Abstract
In this paper, we present a mathematical model with experimental support of how several key parameters govern the adsorption of active retrovirus particles onto the surface of adherent cells. These parameters, including time of adsorption, volume of virus, and the number, size, and type of target cells, as well as the intrinsic properties of the virus, diffusion coefficient, and half-life (t1/2), have been incorporated into a mathematical expression that describes the rate at which active virus particles adsorb to the cell surface. From this expression, we have obtained estimates of Cvo, the starting concentration of active retrovirus particles. In contrast to titer, Cvo is independent of the specific conditions of the assay. The relatively slow diffusion (D = 2 × 10−8 cm2/s) and rapid decay (t1/2 = 6 to 7 h) of retrovirus particles explain why Cvo values are significantly higher than titer values. Values of Cvo also indicate that the number of defective particles in a retrovirus stock is much lower than previously thought, which has implications especially for the use of retroviruses for in vivo gene therapy. With this expression, we have also computed AVC (active viruses/cell), the number of active retrovirus particles that would adsorb per cell during a given adsorption time. In contrast to multiplicity of infection, which is based on titer and is subject to the same inaccuracies, AVC is based on the physicochemical parameters of the transduction assay and so is a more reliable alternative.
Recombinant retroviruses are promising vehicles for the transfer of genes into mammalian cells for the purpose of gene therapy and have been tested clinically for the treatment of a variety of diseases, including cancer and AIDS (8). As the number of clinical studies increases, the quantitation of stocks of retrovirus and comparisons between different laboratories and clinical trials will become increasingly important. This task can be facilitated by a quantitative understanding of the steps of retrovirus-mediated gene transfer. These analyses can provide insight into the effects that key physicochemical factors have on transduction and suggest new strategies to improve the efficiency of retrovirus-mediated gene transfer.
Stocks of recombinant retroviruses are typically quantitated by measuring titer, the number of gene transfer events per unit volume of retrovirus solution. To determine titer, the virus stock is first diluted a few 1,000-fold and then used to transduce target cells. The titer, expressed as the number of CFU/milliliter, is the number of colonies of transduced cells multiplied by the dilution factor and divided by the volume of retrovirus applied to the target cells. The visualization and quantitation of transduced cells are achieved by the use of retrovirus vectors that carry reporter genes, such as the lacZ gene, or antibiotic resistance genes, such as the neo gene.
Although titer is a conventional measure of retrovirus bioactivity, it suffers from certain problems and inconsistencies. The number of gene transfer events depends on the specific conditions of the assay, and standardized conditions have not been established. Numerous factors can influence titer, including the time of exposure of cells to the virus, the number and type of target cells, the volume of the virus-containing medium, and the half-life (t1/2) of retrovirus particles. Thus, titer reflects the number of gene transfer events under a highly specific set of conditions. With these caveats, it is clear that titer is not an absolute measure of the concentration of active retrovirus particles. Compounding these problems is the fact that values for titer are incorporated into the calculation of the multiplicity of infection (MOI), the expected number of gene transfer events per cell. Even though MOI has been shown to be an unreliable and inaccurate predictor of the transduction process (15), it is still commonly used to predict the extent to which a cell population is genetically modified. Clearly, a more accurate and reliable quantitative measure of retrovirus activity is needed to help standardize stocks of virus as well as predict the potency with which these virus stocks can deliver genes to a target cell population.
Here, we present experimental data and mathematical analyses of a critical step in retrovirus-mediated gene transfer, the diffusion and adsorption of virus particles on target cells. We show that the adsorption of these relatively large virus particles is diffusion limited. Moreover, transduction is also limited by the fact that virus particles lose bioactivity with time (t1/2 = 6 to 7 h). Thus, only a small fraction (∼10%) of the total number of active virus particles in a stock are able to diffuse, adsorb, and successfully transduce a target cell before the particles lose their biological activity. These results demonstrate that, contrary to widespread perception, many of the particles in a stock of recombinant retroviruses are initially active particles. Stocks do not contain large numbers of defective particles. Rather, the combined limitations of slow diffusion and virus decay limit the ability of particles to successfully infect a cell. We have incorporated these physicochemical parameters of diffusivity and half-life, along with the number and size of the target cells and the time of adsorption, into a mathematical model that can be used to calculate the initial concentration of active retrovirus particles in a starting stock (Cvo). Since Cvo is independent of the conditions of the transduction assay, it is more reliable and may aid in the standardized comparisons of the potency of stocks of virus. Moreover, with this model, we present an alternative to MOI that is more reliable at predicting the expected number of gene transfer events in a population of adherent cells.
MATERIALS AND METHODS
Cell culture.
Human fibroblasts (HuFb) were isolated from newborn human foreskins. Briefly, the tissue was trimmed to remove the fat and muscle layers underlying the dermis and then repeatedly rinsed in sterile phosphate-buffered saline (PBS). The tissue was subsequently cut into small pieces (0.2 × 0.2 cm2) and placed on the tissue culture plate, with the dermal side contacting the tissue culture plate. After allowing the skin pieces to dry to the tissue culture plate for approximately 45 min, 10 ml of Dulbecco's modified Eagle's medium (Gibco BRL, Gaithersburg, Md.) containing 20% fetal bovine serum (HyClone Laboratories, Inc., Logan, Utah), 100 U of penicillin, and 100 μg of streptomycin (Gibco BRL) per ml was added to the tissue culture plate and placed in an incubator at 37°C with 10% CO2. The medium was changed every 3 to 4 days. Seven to 10 days later, fibroblasts migrated out from the dermis onto the surface of the tissue culture plate. Thereafter, the cells were passed every week when they reached confluence.
NIH 3T3 cells and lacZ virus-producing cell lines were cultured in Dulbecco's modified Eagle's medium (Gibco BRL) with 10% bovine calf serum (HyClone Laboratories, Inc.), 100 U of penicillin, and 100 μg of streptomycin (Gibco BRL) per ml at 37°C with 10% CO2. lacZ virus-containing medium was harvested from confluent cultures of an amphotropic packaging cell line (ΨCRIP) (15). Fresh medium (10 ml in a 10-cm tissue culture dish) was added to the cells 24 h before the virus was harvested. The lacZ virus-containing medium was filtered through 0.45-μm-pore-size filters (Gelman Sciences, Ann Arbor, Mich.), aliquoted, and stored at −80°C until use.
Transduction assay.
Tenfold serial dilutions of the lacZ virus stocks were made in cell culture medium. Polybrene was added at a concentration of 8 μg/ml and 2 ml of virus was added to the target cells plated in six-well plates (Costar Corp., Cambridge, Mass.) the previous day. At the end of exposure of cells to the virus (time as indicated in each experiment), the virus-containing medium was removed, fresh medium was added, and the cells were allowed to grow for 48 h. Subsequently, the cells were fixed and stained for β-galactosidase activity with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). Briefly, cells were washed with PBS and fixed in PBS containing 0.5% glutaraldehyde for 10 min at room temperature. Plates were washed with PBS containing 1 mM MgCl2 and stained for lacZ activity by incubation in a solution containing PBS, 1 mM MgCl2, 3.3 mM K3Fe(CN)6, 3.3 mM K4Fe(CN)6 · 3H2O, and 1 mg of X-Gal per ml overnight at 37°C. The reaction mixture was removed, and the cells were washed with PBS and air dried. Colonies of lacZ+ cells were counted with the aid of a dissecting microscope.
Measurements of target cell surface area.
The average size of the target cells (NIH 3T3 and HuFb) was determined experimentally by fluorescence microscopy. Cells were plated in six-well plates, and the next day they were stained with 10 μmol of the cytoplasmic dye CellTracker Orange CMTMR (5-(and-6)-(((4-chloromethyl)benzoyl)amino)tetramethylrhodamine) for 60 min at 37°C (Molecular Probes, Eugene, Oreg.). The CellTracker probe allows labeling of viable cells for at least 24 h after loading and often through several cell divisions. The cells were viewed with a fluorescence microscope (Nikon Diaphot). Forty areas were randomly selected in six identical wells for image analysis. In each area, several cells were selected at random and the surface area was determined by delineating the perimeter of each cell with the computer software Metamorph (Universal Imaging Corp., West Chester, Pa.). When cells were close to each other so that their borders could not be easily distinguished, they were excluded from the analysis. More than 100 cells were used and the average surface area was calculated.
Quantitation of retrovirus adsorption by an ELISA for p30 capsid protein.
Target cells (NIH 3T3 and HuFb) were grown to confluence in six-well plates, and 1 ml of undiluted virus was added at t = 0. At various times, the virus was removed, centrifuged to remove debris, and stored at −80°C. The concentration of virus particles was measured by an enzyme-linked immunosorbent assay (ELISA) for the p30 capsid protein, as previously described (9). Briefly, the wells of 96-well ELISA plates (Fisher Scientific, Agawam, Mass.) were coated with 100 μl of a p30 antibody (10 μg/ml) (purified from the supernatant harvested from the CRL-1912 hybridoma cell line [American Type Culture Collection, Rockville, Md.]) per ml with overnight incubation at 4°C. The next day, nonspecific binding sites were blocked with 200 μl of BLOTTO blocker in Tris-buffered saline (Pierce, Rockford, Ill.) for 30 min at 37°C. The samples containing the lacZ virus were thawed at 37°C and boiled for 5 min to expose the capsid protein. The denatured particles were then incubated (at 100 μl per well) for 1 h at 37°C. After washing with PBS–0.05% Tween 20, a secondary antibody, goat polyclonal anti-p30 antibody (78S221; Quality Biotech, Camden, N.J.), was added at a 1:300 dilution in BLOTTO (100 μl per well) for 1 h at 37°C. This was followed by washing with PBS–0.05% Tween 20 and incubation for 1 h at 37°C with 100 μl of a horseradish peroxidase-conjugated rabbit anti-goat immunoglobulin G polyclonal antibody (Zymed Laboratories, South San Francisco, Calif.) diluted 1:5,000 in BLOTTO. The substrate (10 mg of o-phenylenediamine dihydrochloride and 10 μl of H2O2 in 25 ml of substrate buffer containing 5.1 mg of citric acid mono-hydrate per ml and 13.78 mg of Na2HPO4 · 7H2O per ml in distilled water) was added (100 μl per well), and the reaction was allowed to proceed for 10 min before the addition of 50 μl of 8 N H2SO4 (stop solution) per well. The optical density was read at 490 to 650 nm with an ELISA plate reader (ThermoMax plate reader; Molecular Devices, Menlo Park, Calif.).
RESULTS
Adsorption of retrovirus particles is diffusion limited.
Valentine and Allison used the theory of Brownian motion to develop a model for the adsorption of virus particles on flat surfaces (24). In their model, the fraction of adsorbed viruses, fads, is given as a function of time, t, the diffusion coefficient of virus particles is D, and the depth of the virus-containing medium is l. The fraction of adsorbed viruses is defined as the ratio of the number of adsorbed viruses, N, to the total number of virus particles in solution, which is the product of virus concentration, Cvo, times the volume of the virus-containing medium, V. Mathematically, the fraction of adsorbed viruses is as follows:
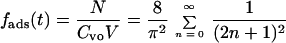 |
1 |
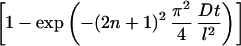 |
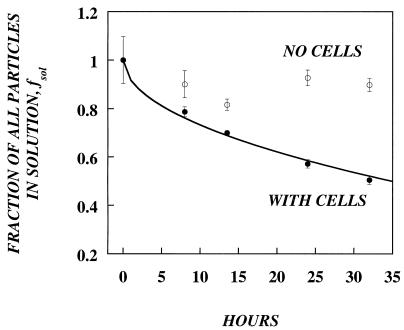
In order to analyze the validity of this model in a retrovirus-target cell system, we measured the adsorption of retrovirus to fibroblasts with an ELISA for the p30 capsid protein. Retrovirus-containing medium was incubated at 37°C in tissue culture dishes with or without a confluent monolayer of NIH 3T3 cells. At various times, aliquots were removed and assayed for levels of p30 (Fig. 1). Without cells, the amount of p30 remained relatively constant for the duration of the experiment, suggesting that nonspecific binding and degradation of p30 were minimal. In the presence of cells, the fraction of retrovirus particles that remained in solution, fsol, decreased with time and reached almost 50% in 33 h. With nonlinear regression analysis to fit equation 1 to the fraction of adsorbed data (fads = 1 − fsol), we estimated a diffusion coefficient (D) for retroviral particles. The best-fit value of D was 1.74 × 10−8 cm2/s, which is close to the diffusion coefficient of other similar-size retroviruses (2 × 10−8 cm2/s) (21).
FIG. 1.
Kinetics of retrovirus adsorption. lacZ retrovirus with Polybrene was added either to an empty plate (open circles) or to a plate confluent with NIH 3T3 fibroblasts (solid circles). Adsorption was monitored with an ELISA that measured the amount of p30 remaining in the medium. The diffusion coefficient was calculated by fitting the experimental data to the Valentine and Allison equation (line) by nonlinear regression analysis. The best-fit value, D = 1.74 × 10−8 cm2/s.
Retrovirus particles are intrinsically unstable.
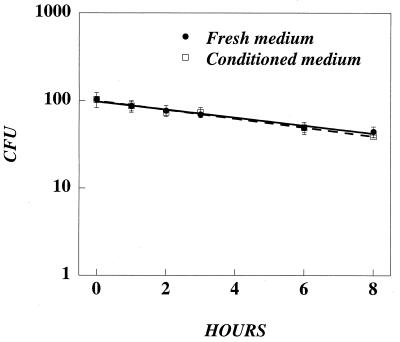
Although equation 1 fits well with the adsorption data, it is unable to predict the number of active retrovirus particles that are adsorbed over time because it does not take into account the instability of retrovirus particles. Previous studies have shown that retroviruses lose activity with time. For several retroviruses used for gene therapy applications, the half-life has been calculated to be 5 to 8 h (7, 11, 12, 18). For other retroviruses, the half-life has been reported to be 3 to 9 h (19). Each of these studies measured half-life in undiluted stocks of virus that also contained conditioned medium produced by the packaging cell line. To determine if decay was intrinsic to the virus particle or was affected by the presence of conditioned medium, we compared the stability at 37°C of undiluted versus diluted lacZ virus (diluted 1:100 in fresh medium). At various time points, aliquots were removed, further diluted in fresh medium, and used to transduce NIH 3T3 cells. Undiluted and diluted lacZ virus declined with nearly identical kinetics (t1/2 = ∼6.5 h) (Fig. 2), suggesting that decay is intrinsic to the particles and is not influenced by conditioned medium.
FIG. 2.
Recombinant retroviruses decay with a half-life of ∼6.5 h in fresh and conditioned media. Undiluted (solid circles) or diluted (1:100) lacZ retrovirus (open circles) was incubated at 37°C. At various times, 0.5-ml aliquots were taken and frozen. To test the activity of the virus, NIH 3T3 cells were plated at 6.0 × 104 cells per well in six-well plates and the next day were transduced with 1.0 ml of lacZ retrovirus for 4.0 h at a final dilution of 1:500 in the presence of Polybrene. Two days later, the number of gene transfer events (CFU) was measured by X-Gal staining.
We modified equation 1 to account for decay, assuming decay is a first-order reaction; thus the fraction of adsorbed retrovirus particles which are active, factive, is given as
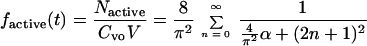 |
2 |
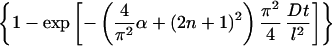 |
The only difference is the term α, which is equal to the rate of decay, kdv, divided by the rate of diffusion, D/l2. With very long adsorption times, the exponential term in equation 2 tends to be 0, and the fraction of adsorbed active particles reaches a constant whose value is ≤1. If a virus is stable and does not decay, then α = 0, equation 2 becomes identical to equation 1, and for very long times the fraction of adsorbed particles asymptotically approaches a value of 1. If, however, a virus decays, only a fraction of the virus particles will then be active by the time they diffuse and adsorb on the cell surface. At very long times, the fraction of adsorbed particles asymptotically approaches a value, <1, that depends on the rate of decay relative to the rate of diffusion as expressed in the ratio α.
Decay limits the number of virus particles that have activity when they adsorb.
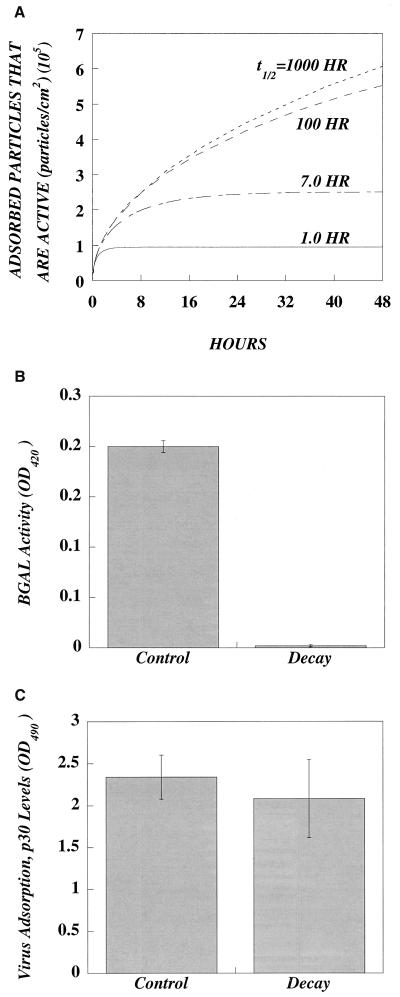
To reveal the relationship between decay and the adsorption of active particles, we simulated the adsorption of viruses of differing decay rates or half-lives with equation 2 (Fig. 3). Adsorption to a confluent layer of cells in a six-well plate with 1 ml (l, ∼1.0 mm) of virus-containing medium was simulated. At the start, the virus concentration was 107 virus particles/ml and all virus particles were active. As shown in Fig. 3, the number of adsorbed particles that are active decreases as the half-life decreases. Although all were active at the start, viruses with short half-lives lose activity during the time it takes to diffuse to the cell surface.
FIG. 3.
Active and inactive particles adsorb, but virus decay limits the number of adsorbed active particles. (A) Simulation of the effect of various half-lives on the adsorption of active retrovirus with equation 2. Confluent monolayers of cells in a six-well plate were exposed for 48 h to 1.0 ml of retrovirus with an initial concentration of 107 active particles per ml and different half-lives. Both the rate of adsorption of active particles and the steady-state levels decrease as half-life decreases. (B) Decay of virus activity has minimal effect on virus adsorption. lacZ virus was divided into two aliquots; one was decayed by incubation for 24 h at 37°C, and the other was an untreated control. (C) To measure adsorption, NIH 3T3 cells (1.5 × 105 per well) were plated in a six-well dish. After 72 h, the medium was replaced with the control or with decayed virus sample containing 8 μg of Polybrene per ml. After 2 h, the virus was removed, the cells were lysed, and the lysate was analyzed for the presence of p30 capsid protein via ELISA.
It also follows that if significant numbers of viruses are losing activity, those viruses which do successfully infect a cell represent only a fraction of the total number of active particles in the starting stock. With the data from Fig. 3, we calculated the fraction of the initial virus particle number that would have activity after a 24-h adsorption. This fraction decreases with decreasing half-life (i.e., 41%, t1/2 = 1,000 h; 39%, t1/2 = 100 h; 23%, t1/2 = 7 h; and 9%, t1/2 = 1 h). Thus, for a typical retrovirus with a half-life of 7 h, this simulation shows that for every 5 particles that adsorb during the 24 h, at the most only ∼1 will have activity when it adsorbs, even if all particles were active at the start of transduction. Conversely, for each successful gene transfer event there must be, at least, ∼4 other virus particles that were active in the initial stock at the start of the transduction assay.
Although decay limits the number of adsorbed particles that are active, it was unclear whether active and inactive particles adsorb with the same efficiency. To determine if decay influences adsorption, we thawed a stock of virus and divided it into two aliquots. The control aliquot was refrozen immediately, while the decay aliquot was refrozen after incubation for 24 h at 37°C. The bioactivity of the decayed stock was about 100 times lower than that of the control (Fig. 3B). We then performed 2-h adsorption experiments on NIH 3T3 cells with these same samples (Fig. 3C). The control and decayed stocks yielded comparable levels of p30 adsorption, suggesting that the loss of virus activity does not impair the adsorption of the particle.
Typical volumes of virus are in excess and do not limit the number of virus particles that have activity when they adsorb.
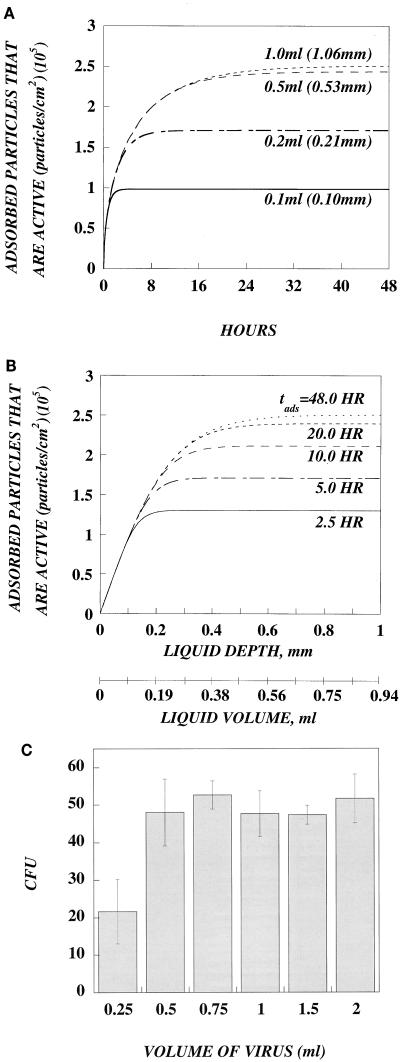
Given the limitations caused by diffusional resistances and decay, the depth of the virus-containing medium should play a role in viral adsorption. We used equation 2 to simulate the effects of various depths on the number of adsorbed, active retrovirus particles. As shown in Fig. 4A, there is a critical depth beyond which the number of adsorbed active particles remains relatively unchanged. We define the critical depth as the depth at which the number of adsorbed active particles reaches 95% of the maximum (plateau) value. Since the distance that a particle travels increases with time (l = √2Dt), the critical depth increases with the time of adsorption (i.e., 0.16 mm, tads = 2.5 h; 0.23 mm, tads = 5 h; 0.33 mm, tads = 10 h; 0.4 mm, tads = 20 h; and 0.45 mm, tads = 48 h) (Fig. 4B). The largest critical value corresponds to the distance that a virus particle travels within approximately two half-lives (in this simulation, t1/2 = 7 h). Therefore, for volumes of 0.5 ml and larger (typically used in a six-well plate), adsorption of active particles does not increase with increasing volume. Although an increase in volume above 0.5 ml provides more total active particles, the distance they must diffuse to reach the cell surface is also increased. As a result, most of the additional particles lose activity before they are adsorbed. For exceedingly small volumes, the situation is different. At low volumes (<0.2 ml), the diffusional distance to the target cells is short, and thus increasing volume does result in the adsorption of more active particles. However, these small volumes are rarely used. Typical transductions are not limited by the volume of virus.
FIG. 4.
Effect of various volumes on the adsorption of active retrovirus. (A) Simulation of the effect of various volumes on the adsorption of active retrovirus with equation 2. Confluent monolayers of cells in a six-well plate were incubated for 48 h with different volumes of retrovirus with an initial concentration of 107 active particles per ml (t1/2 = 7.0 h). This simulation shows that for volumes which are typically used (0.5 and 1.0 ml), there is no significant difference in the number of adsorbed active particles. Beyond a critical volume, there is no significant increase in the number of adsorbed active particles. Differences only occur for exceedingly small volumes that are rarely used in six-well plates (0.2 and 0.1 ml). (B) Data from the same simulation plotted as a function of liquid depth or volume for different adsorption times to more accurately illustrate the threshold volume. The critical volume varies for different adsorption times, because the particles have additional time to diffuse greater distances. (C) The number of infectious events does not increase for volumes of retrovirus above the critical volume. NIH 3T3 cells (8.0 × 104 cells/well; six-well plate) were transduced for 2.5 h the next day with different volumes of lacZ retrovirus diluted 1:500. Two days later, the number of CFU was measured by X-Gal staining. Increasing the volume of retrovirus fourfold from 0.5 to 2.0 ml did not increase the number of CFU, in agreement with the predictions of the mathematical model. A decrease in CFU occurred only with 0.25 ml of virus, a volume below the threshold volume.
This simulation of varying volumes shows that error can be introduced into the computation of titer. Titer (CFU/milliliter) is the number of CFU times the dilution factor of the virus stock, divided by the volume of virus used in the transduction assay. If the liquid depth of the virus is above the critical value, CFU is not linearly dependent on volume and error can be introduced into titer if CFU is normalized to volume. For example, if 2.0 ml of virus-containing medium is used in a six-well plate, then the titer may be underestimated by at least fourfold, since the number of CFU should be the same when the volume is between 0.5 and 2.0 ml.
To test this prediction, transduction experiments were performed with different volumes of virus (0.25 to 2 ml), and the target cells were exposed to the virus for 2.5 h (the same conditions as in our simulation) to avoid evaporation of the medium. As shown in Fig. 4C, the number of CFU was unchanged for volumes of 0.5 ml or greater, in agreement with the prediction of equation 2 (Fig. 4A and B). A decrease in the number of CFU only occurred with 0.25 ml, a volume of virus below the critical volume.
The number of gene transfer events is proportional to cell density.
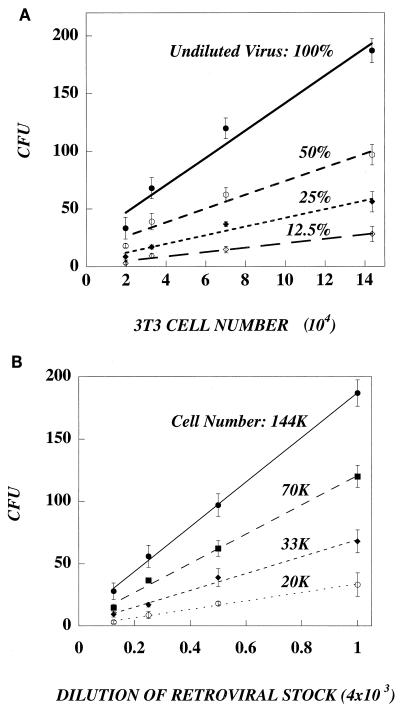
An additional complexity in retrovirus-mediated gene transfer is the fact that the target cells are never infected at confluence. Rather, the cells are plated at subconfluent densities in order to promote cell division, a requirement for successful retrovirus transduction (14). To test the relationship between cell density and transduction, we transduced varying numbers of NIH 3T3 cells with a range of lacZ virus concentrations for 4 h to minimize changes in cell density (Fig. 5). Not surprisingly, the number of CFU is proportional to virus concentration, with less-diluted virus stocks producing more CFU with all cell densities tested (Fig. 5A). Figure 5B also shows that CFU is proportional to cell density. For all virus concentrations, the number of CFU is larger when the cell density is increased. The probability of a virus contacting a cell is increased as the dish is covered with more cells. Linearity begins to break down at the higher cell densities, presumably because transduction is influenced by changes in the rate of cell division. In a typical transduction assay, cell density is not standardized between labs, and it is clear from Fig. 5 that cell density can significantly alter the values of titer. There was a sixfold difference in the number of CFU when the same virus concentration was assayed on 2 × 104 versus 14 × 104 cells.
FIG. 5.
The number of gene transfer events is proportional to the concentration of retrovirus and the number of target cells. NIH 3T3 cells were plated at various densities (1.0 × 104, 2.0 × 104, 4.0 × 104, and 8.0 × 104 cells/well; six-well plate) and were transduced the next day (3.5 h) with 1.0 ml of lacZ retrovirus of different dilutions (1:250, 1:500, 1:1,000, and 1:2,000) in the presence of Polybrene. At the start of transduction, the number of target cells was counted in parallel wells. Two days later, the number of CFU was measured by X-Gal staining. The number of CFU is plotted as a function of virus dilution (A). CFU increases as virus concentration increases. The number of CFU is plotted as a function of cell number at the start of transduction (B). The number of CFU increases as cell number increases.
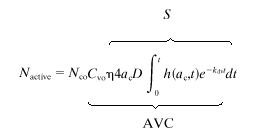
Equations 1 and 2 are valid only when target cells cover the entire surface of the dish, but standard transductions are performed on subconfluent cultures. In this surface topography, viruses adsorb on adsorbent patches (target cells) sparsely distributed on a nonadsorbent surface, thereby requiring a different mathematical treatment to describe the particle flux. Since the cell densities typically used in transduction experiments are low, the distance between target cells is on the order of a few cell diameters. In this case, it is reasonable to assume that the diffusion of virus particles onto one cell does not affect the diffusion onto a neighboring cell. We approximated this problem as equivalent to the diffusion of virus particles onto the surface of a disk-shaped target cell lying on a flat plate, a problem previously solved for the flux of current at a stationary disk electrode (22). Therefore, to derive an equation that estimates the number of active virus particles, Nactive, adsorbed on a dish of subconfluent cells, we modified the equation of Shoup and Szabo (22) to account for virus decay as follows:
 |
3 |
where AVC is the number of adsorbed active retroviral particles per target cell, Nco is the total number of cells at the start of transduction, ac is the average radius of the target cells, D is the diffusion coefficient of the virus, Cvo is the starting concentration of active retrovirus particles, and S is the number of active virus particles adsorbed on a cell relative to a starting virus concentration. The function h(t) was derived by Shoup and Szabo and reflects the flux of particles on each cell at time t, over the steady-state particle flux (22). Since transduction is a multistep process (adsorption through gene expression) and not all steps proceed with 100% efficiency, we introduced the term η, the overall efficiency of ≤1. For NIH 3T3 cells that are known to have high transduction efficiency, we set η = 1.
From equation 3, it can be seen that most of the variables are easily defined or controlled when setting up a transduction. For a range of commonly used adsorption times and several target cell radii, we have provided numerical values for the integral of equation 3 (Table 1). One term that needs to be measured is ac, the average radius of the target cells. To compute the radius, we measured the area of 100 NIH 3T3 cells in 40 randomly chosen fields. The average area of a single cell was 152.7 ± 48.8 μm2, yielding an average radius of approximately 7 μm. For short adsorption times (2 to 4 h), the total area occupied by all cells remains approximately constant. The other unknown term of equation 3 is Cvo, the concentration of active retrovirus particles in the medium at the start of transduction. This term can be calculated with equation 3, and then the concentration of active retroviruses in the undiluted virus stock is obtained by multiplying Cvo by the dilution factor applied when setting up the assay to measure CFU. Alternatively, Cvo can be calculated by dividing the slope of the linear curves (CFU versus Nco) by the function S (see equation 3).
TABLE 1.
Numerical values of the integral in equation 3 for various times of exposure of cells to the virus and target cell radii, ac
| t (h) |
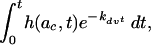 where ac (μm) is where ac (μm) is
|
|||
|---|---|---|---|---|
| 7 | 10 | 15 | 20 | |
| 0.5 | 0.533 | 0.553 | 0.586 | 0.622 |
| 1.0 | 1.015 | 1.042 | 1.088 | 1.135 |
| 1.5 | 1.469 | 1.502 | 1.557 | 1.613 |
| 2.0 | 1.900 | 1.937 | 1.999 | 2.063 |
| 2.5 | 2.309 | 2.349 | 2.418 | 2.487 |
| 3.0 | 2.697 | 2.741 | 2.815 | 2.889 |
| 3.5 | 3.066 | 3.112 | 3.191 | 3.270 |
| 4.0 | 3.416 | 3.465 | 3.548 | 3.631 |
| 4.5 | 3.750 | 3.801 | 3.887 | 3.974 |
| 5.0 | 4.067 | 4.120 | 4.209 | 4.299 |
| 5.5 | 4.368 | 4.423 | 4.515 | 4.609 |
| 6.0 | 4.655 | 4.711 | 4.806 | 4.902 |
| 6.5 | 4.927 | 4.986 | 5.083 | 5.181 |
| 7.0 | 5.187 | 5.246 | 5.346 | 5.447 |
| 7.5 | 5.434 | 5.494 | 5.596 | 5.699 |
| 8.0 | 5.668 | 5.730 | 5.834 | 5.939 |
Equation 3 predicts that the number of CFU is linearly proportional to cell density. As shown in Fig. 5, this is true over a range of cell densities, but it does not hold at very high cell densities. Therefore, for each cell type, it is important to determine the range of target cell numbers for which the number of CFU is linear. With the slopes of all four curves in Fig. 5B, we determined Cvo as the ratio slope/S. The average value of Cvo was (6.25 ± 0.85) × 105 active retroviral particles/ml when assayed on NIH 3T3 cells.
Cvo is not the same as titer. As shown in Fig. 5, values of titer would change sixfold, depending on how many cells were used in the transduction assay. The same lacZ virus stock has a titer of 8.3 × 103 CFU/ml when assayed on the lowest cell number or 4.7 × 104 CFU/ml when assayed on the highest cell number. In contrast to titer, Cvo was independent of target cell number. Also of interest is the fact that values of titer range from 1 to about 8% of the concentration of active virus particles, Cvo. This suggests that the initial concentration of active retrovirus particles is much higher than the number of particles that successfully produce a gene transfer event (12).
RTE of target cells.
In equation 3, all postadsorption steps are lumped into the term η; however, this factor may vary between target cell types. To compare the transducibility of different cell types, we used the same lacZ stock to transduce varying numbers of either NIH 3T3 cells or diploid human fibroblasts. As shown in Fig. 6, values of CFU were lower on human fibroblasts than on NIH 3T3 cells, even though human fibroblasts were slightly larger than NIH 3T3 cells, with an average surface area of 285 μm2 and an average radius of 10 μm. In the range of cell densities where the number of CFU is linearly proportional to the number of target cells, we can write equation 3 for both cell types and then divide by parts to obtain an expression for the ratio of efficiencies or the relative transduction efficiency (RTE), as a function of the ratio of the number of gene transfer events and the ratios of the number and radii of the target cells:
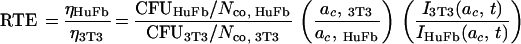 |
4 |
where I is the integral as shown in equation 3 (for numerical values, see Table 1). Alternatively, the ratio of lumped efficiencies can be written as a function of the slopes of the linear curves (from equation 4):
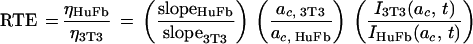 |
5 |
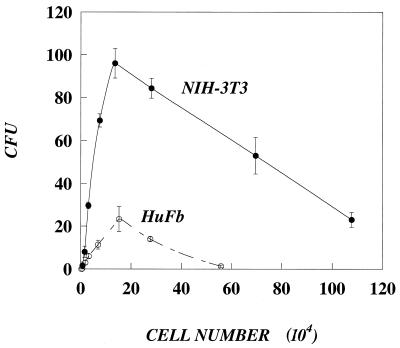
The integral, I, is a weak function of the cell radius. The ratio of the integrals may be significant only for short adsorption times (Table 1), but for longer times (e.g., 4 to 8 h), it is only slightly greater than one and, therefore, it does not contribute significantly to the value of RTE. With equation 5 and the slopes of the linear parts of the curves in Fig. 6, we calculated the lumped efficiency of transduction of human fibroblasts to be 15% of the efficiency of NIH 3T3 cells.
FIG. 6.
Values of CFU of the same retrovirus stock are different on NIH 3T3 cells versus HuFb. Cells were plated at various densities and were transduced the next day (3.5 h) with 1.0 ml of lacZ retrovirus (diluted 1:250). At the start of transduction, the number of target cells was counted in parallel wells. Two days later, the number of CFU was measured by X-Gal staining. The number of CFU of the same virus stock is lower on human fibroblasts; however, for both cell types there is a range of cell densities where the number of CFU is linearly proportional to cell number.
AVC: an alternative to MOI.
The MOI (the number of infectious viruses per cell) is typically used to predict the extent to which a cell population is transduced. However, since titer is used to compute MOI, it suffers from the same lack of standardization and accuracy. As an alternative, we propose AVC, the number of active retrovirus particles that would adsorb per cell during a given adsorption time. According to equation 3, the only variables needed to calculate AVC are Cvo and η. For example, if NIH 3T3 cells (ac = 7 × 10−4cm) (η = 1) are infected for 4 h with a viral stock with 106 active particles/ml (i.e., Cvo = 106 active particles/ml) and the virus has a 7-h half-life, the expected number of adsorbed active particles will be 0.6 particles/cell. Since the transduction efficiency η is probably less than one (η ≤ 1), the value of AVC is an upper limit. For cell targets other than NIH 3T3 cells, this analysis requires the measurement of RTE.
DISCUSSION
Although titer is a widely used quantitative measure of the bioactivity of a stock of recombinant retrovirus, the key parameters of the transduction assay used to compute titer have not been standardized. Titer is a measure of the number of retroviruses that successfully transduce target cells under a very specific set of conditions, and thus rigorous comparisons between experiments, laboratories, and clinical trials are difficult. Our data and other published studies have shown significant problems with titer (15). In this study, we show that titer can vary (i) 6-fold, depending on the number of target cells; (ii) ∼6- to 7-fold, depending on the target cell type (NIH 3T3 cells versus human fibroblasts); (iii) 3- to 4-fold, depending on the adsorption time (2 versus 24 h); and (iv) 4-fold, depending on the volume of virus used in the transduction assay. In this paper, we present a mathematical model with experimental support of how certain physicochemical parameters of the transduction assay (time of adsorption, volume of virus, target cell number, and target cell size), as well as the intrinsic properties of virus particles themselves (diffusion coefficient and half-life), influence the adsorption of active viruses and the measurement of the activity of a virus stock. From these analyses, we have derived Cvo, the starting concentration of active particles in a stock of retrovirus; AVC (active viruses/cell), the predicted number of active particles that would be adsorbed per cell in a given adsorption time; and RTE, the relative transduction efficiency of one cell type versus another. Since these expressions standardize the key parameters of the transduction assay, they are more reliable than titer (and MOI) and should aid in the quantitative comparison of data.
To compute Cvo from equation 3 requires input of the time of adsorption, cell number, and cell radius as well as the number of CFU from the transduction assay. To be accurate, the number of CFU must be linearly proportional to cell number and our data show that this is true over a range of cell densities. However, this range can vary between cell types; as cell numbers were increased beyond this range, the number of CFU declined, possibly due to a decrease in the rate of cell growth, which is known to influence transduction (2, 14). Equation 3 assumes that every particle that adsorbs on the cell surface will result in a successful gene transfer event; however, since the probability of each step after adsorption is ≤1 (parameter η in equation 3), values of Cvo are a lower limit. Nevertheless, Cvo, the concentration of active viruses at the start of transduction, is significantly higher than titer. This is explained by the fact that retrovirus-mediated gene transfer is limited by the slow diffusivity and decay of the virus particles, consistent with the conclusions of other investigators (5, 6). In one study, transduction was improved by flowing virus through a porous membrane with attached cells (6), which suggested that the stock contains additional active particles if they are brought to the target cells fast enough. In another study, the same virus stock was used to transduce multiple dishes of target cells in a series. The number of transduced cells changed only slightly with each successive dish (23), which suggested that the number of active viruses is greater than what can be determined by a single measurement of titer. Our simulation data show that transduction is independent of volume for liquid depths above 500 μm. Those viruses located more than 500 μm above the cells do not contribute appreciably to the number of gene transfer events, in agreement with our previous report (15) and those of other investigators (5, 6). Retroviral particles travel by diffusion approximately 300 μm in one half-life (t1/2 = 7 h). Taken together, these results suggest that retroviral stocks contain a significant fraction of particles that are active and could transduce a cell if they reached the cell surface before they decay.
Since Cvo is higher than titer, this indicates that a significantly larger fraction of virus particles in a stock are active than has been previously thought. Based on measurements of titer, it has been widely believed that only a very small fraction of the particles in a stock are active and the vast majority are inactive defective particles. It has been estimated that between 0.1 and 1% of the particles are active (20). In our study, we show that the limitations of virus diffusion and decay prevent many of the active virus particles in a stock from successfully transducing a cell. These particles are not inherently defective at the start of infection but rather lose activity before they can diffuse, adsorb, and successfully transduce a cell. The values of Cvo that we calculate in this paper are at least 10-fold higher than those of titer, and thus stocks of virus have many more active particles than previously thought. We do not assert that all particles are active in a stock of retrovirus. During production of the virus by the packaging cell line, virus decay does occur and defective particles do accumulate in the stock. Based on the kinetics of production and decay, we estimated in a previous study that, at most, ∼38% of the particles are active in a typical stock produced at 37°C over 24 h (12). The remainder of the particles are truly defective because they have decayed during the production process. In addition, the proportion of defective particles may be somewhat higher due to the possibility that some particles were defective at birth as a result of errors in assembly. Strategies that improve gene transfer, such as convective devices (6) and centrifugation (4), do so because they deliver more active virus particles to the cell surface in the shortest possible time. Good estimates of the true number of active virus particles in a stock, as provided by Cvo, can help determine the effectiveness of these and similar strategies as well as provide an estimate to the maximum possible benefit achievable with these approaches.
In addition to limitations due to slow diffusivity and decay, the efficiency of the various steps following the adsorption of an active virus (i.e., internalization, intracellular processing, integration, etc.) is not known and may vary between cell types. Therefore, we introduced the term η into equation 3 to cover these efficiencies. In this paper, we assume that η = 1 for NIH 3T3 cells because this cell type is highly infectable with recombinant retroviruses. However, this efficiency is probably <1, and thus it is likely that an active virus particle which diffuses and adsorbs on the cell surface may not lead to a successful transduction event, because it may lose activity at one of the many subsequent steps. Nevertheless, the particle was active (not defective) in the starting virus stock and was active when it adsorbed. If η < 1, then the proportion of active virus in a virus stock is even higher than the estimates provided in this paper. Thus, our estimates of Cvo, the concentration of active virus in the starting stock, is a lower limit.
The fact that titer does not accurately reflect the true number of active particles in a retrovirus stock has important implications for in vivo gene transfer. The related lentivirus vectors are being used for in vivo gene transfer, and the potency of these stocks is measured as a titer (16). Since titer significantly underestimates the number of active particles, it will be difficult to accurately quantitate the true potency of these stocks after in vivo injection.
We used our analysis to calculate RTE, the relative transduction efficiency of various cell types with respect to a standard cell type, such as NIH 3T3 cells. Equation 5 shows that the transduction efficiency—the probability of successful gene transfer following virus adsorption—is not just the simple ratio of the numbers of CFU on both cell types; rather, it must also incorporate the number and radii of each cell type. Comparison of the RTE of different cell types is accurate only when CFU is linearly proportional to cell number. The RTE of human fibroblasts, which have radii slightly larger than NIH 3T3 cells, was calculated to be 15% of the efficiency of NIH 3T3 cells, suggesting that, at most, one of six active viral particles that adsorb on human fibroblasts will lead to a successful gene transfer event. Since the flux of particles to a cell with a larger radius should be increased, the difference in RTE must be attributable to differences in the efficiency of any of the numerous processes that lead to successful integration and transgene expression that we have lumped into the term η. For example, NIH 3T3 cells divide more rapidly (in ∼12 h) than human fibroblasts (∼24 h) and since retroviruses decay after internalization with a half-life of about 6 h (1, 3), the additional time through the cell cycle could be one explanation for the decrease in transduction efficiency of human fibroblasts. Also, differences in the transduction efficiency of cell types have been attributed to differences in receptor number (17). By normalizing the physical parameters of the transduction assay, measurements of RTE should help provide more accurate comparisons of the transduction efficiencies of different cell types and aid biochemical investigations into the mechanisms of these differences.
We also computed AVC, the predicted number of active particles that would be adsorbed per cell in a given adsorption time, and propose this as an alternative to MOI. MOI, an expression based on titer, is often used to determine the probability of infection of a population of target cells by DNA and RNA viruses. However, recent experimental results have shown that increasing the number of retrovirus particles per target cell, either by increasing the volume of retrovirus solution (references 5 and 15 and data in this communication) or by decreasing the number of target cells (10, 15), did not result in increased levels of transduction, suggesting that MOI is not an appropriate measure of retrovirus activity (15). Equation 3 shows that the average number of active particles that adsorb on a target cell depends on the time of adsorption, the concentration of virus (not the total number of virus particles added), the radius of the target cell, the diffusion coefficient, and the half-life of the virus. Because AVC describes the number of active particles that adsorb per cell, it is useful for predicting the extent to which a population of cells should be genetically modified. If actual gene transfer efficiency is less than AVC, this would indicate a problem with one or more steps in the gene transfer process. For example, we recently reported that proteoglycans secreted by the packaging cell line limit gene transfer at the highest doses of virus (13). This would be apparent as a significant deviation from AVC at high, but not at low, virus doses (because proteoglycan concentration is reduced by dilution). Likewise, AVC helps to provide a measure of how well the transduction protocol is able to effect gene transfer. The higher the values of AVC, the more effective the transduction protocol. Because AVC is grounded in the physical parameters of the transduction assay, it should be a more reliable predictor of gene transfer than the currently used MOI.
ACKNOWLEDGMENTS
This work was supported in part by the NIH (HD-28528), NSF (BS 9800617), and the Shriners Hospital for Children.
REFERENCES
- 1.Andreadis S, Brott D A, Fuller A O, Palsson B O. Moloney murine leukemia virus-derived retroviral vectors decay intracellularly with a half-life in the range of 5.5 to 7.5 hours. J Virol. 1997;71:7541–7548. doi: 10.1128/jvi.71.10.7541-7548.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andreadis S, Fuller A O, Palsson B O. Cell cycle dependence of retroviral transduction: an issue of overlapping time scales. Biotechnol Bioeng. 1998;58:272–281. doi: 10.1002/(sici)1097-0290(19980420)58:2/3<272::aid-bit23>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 3.Andreadis S, Palsson B O. Kinetics of retrovirus mediated gene transfer: the importance of the intracellular half-life of retroviruses. J Theor Biol. 1996;182:1–20. doi: 10.1006/jtbi.1996.0140. [DOI] [PubMed] [Google Scholar]
- 4.Bahnson A B, Dunigan J T, Baysal B E, Mohney T, Atchison R W, Nimgaonkar M T, Ball E D, Barranger J A. Centrifugal enhancement of retroviral mediated gene transfer. J Virol Methods. 1995;54:131–143. doi: 10.1016/0166-0934(95)00035-s. [DOI] [PubMed] [Google Scholar]
- 5.Chuck A S, Clarke M F, Palsson B O. Retroviral infection is limited by Brownian motion. Hum Gene Ther. 1996;7:1527–1534. doi: 10.1089/hum.1996.7.13-1527. [DOI] [PubMed] [Google Scholar]
- 6.Chuck A S, Palsson B O. Consistent and high rates of gene transfer can be obtained using flow-through transduction over a wide range of retroviral titers. Hum Gene Ther. 1996;7:743–750. doi: 10.1089/hum.1996.7.6-743. [DOI] [PubMed] [Google Scholar]
- 7.Chuck A S, Palsson B O. Membrane adsorption characteristics determine the kinetics of flow-through transductions. Biotechnol Bioeng. 1996;51:260–270. doi: 10.1002/(SICI)1097-0290(19960805)51:3<260::AID-BIT2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 8.Crystal R G. Transfer of genes to humans: early lessons and obstacles to success. Science. 1995;270:404–410. doi: 10.1126/science.270.5235.404. [DOI] [PubMed] [Google Scholar]
- 9.Forestell S P, Bohnlein E, Rigg R J. Retroviral end-point titer is not predictive of gene transfer efficiency: implications for vector production. Gene Ther. 1995;2:723–730. [PubMed] [Google Scholar]
- 10.Kahn M L, Lee S W, Dichek D A. Optimization of retroviral vector-mediated gene transfer into endothelial cells in vitro. Circ Res. 1992;71:1508–1517. doi: 10.1161/01.res.71.6.1508. [DOI] [PubMed] [Google Scholar]
- 11.Kotani H, Newton P B I, Zhang S, Chiang Y L, Otto E, Weaver L, Blaese R M, Anderson W F, McGarrity G J. Improved methods of retroviral vector transduction and production for gene therapy. Hum Gene Ther. 1994;5:19–28. doi: 10.1089/hum.1994.5.1-19. [DOI] [PubMed] [Google Scholar]
- 12.LeDoux J M, Davis H E, Yarmush M L, Morgan J R. Kinetics of retrovirus production and decay. Biotechnol Bioeng. 1999;63:654–662. [PubMed] [Google Scholar]
- 13.LeDoux J M, Morgan J R, Yarmush M L. Proteoglycans secreted by packaging cell lines inhibit retrovirus infection. J Virol. 1996;70:6468–6473. doi: 10.1128/jvi.70.9.6468-6473.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller D G, Adam M A, Miller A D. Gene transfer by retrovirus vectors occurs only in cells that are actively replicating at the time of infection. Mol Cell Biol. 1990;10:4239–4242. doi: 10.1128/mcb.10.8.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morgan J R, LeDoux J M, Snow R G, Tompkins R G, Yarmush M L. Retrovirus infection: effect of time and target cell number. J Virol. 1995;69:6994–7000. doi: 10.1128/jvi.69.11.6994-7000.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 17.Orlic D, Girard L J, Jordan C T, Anderson S M, Cline A P, Bodine D M. The level of mRNA encoding the amphotropic retrovirus receptor in mouse and human hematopoietic stem cells is low and correlates with the efficiency of retrovirus transduction. Proc Natl Acad Sci USA. 1996;93:11097–11102. doi: 10.1073/pnas.93.20.11097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paul R W, Morris D, Hess B W, Dunn J, Overell R W. Increased viral titer through concentration of viral harvests from retroviral packaging cell lines. Hum Gene Ther. 1993;4:609–615. doi: 10.1089/hum.1993.4.5-609. [DOI] [PubMed] [Google Scholar]
- 19.Prince A M. Quantitative studies on Rous sarcoma virus. V. An analysis of the mechanism of virulence of the Bryan “high titer” strain of RSV. Virology. 1960;11:371–399. doi: 10.1016/0042-6822(60)90082-9. [DOI] [PubMed] [Google Scholar]
- 20.Rein A L, Gerwin B I, Bassin R H, Schwarm L, Schidlovsky G. A replication-defective variant of Moloney murine leukemia virus. I. Biological characterization. J Virol. 1978;25:146–156. doi: 10.1128/jvi.25.1.146-156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salmeen I, Rimai L, Luftig R B, Liebes L, Retzer E, Rich M, McCormick J J. Hydrodynamic diameters of murine mammary, Rous sarcoma, and feline leukemia RNA tumor viruses: studies by laser beat frequency light-scattering spectroscopy and electron microscopy. J Virol. 1976;17:584–596. doi: 10.1128/jvi.17.2.584-596.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shoup D, Szabo A. Chronoamperometric current at finite disk electrodes. J Electroanal Chem. 1982;140:237–245. [Google Scholar]
- 23.Tavoloni N. A simple procedure to determine the biological titer of recombinant retroviral vectors. Gene Ther. 1997;4:150–155. doi: 10.1038/sj.gt.3300370. [DOI] [PubMed] [Google Scholar]
- 24.Valentine R C, Allison A C. Virus particle adsorption. I. Theory of adsorption and experiments on the attachment of particles to non-biological surfaces. Biochim Biophys Acta. 1959;34:10–23. doi: 10.1016/0006-3002(59)90228-8. [DOI] [PubMed] [Google Scholar]