Abstract
The African swine fever virus (ASFV) genome contains a gene, 9GL, with similarity to yeast ERV1 and ALR genes. ERV1 has been shown to function in oxidative phosphorylation and in cell growth, while ALR has hepatotrophic activity. 9GL encodes a protein of 119 amino acids and was highly conserved at both nucleotide and amino acid levels among all ASFV field isolates examined. Monospecific rabbit polyclonal antibody produced to a glutathione S-transferase–9GL fusion protein specifically immunoprecipitated a 14-kDa protein from macrophage cell cultures infected with the ASFV isolate Malawi Lil-20/1 (MAL). Time course analysis and viral DNA synthesis inhibitor experiments indicated that p14 was a late viral protein. A 9GL gene deletion mutant of MAL (Δ9GL), exhibited a growth defect in macrophages of approximately 2 log10 units and had a small-plaque phenotype compared to either a revertant (9GL-R) or the parental virus. 9GL affected normal virion maturation; virions containing acentric nucleoid structures comprised 90 to 99% of all virions observed in Δ9GL-infected macrophages. The Δ9GL virus was markedly attenuated in swine. In contrast to 9GL-R infection, where mortality was 100%, all Δ9GL-infected animals survived infection. With the exception of a transient fever response in some animals, Δ9GL-infected animals remained clinically normal and exhibited significant 100- to 10,000-fold reductions in viremia titers. All pigs previously infected with Δ9GL survived infection when subsequently challenged with a lethal dose of virulent parental MAL. Thus, ASFV 9GL gene deletion mutants may prove useful as live-attenuated ASF vaccines.
African swine fever (ASF) is a highly lethal hemorrhagic disease of domestic pigs for which there is no vaccine or disease control strategy other than animal slaughter (27, 60). The causative agent, African swine fever virus (ASFV), is a large, enveloped, double-stranded DNA virus which is the sole member of the newly named Asfarviridae family (L. K. Dixon et al., personal communication). ASFV is the only known DNA arbovirus (7, 11). Perpetuation and transmission of this virus in nature involves the cycling of virus between two highly adapted hosts, Ornithodoros ticks and wild-pig populations (warthogs and bushpigs) in sub-Saharan Africa (48, 49, 59, 64).
Although the icosahedral morphology of the ASFV virion resembles those of iridoviruses, both the genomic organization of ASFV, which includes a central conserved genomic region with more variable terminal regions, inverted terminal repeats, and terminal cross-links, and its cytoplasmic replication strategy indicate a close relationship with the Poxviridae (23, 47, 53). ASFV is genetically complex; its genome of 170 to 190 kbp encodes 160 or more open reading frames (ORF), and approximately 100 proteins have been observed in virus-infected cells (4, 11, 15, 16, 52, 55). Sequencing and genetic analysis of the complete viral genome have led to the prediction and/or identification of genes encoding virion structural proteins and other proteins with functions involving viral virulence and/or host range, RNA transcription and modification, nucleotide metabolism, DNA replication, protein processing (13, 65–67; Z. Lu, G. K. Kutish, J. G. Neilan, L. Zsak, and D. L. Rock, unpublished data). However, the functions of most ASFV genes involved in viral virulence and/or host range, even those with known cellular homologues, remain unknown.
An ASFV gene with similarity to the ERV1 (essential for respiration and viability) gene family (35) has been identified in the pathogenic ASFV isolate Malawi Lil-20/1 (ORF, 9GL) and a cell culture-adapted European virus, BA71V (ORF, pB119L) (65; Lu et al., unpublished data).
The prototype of the ERV1 gene family, scERV1, was first identified in Saccharomyces cerevisiae, where it was shown to be essential for oxidative phosphorylation, maintenance of mitochondrial genomes, and cell cycle division (35). In yeast, the gene, which is expressed as a low-abundance spliced transcript, has a complex expression pattern that is highly dependent on physiological growth conditions (35, 38, 54). How this mitochondria-associated protein functions in these highly significant biological processes is not understood.
Similar ERV1 homologues, encoding a protein referred to as augmenter of liver regeneration (ALR), or hepatopoietin, have been identified in humans, mice, and rats (24, 37, 61). These genes, which appear to be preferentially expressed in the testes and liver, encode cytosolic proteins that function as growth factors in regeneration of the liver, where they stimulate hepatocyte proliferation, and in some yet-undefined aspect of spermatogenesis (21, 24, 54, 61). Although the protein's mechanism of action is currently unknown, the recent identification of a specific receptor for the ALR protein on hepatocytes suggests that it may function by a receptor-mediated signaling pathway (61).
ERV1 homologues appear to be widespread in nature, present in organisms as diverse as higher plants, roundworms, insects, and protozoa. Interestingly, in addition to ASFV, ERV1 gene homologues are also present in a number of other cytoplasmic DNA viruses, including vaccinia virus, Melanoplus sanguinipes entomopox virus, fish lymphocystis disease virus, and chlorella virus (3).
Here we have examined the function of the ASFV ERV1 homologue, 9GL, in macrophage cell cultures and the swine host using a recombinant 9GL gene deletion mutant. Our data indicate that 9GL affects virion maturation and viral growth in macrophage cell cultures and viral virulence in swine.
MATERIALS AND METHODS
Cell culture and viruses.
Primary porcine blood macrophage cell cultures (PBMC) were prepared from defibrinated swine blood as previously described (20). Briefly, heparin-treated swine blood was incubated at 37°C for 1 h to allow sedimentation of the erythrocyte fraction. Mononuclear leukocytes were separated by flotation over a Ficoll-Paque (Pharmacia, Piscataway, N.J.) density gradient (specific gravity, 1.079). The monocyte/macrophage cell fraction was cultured in plastic Primaria (Falcon; Becton Dickinson Labware, Franklin Lakes, N.J.) tissue culture flasks containing RPMI 1640 medium with 30% L929 supernatant and 20% fetal bovine serum for 48 h (37°C under 5% CO2). Adherent cells were detached from the plastic by using 10 mM EDTA in phosphate-buffered saline and were then reseeded into Primaria T25 6- or 96-well dishes at a density of 5 × 106 cells per ml for use in assays 24 h later.
A European Vero cell culture-adapted ASFV strain, BA71V, was provided by J. M. Escribano (INIA, Madrid, Spain). Highly purified BA71V was obtained by Percoll equilibrium centrifugation as described by Carrascosa et al. (8). ASFV isolates used in this study were as follows: pathogenic tick isolates Chiredzi/83/1, Crocodile/96/1, Crocodile/96/3, Fairfield/96/1, Malawi Lil-20/1, Pretoriuskop/96/4, Pretoriuskop/96/5, and Wildebeeslaagte/96/1; pathogenic pig isolates Cameroon, European-70 (E70), European-75 (E75), Haiti 811, Killean III, Kimakia, La Granja, Lisbon 60, Spencer, Tengani, Victoria Falls, and Zaire; warthog isolate Uganda 61; and bush pig isolate Lee. Chiredzi/83/1 (28) and Malawi Lil-20/1 (26) were isolated from Ornithodoros sp. ticks in 1983. Crocodile/96/1, Crocodile/96/3, Fairfield/96/1, Pretoriuskop/96/1, Pretoriuskop/96/4, and Wildebeeslaagte/96/1 were isolated from Ornithodoros porcinus porcinus ticks collected from the Republic of South Africa in 1996. All other isolates were obtained from the Plum Island Animal Disease Center reference collection (25, 32, 63).
DNA manipulation, PCR cloning, and sequencing.
The 9GL gene was amplified by PCR from purified viral DNA by using a mixture of oligonucleotide primers based on the BA71V (65) and Malawi Lil-20/1 gene sequences (Lu et al., unpublished data): forward primers 5′-TCCTCCAAACTCCGGGAGACGT-3′ and 5′-GTGATGATGTAATGAATAGGAA-3′ and reverse primers 5′-CCTCTATATGGAATGAGATTTTTC-3′ and 5′-ACATTAAGGACCATGACACAGATACT-3′. Amplified products were cloned into the TA cloning vector pCR 2.1 (Invitrogen, San Diego, Calif.). Two to four PCR clones derived from independent amplifications of each isolate were sequenced completely with M13 forward and reverse primers by using an Applied Biosystems PRISM 377 automated DNA sequencer (Perkin-Elmer, Foster City, Calif.). The ABI sequence software (version 3.3) was used for lane tracking and trace extraction. The chromatogram traces were base called with Phred (17), which also produced a quality file containing a predicted probability of error at each base position. The sequences were assembled with Phrap by using the quality files and default settings to produce a consensus sequence for each ASFV isolate (31).
9GL protein expression.
The 9GL ORF was amplified by PCR and cloned into the expression vector pGEX-2T (Pharmacia) by using Malawi Lil-20/1 genomic DNA as the template. Primer pairs, which created BamHI and EcoRI sites at the 5′ and 3′ ends of the ORF respectively, were as follows: forward primer, 5′-GGGGGGATCCATGTTGCATTGGGGACCTAA-3′; reverse primer, 5′-GGGGGAATTCTTATAGAGATGACCAGGCTCCA-3′. Selected 9GL clones were confirmed by DNA sequencing. The glutathione S-transferase (GST)-9GL fusion protein was prepared and purified on Glutathione Sepharose beads as described previously (45). New Zealand White rabbits were inoculated subcutaneously with 1 mg of Glutathione Sepharose-bound GST fusion protein in Freund's complete adjuvant and boosted five times, at 2-week intervals, with the fusion protein extract emulsified in incomplete Freund's adjuvant. The resulting immune serum specifically immunoprecipitated l-[35S]methionine-labeled in vitro-translated 9GL (data not shown).
Primary swine macrophage cell cultures were infected with Malawi Lil-20/1, pulse-radiolabeled for 2-h intervals at various times postinfection with l-[35S]methionine in methionine-deficient RPMI medium, and immunoprecipitated with 9GL-monospecific antibodies as previously described (1). Western blot (immunoblot) analyses of highly purified ASFV virions with 9GL antiserum and antisera against ASFV structural proteins p30 (22) and p54 (50) were performed as previously described (1).
Construction of the recombinant ASFV Δ9GL and its revertant 9GL-R.
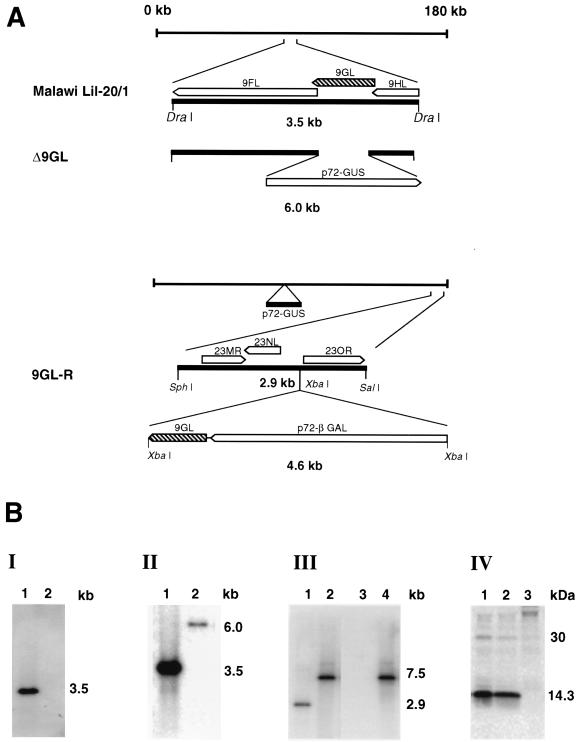
Recombinant ASFVs were generated by homologous recombination between parental ASFV genomes and recombination transfer vectors following infection and transfection of swine macrophage cell cultures (44, 66). Flanking genomic regions, which included portions of 9GL mapping to the left (1.2 kbp) and right (1.15 kbp) of the gene, were amplified by PCR using Malawi Lil-20/1 genomic DNA as a template. The left flanking region was amplified by using a primer pair (forward primer, 5′-GTCGACGTTCCACCCAGGATCTATAACGT-3′; reverse primer, 5′-CTCGAGTATCCGCGGAGTACTGGACCTTCGCGTTT-3′) that introduced XhoI and SalI restriction sites at the 5′ and 3′ ends of the fragment, respectively. The right flanking region was amplified by using a primer pair (forward primer 5′-CTCGAGTGCATGGCAGCGACTCGATA-3′ and reverse primer 5′-GTCGACACAACTCAGCATGCTGCATCT-3′) that introduced SalI and XhoI restriction sites at the 5′ and 3′ ends of the fragment, respectively. Amplified fragments were digested with SalI/XhoI enzymes, cloned into pCR 2.1 to produce pT9GL, and sequenced to verify ASFV sequences. A reporter gene cassette containing the β-glucuronidase (GUS) gene with the ASFV p72 late gene promoter, p72GUS (45), was inserted into SalI-digested pT9GL to yield the transfer vector p72GUSΔ9GL. This construction created an 82-nucleotide deletion in the 9GL ORF (nucleotides 154 to 236) (see Fig. 4). Macrophage cell cultures were infected with Malawi Lil-20/1 and transfected with p72GUSΔ9GL. Recombinant viruses representing independent primary plaques were purified to homogeneity by plaque assay and verified as products of a double-crossover recombination event by using PCR and Southern blot analysis as described previously (44, 66). A recombinant, Δ9GL, was selected for further analysis.
FIG. 4.
Characterization of an ASFV 9GL gene deletion mutant, Δ9GL, and its revertant, 9GL-R. (A) Diagram of Malawi Lil-20/1 and recombinant viruses Δ9GL and 9GL-R. (B) (I and II) Southern blot analysis of DraI-digested parental and Δ9GL viral DNAs (lanes 1 and 2, respectively) probed with deleted 9GL sequences (panel I) or genomic regions flanking the deletion (panel II). (III) Southern blot analysis of SphI/SalI-digested DNAs from Δ9GL (lanes 1 and 3) and its revertant, 9GL-R (lanes 2 and 4), probed with the righthand terminal region (lanes 1 and 2) and a 9GL gene probe (lanes 3 and 4). (IV) Immunoprecipitation analysis of lysates from macrophage cell cultures infected with the parental, revertant, and mutant viruses (lanes 1, 2, and 3, respectively) by using monospecific anti-9GL serum.
A Δ9GL revertant virus, 9GL-R, was constructed by restoring 9GL to the Δ9GL genome by similar methods. A DNA fragment containing the complete 9GL gene and its upstream promoter sequences was amplified by PCR using primer pairs containing NotI restriction endonuclease sites mapping 205 nucleotides upstream and 74 nucleotides downstream of the 9GL ORF (forward primer, 5′-AATAGCGGCCGCGCTGCCCATTGTTTTGGAAATGCCT-3′; reverse primer, 5′-AATAGCGGCCGCCTCCGGGAGACGTTTGTTTTATCCA-3′). The resulting PCR product was subsequently digested with NotI and cloned into a unique NotI site of a transfer vector, pNL1GAL, which was designed for inserting genes into a nonessential area of the right variable region of the Malawi Lil-20/1 genome (2). This transfer vector, pNL1GAL-9GL, which contained a β-galactosidase (GAL) reporter gene, was then used together with Δ9GL virus for transfection and infection. Putative recombinant GAL-positive viruses were purified by plaque assay on macrophage cell cultures and analyzed as described above.
Ultrastructural analysis of Δ9GL-infected macrophage cell cultures and purified virus preparations.
Porcine macrophage cell cultures were either mock infected or infected with ASFV Malawi Lil-20/1 (multiplicity of infection [MOI] = 5) or Δ9GL (MOI = 5) and were harvested at 18, 36, 72, and 96 h postinfection (HPI) by gently scraping the adherent macrophages with a rubber policeman. The cells were centrifuged and processed for transmission electron microscopy as previously described (44). Plastic thin sections were collected on Formvar-coated, carbon-stabilized slot grids (Electron Microscopy Sciences, Fort Washington, Pa.). The occurrence, frequency, and distribution of developing and mature virions were determined, and statistical differences were assessed by the Student t test and chi-square analysis.
To estimate the relative numbers of Δ9GL and 9GL-R virions in infected macrophages, sucrose gradient-purified virus was prepared. Ten 150-cm2 flasks of PBMC were infected (MOI = 10) and harvested at the time of maximum cytopathic effect (36 HPI for 9GL-R-infected cells and 72 HPI for Δ9GL-infected cells). Δ9GL and 9GL-R virions were purified twice on 30 and 60% sucrose step gradients. Aliquots of the purified virus suspensions were titrated as described elsewhere (46). Semipurified virus was fixed with a solution containing 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) for 2 h at 4°C and further processed with 2% osmium tetroxide for 2 h at 4°C followed by 2% aqueous uranyl acetate overnight at 4°C. The virus preparation was dehydrated in ethanol and embedded in Spurr's resin. Virions in thin sections were counted and scored from 25 randomly chosen fields, each outlined by the bars of a 300-mesh grid.
IEM.
Immunoelectron microscopy (IEM) was performed by using fixed and sectioned material, fixed and sectioned virions, and unfixed, purified virions. Two fixatives, periodate-lysine-paraformaldehyde (PLP; Nakane) and 4% freshly prepared paraformaldehyde, and three embedding media (Lowicryl K4M, London Resin White, and Unicryl) were used for IEM of sectioned material. Cells were infected with Malawi Lil-20/1 and processed with the above fixatives and embedding media as previously described (5). The number of gold grains associated with 50 virions was determined. Purified BA71V virions were adsorbed to Formvar-coated nickel grids. For IEM of sectioned material, grids were quenched with 50 mM glycine and were blocked with 5% normal goat serum (NGS) and 1% cold fish gelatin in 20 mM Tris-HCl–0.5 M NaCl. Monospecific rabbit antisera to the ASFV 9GL protein and the ASFV virion structural proteins p54 and p11.6 (5, 50), as well as preimmune rabbit antiserum, were diluted 1:20 to 1:200 in 20 mM Tris-HCl–0.5 M NaCl containing 1% NGS and 1% gelatin and were incubated with grids for 30 min at room temperature. Washing and incubation with a 1:20 dilution of protein A labeled with 10 nm of colloidal gold (Amersham) were performed as previously described (5).
Animal infections.
Yorkshire pigs (30 to 35 kg) were inoculated intramuscularly with either 102 50% tissue culture infective doses (TCID50) of the revertant virus 9GL-R or 102, 104, or 106 TCID50 of the null mutant Δ9GL. A dose of 102 TCID50 of Malawi Lil-20/1 represents a challenge of 10 to 100 100% lethal doses (LD100) (67). Clinical signs of ASF (fever [a rectal temperature greater than or equal to 104°F], anorexia, lethargy, shivering, cyanosis, and recumbency) were monitored daily. Blood samples were collected every other day for 30 days postinfection (DPI). Virus isolation and titration of ASFV in blood samples were performed as previously described (46). Virus titers were calculated by the method of Spearman-Karber and expressed as TCID50 (18).
Nucleotide sequence accession numbers.
The 9GL gene sequences of the isolates used in this study were assigned GenBank accession no. AF081160 (Cameroon), AF081161 (Chiredzi/83/1), AF081162 (Crocodile/96/1), AF081163 (Crocodile/96/3), AF081164 (European-70), AF081165 (European-75), AF081166 (Haiti 811), AF081167 (Fairfield/96/1), AF081168 (Killean III), AF081169 (Kimakia), AF081170 (La Granja), AF081171 (Lee), AF081172 (Lisbon 60), AF081173 (Wildebeeslaagte/96/1), AF081174 (Malawi Lil-20/1), AF081175 (Pretoriuskop/96/4), AF081176 (Pretoriuskop/96/5), AF081177 (Spencer), AF081178 (Tengani), AF081179 (Uganda 61), AF081180 (Victoria Falls), and AF081181 (Zaire).
RESULTS
9GL is similar to yeast ERV1 and rat ALR proteins.
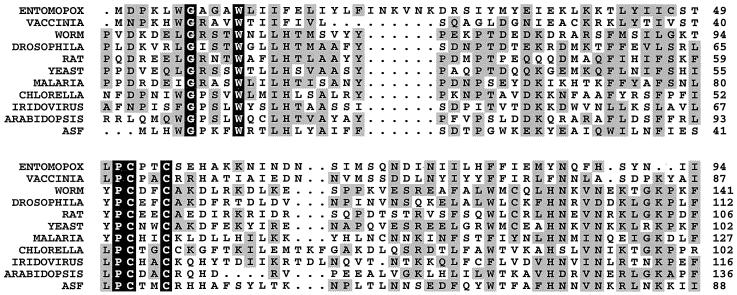
The 9GL protein has significant similarity to the yeast ERV1 and rat ALR proteins (27% identity and 44% homology over 99 residues; FASTA score, 146; z score, 15.5) and slightly less similarity to the ERV1 and ALR homologues found in higher plants (Arabidopsis spp. and corn), roundworms (Caenorhabditis elegans and Schistosoma mansoni), insects (Drosophila melanogaster), and protozoa (Plasmodium falciparum and Trypanosoma brucei), as well as other mammals (humans, mice, pigs, and guinea pigs) (Fig. 1). The 9GL ORF is also similar to genes found in other viruses: an unnamed ORF in the iridovirus fish lymphocystis virus (27% identity over 98 residues; FASTA score, 153; z score, 11.1); the A467R ORF in the chlorella virus; the E10 vaccinia homologue in variola and fowlpox viruses; the MCU40 ORF in molluscum contagiosum virus; and the MSV093 ORF in M. sanguinipes entomopox virus. The viral genes tend to be shorter than the eukaryotic homologues; however, there are no unique motifs which distinguish the viral from the eukaryotic genes. All of these genes encode a highly conserved 96-residue core region (Fig. 1). Also, they all contain two highly conserved motifs: Gly-X-X-X-Trp at residue 4 and Pro-Cys-X-X-Cys at residue 43 of 9GL. The pair of conserved cysteine residues is similar to the glutaredoxin and thioredoxin redox active center motif (Prosite PS00194); however, the glycine-tryptophan motif is unique to 9GL and ERV1 homologues.
FIG. 1.
Alignment of the predicted amino acid sequence of the ASFV Malawi Lil-20/1 ORF 9GL product (ASF) with the yeast ERV1 and rat ALR motif-containing proteins. Homologues shown are as follows (with the GenBank accession number of the nucleotide sequence given in parentheses following each): ENTOMOPOX, M. sanguinipes entomopox virus ORF MSV093 product (AF063866); VACCINIA, vaccinia virus ORF E10R product (M35027); WORM, C. elegans gene product F56C11.3 (AF043697); DROSOPHILA, D. melanogaster homologue (AA696037); RAT, Rattus norvegicus ALR (D30735); YEAST, S. cerevisiae ERV1 (X60722); MALARIA, P. falciparum homologue (AL031745); CHLORELLA, chlorella virus ORF A465R product (U42580); IRIDOVIRUS, fish lymphocystis disease virus homologue (L63545); ARABIDOPSIS, Arabidopsis thaliana (AC003058). The consensus sequence was constructed with the Clustal W computer program (58) by using the Dayhoff PAM 250 symbol comparison table with a 0.5 cutoff value. Invariant amino acids in all proteins are shown on a solid background, while conservative substitutions are shaded. Periods in the sequence denote gaps introduced by the alignment program.
9GL is highly conserved among ASFV isolates.
The degree of 9GL gene conservation among pathogenic ASFV isolates was examined by amplifying the gene from 22 viruses, representing African, European, and Caribbean isolates from both pig and tick sources. The 9GL ORF from each virus was amplified by PCR, cloned, and sequenced completely. All isolates encoded a 119-amino-acid 9GL protein with a predicted molecular mass of 14.4 kDa and a pI of 8.6. At the amino acid level, 9GL proteins were 92 to 100% identical to each other. Two groups were identical at the amino acid level (Fig. 2): (i) an E70X group with Cameroon, E70, E75, Haiti, La Granja, Lee, Lisbon 60, Spencer, Uganda, Zaire, and the BA71V ORF pB119L (GenBank accession no. U18466) product and (ii) a PR4X group with Chiredzi/83/1, Crocodile/96/1, Crocodile/96/3, Pretoriuskop/96/4, Pretoriuskop/96/5, and Wildebeeslaagte/96/1. While the Malawi Lil-20/1 and Killean III isolates were the most different from the other isolates, the differences between amino acid sequences were not sufficiently significant to allow the construction of a fully resolved, well-ordered phylogenetic tree (amino Poisson distance estimate by using a neighbor-joining branch length test; chi-square = 8.9, df = 7, P = 0.26) or a cluster test with 1,000 bootstrap samples (56, 57). There were 10 polymorphic amino acid sites, with 8 occurring in the amino-terminal half, and most were conservative substitutions (PAM 250 cutoff, 0.4) with an 0.01 average change per residue.
FIG. 2.
Alignment of the predicted amino acid sequences encoded by 9GL homologues in ASFV isolates. MAL, Malawi Lil-20/1; KIL, Killean III; KIM, Kimakia; VI, Victoria Falls; TE, Tengani; K1, Fairfield/96/1. Isolates with identical amino acid sequences are represented as E70X (E70, Cameroon, E75, Haiti 811, La Granja, Lee, Lisbon 60, Uganda 61, Spencer, Zaire, and BA71V [GenBank accession no. U18466]) and PR4X (Pretoriuskop/96/4, Pretoriuskop/96/5, Wildebeeslaagte/96/1, Crocodile/96/1, Crocodile/96/3, and Chiredzi/83/1). The consensus sequence was constructed with the Genetics Computer Group (GCG; Madison, Wis.) program Pileup (12) by using the Dayhoff PAM 250 symbol comparison table with a 0.4 cutoff value. Residue differences are shown above the consensus sequence. Asterisks denote amino acid residues conserved in the ERV1-ALR family of proteins.
At the nucleotide level the 9GL genes were 94 to 100% identical to each other. ASFV isolates within the following groups were identical to each other at the nucleotide level: (i) BA71V, Cameroon, E70, E75, Haiti 811, La Granja, Lisbon 60, Uganda, and Zaire; (ii) Lee and Spencer; (iii) Wildebeeslaagte/96/1 and Crocodile/96/1; (iv) Pretoriuskop/96/4 and Pretoriuskop/96/5.
The 9GL gene sequences from the 23 ASFV isolates contained 35 polymorphic nucleotide sites with 99 changes for a 5.67 corrected average transition/transversion ratio and 0.01 average change per nucleotide position. The ratio of synonymous to nonsynonymous codon usage, 6.4, was moderately high and consistent with the high amino acid similarity between the isolates. At the nucleotide level, in contrast to the amino acid comparison, there was evidence of differences between Malawi Lil-20/1, Killean III, and the remaining isolates (Kimura two-parameter distance estimate with branch length test and mutation rate constancy test; chi-square = 28.77, df = 10, P = 0.001). These data indicate that ORF 9GL is highly conserved among a diverse group of pathogenic ASFV isolates, including both pig and tick isolates.
9GL encodes a late nonstructural protein, p14.
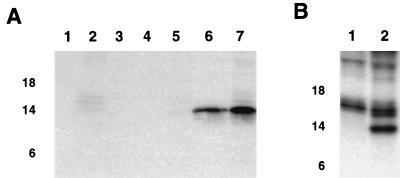
Radioimmunoprecipitation experiments using an anti-9GL monospecific serum were undertaken to analyze 9GL protein expression in ASFV-infected macrophage cell cultures. Anti-9GL serum specifically recognized a protein of 14 kDa (p14) in infected macrophage cell extracts at 7 to 9 HPI, with increasing levels present at later time points in the infection cycle (Fig. 3A). Expression was not detected in infected macrophage cell cultures treated with 1-β-d-arabinofuranosylcytosine (ara-C), a drug that blocks DNA replication and late viral protein synthesis (16) (Fig. 3B). These data demonstrate that p14 is a late viral protein.
FIG. 3.
Expression of p14 in ASFV-infected cell cultures. (A) Cell extracts of mock-infected (lane 1) and Malawi Lil-20/1-infected (lanes 2 to 7) swine macrophage cell cultures were pulse-labeled at 1 to 3 (lane 3), 3 to 5 (lane 4), 5 to 7 (lane 5), 7 to 9 (lane 6), and 9 to 18 (lanes 2 and 7) HPI and immunoprecipitated with anti-9GL monospecific serum (lanes 1 and 3 to 7) or a preimmune serum (lane 2). (B) Extracts of infected cells at 16 HPI maintained in the presence (lane 1) or absence (lane 2) of ara-C were immunoprecipitated with anti-9GL serum. Positions of the molecular size markers in kilodaltons are given on the left.
To determine if p14 was a virion structural protein, Western blot analysis of purified ASFV virions and IEM of virus-infected cells and purified virions were performed by using anti-9GL monospecific serum. ASFV p14 was not detected in Western blots of purified virions, whereas the known structural proteins p30 and p54 were shown to be present. In IEM experiments of infected macrophages incubated with either anti-9GL monospecific serum or preimmune serum, no specific labeling of virions was observed. Specific labeling was, however, detected in sections that were incubated with antiserum to the known structural proteins p11.6 and p54. In addition, no specific immunolabeling of purified ASF virions was detected (with preimmune serum, 1.18 ± 0.5 gold particles per virion; with 9GL antiserum, 0.92 ± 0.4 gold particles per virion). Taken together, these data suggest that p14 is not a virion structural protein.
Construction and analysis of the ASFV 9GL gene deletion mutant, Δ9GL, and its revertant, 9GL-R.
An ASFV 9GL gene deletion mutant, Δ9GL, and its revertant, 9GL-R, were constructed from the pathogenic African isolate Malawi Lil-20/1 and Δ9GL, respectively, as described in Materials and Methods. To construct Δ9GL, an 82-bp region within the 9GL ORF (Fig. 4A) was deleted from Malawi Lil-20/1 and replaced with a 2.4-kb p72GUS reporter gene cassette. The revertant virus, 9GL-R, was constructed by inserting the 9GL ORF, together with its upstream promoter elements and a 72-bp region downstream of its translation termination site, into the right variable region of Δ9GL (Fig. 4A). Genomic DNAs from parental, mutant, and revertant viruses were analyzed by Southern blotting and PCR. Viral DNAs, purified from infected macrophage cell cultures, were digested with DraI, gel electrophoresed, Southern blotted, and hybridized with 32P-labeled DNA probes. As expected, a gene probe specific for the deleted portion of 9GL failed to hybridize with genomic DNA from Δ9GL (Fig. 4B, I, lane 2). A predicted novel DraI fragment of 6.0 kb was observed for Δ9GL when it was hybridized with a probe for the parental 3.5-kb DraI fragment from this region (Fig. 4B, II, lane 2). By using PCR, the deleted 9GL, gene sequences were not detected in DNA preparations prepared from Δ9GL virus stocks (data not shown), and p14 was not detected by radioimmunoprecipitation analysis in Δ9GL-infected macrophage cell cultures (Fig. 4B, IV, lane 3).
The revertant virus 9GL-R exhibited the expected genomic structure. Hybridization with a right variable region probe (the 23-NL gene) demonstrated that the parental 2.9-kb SphI/SalI fragment (Fig. 4B, III, lanes 1 and 2) was replaced by a 7.5-kb fragment which resulted from the insertion of the 4.6-kb 9GL-72βGAL cassette. Restoration of 9GL in the revertant was confirmed by hybridizing the blot with a probe specific for 9GL sequences deleted from Δ9GL. As expected, the terminal 7.5-kb SphI/SalI fragment of 9GL-R hybridized with the probe (Fig. 4B, III, lane 4). Comparable levels of p14 were observed in macrophages infected with Malawi Lil-20/1 and 9GL-R (Fig. 4B, IV, lanes 1 and 2), indicating functional restoration of 9GL in 9GL-R.
9GL affects growth of ASFV in porcine macrophage cell cultures in vitro.
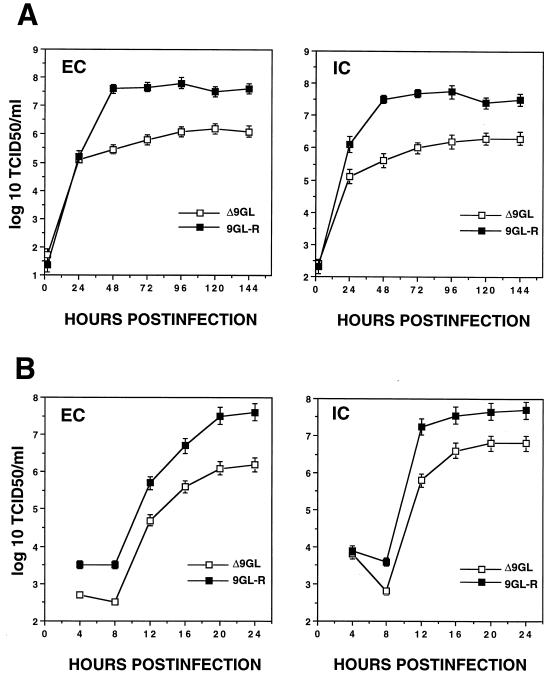
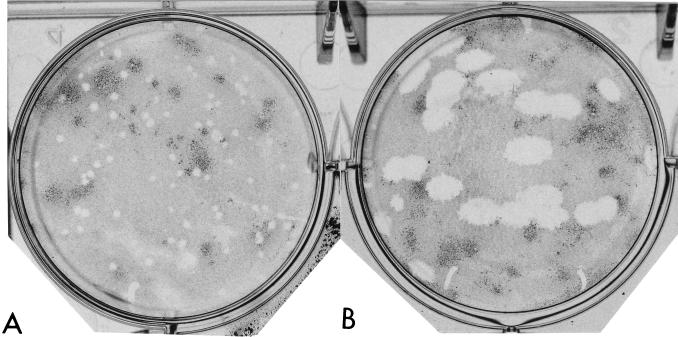
Growth characteristics of Δ9GL were compared to those of 9GL-R by infecting primary swine macrophage cell cultures at low and high MOIs (0.1 and 20, respectively) and determining titers of both cell-associated and extracellular virus at various times postinfection (Fig. 5). Δ9GL exhibited a significant growth defect in macrophage cell cultures. Extra- and intracellular virus titers for Δ9GL-infected cultures were reduced approximately 90 to 99% from those observed for 9GL-R. Consistent with these growth curve data, the plaque size of Δ9GL on macrophage cell cultures was reduced significantly, by approximately 93% (Fig. 6).
FIG. 5.
Growth characteristics of ASFV Malawi Δ9GL and the revertant, 9GL-R, in swine macrophage cell cultures. Porcine macrophage cell cultures were infected at an MOI of 0.1 (A) or 20 (B) with Δ9GL or 9GL-R. At indicated times postinfection, duplicate samples were collected and titrated for extracellular (EC) and intracellular (IC) virus yields. Data are means and standard errors of results from two independent experiments.
FIG. 6.
Plaque formation of Malawi Lil-20/1 and Δ9GL on primary porcine macrophage cell cultures. Porcine macrophage cell cultures were infected with 100 PFU of Δ9GL (A) or Malawi Lil-20/1 (B), overlaid with 0.5% agarose, and incubated at 37°C for 5 days; then they were fixed with a 4% phosphate-buffered saline-paraformaldehyde solution prior to staining with 0.1% crystal violet.
9GL affects ASFV virion maturation in infected macrophages.
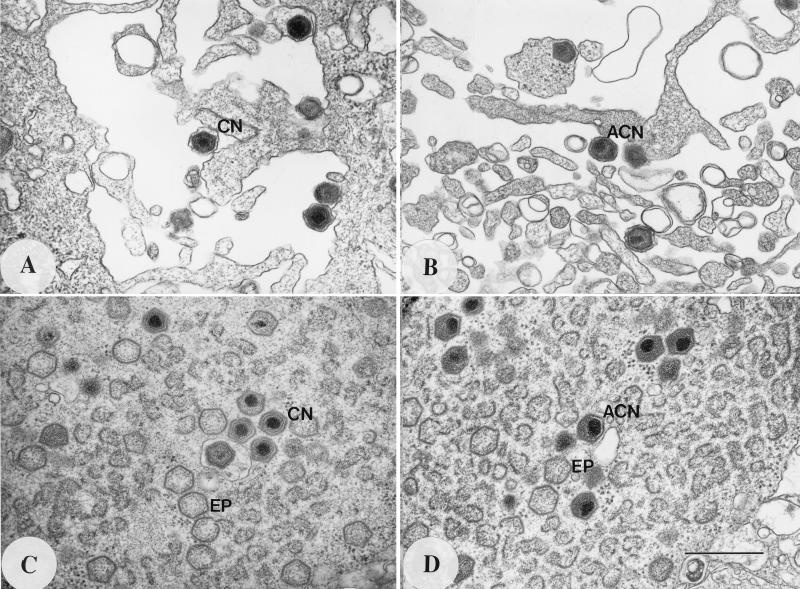
Δ9GL-infected macrophages were examined ultrastructurally to further define the growth defect. These data are shown in Fig. 7 and Table 1. Δ9GL-infected macrophages contained a large number of virus particles containing acentric nucleoids (ACN) (Fig. 7B and D) that resembled intermediate structures in normal virion morphogenesis (6). In these ACN particles, which represented 86 to 95% of all nucleoid-containing particles in the cell, the nucleoid was less condensed and spherical and was spread over two vertices of the viral icosahedral shell. These particles lacked a clearly defined core shell within the icosahedron, and an electron lucent region of varying width separated the nucleoid structure from the outer icosahedron shell. In contrast, 69 to 88% of the virions in Malawi Lil-20/1- or 9GL-R-infected cells contained concentric nucleoids (CN) and exhibited normal mature virion morphology (Fig. 7A and C). The ACN-to-CN ratio of Δ9GL particles in virus factories remained constant (approximately 9:1) at later times postinfection (24, 48, and 72 HPI), indicating that this observation was not the result of a delay in Δ9GL virus maturation (Table 1, experiment 3). Overall, the number of nucleoid-containing particles in 50 Δ9GL-infected cells (n = 175 particles) was reduced by approximately 70% from the number present in 50 cells infected with 9GL-R (n = 557 particles) (Table 1, experiment 2). Unlike parental virus, where budding virions exhibited normal CN particle morphology, approximately 90% of all Δ9GL budding virions showed ACN particle morphology (Fig. 7A and B; Table 1).
FIG. 7.
Budding virions and virus factories from Malawi Lil-20/1- and Δ9GL-infected macrophages at 16 HPI. (A and C) Malawi Lil-20/1-infected macrophages; (B and D) Δ9GL-infected macrophages. EP, empty particle. Magnification, ×42,536. Bar, 0.5 μm.
TABLE 1.
Virion morphology in Malawi Lil-20/1- and Δ9GL-infected macrophage cell cultures and purified virus preparations
| Virus | Cell culture
|
Purified virusa
|
||||
|---|---|---|---|---|---|---|
| No. of infected cells | No. with ACN/total no. (%) | No. with CN/total no. (%) | Titer (log10 TCID50/ml) | No. with ACN/total no. (%) | No. with CN/total no. (%) | |
| Expt 1 | ||||||
| MAL | 121/388 (31) [B = 0/111 (0)]b | 267/388 (69) [B = 111/111 (100)] | ||||
| Δ9GL | 395/416 (95) [B = 60/67 (90)] | 21/416 (5) [B = 7/67 (10)] | ||||
| Expt 2 | ||||||
| 9GL-R | 50 | 66/557 (12) | 491/557 (88) | 9.3 | 59/1,350 (4) | 1,291/1,350 (96) |
| Δ9GL | 50 | 163/175 (93) | 12/175 (7) | 7.3 | 110/137 (80) | 27/137 (20) |
| Expt 3 (Δ9GL) | ||||||
| 24 HPI | 25 | 81/90 (90) | 9/90 (10) | |||
| 48 HPI | 25 | 39/43 (90) | 4/43 (10) | |||
| 72 HPI | 25 | 114/133 (86) | 19/133 (14) | 7.3 | 144/164 (88) | 20/164 (12) |
Viruses in 25 random fields were counted and scored.
B, particles budding from the cell membrane.
To confirm the above observation that the total number of nucleoid-containing virions was reduced in Δ9GL-infected cells, virions were purified from Δ9GL- and 9GL-R-infected cells and counted in thin sections by using an electron microscope as described in Materials and Methods. Compared to 9GL-R-infected cells (n = 1,350 virions), the number of Δ9GL virions (n = 137 virions) was reduced by approximately 90%, and of these, approximately 85% were ACN particles (Table 1, experiment 2). Δ9GL exhibits a 2.0-log10-unit growth defect in macrophage cell cultures (Fig. 5). These data indicate that 1.0 log10 unit of the defect is due to a reduction in the total number of virions produced in the infected cells. If ACN particles are not infectious, the ACN-to-CN particle ratio (9:1) for Δ9GL could account for the additional 1.0-log10-unit reduction in infectivity.
9GL affects viral virulence in domestic swine.
The observed growth defect for Δ9GL in macrophages, the major target cell for the virus in vivo, suggested that 9GL may affect viral virulence in domestic swine. To study the role of 9GL in pig virulence, Yorkshire pigs were inoculated intramuscularly with 102 TCID50 of the revertant virus, 9GL-R, and either 102, 104, or 106 TCID50 of the 9GL null mutant, Δ9GL. A dose of 102 TCID50 of Malawi Lil-20/1 represents a challenge of between 10 and 100 LD100. Results of these experiments are shown in Table 2. In contrast to 9GL-R, where mortality was 100%, all Δ9GL-infected animals survived infection, even when infected with doses that should represent 10,000 to 100,000 LD100. The disease pattern observed for 9GL-R-infected animals contrasted markedly with that seen following infection with Δ9GL. With the exception of a transient, and significantly lower, fever response (P = 0.0001), Δ9GL-infected animals remained clinically normal following infection, whereas animals infected with 9GL-R presented with clinical signs of ASF on 4 to 6 DPI, and these symptoms progressed until death in all cases. Both mean and maximum viremia titers in Δ9GL-infected animals were significantly reduced by 100- to 1,000-fold from control values (P = 0.0001 to 0.009) during this acute disease period. The duration of viremia in Δ9GL-infected pigs ranged from 23 to 26 DPI. Thus, deletion of 9GL from Malawi Lil-20/1 markedly attenuated the virus in swine.
TABLE 2.
Swine survival, fever response, and viremia following infection with Malawi Δ9GL on 9GL-R
| Groupa | No. surviving/total no. | Days to death (±SE) | Feverb
|
Viremiab
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Days to onset (±SE) | No. of days of fever (±SE) | Mean temp (°F) (±SE) | Days to onset (±SE) | Days of viremia (±SE) | Titer (log10 TCID50/ml)
|
||||
| Mean | Maximum | ||||||||
| 9GL-R/102(n = 4) | 0/4 | 7.5 (±0.8) | 4.5 (±0.5) | 2.5 (±0.7) | 105.9 (±0.2) | 5.0 (±0.0) | 2.5 (±0.5) | 8.0 (±0.3) | 8.1 (±0.3) |
| Δ9GL/102(n = 4) | 4/4 | 6.5 (±2.0) | 3.8 (±1.9) | 104.1* (±0.1) | 6.0 (±1.3) | 24.0 (±1.2) | 5.2† (±0.1) | 6.3‡ (±0.1) | |
| Δ9GL/104(n = 4) | 4/4 | 8.3 (±0.3) | 1.3 (±0.3) | 104.1* (±0.1) | 5.5 (±0.5) | 26.0 (±2.0) | 5.5† (±0.1) | 6.8‡ (±0.4) | |
| Δ9GL/106(n = 4) | 4/4 | 6.3 (±1.9) | 2.5 (±1.2) | 104.1* (±0.1) | 4.0 (±1.5) | 23.3 (±2.9) | 5.2† (±0.8) | 6.7‡ (±0.8) | |
Virus/number of 50% tissue culture infective doses (number of animals in group).
Δ9GL values significantly different from those of the Malawi 9GL-R group are indicated as follows: *, P = 0.0001; †, P = 0.0001; ‡, P = 0.009.
At 42 DPI, convalescent Δ9GL-infected animals were challenged with 104 TCID50 of parental Malawi Lil-20/1 to assess the level of immunity conferred by prior infection with Δ9GL. Data from this experiment are shown in Table 3. Following challenge, animals in all three Δ9GL dose groups survived infection and remained clinically normal, indicating that a solid level of homologous protective immunity had been induced by Δ9GL infection. With the exception of a single animal in each group, animals which received 104 or 106 TCID50 of Δ9GL did not develop fever or detectable viremia following Malawi Lil-20/1 challenge. Pigs infected with the lower, 102-TCID50 dose of Δ9GL responded with a transient fever and low-level viremia following challenge infection.
TABLE 3.
Survival, fever response, and viremia following challenge of Δ9GL-immunized pigs with Malawi Lil-20/1a
| Groupb | No. surviving/total no. | Fever
|
Viremia
|
||||
|---|---|---|---|---|---|---|---|
| Days to onset (no. of animals with fever) | Duration (no. of days) | Maximum temp (°F) | Days to onset (no. of viremic animals) | Duration (no. of days) | Maximum titer (log10 TCID50/ml) | ||
| Δ9GL/106(n = 4) | 4/4 | 3.0 (1) | 4.0 | 106.6 | 5.0 (1) | 23.0 | 5.5 |
| Δ9GL/104(n = 4) | 4/4 | 5.0 (1) | 2.0 | 106.2 | 5.0 ± 0.0 (2) | 13.0 ± 4.0 | 4.0 ± 0.5 |
| Δ9GL/102(n = 4) | 4/4 | 8.0 ± 2.5 (4) | 7.2 ± 2.8 | 106.0 ± 0.7 | 6.0 ± 1.0 (4) | 7.5 ± 4.3 | 3.5 ± 0.2 |
Δ9GL-immunized pigs were challenged intramuscularly with 104 TCID50 of Malawi Lil-20/1 at 42 DPI.
Virus/number of 50% tissue culture infective doses (number of animals in group).
DISCUSSION
Here, we have shown that the ASFV ERV1/ALR gene family homologue, 9GL, is highly conserved among virus isolates and that it encodes a late nonstructural viral protein of 14 kDa (p14) which affects virion maturation and viral growth in swine macrophages in vitro and viral virulence in swine.
Overall, our data suggest that the 2.0-log10-unit Δ9GL growth defect in macrophages is due to a 1.0-log10-unit reduction in the overall number of virus particles in infected cells and to a 1.0-log10-unit reduction in the infectivity of the particles actually produced (Table 1). Approximately 90% of the Δ9GL virions in virus factories and budding from infected-cell membranes contained an ACN structure that resembled the intermediate structures observed during ASFV virion maturation (6). While the infectivity of these intermediate ASFV particles is not known, the above data which indicate a 1.0-log10-unit reduction in Δ9GL infectivity suggest that they are not infectious. If this is the case, then only 1 Δ9GL virion in 10 is a mature, infectious particle.
While clearly important, p14 is nonessential for production of mature infectious virions. Thus, the protein does not perform an obligate function in virion maturation and/or assembly. Rather, it appears to affect the overall efficiency of the virion assembly process, either directly or indirectly. Using IEM and Western blotting with anti-9GL antiserum, we were unable to detect p14 in virions. This suggests a nonstructural function for the protein in efficient virion assembly. As is the case with scERV1 in yeast and with other ERV1 homologues, our data suggest that p14 may be a low-abundance protein in infected cells (24, 29, 35, 36).
Local energy production in the ASFV-infected cell appears to be important for virus particle assembly. Actively respiring mitochondria migrate to and cluster in proximity to viral factories containing assembling particles in infected Vero cells and macrophages (51; T. G. Burrage, L. Zsak, J. G. Neilan, and D. L. Rock, unpublished data). It is also known that ASFV infection results in induction of mitochondrial stress-response proteins p74 and cpn60 (51). Given that scERV1 is indispensable for oxidative phosphorylation and for the maintenance of mitochondrial genomes in yeast (35). It is possible that p14 may perform some mitochondria-related function in the infected cell that mediates virus-induced stress, thus allowing sufficient energy production for efficient virion assembly and maturation.
The p14 protein does contain conserved cysteine residues indicative of redox-active centers found in glutaredoxin and thioredoxin (10, 14, 30). These enzymes are multifunctional, performing roles in DNA replication, protein synthesis, protein folding, and photosynthesis (30). Thus, p14 could conceivably perform an important redox function that is critical to efficient virion assembly and maturation.
The Malawi Lil-20/1 9GL deletion mutant Δ9GL was markedly attenuated in swine (Table 2). This represents the first complete attenuation of Malawi Lil-20/1 or any other pathogenic African ASFV isolate by engineered gene deletion. The actual mechanism(s) underlying Δ9GL attenuation in the host is unknown. The behavior of the mutant virus in macrophage cell cultures and the gene's homology with ERV1 does, however, suggest several possibilities, including a possible growth defect and viral interference in vivo, that might account for the attenuated phenotype in swine.
ASFV targets cells of the mononuclear-phagocytic system, including highly differentiated fixed-tissue macrophages; affected tissues show extensive damage following infection with highly virulent virus isolates (9, 33, 34, 40, 43). ASFV isolates of lesser virulence also infect these cells, but the degree of tissue involvement and the resulting tissue damage are much less severe (27, 40, 41). The abilities of ASFV to replicate efficiently and induce cytopathology in macrophages in vivo are likely to be critical factors in ASFV virulence. Attenuation of Δ9GL may be the direct result of a macrophage growth defect in vivo. The Δ9GL macrophage growth defect in vitro and the reduced viremia titers (100- to 1000-fold) for Δ9GL-infected animals are consistent with this. A viral growth defect would slow generalization of infection and reduce lymphoid tissue damage, thus permitting an appropriate protective host immune response to be mounted.
Approximately 90% of the Δ9GL virions produced by infected macrophages contain an ACN, and they resemble the intermediate particles seen during virion morphogenesis (6). Our data discussed above suggest that these particles may be noninfectious. This high ratio of defective to infectious Δ9GL particles could conceivably result in viral interference where defective particles are interfering in some way with efficient replication of the few mature, fully infectious particles. In addition, the large excess of defective ASF virions could actually be serving as immunizing antigens, accelerating the induction of a protective host immune response in the face of acute viral infection.
Interestingly, the rat ERV1 homolog (ALR) is thought to function in liver regeneration in part by regional regulation of liver-resident natural killer (NK) cells. In vivo administration of ALR induced inhibition of NK cell cytotoxic activities in liver by an unknown intermediary mechanism (19). An NK cell response represents one of the earliest host responses to viral infection, and it may be important in the early stages of ASFV infection (42). If p14 functioned as an inhibitor of NK cell activity, either directly or indirectly, it would accelerate viral spread during the early, critical phases of infection.
The possibilities for Δ9GL attenuation discussed above are not mutually exclusive. The marked attenuation of the mutant in animals may result from a combination of these mechanisms.
Significantly, all Δ9GL-infected animals were protected when challenged with parental virulent Malawi Lil-20/1 at 42 DPI. However, the dose of Δ9GL used for infection did affect the level of immunity conferred (Table 3). Animals in the two higher-dose Δ9GL groups (104 and 106 TCID50) exhibited a solid level of homologous protective immunity similar to that reported for animals recovering from infection with pathogenic ASFV isolates (25, 39, 62). Only two of eight animals had a fever response, and viremia was detected in only three of the eight. In contrast, all animals in the lowest-dose Δ9GL group (102 TCID50) had significant fever responses and viremia following challenge. This apparent difference in immune status appears to be unrelated to initial Δ9GL infection. Similar infection patterns were observed in all three dose groups (Table 2).
ASFV 9GL gene deletion mutants will be a powerful tool for defining the number of different ASFV antigenic types that a vaccine must protect against. For the first time, it is possible to reliably attenuate highly virulent African field isolates for use in cross-protection experiments in pigs. An understanding of ASFV cross-protective immunity will be critical for developing a broadly protective ASFV vaccination strategy.
Overall, these data indicate that ASFV 9GL gene deletion mutants are excellent candidates for possible use as live-attenuated ASF vaccines.
ACKNOWLEDGMENTS
We thank Aniko Zsak, George Smoliga, and the PIADC animal care staff for excellent technical assistance and Steven Kleiboeker, Glen Scoles, and Stefan Swanepoel for providing African tick isolates of ASFV.
REFERENCES
- 1.Afonso C L, Alcaraz C, Brun A, Sussman M D, Onisk D V, Escribano J M, Rock D L. Characterization of p30, a highly antigenic membrane and secreted protein of African swine fever virus. Virology. 1992;189:368–373. doi: 10.1016/0042-6822(92)90718-5. [DOI] [PubMed] [Google Scholar]
- 2.Afonso C L, Zsak L, Carrillo C, Borca M V, Rock D L. African swine fever virus NL gene is not required for virus virulence. J Gen Virol. 1998;79:2543–2547. doi: 10.1099/0022-1317-79-10-2543. [DOI] [PubMed] [Google Scholar]
- 3.Afonso C L, Tulman E R, Lu Z, Oma E, Kutish G F, Rock D L. The genome of Melanoplus sanguinipes entomopoxvirus. J Virol. 1999;73:533–552. doi: 10.1128/jvi.73.1.533-552.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alcaraz C, Pasamontes B, Ruiz G F, Escribano J M. African swine fever virus-induced proteins on the plasma membranes of infected cells. Virology. 1989;168:406–408. doi: 10.1016/0042-6822(89)90283-3. [DOI] [PubMed] [Google Scholar]
- 5.Borca M V, Irusta P M, Kutish G F, Carrillo C, Afonso C L, Burrage T, Neilan J G, Rock D L. A structural DNA binding protein of African swine fever virus with similarity to bacterial histone-like proteins. Arch Virol. 1996;141:301–313. doi: 10.1007/BF01718401. [DOI] [PubMed] [Google Scholar]
- 6.Brookes S M, Hyatt A D, Wise T, Parkhouse R M E. Intracellular virus DNA distribution and the acquisition of the nucleoprotein core during African swine fever virus particle assembly: ultrastructural in situ hybridisation and DNase-gold labelling. Virology. 1998;249:175–188. doi: 10.1006/viro.1998.9308. [DOI] [PubMed] [Google Scholar]
- 7.Brown F. The classification and nomenclature of viruses: summary of results of meetings of the International Committee on Taxonomy of Viruses in Sendai, September 1984. Intervirology. 1986;25:141–143. doi: 10.1159/000150091. [DOI] [PubMed] [Google Scholar]
- 8.Carrascosa A L, del Val M, Santarén J F, Viñuela E. Purification and properties of African swine fever virus. J Virol. 1985;54:337–344. doi: 10.1128/jvi.54.2.337-344.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colgrove G S, Haelterman E O, Coggins L. Pathogenesis of African swine fever in young pigs. Am J Vet Res. 1969;30:1343–1359. [PubMed] [Google Scholar]
- 10.Coppock D L, Cina-Poppe D, Gilleran S. The quiescin Q6 gene families: thioredoxin and ERV1. Genomics. 1998;54:460–468. doi: 10.1006/geno.1998.5605. [DOI] [PubMed] [Google Scholar]
- 11.Costa J V. African swine fever virus. In: Darai G, editor. Molecular biology of iridoviruses. Norwell, Mass: Kluwer Academic Publishers; 1990. pp. 247–270. [Google Scholar]
- 12.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dixon L K, Twigg S R F, Baylis S A, Vydelingum S, Bristow C, Hammond J M, Smith G L. Nucleotide sequence of a 55 kbp region from the right end of the genome of a pathogenic African swine fever virus isolate (Malawi LIL20/1) J Gen Virol. 1994;75:1655–1684. doi: 10.1099/0022-1317-75-7-1655. [DOI] [PubMed] [Google Scholar]
- 14.Ellis L B M, Saurugger P, Woodward C. Identification of the three-dimensional thioredoxin motif: related structure in the ORF3 protein of the Staphylococcus aureus mer operon. Biochemistry. 1992;31:4882–4891. doi: 10.1021/bi00135a020. [DOI] [PubMed] [Google Scholar]
- 15.Escribano J M, Tabarés E. Proteins specified by African swine fever virus. V. Identification of immediate early, early and late proteins. Arch Virol. 1987;92:221–238. doi: 10.1007/BF01317479. [DOI] [PubMed] [Google Scholar]
- 16.Estevez A, Marquez M I, Costa J V. Two-dimensional analysis of African swine fever virus proteins and proteins induced in infected cells. Virology. 1986;152:192–206. doi: 10.1016/0042-6822(86)90384-3. [DOI] [PubMed] [Google Scholar]
- 17.Ewing B, Hillier L, Wendl M C, Green P. Base calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 18.Finney D J. Statistical methods in biological assays. 2nd ed. New York, N.Y: Macmillan Publishing Co.; 1978. pp. 394–398. [Google Scholar]
- 19.Francavilla A, Vujanovic N L, Polimeno L, Azzarone A, Iacobellis A, Deleo A, Hagiya M, Whiteside T L, Starzl T E. The in vivo effect of hepatotrophic factors augmenter of liver regeneration, hepatocyte growth factor, and insulin-like growth factor-II on liver natural killer cell functions. Hepatology. 1997;25:411–415. doi: 10.1002/hep.510250225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Genovesi E V, Villinger F, Gerstner D J, Whyard T C, Knudsen R C. Effect of macrophage-specific colony-stimulating factor (CSF-1) on swine monocyte/macrophage susceptibility to in vitro infection by African swine fever virus. Vet Microbiol. 1990;25:153–176. doi: 10.1016/0378-1135(90)90074-6. [DOI] [PubMed] [Google Scholar]
- 21.Giorda R, Hagiya M, Seki T, Shimonishi M, Sakai H, Michaelson J, Francavilla A, Starzl T E, Trucco M. Analysis of the structure and expression of the augumenter of liver regeneration (ALR) gene. Mol Med. 1996;2:97–108. [PMC free article] [PubMed] [Google Scholar]
- 22.Gómez-Puertas P, Rodríguez F, Ovideo J M, Ramiro-Ibáñez F, Ruiz-Gonzalvo F, Alonso C, Escribano J M. Neutralizing antibodies to different proteins of African swine fever virus inhibit both virus attachment and internalization. J Virol. 1996;70:5689–5694. doi: 10.1128/jvi.70.8.5689-5694.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.González A, Talavera A, Almendral J M, Viñuela E. Hairpin loop structure of African swine fever virus DNA. Nucleic Acids Res. 1986;14:6835–6844. doi: 10.1093/nar/14.17.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hagiya M, Francavilla A, Polimeno L, Ihara I, Sakai H, Seki T, Shimonishi M, Porter K A, Starzl T E. Cloning and sequence analysis of the rat augmenter of liver regeneration (ALR) gene: expression of biologically active recombinant ALR and demonstration of tissue distribution. Proc Natl Acad Sci USA. 1994;91:8142–8146. doi: 10.1073/pnas.91.17.8142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamdy F M, Dardiri A H. Clinical and immunologic responses of pigs to African swine fever virus isolated from the Western hemisphere. Am J Vet Res. 1984;45:711–714. [PubMed] [Google Scholar]
- 26.Haresnape J M, Wilkinson P J, Mellor P S. Isolation of African swine fever virus from ticks of the Ornithodoros moubata complex (Ixodoidea: Argasidae) collected within the African swine fever enzootic area of Malawi. Epidemiol Infect. 1988;101:173–185. doi: 10.1017/s0950268800029332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hess W R. African swine fever: a reassessment. Adv Vet Sci Comp Med. 1982;25:39–69. [PubMed] [Google Scholar]
- 28.Hess W R, Endris R G, Lousa A, Caiado J M. Clearance of African swine fever virus from infected tick (Acari) colonies. J Med Entomol. 1989;26:314–317. doi: 10.1093/jmedent/26.4.314. [DOI] [PubMed] [Google Scholar]
- 29.Hofhaus G, Stein G, Polimeno L, Francavilla A, Lisowsky T. Highly divergent amino termini of the homologous human ALR and yeast scERV1 gene products define species specific differences in cellular localization. Eur J Cell Biol. 1999;78:349–356. doi: 10.1016/S0171-9335(99)80069-7. [DOI] [PubMed] [Google Scholar]
- 30.Holmgren A. Thioredoxin. Annu Rev Biochem. 1985;54:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- 31.Kleiboeker S B, Kutish G F, Neilan J G, Lu Z, Zsak L, Rock D L. A conserved African swine fever virus right variable region gene, l11L, is nonessential for growth in vitro and virulence in domestic swine. J Gen Virol. 1998;79:1189–1195. doi: 10.1099/0022-1317-79-5-1189. [DOI] [PubMed] [Google Scholar]
- 32.Knudsen R C, Genovesi E V, Whyard T C. In vitro immune serum-mediated protection of pig monocytes against African swine fever virus. Am J Vet Res. 1987;48:1067–1071. [PubMed] [Google Scholar]
- 33.Konno S, Taylor W D, Dardiri A H. Acute African swine fever. Proliferative phase in lymphoreticular tissue and the reticuloendothelial system. Cornell Vet. 1971;61:71–84. [PubMed] [Google Scholar]
- 34.Konno S, Taylor W D, Hess W R, Heuschele W P. Liver pathology in African swine fever. Cornell Vet. 1971;61:125–150. [PubMed] [Google Scholar]
- 35.Lisowsky T. Dual function of a new nuclear gene for oxidative phosphorylation and vegetative growth in yeast. Mol Gen Genet. 1992;232:58–64. doi: 10.1007/BF00299137. [DOI] [PubMed] [Google Scholar]
- 36.Lisowsky T. ERV1 is involved in the cell-division cycle and the maintenance of mitochondrial genomes in Saccharomyces cerevisiae. Curr Genet. 1994;26:15–20. doi: 10.1007/BF00326299. [DOI] [PubMed] [Google Scholar]
- 37.Lisowsky T, Weinstatsaslow D L, Barton N, Reeders S T, Schneider M C. A new human gene located in the PKD1 region of chromosome 16 is a functional homologue to ERV1 of yeast. Genomics. 1995;29:690–697. doi: 10.1006/geno.1995.9950. [DOI] [PubMed] [Google Scholar]
- 38.Lisowsky T. Removal of an intron with unique 3′ branch site creates an amino-terminal protein sequence directing the scERV1 gene product to mitochondria. Yeast. 1996;12:1501–1510. doi: 10.1002/(sici)1097-0061(199612)12:15<1501::aid-yea40>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 39.Mebus C A, Dardiri A H. Western hemisphere isolates of African swine fever virus: asymptomatic carriers and resistance to challenge inoculation. Am J Vet Res. 1980;41:1867–1869. [PubMed] [Google Scholar]
- 40.Mebus C A. African swine fever. Adv Virus Res. 1988;35:251–269. doi: 10.1016/s0065-3527(08)60714-9. [DOI] [PubMed] [Google Scholar]
- 41.Mebus C A, McVicar J W, Dardiri A H. Comparison of the pathology of high and low virulence African swine fever virus infections. In: Wilkinson P J, editor. Proceedings of CEC/FAO Expert Consultation in African Swine Fever Research. Luxembourg: Commission of the European Communities; 1981. pp. 183–194. [Google Scholar]
- 42.Mendoza C, Videgain S P, Alonso F. Inhibition of natural killer activity in porcine mononuclear cells by African swine fever virus. Res Vet Sci. 1991;51:317–321. doi: 10.1016/0034-5288(91)90084-2. [DOI] [PubMed] [Google Scholar]
- 43.Moulton J, Coggins L. Comparison of lesions in acute and chronic African swine fever. Cornell Vet. 1968;58:364–388. [PubMed] [Google Scholar]
- 44.Neilan J G, Lu Z, Kutish G F, Zsak L, Burrage T G, Borca M V, Carrillo C, Rock D L. A BIR motif containing gene of African swine fever virus, 4CL, is nonessential for growth in vitro and viral virulence. Virology. 1997;230:252–264. doi: 10.1006/viro.1997.8481. [DOI] [PubMed] [Google Scholar]
- 45.Neilan J G, Lu Z, Kutish G F, Zsak L, Lewis T L, Rock D L. A conserved African swine fever virus IκB homolog, 5EL, is nonessential for growth in vitro and virulence in domestic pigs. Virology. 1997;235:377–385. doi: 10.1006/viro.1997.8693. [DOI] [PubMed] [Google Scholar]
- 46.Onisk D V, Borca M V, Kutish G, Kramer E, Irusta P, Rock D L. Passively transferred African swine fever virus antibodies protect swine against lethal infection. Virology. 1994;198:350–354. doi: 10.1006/viro.1994.1040. [DOI] [PubMed] [Google Scholar]
- 47.Ortin J, Enjuanes L, Viñuela E. Cross-links in African swine fever virus DNA. J Virol. 1979;31:579–583. doi: 10.1128/jvi.31.3.579-583.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Plowright W, Parker J, Pierce M A. The epizootiology of African swine fever in Africa. Vet Rec. 1969;85:668–674. [PubMed] [Google Scholar]
- 49.Plowright W, Parker J, Pierce M A. African swine fever virus in ticks (Ornithodoros moubata, Murray) collected from animal burrows in Tanzania. Nature (London) 1969;221:1071–1073. doi: 10.1038/2211071a0. [DOI] [PubMed] [Google Scholar]
- 50.Rodriguez F, Alcaraz C, Eiras A, Yáñez R J, Rodriguez J M, Alonso C, Rodriguez J F, Escribano J M. Characterization and molecular basis of heterogeneity of the African swine fever virus envelope protein p54. J Virol. 1994;68:7244–7252. doi: 10.1128/jvi.68.11.7244-7252.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rojo G, Chamorro M, Salas M L, Vinuela E, Cuezva J M, Salas J. Migration of mitochondria to viral assembly sites in African swine fever virus-infected cells. J Virol. 1998;72:7583–7588. doi: 10.1128/jvi.72.9.7583-7588.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Santarén J F, Viñuela E. African swine fever virus-induced polypeptides in Vero cells. Virus Res. 1986;5:391–405. doi: 10.1016/0168-1702(86)90031-6. [DOI] [PubMed] [Google Scholar]
- 53.Sogo J M, Almendral J M, Talavera A, Viñuela E. Terminal and internal inverted repetitions in African swine fever virus DNA. Virology. 1984;133:271–275. doi: 10.1016/0042-6822(84)90394-5. [DOI] [PubMed] [Google Scholar]
- 54.Stein G, Lisowsky T. Functional comparison of the yeast scERV2 genes. Yeast. 1998;14:171–180. doi: 10.1002/(SICI)1097-0061(19980130)14:2<171::AID-YEA209>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 55.Tabarés E. Characterization of African swine fever virus proteins. In: Becker Y, editor. African swine fever. Boston, Mass: Martinus Nijhoff; 1987. pp. 51–61. [Google Scholar]
- 56.Takezaki N, Rzhetsky A, Nei M. Phylogenetic test of the molecular clock and linearized trees. Mol Biol Evol. 1995;12:823–833. doi: 10.1093/oxfordjournals.molbev.a040259. [DOI] [PubMed] [Google Scholar]
- 57.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 58.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thomson G R, Gainaru M, Lewis A, Biggs H, Nevill E, Van Der Pypekamp M, Gerbes L, Esterhuysen J, Bengis R, Bezuidenhout D, Condy J. The relationship between ASFV, the warthog and Ornithodoros species in southern Africa. In: Wilkinson P J, editor. Proceedings of CEC/FAO Research Seminar. ASF, EUR 8466 EN. Luxembourg: Commission of the European Communities; 1983. pp. 85–100. [Google Scholar]
- 60.Viñuela E. African swine fever. Curr Top Microbiol Immunol. 1985;116:151–170. doi: 10.1007/978-3-642-70280-8_8. [DOI] [PubMed] [Google Scholar]
- 61.Wang G, Yang X-M, Zhang Y, Wang Q-M, Chen H, Wei H, Xing G-C, Xie L, Hu Z-Y, Zhang C, Fang D-C, Wu C-T, He F-C. Identification and characterization of receptor for mammalian hepatopoietin that is homologous to yeast ERV1. J Biol Chem. 1999;274:11469–11472. doi: 10.1074/jbc.274.17.11469. [DOI] [PubMed] [Google Scholar]
- 62.Wardley R C, Norley S G, Martins C V, Lawman M J. The host response to African swine fever virus. Prog Med Virol. 1987;34:180–192. [PubMed] [Google Scholar]
- 63.Wesley R D, Pan I C. African swine fever DNA: restriction endonuclease cleavage patterns of wild-type, Vero cell-adapted and plaque-purified virus. J Gen Virol. 1982;63:383–391. doi: 10.1099/0022-1317-63-2-383. [DOI] [PubMed] [Google Scholar]
- 64.Wilkinson P J. African swine fever virus. In: Pensaert M B, editor. Virus infections of porcines. Amsterdam, The Netherlands: Elsevier Science Publishers; 1989. pp. 17–35. [Google Scholar]
- 65.Yáñez R J, Rodríguez J M, Nogal M L, Yuste L, Enríquez C, Rodriguez J F, Viñuela E. Analysis of the complete nucleotide sequence of African swine fever virus. Virology. 1995;208:249–278. doi: 10.1006/viro.1995.1149. [DOI] [PubMed] [Google Scholar]
- 66.Zsak L, Lu Z, Kutish G F, Neilan J G, Rock D L. An African swine fever virus virulence-associated gene NL-S with similarity to the herpes simplex virus ICP34.5 gene. J Virol. 1996;70:8865–8871. doi: 10.1128/jvi.70.12.8865-8871.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zsak L, Caler E, Lu Z, Kutish G F, Neilan J G, Rock D L. A nonessential African swine fever virus gene UK is a significant virulence determinant in domestic swine. J Virol. 1998;72:1028–1035. doi: 10.1128/jvi.72.2.1028-1035.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]