledges support by the Vetenskapsradet Postdoctoral Fellowship.
References
- [1].Gerstberger S, Hafner M, Tuschl T, A census of human RNA-binding proteins, Nature Publishing Group, 2014. 829–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Castello A, Hentze MW, Preiss T, Metabolic enzymes enjoying new partnerships as RNA-binding proteins, Trends Endocrinol. Metab 26 (2015) 746–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kim Y. et al. , PKR is activated by cellular dsRNAs during mitosis and acts as a mitotic regulator, Genes Dev. 28 (2014) 1310–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ray D. et al. , A compendium of RNA-binding motifs for decoding gene regulation, Nature 499 (2013) 172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kishore S, Luber S, Zavolan M, Deciphering the role of RNA-binding proteins in the post-transcriptional control of gene expression, Brief Funct. Genom 9 (2010) 391–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Van Nostrand EL et al. , Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP), Nat. Methods 13 (2016) 508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Licatalosi DD et al. , HITS-CLIP yields genome-wide insights into brain alternative RNA processing, Nature 456 (2008) 464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Schneider C, Kudla G, Wlotzka W, Tuck A, Tollervey D, Transcriptome-wide analysis of exosome targets, Mol. Cell 48 (2012) 422–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hendrickson G, Kelley DR, Tenen D, Bernstein B, Rinn JL, Widespread RNA binding by chromatin-associated proteins, Genome Biol. 17 (2016) 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hafner M. et al. , Transcriptome-wide identification of RNA-binding protein and MicroRNA target sites by PAR-CLIP, Cell 141 (2010) 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ascano M, Hafner M, Cekan P, Gerstberger S, Tuschl T, Identification of RNA-protein interaction networks using PAR-CLIP, Wiley Interdiscip. Rev. RNA 3 (2012) 159–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kramer K. et al. , Photo-cross-linking and high-resolution mass spectrometry for assignment of RNA-binding sites in RNA-binding proteins, Nat. Methods 11 (2014) 1064–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kishore S. et al. , A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins, Nat. Methods 8 (2011) 559–564. [DOI] [PubMed] [Google Scholar]
- [14].Friedersdorf MB, Keene JD, Advancing the functional utility of PAR-CLIP by quantifying background binding to mRNAs and lncRNAs, Genome Biol. 15 (2014) R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mili S, Steitz JA, Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses, RNA 10 (2004) 1692–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lau NC, Lim LP, Weinstein EG, Bartel DP, An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans, Science 294 (2001) 858–862. [DOI] [PubMed] [Google Scholar]
- [17].Hafner M. et al. , RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries, RNA 17 (2011) 1697–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hafner M. et al. , Barcoded cDNA library preparation for small RNA profiling by next-generation sequencing, Methods 58 (2012) 164–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hafner M. et al. , Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing, Methods 44 (2008) 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells, 16 (2009) 45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Dutka T, Sarshad AA, Hafner M, Field Guidelines for Genetic Experimental Designs in High-Throughput Sequencing 261–289, Springer International Publishing, 2016, 10.1007/978-3-319-31350-4_11. [DOI] [Google Scholar]