Abstract
The receptors for entry of herpes simplex viruses 1 and 2 (HSV-1 and -2), widely expressed in human cell lines, are members of a subset of the immunoglobulin superfamily exemplified by herpesvirus entry mediator C (HveC) and the herpesvirus immunoglobulin-like receptor (HIgR). This report focuses on two members of this subset, herpesvirus entry mediator B (HveB), recently designated nectin2/PRR2α, and its splice variant isoform, nectin2/PRR2δ. Nectin2α and -δ share the ectodomain but differ in the transmembrane and cytoplasmic regions. HveB was reported to enable entry of HSV-1 carrying mutations in glycoprotein D (gD) and of HSV-2, but not of wild-type (wt) HSV-1. We report that (i) both nectin2α and -δ served as receptors for the entry of HSV-1 mutant viruses HSV-1(U10) and -(U21) and AP7r that carry the Leu25Pro substitution in gD but not for HSV-1 mutants U30 and R5000 that carry the Ser140 or Ala185 substitution in gD. All of these mutants were able to overcome the block to entry mediated by expression of wt gD. (ii) Infection of cells expressing nectin2α or -δ required exposure to multiplicities of infection about 100-fold higher than those required to infect cells expressing HveC or HIgR. (iii) gD from HSV-1(U21) bound in vitro soluble forms of nectin2. The association was weaker than that to the soluble form of HveC/HIgR. Binding of wt HSV-1 gD to soluble nectin2 was not detectable. (iv) A major region of nectin2 functional in virus entry mapped to the V domain, located at the N terminus.
The process of herpes simplex virus (HSV) entry into cells begins with low-affinity attachment to heparan sulfate glycosaminoglycans (30). This interaction greatly enhances virus entry, as infectivity is reduced in cells devoid of glycosaminoglycans (3). Human herpesvirus mediator A (HveA), a novel member of the tumor necrosis factor receptor family, was the first cellular mediator shown to be required for HSV entry into human cells (25). Antibodies to HveA blocked HSV infectivity in cells transfected with the mediator or in T lymphocytes but not universally in the cell lines currently used in HSV studies. The narrow distribution and usage of HveA, together with its activity restricted only to some HSV strains, led to the further search for cellular proteins necessary for HSV entry into human cells. Recently, the members of a cluster of the immunoglobulin (Ig) superfamily were identified as the HSV receptors widely expressed in human cell lines like HEp-2, HeLa, and human fibroblasts (8, 13). The cluster includes three major groups of human proteins, each of which has known splice variant isoforms. For all of the members, six conserved cysteines define the same basic structure made of one V and two C2 domains (11, 21). They are (i) HveC, previously known as PRR1, for poliovirus receptor-related 1 (13, 21), and its splice variant isoform HIgR, for herpesvirus Ig-like receptor (8); (ii) two molecules originally designated PRR2α and -δ, also splice variant isoforms (11), the former redesignated HveB (32); and (iii) the poliovirus receptor (23), also designated CD155 or herpesvirus entry mediator D (HveD) (13), of which four isoforms are known. HveC and HIgR share the ectodomain and differ in the transmembrane and cytoplasmic regions but cannot be differentiated with respect to their function as mediators of HSV entry (8). They enable entry of all of the HSV type 1 (HSV-1) and HSV-2 strains tested, as well as of bovine herpesvirus 1 (BHV-1) and porcine pseudorabies virus (PRV) (8, 13). The V domain contains the major functional region in HSV-1 entry and interacts physically with glycoprotein D (gD) (7, 19), the envelope glycoprotein known to mediate HSV entry into cells by interaction with the receptor (8, 18).
In contrast to HveC and HIgR, HveB/PRR2α was reported to mediate the entry of a restricted number of HSV strains, namely, HSV-1-rid1, -rid2, and ANG and HSV-2 (32), while its murine homologue mediates the entry of PRV (29). Human PRR2α and PRR2δ, two isoforms with the same ectodomain and different transmembrane and cytoplasmic regions (11), are homophilic adhesion molecules expressed specifically at interendothelial junctions (20). Recently, they were found to be recruited to adherens junctions via afadin through PDZ domains and were renamed nectin2/PRR2 (31). We note that for a cellular protein with viral receptor activity, it is an established procedure to maintain the name based on the cellular function (thus, for example, the receptors or coreceptors for human immunodeficiency virus and adenoviruses maintain the designation CD4, chemokine receptors 5 and 4, or integrins). According to this rule, PRR2α/HveB and PRR2δ are designated nectin2α and -δ below.
Cells which express wild-type (wt) HSV-1 gD (wtgD1) constitutively are resistant to infection with wt HSV-1 (5, 16). The block to infection is overcome by spontaneous gD-unrestricted mutants selected for the ability to enter cells expressing HSV-1 gD, namely, HSV-1(U10), -(U21), -(U30), -rid1, and -rid2 (4, 6, 9), as well as by a mutant resistant to monoclonal antibody (MAb) AP7 (AP7r virus) (24); by R5000 (27), a virus able to infect resistant clones of thymidine kinase-deficient baby hamster kidney (BHKtk−) cells, or by the ANG strain (9). The amino acid substitutions in gD that enable these viruses to overcome the block to infection mediated by HSV-1 gD (wtgD1) map at three specific loci, amino acids 25 (U10, U21, AP7r virus, and ANG), 27 (rid1, rid2, and ANG), 140 (R5000), and 185 (U21 and U30). The phenomenon of restriction to infection mediated by expression of wtgD1 predicted that constitutive expression of wtgD1 sequesters the major receptor normally employed by the wt virus for entry into cells but allows entry of the HSV-1 gD-unrestricted mutants through one or more secondary receptors and that the interaction between secondary receptors and mutant forms of gD would be at low affinity (4, 5).
The objective of the present study was to determine whether both isoforms of nectin2, α and δ, mediate entry of HSV mutants, to identify the mutations in gD-unrestricted mutants that enable nectin2/PRR2 to serve as a receptor for these viruses, to locate the region of nectin2 functional in virus entry, and to determine if mutant gD interacts physically with nectin2. So far, it has been reported that wtgD1 binds to HveC and to HveA with high affinity and that mutant gD from strain rid1 binds both HveC and HveA with even higher affinity (18); binding of mutant gD to HveB has not been investigated.
We report that (i) both isoforms of nectin2, α and δ, enabled entry of some HSV-1 mutants and recombinants carrying the L25P substitution in gD but not other substitutions, (ii) a major functional region for entry was located in the V domain positioned at the N terminus, (iii) nectin2/PRR2α and -δ were much less efficient at mediating entry of the unrestricted mutant HSV-1(U21) than were HveC and HIgR, and (iv) gD from HSV-1(U21) (gDU21) interacted physically with the soluble form of nectin2. The association was weaker than that of gD from the wt virus to soluble HveC/HIgR.
MATERIALS AND METHODS
Cells and viruses.
All cells were grown in Dulbecco's modified Eagle medium supplemented with 5% fetal calf serum. J1.1-2 cells were previously described (8). HSV-1(F), HSV-2(G), HSV-1(MP), Sc-16, KOS, HFEM, and BHV-1 were also previously described (12, 14, 15, 17, 28, 33), as were HSV-1(U21), (U10), and (U30), their marker transfer and marker rescue recombinants (4, 6). The mutants resistant to MAbs AP7, AP12, LP2, LP14, R5000, and R5001 have been described earlier (24, 27). Viruses were grown and titrated by plaque assay in Vero cells. Virus infectivity in nectin2 cells was detected by immunostaining or β-galactosidase (β-gal) expression, by reading the optical density (OD) at 405 nm (25), or by light microscopy observation of β-gal-expressing cells.
Construction of cells expressing nectin2α and -δ.
Cells expressing nectin2α and -δ were obtained by transfection of pLX2S or pLX2L containing full-length nectin2α or nectin2δ cDNAs from TF-1 cells cloned in the pLXSN vector (20). Stable transformants of J1.1-2 cells expressing nectin2α or -δ were obtained by G418 neomycin selection, staining with MAb R2.477, and enrichment by cell sorting in a Becton Dickinson Vantage cell sorter, according to nectin2 expression. For construction of nectin2α and -δ cells harboring α27p-LacZ, pCEPα27p-LacZ was generated as follows. The lacZ gene was removed from pON832 (gift of E. Mocarski, Stanford University) by BamHI digestion and cloned into the BamHI site downstream of the HSV-1 α27 promoter in pRB3053, generating pRB3053LacZ. The cassette containing the α27 promoter-lacZ fusion was cloned into pCEP4 (Invitrogen) devoid of the SalI cassette, containing the cytomegalovirus promoter, the multicloning site, and the simian virus 40 polyadenylation signal. Stable transformants of nectin2α or -δ cells harboring pCEPα27p-LacZ were derived by hygromycin (250 μg/ml) selection.
Antibodies.
MAbs R2.477 and R2.525 were submitted to the VI International Workshop on Human Leukocyte Differentiation Antigen (22). MAb BC12 was purchased from Diaclone. MAb R1.302, directed to HIgR/PRR1, was previously described (22). MAb LP1 to α-TIF and MAb 1240 to BHV-1 gB were gifts from T. Minson (Cambridge University) and M. Ackermann (University of Zurich), respectively. Rabbit polyclonal antiserum to gM was previously described (2). Purified mouse IgGs, goat polyclonal sera against the Fc fragment of human IgG, and alkaline phosphatase-conjugated anti-mouse and anti-rabbit antibodies were from Sigma. Biotinylated anti-mouse and anti-rabbit secondary antibodies (ABC kit) were from Vector Laboratories.
Construction, production, and purification of soluble forms of nectin2.
The entire extracellular domain of nectin2 (amino acids 1 to 347) was amplified by PCR with primers SBPRR2.5 (AATT TAGA TATC ATGG CCCG GGCC GCTG CCCT C) and SPRR2.3n (TTAT ACTT GCGG CCGC TCGG ACAA AGAT GACC TGCG C) as previously described (20). The V domain of nectin2 (amino acids 1 to 160) was amplified with primers SBPRR2.5 and CFLR2V (GTTG CGGC CGCT ATGA CTCT GAGC CAGG TCAT C). The PCR products were cloned in frame with the Fc fragment of the human IgG1 sequence by using the Cos Fc Link vector (SmithKline Beecham Pharmaceuticals, King of Prussia, Pa.) to produce Fc-tagged soluble forms, designated VCC2-Fc and V2-Fc, respectively. For production, transfections of COS-1 cells were carried out with Fugene 6 (Boehringer Mannheim). The proteins were purified on Affigel protein A. The purity and quality of the proteins were monitored by gel electrophoresis, followed by silver staining (Bio-Rad), as previously described (20). The production of VCC1-Fc, V1-Fc, and CTLA4-Fc was previously described (7).
ELISA.
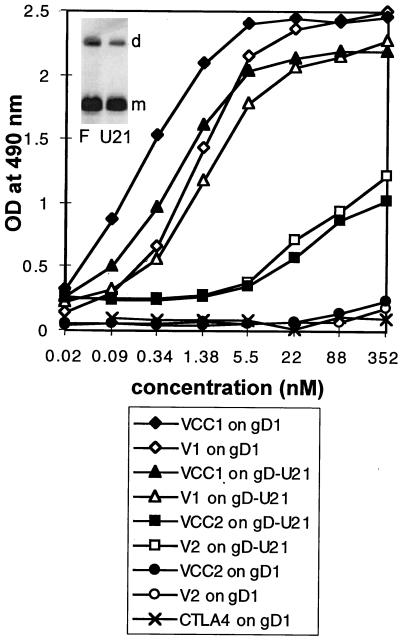
A sandwich enzyme-linked immunosorbent assay (ELISA) for the soluble forms of nectin2/PRR2 was performed with 96-well trays coated with an antibody against the human Fc fragment (Sigma) at 10 μg/ml. After saturation of wells with bovine serum albumin, 10−9 M VCC2-Fc or V2-Fc was reacted with biotinylated MAbs R2.477, R2.525, BC12, and R1.302 (2.5 μg/ml), followed by streptavidin-peroxidase and One Step ABTS (Pierce). gD1 and gDU21 were purified from HSV-1(F)- or HSV-1(U21)-infected BHK cells, and purification was monitored by silver staining. Membranes were obtained from the microsomal fraction of cytoplasm, solubilized in 1% Nonidet P-40 in 20 mM Tris-HCl buffer, and 150 mM NaCl, and protease inhibitors and purified by affinity chromatography to MAb #30 to gD (4) immobilized to Affigel. This resulted in purification practically to homogeneity (see the insert in Fig. 6). Microwell plates were coated with 16 nM gD1 or gDU21 in 100 μl of bicarbonate buffer (15 mM Na2CO3, 35 mM NaHCO3, 0.02% NaN3, pH 9.5) at 4°C overnight and then reacted with the indicated concentrations of the soluble forms of VCC1-Fc, V1-Fc, VCC2-Fc, and V2-Fc diluted in 1% bovine serum albumin–2% NaCl in phosphate-buffered saline. The negative control consisted of CTLA4-Fc. Binding was detected by incubation with anti-human Ig coupled to peroxidase (1:6,000) for 45 min at 37°C, followed by incubation with o-phenylenediamine (Sigma) at 0.5 mg/ml in 2.5 mM citric acid–5 mM Na2HPO4–0.009% H2O2, blocking with H2SO4 1:6 in H2O, and reading of the OD at 490 nm.
FIG. 6.
In vitro binding of gDU21 and wtgD1 to soluble forms of HveC/HIgR (VCC1-Fc and V1-Fc) or nectin2 (VCC2-Fc and V2-Fc) receptors or to CTLA4-Fc as a negative control. Affinity-purified gDU21 or wtgD1 was immobilized on microwells and then reacted with increasing concentrations of the indicated proteins. Binding was detected with anti-human IgG-peroxidase. The insert in the top left corner represents silver staining of affinity-purified gD from HSV-1(F)-infected (F) or HSV-1(U21)-infected (U21) cells. The lower band represents the monomeric form (m), and the upper band represents the dimeric form (d), of gD.
Immunostaining and FACS analysis.
For immunostaining, cells were fixed with ethanol and reacted with a rabbit polyclonal antibody to gM (1:2,000), MAb LP1 to α-TIF (1:1,500), or MAb 1240 to BHV-1 gB (undiluted hybridoma culture medium), followed by appropriate biotinylated or nonbiotinylated secondary antibodies. Fluorescence-activated cell sorter (FACS) analysis was performed as previously described (22) with a FACScan flow cytometer.
Infectivity blocking assays.
For assays of infectivity blocking by MAbs R2.525, BC12, and R2.477, cells grown in 96- or 48-well trays were preincubated with the indicated amounts of purified IgGs from the indicated antibodies in Dulbecco's modified Eagle medium supplemented with heat-inactivated fetal bovine serum for 2 h at 37°C. HSV-1(U21) (50 PFU/cell) was added, and the mixture was incubated for a further 90 min at 37°C. The viral inoculum was removed, and the cells were rinsed twice, overlaid with medium containing the same concentration of antibodies that was present during the preabsorption, and incubated for 16 h at 37°C. Infection was monitored by immunostaining of cells with rabbit polyclonal anti-gM antibodies for HSV-1 strains in nectin2α- and -δ-expressing cells or by β-gal activity measurement in nectin2α- and -δ-expressing cells carrying α27p-LacZ. OD was read in a Bio-Rad ELISA reader. Triplicates were run for each antibody concentration. Data reported are averages of at least two experiments. A 100% value represent the value obtained with infected cells not exposed to antibodies.
RESULTS
gD-unrestricted mutants HSV-1(U10) and -(U21), but not HSV-1(U30), infect cell lines expressing both isoforms of nectin2, α and δ.
In order to construct cell lines expressing nectin2α and -δ, J1.1-2 cells, a derivative of BHKtk− cells resistant to infection with all of the HSV-1 and -2 strains tested, including gD-unrestricted mutants HSV-1(U21), -(U10), and -(U30), were transfected with pLX2S or pLX2L, containing the full-length nectin2α and -δ cDNAs from TF-1 cells, respectively. Neomycin-resistant cells were selected and enriched by cell sorting following staining with MAb R2.477. The extents of expression of nectin2α and -δ were similar, as detected by FACS analysis (data not shown).
In the first series of experiments, we ascertained whether both isoforms of nectin2, α and δ, can serve as receptors for HSV-1(U10), -(U21), and -(U30) mutants. We found that HSV-1(U10) and -(U21) were able to infect cells expressing nectin2α or -δ [Fig. 1, data shown for HSV-1(U21)]. Surprisingly, HSV-1(U30) did not infect nectin2α- or -δ-expressing cells (Fig. 2), despite the fact that, like HSV-1(U10) and -(U21), it was selected for its ability to overcome the resistance to infection of cells expressing wtgD1. HSV-1(U30) can employ HveC and HIgR as primary receptors (8).
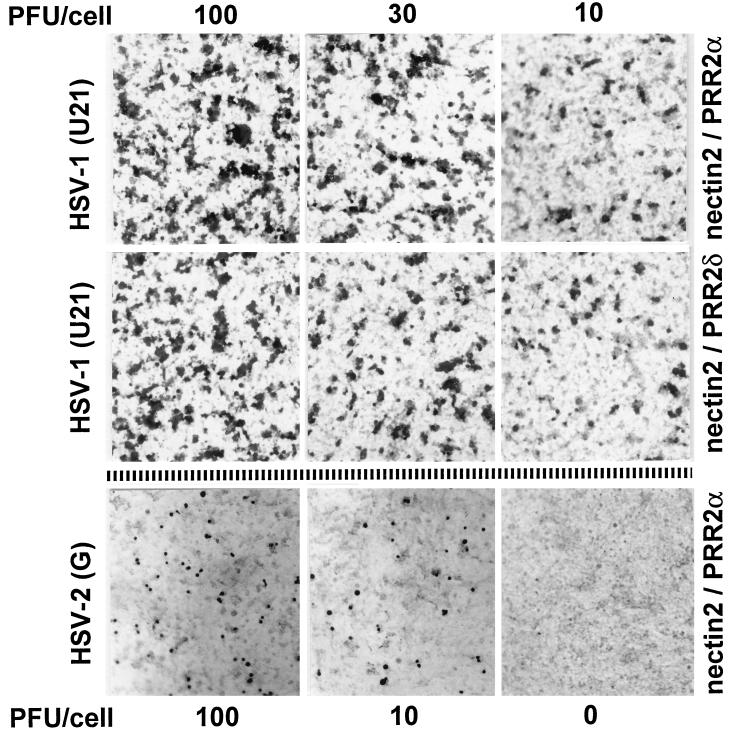
FIG. 1.
Infection of nectin2α (first and third rows) and nectin2δ (second row)-expressing cells with HSV-1(U21) at 100, 30, and 10 PFU/cell and with HSV-2(G) at 100 and 10 PFU/cell. Uninfected nectin2α-expressing cells, bottom right panel. Infection was detected by immunostaining with polyclonal antibodies to gM or MAb LP1 to α-TIF.
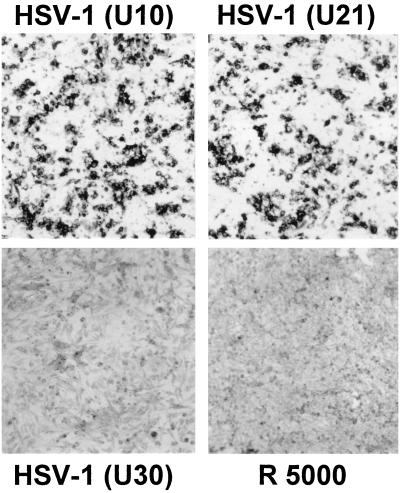
FIG. 2.
Infection of nectin2α-expressing cells with HSV-1(U10) and -(U21) and exposure to HSV-1(U30) and R5000 at 30 PFU/cell. Infection was detected by immunostaining with polyclonal antibodies to gM.
We noticed a striking difference in the multiplicity of infection (MOI) required to enter nectin2α- and -δ-expressing cells compared to that required to enter HveC- or HIgR-expressing cells. Thus, an MOI of 10 PFU/cell resulted in infection of only a small minority of nectin2-expressing cells. When the MOI was increased to 30 and 100 PFU/cell, the number of infected cells increased but remained much below 100% (Fig. 1). This was true for both nectin2α- and -δ-expressing cells. We note that HIgR-expressing cells can be infected at 100% efficiency with MOIs as low as 3 to 5 PFU/cell (8). Infection of nectin2α- and -δ-expressing cells therefore requires about 2 log higher MOIs than infection of HIgR cells. Nectin2α and -δ are cell adhesion molecules that act through homophilic interaction (1, 20). In order to check if the high MOI required to enter nectin2α- and -δ-expressing cells was due to low receptor availability, we infected monolayers of 70% confluent and fully confluent cells in parallel and scored the numbers of infected cells. Subconfluent or confluent cultures expressed very similar amounts of nectin2, as detected by FACS analysis (data not shown). No difference was observed between the amounts of infected cells (data not shown), indicating that homophilic interaction, when present, did not result in subtraction of receptors for the incoming virus. Altogether, the results show that not all of the gD-unrestricted mutants, selected for the ability to overcome the block to infection mediated by wtgD1, were able to infect cells expressing nectin2α or -δ. The finding that both isoforms, α and δ, mediated entry of HSV-1(U21) and -(U10) suggested that the ectodomain of the molecules carries the major functional domain and that differences in the transmembrane and cytoplasmic tails are not crucial for this activity. Nectin2α- or -δ-expressing cells were infected at much lower efficiency than HveC- or HIgR-expressing cells.
It was reported that expression of HveB (nectin2α) in CHO cells, which are partially susceptible to HSV-2 entry, augments infection with HSV-2(G) by about twofold, suggesting that it can serve as a mediator of HSV-2 entry (32). J1.1-2 cells are highly resistant to HSV-2 infection (8) and therefore represent a suitable system with which to address the issue of whether nectin2 may function as a mediator of HSV-2 entry. We found that expression of nectin2α or -δ in J1.1-2 conferred some degree of susceptibility to HSV-2 but with very low efficiency, inasmuch as infection at 100 PFU/cell resulted in a number of infected cells much lower than that observed with HSV-1(U21) (Fig. 1). Due to the very low efficiency of HSV-2 entry into nectin2 cells, HSV-2 was not investigated further.
Several strains of HSV-1, KOS, Sc16, MP, and HFEM, as well as BHV-1, had almost no ability to enter nectin2-expressing cells, in agreement with previous data (32).
Nectin2α- and -δ-expressing cells were almost undistinguishable in all assays, and subsequent results were obtained with both cell lines but are shown mainly for one cell line only.
Nectin2 enables entry of gD mutants carrying the L25P substitution, a mutation that enables mutant viruses to overcome the block to entry into cells expressing wt HSV-1 gD.
HSV-1(U10) gD carries the L25P amino acid substitution. HSV-1(U21) gD carries both the L25P and A185T amino acid substitutions. These viruses also carry other mutations outside of the gD gene (4). In order to ascertain if the ability of HSV-1(U21) and -(U10) to infect nectin2-expressing cells is dependent on the mutations present in gD and not on mutations present outside of the gD gene, nectin2 cells were infected with recombinants RFU21 and RFU10, which were created by marker transfer of the mutant gDs from HSV-1(U21) and HSV-1(U10) into HSV-1(F) and with the marker rescue viruses RsU21 and RsU10, which were created by rescue of mutant gD in HSV-1(U21) or HSV-1(U10) with wtgD1. The mutant virus AP7r, which carries the L25P substitution was included in this assay. Infection (100 and 30 PFU/cell) was monitored by immunostaining of infected cells with rabbit polyclonal antibodies to gM. Representative examples are shown in Fig. 2. The results, summarized in Table 1 as positive or negative, demonstrate that the recombinants of HSV-1(U10) and -(U21) viruses were, indeed, able to infect nectin2 cells and that the AP7r virus was also able to infect nectin2 cells, while the marker rescue viruses RsU21 and RsU10 did not infect nectin2α- and -δ-expressing cells. There was no detectable difference in the number of cells infected with HSV-1(U10) or -(U21) (Fig. 2), indicating that the ability of HSV-1(U21) to employ nectin2 was not dependent on the additional A185T substitution in gD. The results demonstrate that nectin2 serves as a specific receptor for gD mutants carrying the L25P substitution and not for the mutant carrying the A185T substitution and that the A185T substitution does not alter the effect of the L25P substitution. Previously, no recombinant was used in studies in which the rid1, rid2, and ANG strains were shown to infect HveB-expressing CHO cells (32) to unambiguously ascribe the observed phenotype to the mutations in gD. This is relevant in view of the fact that HSV-1(U21) was reported to carry mutations that allow infection of gD-expressing cells also outside the gD gene (4) and also that mutations in gK can partially overcome the gD-mediated restriction to infection (26).
TABLE 1.
Summary of HSV-1 mutants and recombinants able to infect nectin2-expressing cells
| Virus strain
|
Mutation(s) in gD relative to parental strain | Infectivity in nectin2-expressing cells | |
|---|---|---|---|
| Parental | Mutant | ||
| HSV-1 (F) | − | ||
| U21 | L25P, A185T | + | |
| U10 | L25P | + | |
| U30 | A185T | − | |
| RFU21 | L25P, A185T | + | |
| RFU10 | L25P | + | |
| RsU21 | None | − | |
| RsU10 | None | − | |
| R5000 | S140N | − | |
| R5001 | S140N | − | |
| KOS | −a | ||
| rid1 | Q27P | +a | |
| rid2 | Q27R | +a | |
| Sc16 | − | ||
| AP7R | L25P | + | |
| LP2R | S216N | − | |
| LP14R | R16H | − | |
| AP12R | I129T | − | |
| ANG | L25P, Q27R | +a | |
According to reference 32.
We next tested whether nectin2 can serve as a receptor for mutants carrying other substitutions in gD. R5000 and its recombinant R5001 carry a substitution at Ser140 (27). The mutants selected for resistance to MAbs LP14, AP12, and LP2 carry substitutions at amino acids 16, 129, and 216 (24). The results shown in part in Fig. 2 and summarized in Table 1 show that nectin2 could not serve as a receptor for any of these mutants, despite the fact that R5000 and R5001 can infect cells expressing wt HSV-1 gD. We conclude on the basis of the mutants tested that nectin2 can serve as a receptor for gD carrying the L25P substitution. This mutation coincides with one of the mutations which enable HSV-1 to overcome the block to infection mediated by HSV-1 gD. Nectin2 cannot serve as a receptor for viruses carrying mutations in gD at residues 16, 129, 140, 185, and 216. Mutation at residue 185 does not affect the ability to employ nectin2 conferred by the L25P substitution. We note that, relative to HSV-1 gD, HSV-2 gD, which could employ nectin2, although with very low efficiency, is not mutated at amino acid 25 or at amino acid 27, where the substitutions of HSV-1-rid1, -rid2, and ANG are located. However, it carries substitutions nearby, at amino acids 21 and 35 (10), which might account for the weak ability of nectin2 to serve as a receptor for HSV-2.
As no difference was observed between HSV-1(U10) and HSV-1(U21) in the ability to employ nectin2 as a receptor and as the A185T mutation, alone, or in combination, had no additional effect on entry conferred by the L25P substitution, the subsequent studies were performed with HSV-1(U21).
MAbs R2.525 and BC12, but not R2.477, inhibit entry of HSV-1(U21).
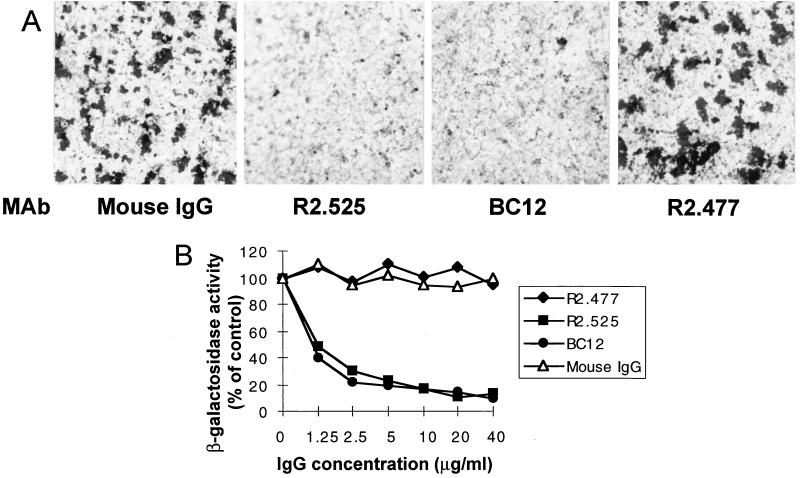
Three MAbs to nectin2α and -δ were assayed for the ability to block the entry of HSV-1(U21). They are MAbs R2.525 and R2.477, derived previously in one of our laboratories (22), and BC12 (Diaclone). In order to quantify virus entry, derivatives of nectin2α and -δ cells containing the lacZ gene under the control of the α27 promoter (α27p-LacZ) were constructed. Nectin2α and -δ cells, whether carrying α27p-LacZ or not, were incubated with increasing concentrations of MAbs for 2 h at 37°C prior to infection with HSV-1(U21). Cells were fixed 16 h later, and virus infection was monitored by immunoperoxidase staining with a polyclonal antibody to gM or by assay of β-gal activity. The results in Fig. 3A and B show that MAbs R2.525 and BC12 inhibited HSV-1(U21) entry in a dose-dependent fashion. Inhibition was about 90% at 20 μg/ml. By contrast, MAb R2.477 did not inhibit virion infectivity (data are shown for nectin2α). These results suggest that MAbs R2.525 and BC12, but not R2.477, bind or affect the nectin2 site functional in HSV-1(U21) entry.
FIG. 3.
Block of HSV-1(U21) entry by MAbs to nectin2. (A) Nectin2α-expressing cells were preincubated with MAb R2.525, BC12, or R2.477 to nectin2 or mouse IgGs (0.16 mg/ml) and then infected with HSV-1(U21). Infection was monitored by immunostaining 16 h later. (B) Nectin2α-expressing cells carrying α27p-LacZ were preincubated with the indicated concentrations of R2.525, BC12, and R2.477 antibodies or mouse IgGs and then infected with HSV-1(U21). Infection was quantitated as β-gal activity. A value of 100% represents the value obtained with infected cells not exposed to antibodies.
MAbs R2.525 and BC12, but not R2.477, bind the V domain of nectin2.
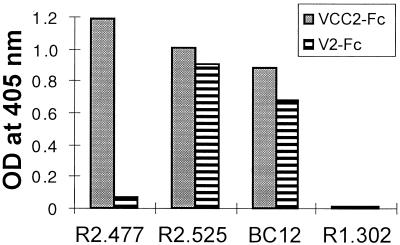
In order to define the location of the nectin2 epitopes recognized by the three antibodies tested above, soluble forms of nectin2 consisting of the entire ectodomain or of the single V domain, designated VCC2-Fc and V2-Fc, were constructed by cloning the domains of interest in the Cos Fc Link vector. The recombinant proteins, VCC2-Fc and V2-Fc, were purified by affinity chromatography to Affigel protein A from the extracellular medium of COS-1-transfected cells. The binding of MAbs R2.525, BC12, and R2.477 to VCC2-Fc and V2-Fc was determined in a sandwich ELISA. Microwells were first coated with anti-human antibodies and thereafter with VCC2-Fc and V2-Fc and then reacted with MAbs R2.525, BC12, and R2.477. Figure 4 shows that all of the MAbs bound VCC2-Fc, which contains the full-length ectodomain. Interestingly, MAbs R2.525 and BC12 had high levels of binding activity to V2-Fc, which contains the single V domain whereas MAb R2.477 did not bind V2-Fc. This demonstrates that the epitope recognized by the first two antibodies is located in the V domain, whereas the epitope recognized by MAb R2.477 appears to be located outside of the V domain but within the ectodomain.
FIG. 4.
ELISA binding of MAbs R2.525, R2.477, and BC12, directed to nectin2, with soluble forms of nectin2 containing the entire ectodomain (VCC2-Fc) or the single V domain (V2-Fc).
Soluble forms of nectin2 containing the single V domain block HSV-1(U21) infectivity.
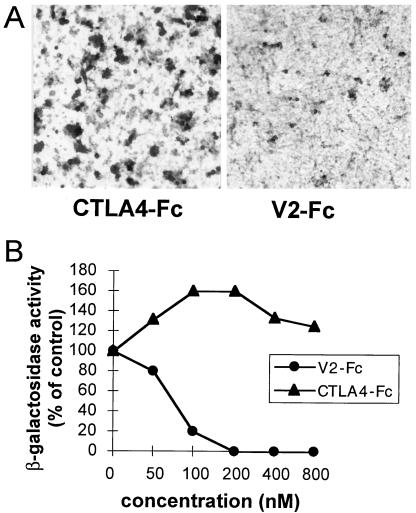
The above results show that two MAbs which inhibit HSV-1(U21) infectivity recognize epitopes located in the V domain, whereas a MAb which does not block virus infectivity does not bind the V domain. This provides evidence that a major nectin2 site functional for HSV-1(U21) entry resides in the V domain. To confirm these data, we tested whether V2-Fc, the soluble form of nectin2 containing the single V domain, competed with the cell-bound full-length receptor and blocked HSV-1(U21) entry. The inhibition assay was performed with nectin2α and -δ-expressing cells and with the respective cells carrying α27p-LacZ. The results in Fig. 5A and B show that V2-Fc inhibited virus infectivity in a dose-dependent manner. Inhibition was complete at 200 nM (the data shown are for nectin2α cells). The results confirm that a major nectin2 region functional in HSV-1(U21) entry is located in the V domain. This feature is similar to that observed with HIgR and HveC and HSV-1 (7, 19).
FIG. 5.
Entry of HSV-1(U21) into nectin2α-expressing cells is competitively blocked by a soluble form of nectin2 carrying the single V domain (V2-Fc). (A) HSV-1(U21) (100 PFU/cell) was preincubated with V2-Fc (200 nM) or CTLA4-Fc and then allowed to adsorb to nectin2α-expressing cells. Infection was monitored by immunoperoxidase staining. (B) Aliquots of HSV-1(U21) were preincubated with V2-Fc or CTLA4-Fc and then allowed to adsorb to nectin2α-expressing cells carrying α27p-LacZ. Infection was quantified as β-gal activity. Each point represents the average of triplicate assays. A value of 100% indicates the OD measured in virus-infected cultures treated with no antibody.
gD from HSV-1(U21) interacts physically with soluble forms of nectin2.
In the final series of experiments, we wanted to ascertain whether a soluble form of nectin2 interacts physically with gD from HSV-1(U21) (gDU21) and whether the binding site resides in the V domain. Previously, binding of gD from the rid1 mutant with HveB was not reported. gDU21 and wtgD1, each purified to homogeneity from lysates of HSV-1(F)- or HSV-1(U21)-infected cells by affinity chromatography (Fig. 6), were immobilized on microtiter wells and then reacted with increasing concentrations of the soluble forms of HIgR or nectin2, designated VCC1-Fc, V1-Fc, VCC2-Fc, and V2-Fc, respectively. Binding was detected by reactivity of anti-human Fc antibodies. The results in Fig. 6 indicate that both wtgD1 and gDU21 bound to VCC1-Fc and V1-Fc with very similar patterns. gDU21 bound VCC2-Fc and V2-Fc. The association was weaker than that to VCC1-Fc and V1-Fc. wtgD1 did not bind VCC2-Fc in the concentration range tested. It was reported that gD from HSV-1-rid1 and ANG had a somewhat higher affinity to HveC than wtgD1 (18); we have not observed a similar increase. We note that gDrid1 is mutated at amino acid 27, whereas gDU21 is mutated at amino acids 25 and 185. Determination of whether the slight changes in association to HveC or HIgR correlate with the different mutations was beyond the scope of the present investigation. Two major conclusions can be drawn from these results. First, a physical interaction occurs between nectin2 and gDU21. Second, the association between soluble nectin2 and gDU21 is weaker than that between wtgD1 or gDU21 and a soluble form of HveC or HIgR.
DISCUSSION
Substitutions at or near residue 25 confer on gD the ability to employ nectin2 as a receptor.
Of the mutants tested in this study, nectin2 could serve as a receptor only for viruses carrying the substitution at amino acid 25. Surprisingly, the mutants HSV-1(U30) and R5000, carrying gD substitutions at amino acids 185 (U30) and 140 (R5000) that enabled entry into cells expressing wt HSV-1 gD, were not able to utilize nectin2, suggesting that these viruses are able to employ still another, as yet unknown, secondary receptor. The results presented earlier (32) suggested an involvement also of amino acid 27. Current results demonstrate that mutant gD interacts physically with soluble forms of nectin2. Remarkably, the mutations at residues 25 and 27 do not greatly affect the ability of HSV-1 gD to interact with HveC or HIgR. This interaction has been finely mapped and involves two gD regions, recognized by group Ia and Ib MAbs to gD, located between amino acids 216 and 234 and 222 and 252, respectively (18). It can be concluded that a domain of gD that enables viral entry via nectin2 is located at or near amino acid 25. The substitutions at residue 25, and possibly 27, appear to confer on gD, which in the wt form seems to be unable to interact with nectin2, some degree of ability to interact with nectin2 while not grossly altering its ability to interact with HveC/HIgR. That HveC and HIgR can tolerate a number of substitutions in gD is indirectly supported by the observation that HveC and HIgR can mediate the entry also of HSV-2, BHV-1, and PRV (8, 13) and of HSV-1 mutants resistant to MAbs LP2, LP14, and AP12. The data suggest that the interaction of mutant gD from HSV-1(U21) with nectin2 involves a domain not involved in the interaction between wt HSV-1 gD and the ectodomain of HveC/HIgR.
Nectin2α and -δ represent low-efficiency receptors for HSV-1(U21).
Evidence for the conclusion that nectin2α and -δ are low-efficiency HSV-1(U21) receptors rests on the observation that an MOI about 100-fold higher was required for HSV-1(U21) to infect cells expressing the nectin2 receptor than to infect cells expressing HveC/HIgR. Our results suggest that homophilic interaction of nectin2 in confluent cultures did not represent a limiting factor in the availability of the nectin2 receptor for the infecting virus, since the yield of infected cells did not change when confluent or subconfluent cell cultures were infected. Nectin2 interacted physically with mutant gD from HSV-1(U21). Interestingly, the association between gDU21 and soluble nectin2 was weaker than that between wtgD1 or gDU21 and soluble HIgR. The weaker association is in keeping with the ability of nectin2 to serve as low-efficiency receptor for HSV-1(U21). The failure to detect an association between wtgD1 and soluble nectin2 is in keeping with the inability of nectin2 to serve as a receptor for HSV-1(F) (32).
A major region of nectin2 functional in the entry of HSV-1 mutants is located in the V domain of the molecule.
Both isoforms of nectin2, α and δ, were able to mediate the entry of HSV-1(U21). The two isoforms share the ectodomain and differ in the transmembrane and cytoplasmic regions. This finding provided the first evidence that the major region of nectin2 functional in HSV entry is located in the ectodomain of the molecule. The functional region was further located to the V domain on the basis of two lines of evidence. First, two MAbs that blocked the entry of virions recognized an epitope located in the V domain. Conversely, a MAb that did not block virion infectivity recognized an epitope located outside of the V domain. Second, a soluble form of the receptor, consisting of the V domain fused to the Fc portion of human IgG could compete with the cell-bound full-length receptor and reduce virion infectivity. In this respect, nectin2 does not differ from HIgR and HveC, whose major region functional in HSV-1 entry resides in the V domain (7, 19), and does not even differ from the poliovirus receptor, whose functional region also resides in the V domain. The extent of divergence between the V domains of HveC/HIgR and nectin2 is 67.1% at the amino acid level. This divergence modulates the binding of two forms of gD differing at residue 25. While HveC and HIgR bind gD from both wt HSV-1 and the HSV-1(U21) and -rid1 mutant strains with high affinity, nectin2 binds gD from HSV-1(U21) with low efficiency but does not appear to bind gD from wt HSV-1 at all.
ACKNOWLEDGMENTS
We thank E. Mocarski (Stanford University), T. Minson (Cambridge University), and M. Ackermann (University of Zurich) for the gifts of plasmid pON832, MAb LP1, and the MAb to BHV-1 gB, respectively.
The studies at the University of Bologna were aided by Progetto Finalizzatto Biotecnologie, by Telethon, and by grants from the Ministry of Education and Research, ex60% and ex40%. The studies at the INSERM U119, Marseille, were aided by INSERM, the Association pour la Recherche Contre le Cancer (ARC), and the Ligue Nationale Francaise Contre le Cancer (LNFCC).
REFERENCES
- 1.Aoki J, Koike S, Asou H, Ise I, Suwa H, Tanaka T, Miyasaka M, Nomoto A. Mouse homolog of poliovirus receptor-related gene 2 product, mPRR2, mediates homophilic cell aggregation. Exp Cell Res. 1997;235:374–384. doi: 10.1006/excr.1997.3685. [DOI] [PubMed] [Google Scholar]
- 2.Baines J D, Roizman B. The UL10 gene of herpes simplex virus 1 encodes a novel viral glycoprotein, gM, which is present in the virion and in the plasma membrane of infected cells. J Virol. 1993;67:1441–1452. doi: 10.1128/jvi.67.3.1441-1452.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banfield B W, Leduc Y, Esford L, Schubert K, Tufaro F. Sequential isolation of proteoglycan synthesis mutants by using herpes simplex virus as a selective agent: evidence for a proteoglycan-independent virus entry pathway. J Virol. 1995;69:3290–3298. doi: 10.1128/jvi.69.6.3290-3298.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandimarti R, Huang T, Roizman B, Campadelli Fiume G. Mapping of herpes simplex virus 1 genes with mutations which overcome host restrictions to infection. Proc Natl Acad Sci USA. 1994;91:5406–5410. doi: 10.1073/pnas.91.12.5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campadelli-Fiume G, Arsenakis M, Farabegoli F, Roizman B. Entry of herpes simplex virus 1 in BJ cells that constitutively express viral glycoprotein D is by endocytosis and results in degradation of the virus. J Virol. 1988;62:159–167. doi: 10.1128/jvi.62.1.159-167.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campadelli-Fiume G, Qi S, Avitabile E, Foà-Tomasi L, Brandimarti R, Roizman B. Glycoprotein D of herpes simplex virus encodes a domain which precludes penetration of cells expressing the glycoprotein by superinfecting herpes simplex virus. J Virol. 1990;64:6070–6079. doi: 10.1128/jvi.64.12.6070-6079.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cocchi F, Lopez M, Menotti L, Aoubala M, Dubreuil P, Campadelli-Fiume G. The V domain of herpesvirus Ig-like receptor (HIgR) contains a major functional region in herpes simplex virus-1 entry into cells and interacts physically with the viral glycoprotein D. Proc Natl Acad Sci USA. 1998;95:15700–15705. doi: 10.1073/pnas.95.26.15700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cocchi F, Menotti L, Mirandola P, Lopez M, Campadelli-Fiume G. The ectodomain of a novel member of the immunoglobulin superfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J Virol. 1998;72:9992–10002. doi: 10.1128/jvi.72.12.9992-10002.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dean H J, Terhune S S, Shieh M T, Susmarski N, Spear P G. Single amino acid substitutions in gD of herpes simplex virus 1 confer resistance to gD-mediated interference and cause cell-type-dependent alterations in infectivity. Virology. 1994;199:67–80. doi: 10.1006/viro.1994.1098. [DOI] [PubMed] [Google Scholar]
- 10.Dolan A, Jamieson F E, Cunningham C, Barnett B C, McGeoch D J. The genome sequence of herpes simplex virus type 2. J Virol. 1998;72:2010–2021. doi: 10.1128/jvi.72.3.2010-2021.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eberlè F, Dubreuil P, Mattei M G, Devilard E, Lopez M. The human PRR2 gene, related to the human poliovirus receptor gene (PVR), is the true homolog of the murine MPH gene. Gene. 1995;159:267–272. doi: 10.1016/0378-1119(95)00180-e. [DOI] [PubMed] [Google Scholar]
- 12.Ejercito P M, Kieff E D, Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 13.Geraghty R J, Krummenacher C, Cohen G H, Eisenberg R J, Spear P G. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 14.Hill T J, Field H J, Blyth W A. Acute and recurrent infection with herpes simplex virus in the mouse: a model for studying latency and recurrent disease. J Gen Virol. 1975;28:341–353. doi: 10.1099/0022-1317-28-3-341. [DOI] [PubMed] [Google Scholar]
- 15.Hoggan M D, Roizman B. The isolation and properties of a variant of herpes simplex producing multinucleated giant cells in monolayer cultures in the presence of antibody. Am J Hyg. 1959;70:208–219. doi: 10.1093/oxfordjournals.aje.a120071. [DOI] [PubMed] [Google Scholar]
- 16.Johnson R M, Spear P G. Herpes simplex virus glycoprotein D mediates interference with herpes simplex virus infection. J Virol. 1989;63:819–827. doi: 10.1128/jvi.63.2.819-827.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kieff E D, Bachenheimer S L, Roizman B. Size, composition, and structure of the deoxyribonucleic acid of herpes simplex virus subtypes 1 and 2. J Virol. 1971;8:125–132. doi: 10.1128/jvi.8.2.125-132.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krummenacher C, Nicola A V, Whitbeck J C, Lou H, Hou W, Lambris J D, Geraghty R J, Spear P G, Cohen G H, Eisenberg R J. Herpes simplex virus glycoprotein D can bind to poliovirus receptor-related protein 1 or herpesvirus entry mediator, two structurally unrelated mediators of virus entry. J Virol. 1998;72:7064–7074. doi: 10.1128/jvi.72.9.7064-7074.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krummenacher C, Rux A H, Whitbeck J C, Ponce-de-Leon M, Lou H, Baribaud I, Hou W, Zou C, Geraghty R J, Spear P G, Eisenberg R J, Cohen G H. The first immunoglobulin-like domain of HveC is sufficient to bind herpes simplex virus gD with full affinity, while the third domain is involved in oligomerization of HveC. J Virol. 1999;73:8127–8137. doi: 10.1128/jvi.73.10.8127-8137.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez M, Aoubala M, Jordier F, Isnardon D, Gomez S, Dubreuil P. The human poliovirus receptor related 2 protein is a new hematopoietic/endothelial homophilic adhesion molecule. Blood. 1998;92:4602–4611. [PubMed] [Google Scholar]
- 21.Lopez M, Eberlè F, Mattei M G, Gabert J, Birg F, Bardin F, Maroc C, Dubreuil P. Complementary DNA characterization and chromosomal localization of a human gene related to the poliovirus receptor-encoding gene. Gene. 1995;155:261–265. doi: 10.1016/0378-1119(94)00842-g. [DOI] [PubMed] [Google Scholar]
- 22.Lopez M, Jordier F, Bardin F, Coulombel L, Chabannon C, Dubreuil P. Identification of a new class of IgG superfamily antigens expressed in hemopoiesis. In: Kishomoto T, et al., editors. Leukocyte typing VI, white cells differentiation antigens. New York, N.Y: Garland Publishing, Inc.; 1997. pp. 1081–1083. [Google Scholar]
- 23.Mendelsohn C L, Wimmer E, Racaniello V R. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell. 1989;56:855–865. doi: 10.1016/0092-8674(89)90690-9. [DOI] [PubMed] [Google Scholar]
- 24.Minson A C, Hodgman T C, Digard P, Hancock D C, Bell S E, Buckmaster E A. An analysis of the biological properties of monoclonal antibodies against glycoprotein D of herpes simplex virus and identification of amino acid substitutions that confer resistance to neutralization. J Gen Virol. 1986;67:1001–1013. doi: 10.1099/0022-1317-67-6-1001. [DOI] [PubMed] [Google Scholar]
- 25.Montgomery R I, Warner M S, Lum B J, Spear P G. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 26.Pertel P E, Spear P G. Partial resistance to gD-mediated interference conferred by mutations affecting herpes simplex virus type 1 gC and gK. J Virol. 1997;71:8024–8028. doi: 10.1128/jvi.71.10.8024-8028.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roller R J, Roizman B. A herpes simplex virus 1 US11-expressing cell line is resistant to herpes simplex virus infection at a step in viral entry mediated by glycoprotein D. J Virol. 1994;68:2830–2839. doi: 10.1128/jvi.68.5.2830-2839.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schröder C H, Stegmann B, Lauppe H F, Kaerner H C. An unusual defective genotype derived from herpes simplex virus strain ANG. Intervirology. 1975;6:270–284. doi: 10.1159/000149481. [DOI] [PubMed] [Google Scholar]
- 29.Shukla D, Rowe C L, Dong Y, Racaniello V R, Spear P G. The murine homolog (Mph) of human herpesvirus entry protein B (HveB) mediates entry of pseudorabies virus but not herpes simplex virus types 1 and 2. J Virol. 1999;73:4493–4497. doi: 10.1128/jvi.73.5.4493-4497.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spear P G, Shieh M T, Herold B C, WuDunn D, Koshy T I. Heparan sulfate glycosaminoglycans as primary cell surface receptors for herpes simplex virus. Adv Exp Med Biol. 1992;313:341–353. doi: 10.1007/978-1-4899-2444-5_33. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi K, Nakanishi H, Miyahara M, Mandai K, Satoh K, Satoh A, Nishioka H, Aoki J, Nomoto A, Mizoguchi A, Takai Y. Nectin/PRR: an immunoglobulin-like cell adhesion molecule recruited to cadherin-based adherens junctions through interaction with Afadin, a PDZ domain-containing protein. J Cell Biol. 1999;145:539–549. doi: 10.1083/jcb.145.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warner M S, Geraghty R J, Martinez W M, Montgomery R I, Whitbeck J C, Xu R, Eisenberg R J, Cohen G H, Spear P G. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by mutants of herpes simplex virus type 1, herpes simplex virus type 2, and pseudorabies virus. Virology. 1998;246:179–189. doi: 10.1006/viro.1998.9218. [DOI] [PubMed] [Google Scholar]
- 33.Wildy P, Russel W C, Horne R W. The morphology of herpes virus. Virology. 1960;12:204–222. doi: 10.1016/0042-6822(60)90195-1. [DOI] [PubMed] [Google Scholar]